Abstract
In ribosomopathies, the Diamond-Blackfan anemia (DBA) or 5q- syndrome, ribosomal protein (RP) genes are affected by mutation or deletion, resulting in bone marrow erythroid hypoplasia. Unbalanced production of ribosomal subunits leading to a limited ribosome cellular content regulates translation at the expense of the master erythroid transcription factor GATA1. In RPS14-deficient cells mimicking 5q- syndrome erythroid defects, we show that the transcript length, codon bias of the coding sequence (CDS) and 3’UTR (untranslated region) structure are the key determinants of translation. In these cells, short transcripts with a structured 3’UTR and high codon adaptation index (CAI) showed a decreased translation efficiency. Quantitative analysis of the whole proteome confirmed that the post-transcriptional changes depended on the transcript characteristics that governed the translation efficiency in conditions of low ribosome availability. In addition, proteins involved in normal erythroid differentiation share most determinants of translation selectivity. Our findings thus indicate that impaired erythroid maturation due to 5q- syndrome may proceed from a translational selectivity at the expense of the erythroid differentiation program, and suggest that an interplay between the CDS and UTR may regulate mRNA translation.
Introduction
A haploinsufficiency or mutation of ribosomal protein (RP) genes causes alterations in the ribosome’s translation capacity; these disorders are known as “ribosomopathies”. Diamond-Blackfan anemias (DBA) are provoked by heterozygous lossof- function mutations in one of 18 different RP genes,1whilst the haploinsufficiency of the RPS14 gene accounts for the erythroid phenotype in 5q- syndrome.2,3 In these diseases, the bone marrow (BM) erythroid lineage is commonly hypoplastic and this has prompted the search for the mechanisms underlying specific dysregulation of translation. Several models have been proposed to explain the observed phenotypes in ribosomopathies.4 In one model, changes in the cellular ribosome concentration relative to mRNA levels may cause changes in the translation efficiency of different classes of transcripts due to the competition for ribosomes among the cellular mRNA content.4 In a second model, specialized ribosomes with a modified RP composition, or with RP or rRNA modifications, could be responsible for changes in the interactions with mRNAs and tissue-selective translation. 5 In a third model, the erythroid transcription factor GATA1 is selectively targeted by translation impairment in DBA or post-translational cleavage by caspase in 5qsyndrome. 6,7 DBA and 5q- myelodysplastic syndromes (MDS) have insufficient globin production leading to excess of free heme, accumulation of reactive oxygen species and cell death.8 Furthermore, free heme stops GATA1 synthesis and its suppression of the heme-regulated inhibitor (HRI) activity and subsequent eIF2a phosphorylation is inefficient to rescue globin translation.9,10
Cellular models of DBA that were developed by expressing a shRNA to RPS19 and of somatic 5q- syndrome through the same silencing mechanism for RPS146,11,12 support a role for the unbalanced production of ribosome subunits in the translational decrease of GATA1 transcript which could be dependent on the structure of its 5'UTR (untranslated region).6,12 A global assessment is thus needed of the rules governing translation specificity when ribosome production is diminished. In our current study, we investigated mechanisms that regulate translation under conditions of limited ribosome availability and assessed their contribution to the normal human erythroid differentiation process. We analyzed the characteristics of transcripts occupied by ribosomes under conditions of RPS14 downregulation in both cell lines and primary cells. Our results indicate that the transcript length, codon usage and 3’UTR structure are key factors governing the translation selectivity.
Methods
Cell culture
Human erythroblasts were derived from CD34+ cord blood progenitors and infected by non-inducible GFP-pLenti X1 vector containing shRPS14 or shSCR. The UT-7/EPO cell line was transduced with a scrambled (SCR) or RPS14 shRNA cloned into a pLKO.1 Tet-On vector, selected with puromycine (1 mg/mL) and induced with doxycycline (0.2 g/mL) for 3 days. Cord blood and other patient samples were obtained from the Centre d’Investigation Clinique Paris Descartes Necker Cochin through the Programme Hospitalier de Recherche Clinique (PHRC MDS- 04; INCa-DGOS-5480; IRB IdF 2753).
Quantitative proteomics and data analysis
Label free quantification (LFQ) proteomic experiments were performed as described previously.13 For data and statistical analyses, the MS data were processed with MaxQuant version 1.5.2.8 using human sequences from the Uniprot-Swiss-prot database (Uniprot, release 2015-02) with a false discovery rate (FDR) below 1% for both peptides and proteins. LFQ results from MaxQuant were imported into Perseus software (version 1.5.1.6). Protein copy numbers per cell were then calculated using the Protein ruler plugin of Perseus by standardization to the total histone MS signal. 14 Raw data were deposited, and processed data are provided in the Online Supplementary Table S1. The abundances of erythroid progenitor, precursor and mature stage proteins were obtained from our two previous studies.13,15
Oligonucleotide microarrays: transcriptome and translatome analysis
Fisher Scientific, Waltham, MA, USA). Differential expression analysis was then carried out using a Student t-test corrected by significance analysis microarrays (SAM). Differentially expressed genes were selected using a P-value cut-off <0.05, a calculated q-value ≤0.05 and a minimum fold-change of >1.5 or <1/1.5. These genes were annotated using the Gene Ontology consortium software (www.geneontology.org/). Cytoscape (v3.2.1) and Enrichment Map plug-in were used to generate networks for gene sets enriched with an FDR <0.1.
Codon usage, upstream open reading frames and structure prediction analysis
The codon adaptation index annotation and relative synonymous codon usage (RSCU) analysis were performed using the CAIcal tool (www.genomes.urv.es/CAIcal/).16
Flow cytometry, western blotting and real-time-quantitative polymerase chain reaction
All antibodies and primers are listed in the Online Supplementary Appendix.
Statistical analysis
The statistical analysis of each plot is described above or in the corresponding figure legend. All grouped data values are presented as a mean±standard deviation (SD) or standard error of the mean (SEM). All boxes and whisker plots of expression data are presented as medians ± interquartile range. P-values were calculated using a two-sided Mann-Whitney U-test, Student t-test or Kruskal- Wallis ANOVA test with GraphPadPrism software (GraphPad Software, San Diego, CA, USA). Gene set enrichment analysis (GSEA) was based on the Kolmogorov-Smirnov test.
Data accessibility
The raw and preprocessed HTA 2.0 microarray data are publicly available at the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database (GEO; GSE108822). The raw and preprocessed proteomic data are available via ProteomeXchange with identifiers PXD008650 and PXD009258. Several published datasets were used: (GEO GSE126523; GSE15061; GSE89183; GSE85864; GSE95854).
