Nucleotide analogs targeting viral RNA polymerase have been proved to be an effective strategy for antiviral treatment and are promising antiviral drugs to combat the current severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic. In this study, we developed a robust in vitro nonradioactive primer extension assay to quantitatively evaluate the efficiency of incorporation of nucleotide analogs by SARS-CoV-2 RNA-dependent RNA polymerase (RdRp). Our results show that many nucleotide analogs can be incorporated into RNA by SARS-CoV-2 RdRp and that the incorporation of some of them leads to chain termination.
KEYWORDS: RNA-dependent RNA polymerase, SARS-CoV-2, discrimination value, favipiravir, nsp12-nsp8-nsp7, nucleotide analogs, primer extension, remdesivir, ribavirin, sofosbuvir
ABSTRACT
Nucleotide analogs targeting viral RNA polymerase have been proved to be an effective strategy for antiviral treatment and are promising antiviral drugs to combat the current severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic. In this study, we developed a robust in vitro nonradioactive primer extension assay to quantitatively evaluate the efficiency of incorporation of nucleotide analogs by SARS-CoV-2 RNA-dependent RNA polymerase (RdRp). Our results show that many nucleotide analogs can be incorporated into RNA by SARS-CoV-2 RdRp and that the incorporation of some of them leads to chain termination. The discrimination values of nucleotide analogs over those of natural nucleotides were measured to evaluate the incorporation efficiency of nucleotide analog by SARS-CoV-2 RdRp. In agreement with the data published in the literature, we found that the incorporation efficiency of remdesivir-TP is higher than that of ATP and incorporation of remdesivir-TP caused delayed chain termination, which can be overcome by higher concentrations of the next nucleotide to be incorporated. Our data also showed that the delayed chain termination pattern caused by remdesivir-TP incorporation is different for different template sequences. Multiple incorporations of remdesivir-TP caused chain termination under our assay conditions. Incorporation of sofosbuvir-TP is very low, suggesting that sofosbuvir may not be very effective in treating SARS-CoV-2 infection. As a comparison, 2′-C-methyl-GTP can be incorporated into RNA efficiently, and the derivative of 2′-C-methyl-GTP may have therapeutic application in treating SARS-CoV-2 infection. This report provides a simple screening method that should be useful for evaluating nucleotide-based drugs targeting SARS-CoV-2 RdRp and for studying the mechanism of action of selected nucleotide analogs.
INTRODUCTION
The ongoing global pandemic of coronavirus disease 2019 (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (1–3), which is a single-stranded, positive-sense RNA virus and has one of the largest genomes (around 30 kb) known among RNA viruses (4). SARS-CoV-2 is continuing its spread across the world, and no effective vaccines are available; thus, the search for safe and effective small-molecule inhibitors that are active against SARS-CoV-2 represents a high research priority.
Nucleoside/nucleotide-based inhibitors targeting viral DNA or RNA polymerase have been proven to be the backbone of antiviral therapies (5–8). Currently, there are over 25 approved therapeutic nucleosides/nucleotides used for the therapy of various viral infections of high medical importance, such as HIV infection (tenofovir, AZT, etc.), hepatitis B (lamivudine), hepatitis C (sofosbuvir), and herpes (acyclovir). The proven success of using nucleoside-based drugs to effectively treat viral infections and the current knowledge of the pathway for developing this class of inhibitors make them promising antiviral agents to combat the current pandemic of COVID-19 (9, 10).
Following intracellular phosphorylation, nucleoside analog 5′-triphosphates (the active form of nucleoside analogs) are competitively incorporated into nascent viral RNA chains and thus inhibit viral replication (5). The mechanism of action varies for different nucleotide analogs. The most common mechanism of action is chain termination caused by the incorporation of nucleotide analogs, resulting in the formation of incomplete viral RNA chains (11, 12). Nucleoside analogs not causing chain termination may also be used as antiviral drugs, such as ribavirin and favipiravir, which induce mutagenesis due to their ambiguous base-pairing properties after being incorporated into viral RNA (13–17).
Like other coronaviruses, SARS-CoV-2 encodes an RNA-dependent-RNA polymerase (RdRp) on the nsp12 gene (18). This protein, which catalyzes RNA synthesis, forms a replication complex with nsp7, nsp8, and other virally encoded proteins and host proteins that are responsible for mRNA synthesis, as well as the synthesis of genomic RNA for progeny viruses (19). The structure of the nsp12-nsp8-nsp7 complex was recently determined using cryo-electron microscopy (20). Additionally, unlike other viruses, SARS-CoV-2 contains an exonuclease gene (nsp14) and can perform proofreading activity to remove the mismatched nucleotides from viral RNA (16, 21, 22). This is probably one of the difficulties in developing nucleotide polymerase inhibitors against SARS-CoV-2, and this proofreading activity of SARS-CoV-2 should be taken into consideration when a nucleoside-based antiviral drug against SARS-CoV-2 is being developed. Nucleotide analogs with different mechanisms of action (chain terminators, delayed terminators, and mutagenic nucleoside analogs) may respond differently to SARS-CoV-2 proofreading activity mediated by nsp14. So far, numerous nucleoside/nucleotide analogs have been described to inhibit SARS-CoV-2, including remdesivir, ribavirin, BCX4430, gemcitabine hydrochloride, β-d-N4-hydroxycytidine, and 6-azauridine (9). The detailed mechanisms of action of those nucleotide analogs against SARS-CoV-2 still need to be addressed.
In this study, we developed a robust in vitro nonradioactive primer extension assay using a fluorescently labeled RNA primer annealed to an RNA template. The incorporation efficiency and chain termination ability of a series of nucleotide analogs were determined using this assay. The method is very valuable for evaluation of the antiviral potential of nucleotide analogs, and the structure-activity information derived from these studies can be used to explore the mechanism of action of given nucleotide analogs and to design nucleotide analogs with better properties.
RESULTS
Expression and purification of SARS-CoV-2 nsp12, nsp7, and nsp8.
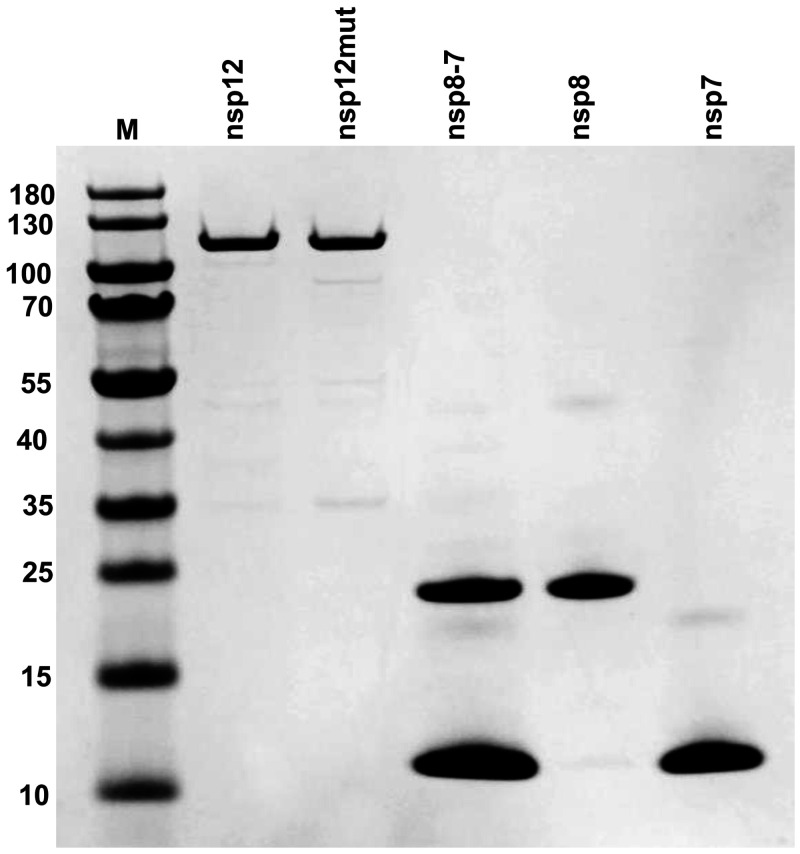
A number of reports have shown that a functional SARS-CoV-2 nsp12-nsp8-nsp7 complex (RdRp) can be expressed and purified from insect cells (23). In this study, nsp12, nsp7, and nsp8 were successfully expressed and purified from Escherichia coli (Fig. 1), and the functional nsp12-nsp8-nsp7 complex was assembled by simply mixing nsp12, nsp7, and nsp8 together in vitro. nsp12 was expressed as a C-terminally His-tagged protein and purified according to the published methods, with modifications (20, 24). nsp7 and nsp8 were expressed as N-terminally His-tagged protein and purified by using nickel agarose resin. It has been reported that a preformed nsp7-nsp8 complex has a better ability to promote nsp12 polymerase activity (19); copurification of nsp7 and nsp8 was also performed by simply mixing cells expressing nsp8 and nsp7 before lysing cells, which allowed nsp7 and nsp8 to form a stable complex during purification. The term “nsp8-7” is used here to represent copurified nsp8 and nsp7. After purification, the identity of the purified proteins was confirmed by mass spectrometry analysis (Fig. S5, S6, and S7).