Results
Limited ribosome availability leads to a translational defect of GATA1
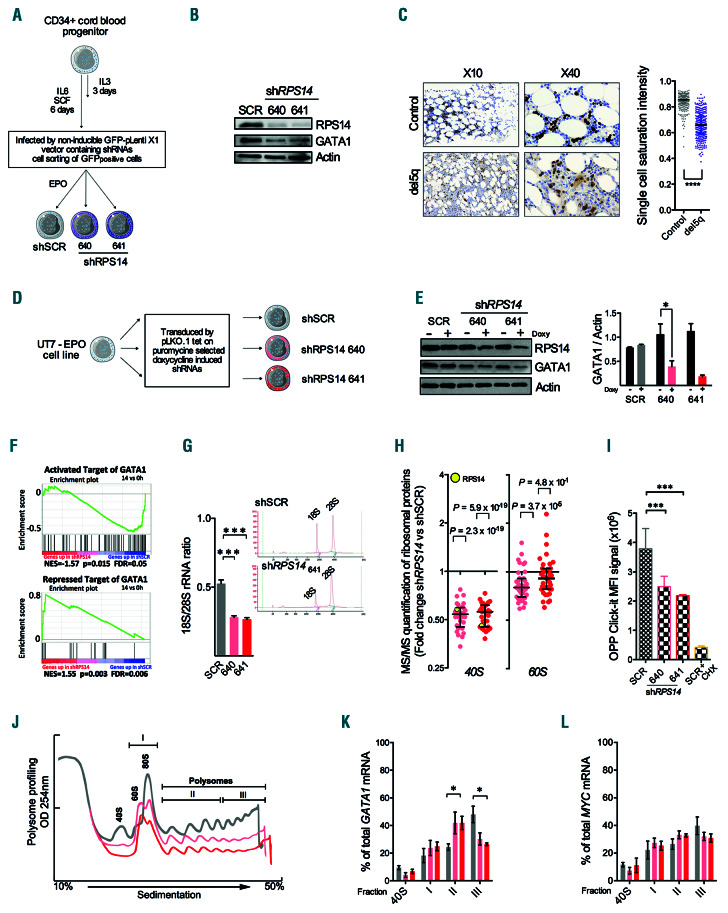
We first investigated the expression of the GATA1 gene in the context of RPS14 downregulation by infecting human primary erythroblasts with a non-inducible pLenti X1 shRPS14 vector (Figure 1A). At 3 days post infection, the expression of GATA1 protein was decreased together with a lower percentage of differentiated glycophorin A (GPA)+ cells (Figure 1B and Online Supplementary Figure S1A). Consistently, immuno-histochemistry analysis of BM biopsy sections and immunofluorescence microscopy of cultured erythroblasts confirmed that GATA1 was less abundant in del(5q) MDS compared to control erythroblasts (Figure 1C and Online Supplementary Figure S1B). By contrast, publicly available transcriptome data for del5q patients indicated that GATA1 transcript levels were normal in MDS with del(5q) (Online Supplementary Figure S1C).17 This suggests that GATA1 gene expression is mainly regulated at a post-transcriptional level.
To investigate this regulatory scenario further, we established stable UT-7/EPO shRPS14 cell lines via an inducible lentiviral vector (Figure 1D). Consistent with our human primary cell model and patient data, the GATA1 protein expression level was found to be decreased without any change in the transcript level (Figures 1E and Online Supplementary Figure S1D). After shRPS14 induction, proliferation was decreased, as evidenced by the accumulation of the cells in G1 phase and the induction of apoptosis, and membrane GPA expression was diminished (Online Supplementary Figure S1E-H). Transcriptome analysis of the UT-7/EPO shRPS14 cell lines indicated a significant modulation of GATA1 target genes including a decreased expression of the majority of activated GATA1 targets and an increased expression of the usually repressed GATA1 targets (Figure 1F, Online Supplementary Table S2 and Online Supplementary Figure S1I-K). As expected, the 18S/28S ratio was diminished (Figure 1G). The quantity of ribosomes per cell was then assessed by label-free mass spectrometry (MS), which enabled an absolute quantification of RP by using histone signals as an internal calibrator.13,14 The quantity of RP of the small subunit 40S, expressed as fold-change (FC) in the copy number per cell, was decreased by 50% in the UT-7/EPO shRPS14 cells (Figure 1H) compared to the shSCR cells (Online Supplementary Table S1). By contrast, the quantity of RP of the large subunit 60S (RPL) was less impacted, thus confirming an unbalanced expression of RP. Consistently, an absolute quantification using O-propargyl- puromycin (OPP)-click-iT® revealed that translation had been globally decreased by half (Figure 1I). The polysome profiling of UT-7/EPO shRPS14 cell lines indicated a strong reduction or absence of the free 40S and a relative increase in the free 60S. The quantity of the entire 80S ribosome was reduced and the height of polysome peaks was lower, revealing that an RPS14 downregulation results in a decreased translating ribosome content (Figure 1J). We collected the sub-fractions corresponding to 40S, 60S, 80S, light and medium polysomes with 2-5 ribosomes on an mRNA, and heavy polysomes that contain >5 ribosomes on an mRNA. Other than 40S, the sub-fractions were pooled as fraction I representing 60S and 80S, fraction II representing light and medium polysomes, and fraction III representing heavy polysomes (Figure 1J). We then compared the abundance of GATA1 and MYCmRNA in each fraction. GATA1 mRNA was less abundant in the heavy polysomes of shRPS14 cells and had shifted to lighter fractions I and II (Figure 1K). This suggests that a majority of GATA1 transcripts carried a lower number of ribosomes. By contrast, the MYC mRNA profile did not vary between conditions in any of the fractions (Figure 1L). We also confirmed the decrease of GATA1 translation in K562 shRPS14 cell lines (Online Supplementary Figure S1L-Q). Taken together, these findings indicate that an RPS14 downregulation induces a decreased ribosome availability leading to selective translation at the expense of GATA1.
Figure 1.
RPS14 downregulation results in limited ribosome availability and reduced GATA1 translation. (A-C) Knockdown of RPS14 by shRNA in human primary erythroblasts from healthy donors. (A) Schematic workflow. (B) RSP14 and GATA1 protein expression determined by western blot. Actin was used as a loading control (n=3). (C) (Left) Representative immunohistochemical images of human bone marrow biopsies stained for GATA1 protein (brown) and counterstained with Hematoxylin & Eosin in del5q patients (n=3) and normal healthy controls (n=3). (Right) A comparison of single cell saturation intensity between patients and healthy controls. A Mann-Whitney test was used to calculate P-value. (D-L) Knockdown of RPS14 in the UT-7/EPO cell line via inducible shRNA (sh640 and sh641). (D) Schematic workflow. (E) GATA1 protein expression determined by western blot. Signals were quantified by normalization to actin (n=3). (F) Transcriptome analysis of UT-7/EPO shRPS14 cell lines. Gene set enrichment analysis (GSEA) shows variations in GATA1 targets. (G) Bioanalyzer quantification of 18S and 28S rRNA expressed as mean ratios±standard error of mean (SEM) (n=3). (H) Quantitative proteomic analysis of ribosomal protein (RP) from whole cell lysates. Data are expressed as a fold change in the copy number per cell between shRPS14 and shSCR conditions in doxycycline-induced cells (n=3). (I) Absolute quantification of translation by quantification of O-propargyl-puromycin (OPP) incorporation per cell by flow cytometry, shown by the mean fluorescence intensity±SEM (n=3). (J) Polysome profiling. Free 40S, free 60S and ribosome 80S (fraction I), light and medium polysomes (fraction II), and heavy polysomes (>5 ribosomes; fraction III) are indicated. (K and L) Relative distribution of GATA1 and MYC mRNA in ribosomal fractions. Quantification values determined by real-time-quantitative polumerase chain reaction are expressed as mean±SEM (n=3). Unpaired two-tailed Student t-test: *P<0.05; **P<0.01; ***P<0.001.
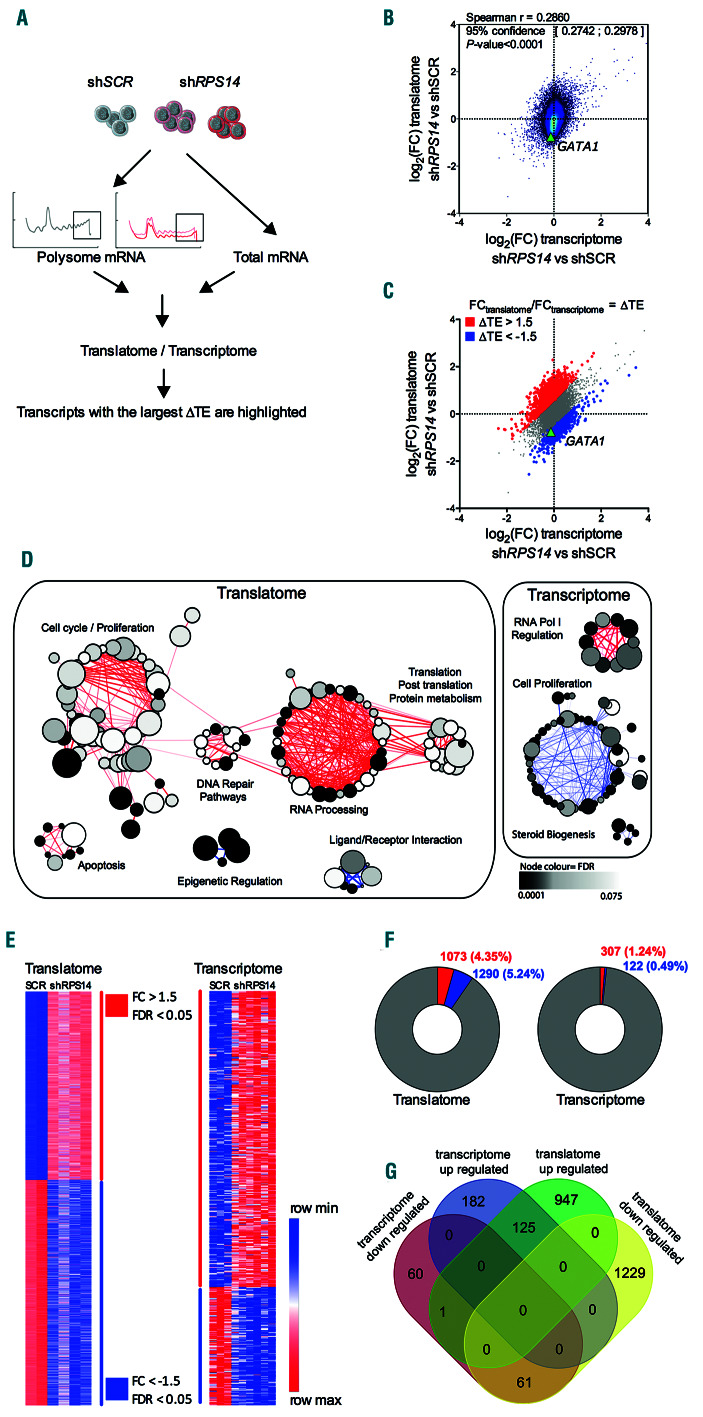
Figure 2.
Selective translation in UT-7/EPO shRPS14 cell lines. (A) Schematic workflow showing RNA extraction from actively translating polysomes (translatome) or from whole cell lysates (transcriptome) in shRPS14 640, shRPS14 641 and shSCR cells to be used for microarray analysis. Comparative analysis of the translatome related to the transcriptome and the determination of transcripts with the largest translational efficiencies variation (ΔTE) are shown. (B) 2D density plot of the log2(foldchange, FC) values obtained for the translatome and transcriptome. A Spearman test (r coefficient with 95% confidence interval and P-value) was used for correlation analysis. (C) Post-transcriptionally regulated components with an (FCtranslatome/FCtranscriptome) ratio >1.5 or <1/1.5 were overlaid as red or blue dots, respectively. (D) Gene set enrichment analysis (GSEA) of differentially expressed genes in the translatome and transcriptome. Enriched cellular programs are indicated using the Enrichment Map App of Cytoscape software (P-value cut-off = 0.005, q-value cut-off = 0.1). Nodes represent the gene sets that were enriched at the top or bottom ranking of the differentially expressed genes. Node size corresponds to the number of genes in the set. Edges indicate overlaps between gene sets and the line thickness indicates the similarity coefficient between gene sets in red for less impacted and blue for downregulated genes. The node color depends on the false discovery rate (FDR) value as shown on the scale. Clusters of nodes were determined with the Autoannotate App and are represented by the circles. (E) Heatmaps representing the significantly up- or downregulated transcripts in the translatome and transcriptome with a P-value cut-off <0.05, a calculated q-value (FDR) in significance analysis microarrays (SAM) ≤0.05 and a minimum fold-change of >1.5 or <1/1.5. The color scale representing the minimum and maximum expression per row is indicated. (F) Numbers of significantly up- or downregulated transcripts shown as doughnuts. (G) Venn diagram showing the overlap between the significantly less impacted or more downregulated transcripts in the translatome and transcriptome.
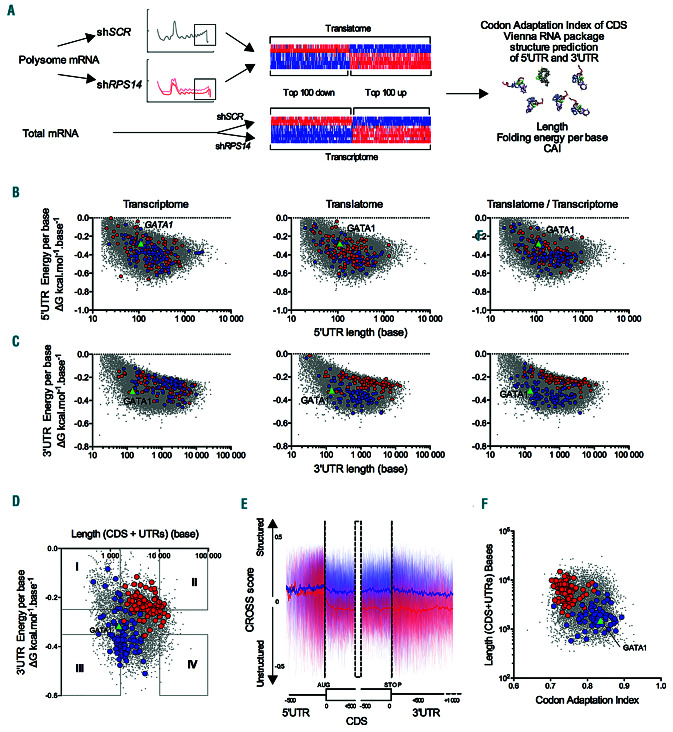
Figure 3.