FIG 1.
nsp12, nsp7, nsp8, and nsp8-7 PAGE protein gel stained with Coomassie blue. nsp12mut contains an active-site mutation (active-site motif SDD changed to SAA). nsp8-7 represents copurified nsp8 and nsp7. M, protein molecular weight markers. The sizes of protein markers (in kilodaltons) are indicated on the left.
nsp12-nsp8-nsp7 complex assembly and activity measurement.
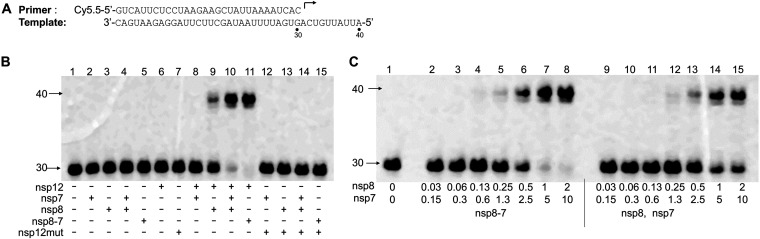
To measure the polymerase activity of the purified enzyme, a primer extension assay employing an RNA template (40-mer RNA corresponding to the sequence of the 3′ end of the SARS-CoV-2 RNA genome) and a fluorescently labeled RNA primer (30-mer) was developed (Fig. 2A). The details of the RNA primer extension method were described previously (25–27). It was reported that nsp7 and nsp8 could form a complex with nsp12, which is essential for RNA synthesis catalyzed by nsp12. In this experiment, different combinations of purified protein (nsp12, nsp7, nsp8, and nsp8-7) were used in the primer extension assay to test the RNA synthesis ability of different combinations (Fig. 2B). The result showed that nsp12, nsp7, or nsp8 alone did not have any measured RNA synthesis ability in the reaction conditions described in this experiment (Fig. 2B, lanes 2, 3, and 6). nsp12 and nsp7 together also did not have RNA polymerase activity (Fig. 2B, lane 8), and nsp12 and nsp8 together had weak RNA polymerase activity (Fig. 2B, lane 9). Maximum polymerase activity was observed in the presence of nsp12, nsp8, and nsp7 together (Fig. 2B, lanes 10 and 11). An active-site mutation (SDD to SAA) of nsp12 (nsp12mut) resulted in no RNA synthesis in the presence of nsp8 and nsp7, which confirms that RNA synthesis is mediated by nsp12. The ability of nsp8-7 to promote nsp12 polymerase was further evaluated through an enzyme dilution assay and compared with that of separately added nsp8 and nsp7 (Fig. 2C). The result showed that nsp8-7 had a better ability to promote nsp12-catalyzed RNA synthesis. nsp8-7 was used in the primer extension assays described in the report, and the nsp8 concentration in nsp8-7 was used to represent the concentration of nsp8-7.
FIG 2.
Analysis of nsp12 polymerase activity using a primer extension assay. (A) RNA primer and template used in this assay. The 30-mer primer (top) contains a fluorescent label (Cy5.5) at the 5′ end, and the arrow indicates the location and direction of primer extension to form a 40-mer product. (B) Analysis of the nsp12 polymerase activity in the presence of nsp7, nsp8, or nsp8-7 (copurified nsp8 and nsp7). The enzymes used in each reaction are indicated at the bottom of the gel. The concentrations for nsp12, nsp12mut, nsp7, nsp8, and nsp8-7 (copurified nsp8 and nsp7) are 50 nM, 50 nM, 10 μM, 2 μM, and 2 μM (2 μM nsp8, 10 μM nsp7), respectively. Different enzymes and P/T (5 nM) were incubated in reaction buffer, and the reactions were initiated by the addition of 100 μM rNTPs, continued at 37°C for 1 h, and then were stopped by addition of stopping solution. The products were separated on denaturing polyacrylamide gels. (C) Primer extension activity using nsp12 and nsp8, nsp7, or nsp8-7 (copurified). nsp12 (50 nM) was used in all the samples. Primer extension reaction was performed as described above. In lanes 2 to 8, copurified nsp8-7 was used, and in lanes 9 to 15, nsp8 and nsp7 were added to the reaction mixture separately. The final concentrations of nsp8 and nsp7 in the reaction mixtures are given under each lane in micromolar units.
Primer extension assay development and optimization.
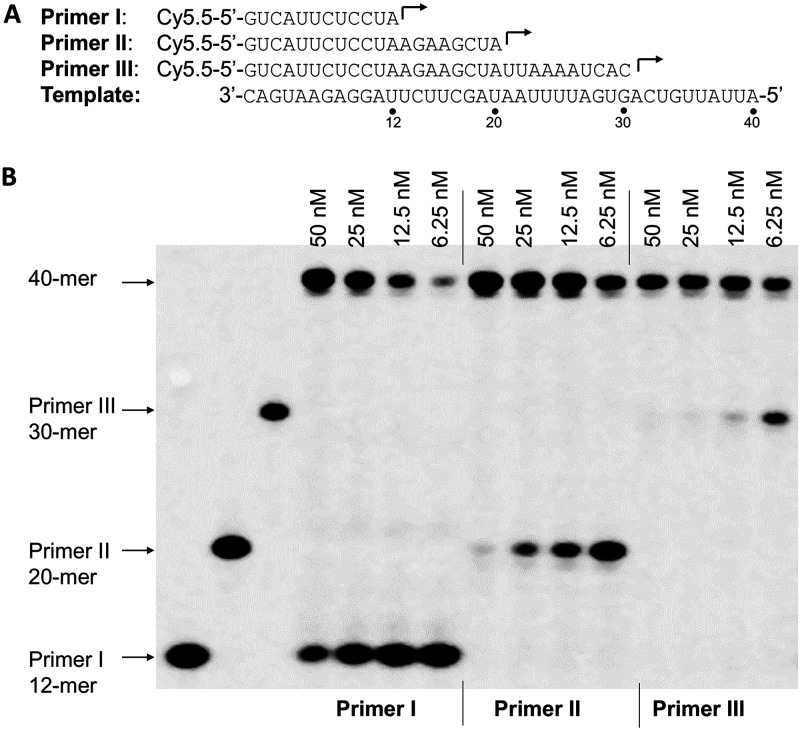
It has been shown that the primer/template (P/T) scaffold could have a major influence on the efficiency of primer extension reaction catalyzed by mitochondrial DNA-dependent RNA polymerase (26). In this study, three RNA primers (primers I, II, and III) complementary to different regions of the 3′ end of the template (40-mer) were used to form three different P/T scaffolds (Fig. 3A). The efficiency of RNA synthesis by SARS-CoV-2 RdRp using three P/T scaffolds was tested in a primer extension assay with a serial dilution of nsp12 and a constant concentration of nsp8-7 (Fig. 3B). The result showed that the SARS-CoV-2 RdRp prefers a longer primer (primer III; 30-mer) for annealing with the RNA template. As a comparison, the efficiency of RNA synthesis using three P/T scaffolds by dengue virus RdRp was similar (data not shown). Primer III (30-mer) (Fig. 3A) was used in the primer extension assays described below.