The codon adaptation index, coding sequence length and thermodynamic characteristics of the 3’UTR in an mRNA govern its translation selectivity under conditions of limited ribosome availability. (A) Schematic representation of the workflow: RNA from actively translating polysomes or whole cell lysates were analyzed using Affymetrix microarrays. The top 100 most expressed (in red) and less expressed (in blue) transcripts were identified by calculating the log2fold change (FC) of the values in shRPS14 compared to shSCR cell lines. The codon adaptation index (CAI) of the coding sequence (CDS) and the thermodynamic characteristics of the untranslated regions (UTR) including the fold energy, length, and energy per base were determined. Comparisons were made of the CDS and UTR characteristics of the top 100 less impacted and downregulated transcripts in the UT-7/EPO shRPS14 translatome and transcriptome. (B and C) Thermodynamic landscape: bi-parametric scatter-plots are shown with the energy per base on the y axis and length on the x axis in a log scale of 5’UTRs (B) and 3’UTR (C). All transcripts described in the University of California Santa Cruz (UCSC) database are represented by gray dots. Red dots denote the top 100 less impacted transcripts and blue dots represent the top 100 more downregulated transcripts in the transcriptome, translatome and FCtranslatome/FCtranscriptome analysis. GATA1 is indicated by a green triangle. (D) Biparametric scatter-plot in a log scale representing the free-energy per base of the 3’UTR and length (CDS+UTR) of each transcript described in the UCSC database, represented as gray dots. Red dots denote the top 100 less impacted transcripts and blue dots indicate the top 100 more downregulated transcripts in FCtranslatome/FCtranscriptome analysis. (E) CROSS global score consensus prediction of the secondary structure profile of the top 100 less impacted (in red) and more downregulated (in blue) impacted transcripts identified by FCtranslatome/FCtranscriptome analysis. Row scores are represented by the light color lines. Mean scores are denoted by the dark color lines. (F) For all human transcripts, the CAI and their respective transcript length (CDS+UTR) are represented as gray dots in a bi-parametric scatter- plot on a log scale. Red dots indicate the top 100 less impacted transcripts and blue dots denote the top 100 more downregulated transcripts in the FCtranslatome/FCtranscriptome analysis. P-values were calculated using a two-sided Mann-Whitney U-test.* P<0.05 ***P<0.001.
Global assessment of translation under limited ribosome availability conditions
To address translation regulation as a whole in the UT- 7/EPO shRPS14 cells, we used Affymetrix HTA 2.0 microarrays to profile the mRNAs present on heavy polysomes referred to as the translatome (Figure 2A). We observed a weak correlation between the transcripts differentially expressed in the translatome and transcriptome of UT-7/EPO shRPS14 compared to UT-7/EPO shSCR cell lines (Figure 2B; Spearman test; r=0.286; P<0.0001; Online Supplementary Table S1). This weak correlation has already been reported in other translatome and transcriptome studies.12,18 The ratio of fold change FCtranslatome/FCtranscriptome enabled a determination of the transcripts with the largest translational efficiency variations (TE) (Figure 2C). GATA1 mRNA was among the notable transcripts with a downregulated TE (Figure 2B and C). GSEA coupled to enrichment map visualization was then used to annotate the differentially enriched biological pathways of the translatome and transcriptome. Using a cytoscape representation (Figure 2D and Online Supplementary Table S3), we observed that the sets of genes less impacted by RP loss in the translatome were clustered into biological pathway annotations that included cell cycle and proliferation, DNA repair and apoptosis, and RNA processing and translation. Conversely, the downregulated gene sets were poorly clustered. The few sets of genes less impacted by RP loss in the transcriptome were found to be involved in RNA polymerase I regulation while the few downregulated gene sets showed an involvement in cell proliferation and steroid biogenesis. GO term enrichment analysis of significantly impacted genes in the translatome and transcriptome confirmed these findings (Online Supplementary Table S3 and Online Supplementary Figure S2).
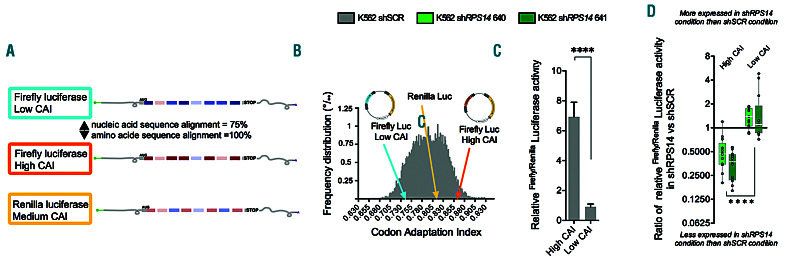
Figure 4.
Validation of codon bias as a determinant of translation selectivity. K562 shRPS14640, shRPS14641 and shSCR cells were used in dual luciferase reporter assays. (A) Schematic representation of the luciferase codon contents. (B) Frequency distribution of the codon adaptation index (CAI) values among all human transcripts. The CAI of firefly and renilla luciferases are indicated. (C) Firefly/renilla luciferase activity in K562 shSCR cells comparing firefly luciferase with a high versus that with a low CAI. (D) Ratios of firefly/renilla luciferase activity in K562 shRPS14 to K562 shSCR comparing firefly luciferase with a high or low CAI (n=13). Medians are shown as white horizontal bars, boxes represent first and third quartiles, and whiskers represent minimal and maximal values. Light green denotes shRPS14640 and dark green indicates shRPS14641. Unpaired two-tailed Student t-test: **P<0.01; ****P<0.0001.
Figure 5.