FIG 3.
Influence of P/T complexes on the efficiency of the primer extension reaction catalyzed by SARS-CoV-2 RdRp. (A) RNA primers (primers I, II, and III) and RNA template used in the experiments whose results are shown in panel B. (B) Primer extension assay using the different P/T complexes from panel A. Four nsp12 concentrations (6.25 nM, 12.5 nM, 25 nM, 50 nM; indicated above the gel), 2 μM nsp8-7 (2 μM nsp8 and 10 μM nsp7), and 5 nM concentrations of different P/T complexes (formed by annealing primer I, II, or III with the template shown in panel A) were used in the assay. The reactions were initiated by the addition of 100 μM rNTP and continued at 37°C for 1 h, and the products were separated on denaturing polyacrylamide gels. Three P/T complexes without nsp12 added were used as negative controls (left three lanes).
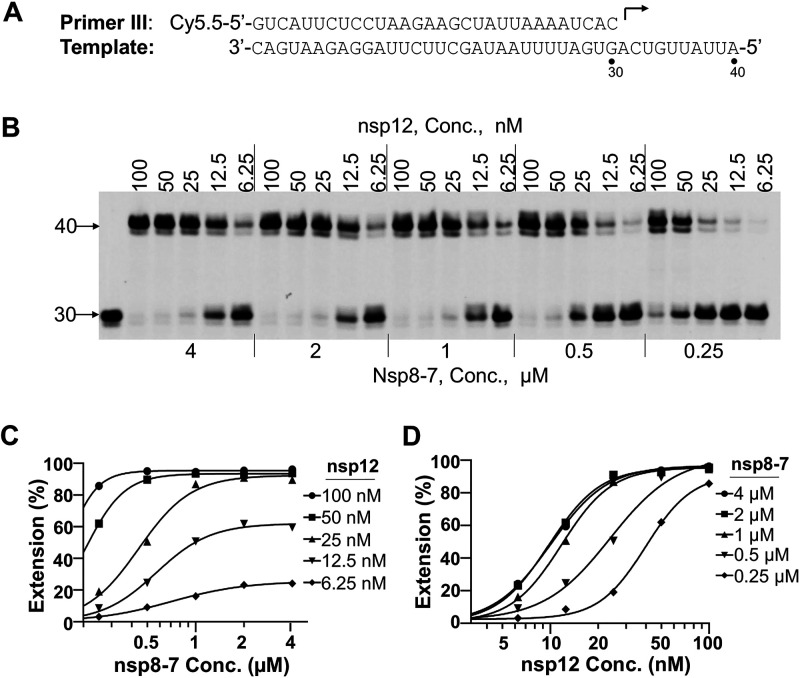
To develop a robust polymerase assay, optimal nsp8-7 and nsp12 concentrations used in the reaction should be carefully determined. An enzyme dilution assay with different concentrations of nsp8-7 and nsp12 was performed (Fig. 4). The concentration of nsp8 was used to represent the concentration of copurified nsp8-7. The result showed that the primer extension products correlated with the concentration of nsp12 (Fig. 4C), and a higher concentration of nsp8-7 had a better ability to promote nsp12 polymerase activity (Fig. 4D). In the primer extension assays described below, 50 nM nsp12 and 2 μM nsp8-7 (2 μM nsp8 and 10 μM nsp7) were used. Under this assay condition, RNA primer (5 nM) can be extended completely while consuming less nsp12 enzyme.
FIG 4.
nsp12 and nsp8-7 concentration optimization. (A) P/T complex used in this assay. (B) Analysis of the primer extension activity in the presence of different concentrations of nsp12 and nsp8-7. Serial dilutions of nsp12 (indicated above the gel), serial dilutions of nsp8-7 (indicated below the gel), and 5 nM P/T were used in this assay. Reactions were initiated by the addition of 100 μM rNTP, carried out at 37°C for 1 h, and then stopped by addition of stopping solution. The products were separated on denaturing PAGE gels. The first lane on the left represents the reaction without rNTP added, which serves as a negative control. The numbers on the left of the gels indicate the locations of the 30-mer primer and 40-mer full-length product. (C) The percentage of full-length products in panel B was plotted against the nsp8-7 concentrations. The results were fitted to sigmoidal dose-response curves using the GraphPad Prism program. (D) The percentage of full-length products in panel B was plotted against the nsp12 concentrations. The results were fitted to sigmoidal dose-response curves using the GraphPad Prism program.
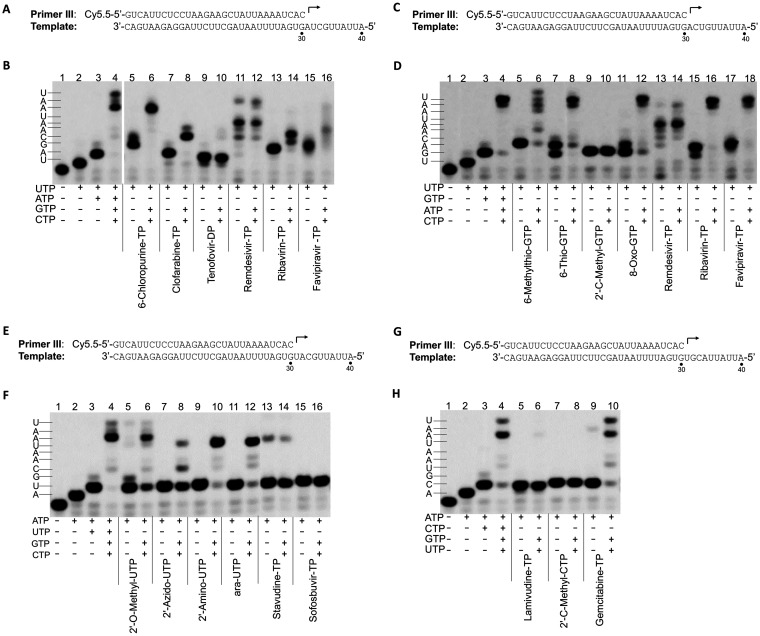
Nucleotide incorporation and chain termination.
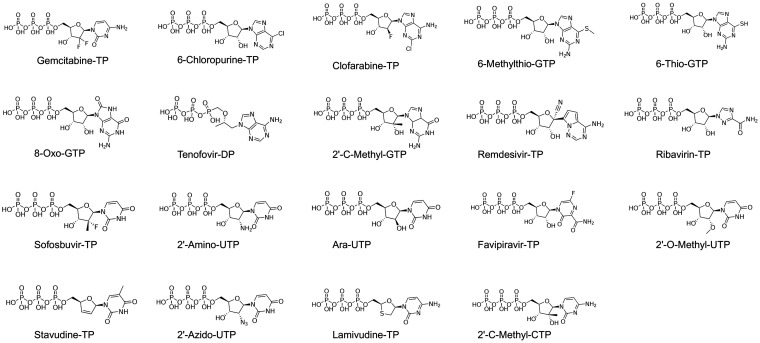
To inhibit viral RNA synthesis or disrupt viral RNA function, a nucleotide analog must be incorporated into newly synthesized viral RNA by RNA polymerase. Several different nucleotide analogs (Fig. 5) that have been used as antiviral drugs or in various applications were selected, and the utilization of those nucleotide analogs by SARS-CoV-2 RdRp and their ability to cause chain termination (one of the major mechanisms of inhibition of viral RNA polymerase) after incorporation were evaluated in the primer extension assay (Fig. 6). The results showed that all the nucleotide analogs tested can be incorporated into RNA by SARS-CoV-2 RdRp. Due to the possibility of misincorporation, the termination ability of those nucleotide analogs may need further investigation.
FIG 5.
Chemical structures of the nucleoside 5′-triphosphate analogs tested in this study. Sofosbuvir-TP and remdesivir-TP are the triphosphate derivatives of sofosbuvir and remdesivir, respectively, although sofosbuvir and remdesivir are prodrugs of active monophosphorylated nucleotides.
FIG 6.
Analysis of incorporation and chain termination effects of nucleotide analogs in primer extension assay catalyzed by SARS-CoV-2 RdRp. (A) P/T complex used in the assay whose results are shown in panel B. The first, second, third, and fourth nucleotides to be incorporated are UTP, ATP, GTP, and CTP, respectively. (B) Analysis of the incorporation and chain termination abilities of the ATP analogs. Incorporation and chain termination of tested nucleotides were measured in two separate assays. For incorporation (lanes 5, 7, 9, 11, 13, and 15), primer extension reactions were initiated by the addition of 0.1 μM UTP and 100 μM ATP analogs, as indicated under each lane. The reactions were performed at 37°C for 30 min and stopped by the addition of stopping solution. For chain termination ability (lanes 6, 8, 10, 12, 14, and 16), primer extension reactions were initiated by the addition of 0.1 μM UTP and 100 μM ATP analogs, as indicated under each lane. The reactions were performed at 37°C for 30 min, 0.1 μM GTP and 0.1 μM CTP were added to the reaction mixtures, and the reactions were continued at 37°C for another 30 min before the addition of stopping solution. The reaction products were resolved by denaturing PAGE. Analysis of incorporation and termination of GTP analogs (C and D), UTP analogs (E and F), and CTP analogs (G and H) were done by using a similar method.
Efficiency of nucleotide analog incorporation by RdRp.
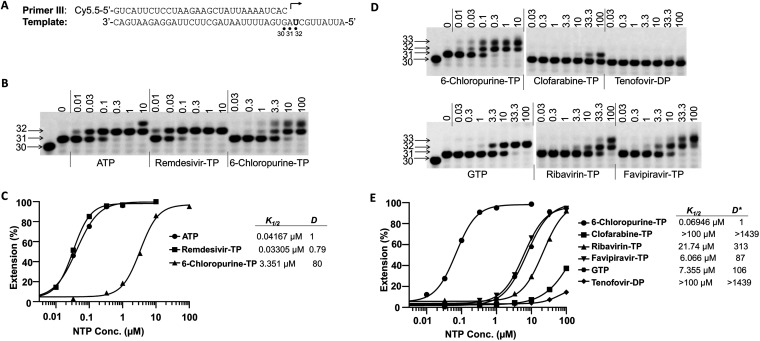
The relative efficiency of incorporation of nucleotide analogs versus natural nucleotides (discrimination value) by viral RNA-dependent RNA polymerase has been used to evaluate the antiviral potential of nucleotide analogs targeting Zika virus and dengue virus RNA-dependent RNA polymerase (25), and it has also been used to evaluate potential mitochondrial toxicity of nucleotide analogs in a primer extension assay by mitochondrial DNA-dependent RNA polymerase (26). Using a similar method, the discrimination values of nucleotide analogs, measured in the SARS-CoV-2 RdRp primer extension assay developed in this study, were employed to evaluate the relative efficiency of incorporation of nucleotide analogs by SARS-CoV-2 RdRp. An initial time course experiment showed that RNA synthesis starting from a preformed RNA/RdRp complex (31-mer) was very fast and was finished within 20 s when 100 μM ribonucleoside triphosphate (rNTP) (Fig. S1B) was used. As a comparison, it took up to 20 min to finish RNA synthesis when no preincubation of RNA and RdRp was performed (Fig. S1C). Those results suggested that the speed-limiting step of RNA synthesis in our assay is the formation of the RNA/RdRp complex.
The efficiency of nucleotide analog incorporation was measured in a single-turnover condition with preformed RNA/RdRp complex as described by Fung et al. (11). To get a quantitative measurement of K1/2 (the nucleotide concentration at which half of the 31-mer product is extended to the 32-mer product), a short reaction time (20 s) after addition of different nucleotide analogs was used. Figure 7A and B show the primer/template design and the results of testing of ATP and two ATP analogs (remdesivir-TP and 6-chloropurine-TP). The measured values of K1/2 for ATP (K1/2, ATP), remdesivir-TP (K1/2, remdesivir-TP), and 6-chloropurine-TP (K1/2, 6-chloropurine-TP) were 0.04167 μM, 0.03305 μM, and 3.351 μM, respectively, and the calculated discrimination values Dremdesivir-TP and D6-chloropurine-TP were 0.79 and 80, respectively (Fig. 7C). This result suggested that remdesivir-TP was incorporated into RNA by SARS-CoV-2 RdRp more efficiently than natural ATP. The efficiency of incorporation of other ATP analogs by SARS-CoV-2 RdRp was much lower than that of ATP. To get a quantitative measurement of K1/2 values, a longer reaction time (15 min) is needed to increase the percentage of incorporation of nucleotide analog by SARS-CoV-2 RdRp. Since the K1/2 of natural ATP is impossible to measure directly under such conditions (it is below the limit of sensitivity of the assay), the K1/2 value of 6-chloropurine-TP was used as a surrogate comparator. A similar strategy has been used to evaluate the efficiency of incorporation of the nucleotide analog by mitochondrial DNA-dependent RNA polymerase (26).
FIG 7.
Measurement of the discrimination values of ATP analogs. (A) Primer and template used to assay ATP analogs, as shown in panels B and D. (B) A representative image of the results of the analysis of Dremdesivir-TP and D6-chloropurine-TP values. nsp12 (50 nM) and nsp8-7 (2 μM) were incubated with 5 nM P/T and 0.1 μM UTP (the first nucleotide to be incorporated) in reaction buffer for 30 min at 37°C and then rapidly mixed with different concentrations (above each lane, in micromolar units) of ATP, remdesivir-TP, or 6-chloropurine-TP. The reactions were continued at 22°C for 20 s before the addition of stopping solution, and the products were resolved by denaturing PAGE. The identity of the tested nucleotide is indicated at the bottom of the gel. The locations of the 30-mer primer and the 31-mer and 32-mer (first and second nucleotide extension products, respectively) are indicated on the left of the gel. (C) Quantitative analysis of ATP, remdesivir-TP, and 6-chloropurine-TP incorporation in the assay whose results are shown in panel B. The incorporation efficiency was evaluated based on the extension of 31-mer to 32-mer products. The measured K1/2 values for ATP, remdesivir-TP, and 6-chloropurine-TP in this experiment were 0.04167 μM, 0.03305 μM, and 3.351 μM, respectively. The discrimination values were calculated as K1/2, ATP analog/K1/2, ATP and are shown on the right of the graph. (D) Representative image of the result of analysis of ATP analogs. Primer extension reactions were performed a described for panel B. After addition of different concentrations of ATP analog, the reactions were continued at 22°C for 15 min before the addition of stopping solution. (E) Quantitative analysis of ATP analogs incorporation in the assay whose results are shown in panel D. K1/2 analysis was as described for panels B and C. The discrimination between ATP analog and 6-chloropurine-TP, D*ATP analog, was calculated as K1/2, ATP analog/K1/2, 6-chloropurine, and the values are shown on the right of the graph. The discrimination between ATP analogs and natural ATP, DcalATP analog, was calculated as D*ATP analog × D6-chloropurine-TP. D6-chloropurine-TP is 78.0 ± 3.5 (average of 2 independent experiments; results of one are shown in panel C). For clofarabine-TP, ribavirin-TP, tenofovir-DP, favipiravir-TP, and GTP (misincorporated as ATP), calculated values of DcalATP analog were >112,242, 24,999 ± 828, >112,242, 7,343 ± 752, and 8,683 ± 600, respectively (averages from 2 independent experiments; results of one are shown here).
Figure 7D and E show the testing of ATP analogs and discrimination value calculation. In this assay, the K1/2 values of several ATP analogs were measured and compared to the K1/2 value of 6-chloropurine-TP, which was used as a reference to calculate the D*ATP analog value (where D*ATP analog = K1/2, ATP analog/K1/2, 6-chloropurine-TP). The discrimination values of the different ATP analogs are summarized in Table 1. Dcal is a calculated discrimination value obtained using the equation DcalATP analog = D*ATP analog × D6-chloropurine. Dcal represents the discrimination of incorporation by RdRp between natural ATP and a tested ATP analog. Misincorporation of GTP base-pairing with uridine in the template was also measured, which can be used as a guideline to evaluate the possibility of nucleotide analog incorporation in the cell. If the incorporation efficiency of an ATP analog is lower than GTP misincorporation efficiency, it probably has less chance to be incorporated in the cell, unless very high intracellular nucleotide analog concentrations can be reached.