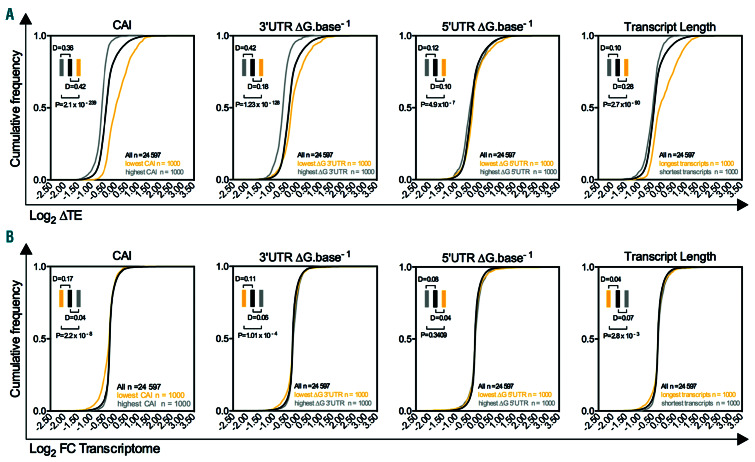
Impact of each mRNA feature on the translation efficiency and transcript stability. (A) Cumulative frequency representation of translation efficiency changes for mRNAs with the highest and lowest codon adaptation index (CAI), the largest and lowest 3’UTR ΔG.base-1 or 5’UTR ΔG.base-1 and the largest and shortest lengths. (B) Cumulative frequency representation of stability changes (Log2 [FC] transcriptome) of transcripts with the highest and lowest CAI, the largest and lowest 3’UTR ΔG.base-1 or 5’UTR ΔG.base-1 and the largest and shortest lengths. Distances were calculated using the Kolmogorov-Smirnov test. Exact P-values were calculated.
The highest and lowest expressed transcripts with an FC >1.5 or <1/1.5 in the translatome and transcriptome were subsequently selected (Figure 2E). A higher number of transcripts of the translatome varied in comparison to transcripts of the whole transcriptome (Figure 2F), with little overlap between the transcripts found to be differentially expressed in the translatome and in the transcriptome (Figure 2G). We then extended our analysis to a published dataset of DBA models targeting RPL5 or RPS19.12 Remarkably, despite differences in cell types, targeted RPs and methods, the TE across the models was highly correlated, particularly when RPS14- or RPS19-targeted cells were compared (Online Supplementary Figure S2B). Since these global expression analyses indicated uncoupling of the translatome and transcriptome, we concluded that a mechanism of selective gene expression regulation was operating at a translational level.
The codon adaptation index, coding sequence length and thermodynamic characteristics of the 3’UTR govern the translation selectivity under conditions of limited ribosome availability
Multiple mechanisms of translation regulation are dependent on the mRNA sequence and structure.19-22 We first focused our analysis on the contribution of UTR to translation selectivity. We took advantage of the availability of thermodynamic parameters of UTR in the University of California Santa Cruz (UCSC) databases to establish the thermodynamic landscape of all the referenced 5’ and 3’UTR in humans.23 Secondary structures in mRNA UTR are characterized by the enthalpy G (fold energy) and the length which are highly correlated (Online Supplementary Figure S3A) and the fold energy per base which relationship to the length is not linear, each of them influencing translation efficiency (Online Supplementary Figure S3B). We compared the 5’UTR and 3’UTR parameters of the 100 transcripts that were the less impacted or the more downregulated in the translatome and transcriptome (Figure 3A). The 5’UTR energy per base was similar between the up- and downregulated mRNAs in the transcriptome and was generally slightly stronger in the mRNAs downregulated on the polysomes (Online Supplementary Figure S3C). The 5’UTR energy per base measure provided a poor separation of the less impacted and the more downregulated transcripts of the translatome while the 3’UTR thermodynamic characteristics were fully distinctive (Figure 3B and C). A strong negative energy per base of the 3’UTR defining a highly structured region and its shortness characterized the transcripts with a lower presence on heavy polysomes, indicating that they were less translated under conditions of RPS14 downregulation (Figure 3C and Online Supplementary Figure S3D). By contrast, the 3’UTR thermodynamic characteristics of the most and the less expressed transcripts in the transcriptome were similar indicating that the regulation took place at a translational level (Figure 3C, left).
Figure 6.
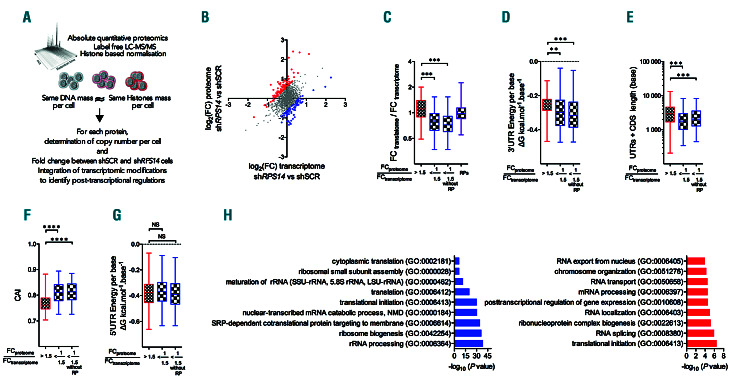
Proteome and transcriptome analyses confirm the identified rules and point to the post-translational regulation of ribosomal protein (RP). (A) Schematic representation of the experimental workflow: absolute quantitative proteomics analysis (label free LC-MS/MS) was performed in shRPS14 640, shRPS14 641 and shSCR UT7/EPO cell lines leading to a determination of the copy number per cell of each protein. Data are expressed as fold-change (FC) of the mean copy number per cell between shRPS14 versus shSCR conditions for three independent experiments. (B) Scatter plots of log2(FC) obtained for the proteome and transcriptome. Post-transcriptionally regulated components with an (FCproteome/FCtranscriptome) ratio >1.5 or <1/1.5 were overlaid as red or blue dots, respectively. (C) ΔTE values determined in the translatome/transcriptome analysis (see Figure 2) of post-transcriptionally regulated components. (D-G) Comparison of the thermodynamic characteristics of the 3’UTR (D), transcript length (E), CAI (F) and 5’UTR (G) between the two groups defined in (B) and the group with an (FCproteome/FCtranscriptome) ratio <1/1.5 minus RP. **P<0.001; ***P<0.001; ****P<0.0001. (H) Gene Ontology enrichment analysis of the most post-transcriptionally up- or downregulated components.
An integrated analysis of the translatome and transcriptome identified the transcripts with the largest TE (Figure 2C). The 3’UTR but not 5’UTR characteristics efficiently distinguished the transcripts with the largest TE from the others (Figure 3B, 3C right, and Online Supplementary Figure S3E). In the thermodynamic landscape, GATA1 transcripts have a short and unstructured 5’UTR and a short and highly structured 3’UTR. The 3’UTR parameters of GATA1 caused it to cluster with the transcripts that were less expressed on the polysomes and had a low TE. Furthermore, the length of the entire transcript encompassing the UTR and CDS was discriminative between transcripts with the largest TE (Online Supplementary Figure S3E). Thus, the transcript length and 3’UTR structure were found to be effective separators of the most and less expressed transcripts and placed GATA1 among the shortest transcripts with a structured 3’UTR (Figure 3D). To confirm the impact of transcript structures on translation outcomes, we used the CROSS method which is based on high-throughput profiling of the RNA structure to calculate the structural profile of an RNA sequence at a single-nucleotide resolution and without sequence length restrictions.24 We determined the structuration propensity score of the 5’UTR, CDS and 3’UTR of 100 transcripts with the most increased or decreased TE. The scores for the 5’UTR region were very similar, whereas those for the CDS and 3’UTR regions were highly discriminative. This suggests that the nucleotides, and therefore codon composition, of the CDS and 3’ sequence were very different between the transcripts with an increased or decreased TE (Figure 3E).
Several prior studies have demonstrated that codon usage is a key determinant of mRNA translation20,25-27 as it modulates ribosome elongation speed, mRNA stability, and co-translational folding of the nascent protein.28-31 As a metric of codon bias, the codon adaptation index (CAI) of each transcript was plotted (Figure 3F). The CAI can range from 0 to 1, with a higher value reflecting the occurrence of more frequent codons that tend to be associated with a faster translation elongation.16 We found from this analysis that the composition of the CDS in optimal codons was completely different in transcripts with an increased or a decreased TE, transcripts with an increased TE having a low CAI, and transcripts with a decreased TE such as GATA1 having a high CAI (Figure 3F). These features governing translation selectivity were confirmed in the K562 shRPS14 cell line model (Online Supplementary Figure S3F). Finally, we extended our analysis to the published transcriptome and translatome datasets of shRPL5 or shRPS19 treated primary human erythroblasts.12 The TE was increased for transcripts with a weak energy per base of the 3’UTR but not the 5’UTR, a long size and a low CAI, as shown in the UT-7 or K562 shRPS14 cell lines (Online Supplementary Figure S3G and H). Targeting of the 40S rather than the 60S subunits appeared to be more deleterious to the translation of mRNAs with structured 5’UTR translation (Online Supplementary Figure S3I). Taken together, these data show that translation selectivity is dependent on the transcript length, 3’UTR structure, and CAI. Furthermore, these data show that translation selectivity is not related to the depletion of one particular RP, but rather to the decrease in ribosome cellular content.
Figure 7.
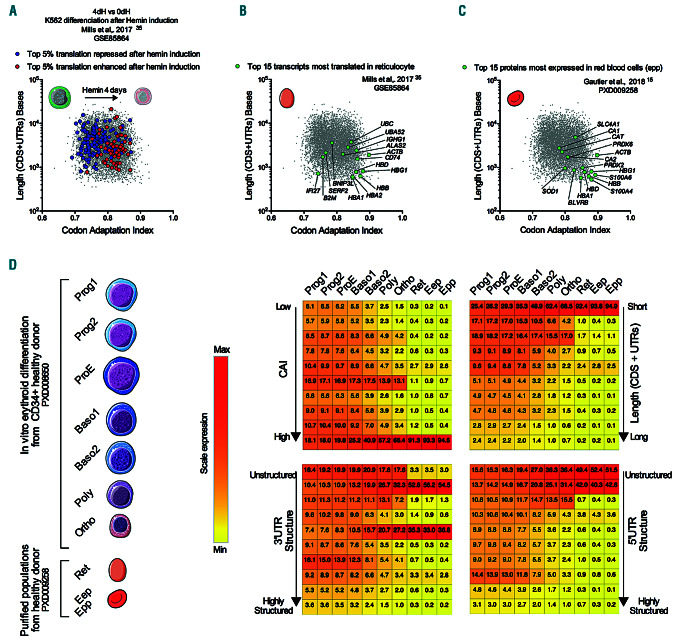
The codon adaptation index, coding sequence length and thermodynamic characteristics of the untranslated regions (UTR) in an mRNA are key determinants of its translation during normal erythropoiesis. (A-C) Bi-parametric scatter plot in a log scale of the codon adaptation index (CAI) and length (coding sequence [CDS]+UTR) of all human transcripts denoted by gray dots. (A) The top five percentile transcripts which are the more or the less translated after hemin induction of differentiation in K562 model (Mills et al.,35 GSE85864) are overlaid in red or blue, respectively. dH: day post hemin. (B) The top 15 transcripts most translated mRNAs in reticulocyte (Mills et al.,35 GSE85864) overlaid in green (C) The top 15 most expressed proteins in red blood cells (Gautier et al.,15 PXD009258) overlaid in green. (D) Dynamic repartition of proteins during erythropoiesis. Heatmaps show repartition of the protein copy numbers ranked according to their transcripts features (CAI, 5’ and 3’UTR structure and total length) in the rows and stages of differentiation in the columns. Each row represents 1/10 of the total scale of the corresponding feature. For example, at the Epp stage 94.5% of the protein cellular content is issued from transcripts with a very high CAI (Top 10 % of CAI). Ret: reticulocytes; Eep: erythrocyte-enriched population; Epp: erythrocyte-purified erythrocytes.
Validation of codon bias as a determinant of translation selectivity
To further validate the contribution of codon bias to translation regulation, we used relative synonymous codon usage (RSCU) as a second metric for codon bias measurement. This analysis confirmed that transcripts with a decreased TE were enriched in optimal codons (Online Supplementary Figure S4A). We then performed luciferase assays using a dual luciferase vector harboring a firefly luciferase (Fluc) with a high CAI and a renilla luciferase (Rluc) with a medium CAI. We also engineered an additional Fluc vector containing the same luciferase amino-acid sequence but with a nucleotide sequence of non-optimal codons to obtain a low CAI compared to the CAI spectra in human transcripts (Figure 4A and B, and Online Supplementary Figure S4B). As expected, the Fluc vector with a low CAI was less expressed than its high CAI counterpart under shSCR conditions (Figure 4C and Online Supplementary Figure S4C). After the induction of shRPS14 640 or shRPS14 641, and normalization to Rluc, the high CAI Fluc was highly repressed but the low CAI Fluc was less affected (Figure 4D and Online Supplementary Figure S4C).
Impact of each mRNA feature on the translation efficiency and transcript stability
To quantify the relative contribution of each characteristic of an mRNA molecule to its translation efficiency, we analyzed the cumulative frequency of the TE. The transcripts with the largest translation upregulation were longer and had a lower CAI. Conversely, the transcripts with the largest translation downregulation were those with a highly structured 3’UTR (Figure 5A). Furthermore, the impact of upstream open reading frames (uORF) on translational regulation appeared to be negligible under conditions of RPS14 downregulation (Online Supplementary Figure S5A). To rule out any effect of the 3’UTR or CAI on mRNA stability,30,32,33 we analyzed the mRNA half-life and decay rate in K562 cells. We found that the CAI and 3’UTR energy per base poorly correlated with mRNA decay and half-life measurement (Online Supplementary Figure S5B). Moreover, our transcriptome analysis did not correlate with either mRNA decay or a reduced mRNA half-life, thus excluding a possible global alteration of decay pathways (Online Supplementary Figure S5C). That the cumulative frequency of the differential expression in transcriptomic analysis did not reveal major changes was a definitive indicator of no changes in RNA decay that are dependent on the 3’UTR or 5’UTR structure or the transcript length. Nevertheless, consistent with the previously reported relationship between codon usage and RNA stability,30 we observed a marginally increased decay in low CAI transcripts (Figure 5B).