TABLE 1.
Discrimination values of nucleotide analogs by SARS-CoV-2 RdRp
| Nucleotide analog | Value (mean ± SD)a |
|
|---|---|---|
| D*analog | Dcalanalog | |
| ATP analogs | ||
| Remdesivir-TP | 0.78 ± 0.02 | |
| 6-Chloropurine-TP | 1 | 78.0 ± 3.5 |
| Clofarabine-TP | >1,439 | >112,242 |
| Ribavirin-TP | 320.5 ± 10.6 | 24,999 ± 828 |
| Tenofovir-DP | >1,439 | >112,242 |
| Favipiravir-TP | 94.1 ± 9.6 | 7,343 ± 752 |
| GTP | 111.3 ± 7.7 | 8,683 ± 600 |
| GTP analogs | ||
| 6-Thio-GTP | 10.6 ± 1.1 | |
| 2′-C-Methyl-GTP | 1 | 61.7 ± 9.0 |
| 6-Methylthio-GTP | 73.2 ± 14.3 | 4,516 ± 884 |
| Oxo-GTP | 37.6 ± 9.7 | 2,317 ± 600 |
| Remdesivir-TP | 1,786.4 ± 173.8 | 110,222 ± 10,726 |
| Favipiravir-TP | >2,319 | >143,089 |
| Ribavirin-TP | >2,319 | >143,089 |
| ATP | 55.5 ± 12.9 | 3,427 ± 797 |
| UTP analogs | ||
| 2′-Amino-UTP | 1 | 75 ± 5 |
| 2′-O-Methyl-UTP | 102.5 ± 5.1 | 7,688 ± 380 |
| 2′-Azido-UTP | 11.1 ± 0.5 | 831 ± 35 |
| ara-UTP | 8.3 ± 0.5 | 625 ± 40 |
| Stavudine-TP | 54.8 ± 6.7 | 4,109 ± 502 |
| Sofosbuvir-TP | 97.4 ± 4.9 | 7,307 ± 368 |
| Ribavirin-TP | 825.8 ± 131.2 | 61,933 ± 9,843 |
| CTP | 68.4 ± 8.6 | 5,131 ± 645 |
D*ATP analog = K1/2 ATP analog/K1/2, 6-chloropurine-TP; DcalATP analog = D*ATP analog × D6-chloropurine-TP; D*GTP analog = K1/2 GTP analog/K1/2, 2′-C-methyl-GTP; DcalGTP analog = D*GTP analog × D2′-C-methyl-GTP; D*UTP analog = K1/2 UTP analog/K1/2, 2′-amino-UTP; DcalUTP analog = D*UTP analog × D2′-amino-UTP. Data are from two independent experiments.
As shown in Table 1, Dcal values for remdesivir-TP, 6-chloropurine-TP, clofarabine-TP, ribavirin-TP, favipiravir-TP, tenofovir-DT, and GTP were 0.78 ± 0.02, 78.0 ± 3.5, >112,242, 24,999 ± 828, 7,343 ± 752, >112,242, and 8,683 ± 600, respectively. Based on comparison with DcalGTP (natural GTP misincorporation as ATP), remdesivir-TP and 6-chloropurine-TP can be incorporated into RNA very efficiently by RdRp; ribavirin-TP and favipiravir-TP showed less ability to be incorporated into RNA by RdRp. Only a small fraction of primer was extended in the presence of clofarabine-TP and tenofovir-DP in the primer extension conditions used in this assay, which suggested that the incorporation efficiency of those two nucleotides is very low. Using a similar strategy, several GTP analogs (Fig. S2) and UTP analogs (Fig. S3) were also tested, and the data are summarized in Table 1. For GTP analogs, 6-thio-GTP and 2′-C-methyl-GTP can be incorporated into RNA very efficiently; the incorporation efficiencies of 6-methylthio-GTP, oxo-GTP, remdesivir-TP, ribavirin-TP, and favipiravir-TP are lower than or close to the misincorporation of ATP base-pairing with cytidine in the template, which suggested that they may not be incorporated into RNA as GTP analogs in the cell. For UTP analogs, 2′-amino-UTP, 2′-azido-UTP and ara-UTP showed higher incorporation efficiency; incorporation efficiency for stavudine-TP, 2′-O-methyl-UTP, sofosbuvir-TP, and ribavirin-TP was very low. Remdesivir-TP and favipiravir-TP were also tested as UTP analogs, and no incorporation was observed under the conditions used in this assay (15 min incubation).
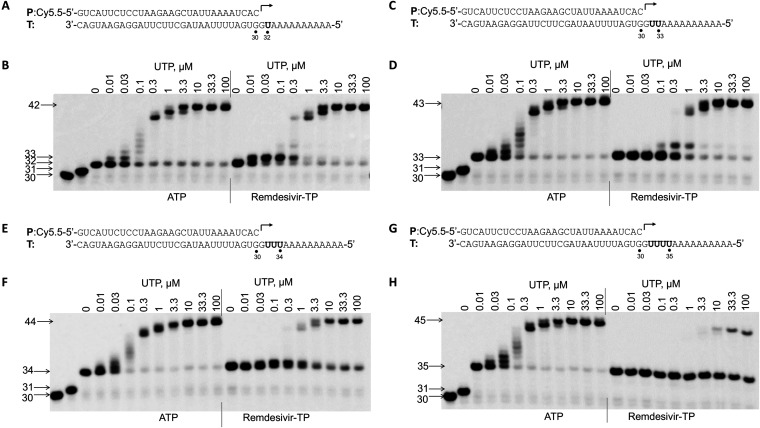
Influence of remdesivir-TP incorporation to RNA synthesis.
It was shown previously that remdesivir-TP could be incorporated into RNA, and this incorporation caused delayed chain termination (23). In this study, the influence of remdesivir-TP incorporation on subsequent nucleotide incorporation during RNA synthesis was tested in a primer extension assay (Fig. 8). To rule out the possibility of premature RNA synthesis caused by RNA sequence variation, a poly(A) sequence was used downstream of uridine (remdesivir-TP incorporation site) in the template (Fig. 8A, C, E, and G). In this study, 4 different templates (shown at the top of each gel image in Fig. 8) were used to test the influence of single, double, triple, and quadruple incorporations of remdesivir-TP on subsequent nucleotide incorporation. After incorporation of ATP or remdesivir-TP, different concentrations of UTP were added to test the RNA synthesis by RdRp. Compared with ATP, single incorporation of remdesivir-TP did not lead to chain termination (Fig. 8B). Interestingly, the incorporation efficiency of UTP after single remdesivir-TP incorporation was increased, as evidenced by a stronger 33-mer band at a low concentration of UTP in the presence of remdesivir-TP compared with that in the presence of ATP (Fig. 8B). Figure 8C and D show that double incorporation of remdesivir-TP also did not lead to chain termination. Figure 8E and F show that triple incorporation of remdesivir-TP did decrease the RNA synthesis efficiency after remdesivir-TP incorporation, as evidenced by the 34-mer band persisting at a high concentration of UTP (Fig. 8F), which suggests a partial termination caused by triple incorporation of remdesivir-TP. Figure 8G and H show that quadruple incorporation of remdesivir-TP greatly decreased the RNA synthesis (Fig. 8H, 35-mer band), which suggests a strong termination effect caused by quadruple incorporation of remdesivir-TP.
FIG 8.
Influence of remdesivir-TP incorporation on incorporation of the next nucleotide. (A) P/T complex used in the experiment whose result is shown in panel B. The bold “U” in the template indicates the position base-pairing with ATP or remdesivir-TP. (B) Influence of single remdesivir-TP incorporation on incorporation of the next nucleotide. nsp12 (50 nM) and nsp8-7 (2 μM) were incubated with 5 nM P/T (A), 0.1 μM CTP (the first nucleotide to be incorporated), and 0.1 μM ATP or remdesivir-TP (the second nucleotide to be incorporated) in reaction buffer for 30 min at 37°C and then rapidly mixed with different concentrations (indicated above the gel, in micromolar units) of UTP. The reactions were continued at 22°C for 20 s before the addition of stopping solution, and the products were resolved by denaturing PAGE. The identity of the tested nucleotide (ATP or remdesivir-TP) is indicated at the bottom of the gel. The locations of the 30-mer primer and the 31-mer (CTP incorporation), 32-mer (single ATP or remdesivir-TP incorporation), and 42-mer (full-length extension products) are indicated on the left of the gel. The influence of double (C and D), triple (E and F), and quadruple (G and H) incorporations of remdesivir-TP on incorporation of the subsequent nucleotide was analyzed using a similar method.