Proteome analyses confirm the identified rules and point to a post-translational regulation of ribosomal protein
We showed in our initial experiments that global translation was diminished per cell (Figure 1I). Therefore, to investigate the impact of translation selectivity on protein expression, we performed an absolute proteomic quantification of our UT7/EPO model and normalized the data using histones to avoid growth difference effects and obtain the copy number per cell for each protein (Figure 6A). Because the number of ribosomes associated with a given mRNA depends on the length of this transcript, analyzing the transcripts present on heavy polysomes could favor the identification of the longest mRNAs. Integrating whole proteome and transcriptome analyses excludes this putative bias and provides an indirect measurement of translation and degradation rates. The log2(FC) of the proteins was plotted to the log2(FC) of the transcripts to identify components that underwent post-transcriptional regulation (Figure 6B and Online Supplementary Table S1). To evaluate post-transcriptional changes, we selected components having expression that was inversely regulated in the proteome and transcriptome with an FCproteome/FCtranscriptome ratio >1.5 or <1/1.5. First, we confirmed that the post-transcriptional changes identified by the proteome analysis were predicted by the TE analysis. At post-transcriptional level, the upregulated components had an increased TE whilst those with downregulated expression had a decreased TE, highlighting that post-transcriptional changes at the proteome level are a direct result of translation selectivity. This also demonstrates that the observed TE values were not only associated with changes in translational occupancy but also with changes in protein quantities (Figure 6C). Furthermore, the post-transcriptionally downregulated components in the proteome were encoded by transcripts with a significantly more structured 3’UTR, a shorter length and a higher CAI than the posttranscriptionally upregulated components (Figures 6D-F), whilst the 5’UTR structure had no impact (Figure 6G).
Interestingly, RP transcripts which mainly harbored a short and unstructured 3’UTR were recognized among the post-transcriptionally downregulated components (Online Supplementary Figure S6A). Removing RP from the analysis increased the concordance between the TE and the protein expression level (Figure 6C-F). These results confirmed that the determinants of translation selectivity predicted by our translatome analysis were relevant. A GO term analysis of the post-transcriptionally regulated components (Figure 6H) revealed that those which were upregulated, were involved in DNA replication, RNA processing and splicing (Figure 6H). These terms overlapped those identified by GSEA and GO analyses of the less impacted transcripts in the translatome (Figure 2D and Online Supplementary Figure S2A). Post-transcriptionally downregulated components were found to be involved in translation, rRNA processing and maturation (Figure 6H). Finally, protein expression is controlled by the rules governing the selection of transcripts on the ribosome. We extended our findings by re-analyzing datasets generated previously in lymphoid cell models carrying mutations in the RPS15 gene.34 Those mutations lead to a decrease in the ribosome half-life and content. We observed in our current analysis that mutant cells had a global protein expression imbalance in favor of proteins whose transcripts had a low CAI and an unstructured 3’UTR (Online Supplementary Figure S6B).
Codon adaptation index, coding sequence length and thermodynamic characteristics of the untranslated regions are key determinants of translation in normal erythropoiesis
Clinical manifestations of ribosomopathies are linked to the cell-specific impact of mutations. Of note, impaired erythropoiesis may at least be partly related to a translation defect of GATA1.6 To gain further insights into the translational regulation that occurs during normal erythroid maturation, we investigated the characteristics of the transcripts in the translatome of the K562 cell line, of the proteins expressed in human erythroblasts at different stages of differentiation and in red blood cells, and of the transcripts in the translatome of healthy donor reticulocytes. 4,13,15,35 We found that a high CAI and a short transcript length characterized the mRNAs that are translated when erythroid differentiation is induced with hemin in K562 cells or in purified reticulocytes (Figure 7A and B). These parameters also characterized the most expressed proteins in red blood cells (Figure 7C). More generally, a high CAI, short transcript length and an unstructured 5’ and 3’ UTR were the characteristics of transcripts corresponding to proteins which show increased expression during the progression of normal erythroid differentiation (Figure 7D and Online Supplementary Figure S7). Our current results thus indicate that the transcripts encoding proteins that accumulate in erythrocytes shared most of the determinants of translation selectivity, which was highlighted by conditions of limited ribosome availability.
Discussion
RP depletion has been recognized as a principal cause of erythroid hypoplasia either in acquired del(5q) MDS or DBA.2,36 The specific impairment of the erythroid lineage has been linked to a decreased representation of GATA1 transcript on polysomes under conditions of an RPS19, RPL5, RPL11 or RPS14 haploinsufficiency.6,11 Our current results indicate that in addition to a reduction in GATA1 mRNA on polysomes, translation as a whole is selective at the expense of erythroid transcripts including globin genes under conditions of low ribosome availability. Consistent with the previous findings of Yang et al.,8 we have observed in our present study that low globin gene translation may account for a disequilibrium in the heme-globin balance, leading to reactive oxygen species (ROS) production and cell death.
Translation efficiency is thought to depend on the thermodynamic properties of a given transcript.23,37 For example, the presence of an IRES in the BAG1 or CSDE1 mRNAs or the length of 5’UTR of the BCAT1 transcript have been implicated in disrupted translation when RPL11 or RPS19 is haploinsufficient.38,39 The structure of the 5’UTR in GATA1 mRNA has been associated with its translation downregulation in the context of RPS19 haploinsufficiency. However, depending on which transcripts were compared with GATA1 mRNA, its 5’UTR may be considered to be either highly structured or unstructured.6,12 Our visualization of the thermodynamic landscape allowed us to establish that, in comparison to all other human 5’UTR sequences, the GATA1 5’UTR is short and unstructured. Our global investigation of all the determinants of translation selectivity in UT-7/EPO and K562 shRPS14 cell line models identified that short mRNAs with a high CAI, a highly structured and short 3’UTR, and to a far lesser extent transcripts with a structured 5’UTR, were specifically less translated. GATA1 mRNA is indeed a short transcript with high CAI, highly structured 3’UTR, but a less structured 5’UTR. We confirmed our present findings using published ribosome profiling data of shRPS19/shRPL5 human primary erythroblasts and proteome data for shRPS15 lymphocytes. 12,34 RPS rather than RPL targeting has been shown to impair the translation of mRNAs with a structured 5’UTR, highlighting the crucial role of the 40S subunit in the initiation and scanning of the 5’UTR.40 Hence, the rules of translation selectivity have been shown to be conserved across the different models of an RP deficiency, demonstrating that translation selectivity has a stronger association with a decrease in the cellular ribosome content than a defect in one particular RP. Interestingly, proteins whose transcripts display most of these characteristics are accumulated during normal erythropoiesis suggesting that such a combination of parameters may allow the expression of a selected proteome in a short time. Consistent with our current results, a high CAI has previously been reported to confer a high elongation speed and a long mRNA half-life, whereas a low CAI has been associated with mRNA decay through the slowing of translation elongation.28-30 The 3’UTR structure and length are also associated with translation repression and decay.32,33 In our current experiments, under limited ribosome availability conditions, we did not observe any decay modifications linked to these two features. Further experiments are thus required to investigate the interplay between CDS and UTR and its role in the control of mRNA stability and translation.