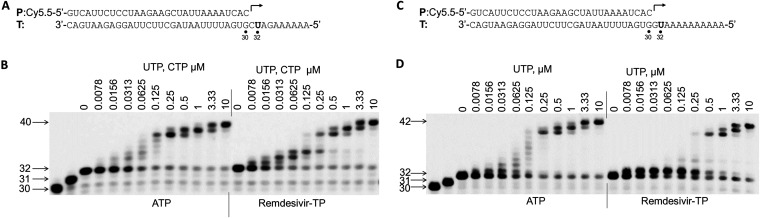
In a paper published by Gordon et al. (23), it was shown that incorporation of remdesivir-TP at position i caused termination of RNA synthesis at position i + 3. Our results shown in Fig. 8B suggest a partial termination at position i + 1 after remdesivir-TP incorporation, as evidenced by the persistence of 33-mer band, and this termination effect can be overcome by higher concentrations of UTP (the next nucleotide to be incorporated). Since the template RNA sequence may have some influence on RNA synthesis, a template having sequence downstream of the RNA synthesis initiation site similar to the template used by Gordon et al. (23) was utilized (Fig. 9A). With this template, a strong termination at position i + 3 was observed after remdesivir-TP incorporation, and this termination can be overcome by higher concentrations of the next nucleotide to be incorporated (Fig. 9B).
FIG 9.
Influence of single remdesivir-TP incorporation on incorporation of the next nucleotide in the presence of different templates. (A) P/T complex used in the experiment whose results are shown in panel B. (B) Influence of single remdesivir-TP incorporation on incorporation of the next nucleotide. nsp12 (50 nM) and nsp8-7 (2 μM) were incubated with 5 nM P/T complex (A), 0.1 μM GTP (the first nucleotide to be incorporated), and 0.1 μM ATP or remdesivir-TP (the second nucleotide to be incorporated) in reaction buffer for 30 min at 37°C and then rapidly mixed with different concentrations (indicated above the gel, in micromolar units) of both UTP and CTP. The reactions were continued at 22°C for 20 s before the addition of stopping solution, and the products were resolved by denaturing PAGE. The identity of the tested nucleotide (ATP or remdesivir-TP) is indicated at the bottom of the gel. The locations of the 30-mer primer and the 31-mer (GTP incorporation), 32-mer (single ATP or remdesivir-TP incorporation), and 40-mer (full-length extension products) are indicated on the left of the gel. (C and D) Influence of single remdesivir-TP incorporation on incorporation of the next nucleotide on a different template was analyzed using the method described for Fig. 8.
DISCUSSION
In this study, SARS-CoV-2 nsp12, nsp8, and nsp7 were successfully constructed, expressed, and purified from E. coli. The nsp12-nsp8-nsp7 complex (RdRp) was assembled in vitro and showed robust RNA synthesis activity. Using purified RdRp, we developed an in vitro nonradioactive primer extension assay and demonstrated that it can be used as a tool to identify nucleotide analog substrates which could be developed into antiviral drugs against SARS-CoV-2. The primer extension assay described in this report can also be used to develop a high-throughput screen assay based on the detection of PPi released from the polymerase reaction to identify nonnucleotide analog polymerase inhibitors against SARS-CoV-2 (25).
It has been shown that many nucleotide analogs currently used as antiviral drugs can be incorporated into RNA by SARS-CoV-2 RNA polymerase (28, 29), which suggests that they have the potential to be developed into antiviral drugs against SARS-CoV-2. As an alternate substrate of RNA polymerase, a nucleotide analog must compete with natural rNTP for incorporation into viral RNA. The relative incorporation efficiency of a nucleotide analog versus natural rNTP (discrimination value) is an important criterion in evaluating the antiviral potential of nucleotide analogs. Like other studies (23), our data showed that remdesivir-TP can be incorporated into RNA as an ATP analog by SARS-CoV-2 RdRp, and the incorporation efficiency is higher than that of natural ATP (Dremdesivir-TP = 0.78 ± 0.02). We also tested remdesivir-TP against C, A, and G in the template, and the result showed that remdesivir-TP can be incorporated as a GTP analog (Fig. S2) with low efficiency and cannot be incorporated as a UTP or CTP analog (Fig. S3 and S4). The efficiency of incorporation of ribavirin-TP and favipiravir-TP as either ATP or GTP analogs measured in our study is very low (even lower than that of GTP or ATP misincorporation), which is in disagreement with the studies done by Ferron et al. (16) and Shannon et al. (17). In those studies, ribavirin-TP and favipiravir-TP were readily incorporated into RNA by SARS-CoV RdRp. One possible explanation for this discrepancy is the stability of the ribavirin-TP and favipiravir-TP used in our assay. Further studies are needed to verify the concentration of ribavirin-TP and favipiravir-TP used in our assay. Sofosbuvir, an approved anti-hepatitis C virus (HCV) drug, has been proposed to be used for treating SARS-CoV-2. Our data show that sofosbuvir-TP can be incorporated into RNA by SARS-CoV-2 RdRp, but the incorporation efficiency is very low, and it probably cannot compete with natural UTP for incorporation by SARS-CoV-2 RdRp in the cell.
Remdesivir has been used as an antiviral drug against SARS-CoV-2, and a number of studies have been performed to study the antiviral mechanism of action of remdesivir (23, 30, 31). It has been shown that incorporation of remdesivir-TP causes delayed chain termination, which in turn blocks viral RNA synthesis. The influence of remdesivir-TP incorporation on RNA synthesis catalyzed by SARS-CoV-2 RdRp was studied using a primer extension assay. Our data showed that single or double incorporation of remdesivir-TP did not lead to immediate chain termination. Quadruple incorporation of remdesivir-TP caused a strong termination. Similar to the data presented by Gordon et al. (23), delayed chain termination was observed due to the incorporation of remdesivir-TP in our assay, but the delayed chain termination pattern is different when templates with different sequences are used. Our data showed that the incorporation efficiency of UTP at position i + 1 after remdesivir-TP incorporation was increased greatly (Fig. 9), which suggested that incorporation of remdesivir-TP may have some influence on the kinetics of subsequent nucleotide incorporation, besides causing delayed chain termination.
Since our data showed that remdesivir-TP can be incorporated into RNA as an ATP analog and a GTP analog (Fig. S2) but cannot be incorporated into RNA as a CTP analog (Fig. S4) or a UTP analog (Fig. S3), the incorporation pattern of remdesivir-TP shown in Fig. 6B, lane 11, and Fig. 6D, lane 13, may suggest that a significant U-G mismatch occurs following remdesivir-TP incorporation. Further studies are needed to understand the influence of remdesivir-TP on the kinetics and fidelity of subsequent nucleotide incorporation and RNA synthesis.
MATERIALS AND METHODS
Chemicals.
ATP, UTP, CTP, and GTP were purchased as 100 mM solutions from Thermo Fisher Scientific (Massachusetts, USA). Urea, taurine, dithiothreitol (DTT), MgCl2, imidazole, and isopropyl-β-d-thiogalactopyranoside (IPTG) were purchased from Bidepharm (Shanghai, China). LB medium, NaCl, and HisPur Ni-nitrilotriacetic acid (NTA) agarose resin were purchased from Thermo Fisher Scientific (Massachusetts, USA). Fluorescently labeled RNA oligonucleotides, as well as unlabeled RNA oligonucleotides, were chemically synthesized and purified by high-performance liquid chromatography (HPLC) by GenScript (Nanjing, China). Stellar competent cells were purchased from TaKaRa (Mountain View, CA, USA). BL21(DE3) cells were purchased from TransGen Biotech (Beijing, China). The pMal-c5X vector was purchased from New England Bioscience (NEB, Ipswich, MA). Tenofovir-DP, 2′-C-methyl-CTP, and 2′-C-methyl-GTP were purchased from Carbosynth (San Diego, CA). 6-Chloropurine-TP, gemcitabine-TP, 6-methyl-thio-GTP, clofarabine-TP, 8-oxo-GTP, 6-thio-GTP, stavudine-TP, and lamivudine-TP were purchased from Jena Bioscience (Jena, Germany). Ribavirin-TP was purchased from Santa Cruz Biotechnology (TX, USA). Favipiravir-TP was purchased from Toronto Research Chemicals (TRC, Toronto, Canada). 2′-Amino-UTP, 2′-O-methyl-UTP, ara-UTP, and 2′-azido-UTP were purchased from Trilink BioTechnologies (San Diego, CA, USA). Sofosbuvir-TP was purchased from MedChemExpress (MCE, New Jersey, USA). Remdesivir-TP was custom synthesized at Wuxi AppTech (Shanghai, China).
nsp12 protein expression and purification.
The SARS-CoV-2 nsp12 gene (corresponding to amino acids 4393 to 5324; UniProt code P0DTD1) was synthesized de novo by GenScript (Nanjing, China) and constructed on the pET22b vector between the NdeI and XhoI sites. nsp12 was expressed with a C-terminal 10-His tag in BL21(DE3) cells at 16°C; 2 mM MgCl2 and 50 μM ZnSO4 were used to supplement the culture during induction. After overnight cultivation, cells were harvested and lysed with a high-pressure homogenizer in buffer containing 25 mM HEPES (pH 7.5), 150 mM NaCl, 4 mM MgCl2, 50 μM ZnSO4, 10% glycerol, 2.5 mM DTT, and 20 mM imidazole. Cell debris was removed by centrifugation at 13,000 rpm. nsp12 was then purified by nickel-affinity chromatography followed by ion-exchange chromatography (using HisTrap FF and Capto HiResQ 5/50 columns, respectively; GE Healthcare, USA). nsp12 eluates with a conductivity of 19 to 23 mS/cm were combined and injected onto a Superdex 200 Increase 10/300 GL column (GE Healthcare, USA) in a buffer containing 25 mM HEPES (pH 7.5), 250 mM NaCl, 1 mM MgCl2, 1 mM tris(2-carboxyethyl)phosphine, and 10% glycerol. Peak fractions were combined, concentrated to 10 μM, and stored at −80°C before enzymatic assay. The nsp12 loss-of-function mutant (SDD-to-SAA mutation; amino acids 5151 to 5153; UniProt code P0DTD1) was prepared in an identical process. Protein identification by mass spectrometry was performed by Biotech-Pack Scientific (Beijing, China).
nsp7 and nsp8 protein expression and purification.
SARS-CoV-2 nsp8 gene (nucleotides 12092 to 12685; strain name, Wuhan-Hu-1; GenBank no. MN908947.3) was synthesized de novo by GenScript (Nanjing, China) and cloned into a pMal-c5X vector under tac promoter control (without a maltose-binding protein [MBP] sequence). Specifically, the entire MBP sequence of pMal-c5X (nucleotides 1527 to 2628) was replaced with the nsp7 gene sequence with the addition of an N-terminal 6-His tag. The SARS-CoV-2 nsp7 gene (nucleotides 11846 to 12091; GenBank no. MN908947.3) was synthesized de novo by GenScript (Nanjing, China) and cloned into the pMal-c5X vector in the same way as nsp8. The expression plasmids were transformed into Stellar competent cells. Protein expression was induced at 16°C overnight night by addition of 0.3 mM IPTG. Cells were harvested, and cell pellets were resuspended in cell lysis buffer (20 mM HEPES [pH 7.5], 10% glycerol, 100 mM NaCl, 0.05% Tween 20, 10 mM DTT, 1 mM MgCl2, 20 mM imidazole, 1× protease inhibitor cocktail). Cell disruption was performed at 4°C for 10 min using a high-pressure homogenizer. The cell extract was clarified by centrifugation at 12,000 rpm for 10 min at 4°C. Individual nsp7 and nsp8 were purified by HisPur Ni-NTA agarose resin separately, and the enzymes were eluted from the resin with elution buffer (20 mM HEPES [pH 7.5], 50 mM NaCl, 300 mM imidazole, 10 mM DTT, 0.01% Tween 20). The eluted enzymes were adjusted to 40% glycerol and stored at −80°C. For copurification of nsp8 and nsp7, nsp8 and nsp7 were expressed separately, and the expression cells were mixed before cell lysis. Protein purification was the same as for individual nsp7 and nsp8 proteins. Under this condition, nsp8 and nsp7 were purified together at once. Protein identification by mass spectrometry was performed by Biotech-Pack Scientific (Beijing, China).
Primer and template annealing.
To generate RNA primer-template complexes, 1 μM fluorescently (Cy5.5) labeled RNA primer and 5 μM unlabeled RNA template were mixed in 50 mM NaCl in deionized water, incubated at 98°C for 10 min, and then slowly cooled to room temperature. The annealed primer-template (P/T) complexes were stored at −20°C before use in the primer extension assay.
Primer extension assay.
The ability of RNA synthesis by purified polymerase was determined in a primer extension reaction using P/T complexes prepared by annealing Cy5.5-labeled RNA primer and unlabeled RNA template (described above). A typical primer extension reaction was performed in a 10-μl reaction mixture containing reaction buffer (20 mM HEPES [pH 7.5], 5 mM MgCl2, 10 mM DTT, 0.01% Tween 20), 5 nM P/T, 50 nM nsp12, and 2 μM nsp8-7 (copurified protein mixture containing 2 μM nsp8 and 10 μM nsp7) unless otherwise specified. The reaction was initiated by the addition of rNTPs at a final concentration of 100 μM, unless otherwise specified, followed by incubation for 1 h at 37°C. The reactions were quenched by the addition of 20 μl stopping solution (8 M urea, 90 mM Tris base, 29 mM taurine, 10 mM EDTA, 0.02% SDS, 0.1% bromophenol blue). The quenched samples were denatured at 95°C for 10 min, and the primer extension products were separated using 10% denaturing polyacrylamide gel electrophoresis (urea-PAGE) in 1× TTE buffer (90 mM Tris base, 29 mM taurine, 0.5 mM EDTA). After electrophoresis, the gels were scanned using an Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, NE). The images were analyzed, and the proper RNA bands were quantified using Image Studio Lite (version 5.2; LI-COR Bioscience, Lincoln, NE). Data were analyzed using GraphPad Prism 7.
Analysis of chain termination ability of nucleotide analogs.
Primer extension reactions were performed as described above. Incorporation and chain termination of tested nucleotides were measured in two separate assays. For the nucleotide analog incorporation assay, P/T complexes (5 nM) and the nsp12-nsp8-nsp7 complex (RdRp) (50 nM nsp12, 2 μM nsp8-7) were incubated with a natural rNTP (the first nucleotide to be incorporated) and tested nucleotide analogs (the second nucleotide to be incorporated), and the reactions were continued at 37°C for 30 min before the addition of stopping solution. For the chain termination assay, nucleotide analogs were incorporated as described above. Then, two natural rNTPs (the third and fourth nucleotides to be incorporated) were added to the reaction mixture, and reactions were continued at 37°C for another 30 min before the addition of stopping solution. The quenched samples were heated at 95°C for 10 min and analyzed by denaturing urea-PAGE as described above. The concentration and identity of nucleotides added for each reaction are described in the legend to Fig. 6.
Measurement of nucleotide analog incorporation efficiency.
Different P/T complexes were designed to test individual analogs using the method described previously (25, 26). To perform the reaction, 5 nM P/T, 50 nM nsp12, and 2 μM nsp8-7 were incubated in reaction buffer in the presence of a 0.1 μM concentration of the first natural ribonucleotide for 30 min at 37°C, and then different concentrations of the nucleotide analogs to be tested were added to the reaction mixtures. The reactions were continued at 22°C for the times indicated in each figure legend and subsequently quenched and analyzed by urea-PAGE as described above. After electrophoresis, the gels were scanned using the Odyssey infrared imaging system. The intensities of the different RNA bands were quantified using Image Studio Lite. The incorporation efficiencies of the different nucleotide analogs were evaluated by measurement of the K1/2 values (the analog triphosphate concentrations resulting in 50% product extension) and the corresponding discrimination values (Danalog, defined as K1/2, analog/K1/2, natural nucleotide when both were measured under the same assay condition), as previously described (11, 25, 26).
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the Bill & Melinda Gates Foundation, Tsinghua University, and Beijing Municipal Government.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N. 2020. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. 2020. Coronavirus disease 2019 (COVID-19): situation report, 88. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 3.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W. 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Y, Liu Q, Guo D. 2020. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol 92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jordheim LP, Durantel D, Zoulim F, Dumontet C. 2013. Advances in the development of nucleoside and nucleotide analogues for cancer and viral diseases. Nat Rev Drug Discov 12:447–464. doi: 10.1038/nrd4010. [DOI] [PubMed] [Google Scholar]
- 6.Quan DJ, Peters MG. 2004. Antiviral therapy: nucleotide and nucleoside analogs. Clin Liver Dis 8:371–385. doi: 10.1016/j.cld.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 7.Seley-Radtke K, Deval J. 2018. Advances in antiviral nucleoside analogues and their prodrugs. SAGE Publications Sage UK, London, England. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pastuch-Gawołek G, Gillner D, Król E, Walczak K, Wandzik I. 2019. Selected nucleos(t)ide-based prescribed drugs and their multi-target activity. Eur J Pharmacol 865:172747. doi: 10.1016/j.ejphar.2019.172747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pruijssers AJ, Denison MR. 2019. Nucleoside analogues for the treatment of coronavirus infections. Curr Opin Virol 35:57–62. doi: 10.1016/j.coviro.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H, Zhou Y, Zhang M, Wang H, Zhao Q, Liu J. 2020. Updated approaches against SARS-CoV-2. Antimicrob Agents Chemother 64:e00483-20. doi: 10.1128/AAC.00483-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fung A, Jin Z, Dyatkina N, Wang G, Beigelman L, Deval J. 2014. Efficiency of incorporation and chain termination determines the inhibition potency of 2′-modified nucleotide analogs against hepatitis C virus polymerase. Antimicrob Agents Chemother 58:3636–3645. doi: 10.1128/AAC.02666-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Clercq E, Neyts J. 2009. Antiviral agents acting as DNA or RNA chain terminators, p 53–84. In Krausslick HG, Bartenschlager R (ed), Antiviral strategies. Springer, New York, NY. [DOI] [PubMed] [Google Scholar]
- 13.Arias A, Thorne L, Goodfellow I. 2014. Favipiravir elicits antiviral mutagenesis during virus replication in vivo. Elife 3:e03679. doi: 10.7554/eLife.03679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baranovich T, Wong S-S, Armstrong J, Marjuki H, Webby RJ, Webster RG, Govorkova EA. 2013. T-705 (favipiravir) induces lethal mutagenesis in influenza A H1N1 viruses in vitro. J Virol 87:3741–3751. doi: 10.1128/JVI.02346-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin Z, Smith LK, Rajwanshi VK, Kim B, Deval J. 2013. The ambiguous base-pairing and high substrate efficiency of T-705 (favipiravir) ribofuranosyl 5′-triphosphate towards influenza A virus polymerase. PLoS One 8:e68347. doi: 10.1371/journal.pone.0068347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferron F, Subissi L, Silveira De Morais AT, Le NTT, Sevajol M, Gluais L, Decroly E, Vonrhein C, Bricogne G, Canard B, Imbert I. 2018. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc Natl Acad Sci U S A 115:E162–E171. doi: 10.1073/pnas.1718806115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shannon A, Selisko B, Le NTT, Huchting J, Touret F, Piorkowski G, Fattorini V, Ferron F, Decroly E, Meier C, Coutard B, Peersen O, Canard B. 2020. Favipiravir strikes the SARS-CoV-2 at its Achilles heel, the RNA polymerase. bioRxiv. doi: 10.1101/2020.05.15.098731. [DOI] [PMC free article] [PubMed]
- 18.Brian D, Baric R. 2005. Coronavirus genome structure and replication, Curr Top Microbiol Immunol 287:1–30. doi: 10.1007/3-540-26765-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subissi L, Posthuma CC, Collet A, Zevenhoven-Dobbe JC, Gorbalenya AE, Decroly E, Snijder EJ, Canard B, Imbert I. 2014. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc Natl Acad Sci U S A 111:E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao Y, Yan L, Huang Y, Liu F, Zhao Y, Cao L, Wang T, Sun Q, Ming Z, Zhang L, Ge J, Zheng L, Zhang Y, Wang H, Zhu Y, Zhu C, Hu T, Hua T, Zhang B, Yang X, Li J, Yang H, Liu Z, Xu W, Guddat LW, Wang Q, Lou Z, Rao Z. 2020. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science 368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bouvet M, Imbert I, Subissi L, Gluais L, Canard B, Decroly E. 2012. RNA 3'-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proc Natl Acad Sci U S A 109:9372–9377. doi: 10.1073/pnas.1201130109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Minskaia E, Hertzig T, Gorbalenya AE, Campanacci V, Cambillau C, Canard B, Ziebuhr J. 2006. Discovery of an RNA virus 3′→ 5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc Natl Acad Sci U S A 103:5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gordon CJ, Tchesnokov EP, Woolner E, Perry JK, Feng JY, Porter DP, Götte M. 2020. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J Biol Chem 295:6785–6797. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirchdoerfer RN, Ward AB. 2019. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat Commun 10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu G, Bluemling GR, Collop P, Hager M, Kuiper D, Gurale BP, Painter GR, De La Rosa A, Kolykhalov AA. 2017. Analysis of ribonucleotide 5′-triphosphate analogs as potential inhibitors of Zika virus RNA-dependent RNA polymerase by using nonradioactive polymerase assays. Antimicrob Agents Chemother 61:e01967-16. doi: 10.1128/AAC.01967-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu G, Bluemling GR, Mao S, Hager M, Gurale BP, Collop P, Kuiper D, Sana K, Painter GR, De La Rosa A, Kolykhalov AA. 2017. Simple in vitro assay to evaluate the incorporation efficiency of ribonucleotide analog 5′-triphosphates into RNA by human mitochondrial DNA-dependent RNA polymerase. Antimicrob Agents Chemother 62:e01830-17. doi: 10.1128/AAC.01830-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sticher ZM, Lu G, Mitchell DG, Marlow J, Moellering L, Bluemling GR, Guthrie DB, Natchus MG, Painter GR, Kolykhalov AA. 2020. Analysis of the potential for N4-hydroxycytidine to inhibit mitochondrial replication and function. Antimicrob Agents Chemother 64:e01719-19. doi: 10.1128/AAC.01719-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jockusch S, Tao C, Li X, Anderson TK, Chien M, Kumar S, Russo JJ, Kirchdoerfer RN, Ju J. 2020. A library of nucleotide analogues terminate RNA synthesis catalyzed by polymerases of coronaviruses causing SARS and COVID-19. bioRxiv. doi: 10.1101/2020.04.23.058776. [DOI] [PMC free article] [PubMed]
- 29.Chien M, Anderson TK, Jockusch S, Tao C, Kumar S, Li X, Russo JJ, Kirchdoerfer RN, Ju J. 2020. Nucleotide analogues as inhibitors of SARS-CoV-2 polymerase. bioRxiv. doi: 10.1101/2020.03.18.997585. [DOI] [PMC free article] [PubMed]
- 30.Gordon CJ, Tchesnokov EP, Feng JY, Porter DP, Götte M. 2020. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J Biol Chem 295:4773–4779. doi: 10.1074/jbc.AC120.013056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tchesnokov EP, Feng JY, Porter DP, Götte M. 2019. Mechanism of inhibition of Ebola virus RNA-dependent RNA polymerase by remdesivir. Viruses 11:326. doi: 10.3390/v11040326. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.