Our current gene expression analysis highlighted pathways involved in cell cycle, proliferation, DNA repair and RNA processing among the transcripts with the highest TE under conditions of reduced translation (Figure 2D). Notably, however, the preferential translation of these transcripts in the context of the global diminution of translation rate (Figure 1J-I) remains an inefficient mechanism of rescuing the cells from death. Understanding these processes may optimize gene expression in some diseases.
Khajuria et al.12 previously reported that transcripts with unstructured 5’UTR were translated more under normal conditions and more impacted by low ribosome availability than mRNAs with a structured 5’UTR. Consistently, we found in our current analyses that the most expressed proteins during normal erythropoiesis are encoded by transcripts with an unstructured 5’UTR, and often with an unstructured 3’UTR (Figure 7D). However, many transcripts with a structured 3’UTR that are impacted under conditions of low ribosome availability also encode erythroid proteins under normal conditions. Hence, RNA binding proteins and miRNA that target the 3’ end of transcripts may play a role in translation selectivity. Other studies have demonstrated the contribution of the 3’UTR length to the regulation of translation.21,41 Highly translated mRNAs have the ability to form a loop through interactions between polyA binding proteins and initiation factors that brings the 5’ and 3’ ends into communication.42 It has also been suggested that ribosomes may move through the 3’UTR to support the recycling and re-initiation of another loop of translation at the 5’UTR.21,43 It would be advantageous for the recycling re-initiation process to have a small distance between the 3’ and 5’ ends.44 In normal conditions, a short and structured 3’UTR may contribute to an increased recycling and a higher translation efficiency by reducing the distance between 3’ and 5’ ends. In agreement with this hypothesis, recent evidence has demonstrated that the depletion of the ribosome recycling/re-initiation protein ABCE1 tended to arrest ribosomes on transcripts with a short and highly structured 3’UTR.45,46 These transcripts, which have similar characteristics to those we found to be preferentially translated during normal erythropoiesis, could be favored by the recycling/re-initiation process. Under conditions of RPS14, RPL5 and RPS19 downregulation, the translation of mRNAs with a short and structured 3’UTR was found to be decreased, suggesting that re-initiation could no longer occur normally and that terminating ribosomes should be released to feed the cellular pool. The overexpression of ribosome rescue factors PELO/HBS1L in RPS19 haplo-insufficient K562 cell line, was reported to restore the hemoglobin levels.35 Whether such a mechanism may compensate for the loss of ribosomes in the disorder caused by an RPS14 haplo-insufficiency will require further investigation.
Several prior yeast and human studies in which the RP genes RPS19, RPL5, RACK1 or RPS26 were mutated or deleted, have also reported that the shortest transcripts were less present on polysomes under conditions of low ribosome availability.47 This is in sharp contrast to observations under normal conditions in a wide range of eukaryotic organisms, in which the shortest transcripts are more efficiently translated than the longest mRNAs.48,49 The translation initiation rate, density of ribosomes on the transcript and protein abundance usually negatively correlates with the CDS length.49 In addition, a high density of ribosomes on short transcripts contributes to the efficiency of their translation.47,48 Recent computational analyses have shown that the recycling/reinitiation process could account for the high density of ribosomes and efficient translation of short mRNAs.44,50 In our current study, the severe defect we observed in the translation of the shortest transcripts could be explained by a diminution of re-initiation. The same reasoning may explain why high CAI transcripts were more impacted by the limited ribosome availability than those with a low CAI. Under normal conditions, it has been shown that a high CAI is an advantage in terms of having access to this process of recycling/re-initiation due to a faster elongation rate.44
In conclusion, the rate of protein synthesis depends on a complex network of regulatory elements that include expression levels of mRNAs, the cellular concentration of ribosomes, the mRNA length, the density of ribosomes, and the initiation and termination rates.4 Our current findings indicate that, when the ribosome concentration becomes a limiting factor, the translation is selective, and is dependent on the mRNA CAI, length and 3’UTR structure. Further investigations are required to better understand how the cellular ribosome concentration modifies translation initiation, translation termination, and ribosome recycling to create the link between the genetic alteration of an RP and impaired translation in erythroid cells.
Supplementary Material
Acknowledgments
The authors want to acknowledge Pr Olivier Kosmider, Dr Narla Mohandas, and Dr Christian Bastard for very helpful discussions. They also want to thank Dr Franck Letourneur from the genom’IC platform, Florent Dumont, bioinformatician funded by the Site de Recherche Intégrée sur le Cancer (SIRIC) CAncer Research for PErsonalized Medicine CARPEM, Alice Rousseau for technical assistance and Marjorie Leduc from the 3P5 proteomic platform of Paris Descartes University.
Funding Statement
Funding: This study was funded by INSERM, by the Institut National du Cancer (INCa) and the Direction Générale de l’Offre de Soins (DGOS) of the French Ministry of Social Affairs and Health through the Programme Hospitalier de Recherche Clinique (PHRC MDS-04: INCa-DGOS_5480). SLG was the recipient of a PhD funding from the Laboratory of Excellence on red cells GR-Ex.
References
- 1.Mirabello L, Khincha PP, Ellis SR, et al. Novel and known ribosomal causes of Diamond- Blackfan anaemia identified through comprehensive genomic characterisation. J Med Genet. 2017;54(6):417-425. [DOI] [PubMed] [Google Scholar]
- 2.Ebert BL, Pretz J, Bosco J, et al. Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature. 2008; 451(7176): 335-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schneider RK, Schenone M, Ferreira MV, et al. Rps14 haploinsufficiency causes a block in erythroid differentiation mediated by S100A8 and S100A9. Nat Med. 2016;22(3): 288-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mills EW, Green R. Ribosomopathies: there’s strength in numbers. Science. 2017; 358(6363). [DOI] [PubMed] [Google Scholar]
- 5.Xue S, Barna M. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat Rev Mol Cell Biol. 2012; 13(6):355-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ludwig LS, Gazda HT, Eng JC, et al. Altered translation of GATA1 in Diamond-Blackfan anemia. Nat Med. 2014;20(7):748-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frisan E, Vandekerckhove J, de Thonel A, et al. Defective nuclear localization of Hsp70 is associated with dyserythropoiesis and GATA-1 cleavage in myelodysplastic syndromes. Blood. 2012;119(6):1532-1542. [DOI] [PubMed] [Google Scholar]
- 8.Yang Z, Keel SB, Shimamura A, et al. Delayed globin synthesis leads to excess heme and the macrocytic anemia of Diamond Blackfan anemia and del(5q) myelodysplastic syndrome. Sci Transl Med. 2016;8(338):338ra67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doty RT, Yan X, Lausted C, et al. Single-cell analyses demonstrate that a heme-GATA1 feedback loop regulates red cell differentiation. Blood. 2019;133(5):457-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rio S, Gastou M, Karboul N, et al. Regulation of globin-heme balance in Diamond-Blackfan anemia by HSP70/GATA1. Blood. 2019;133(12):1358-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilles L, Arslan AD, Marinaccio C, et al. Downregulation of GATA1 drives impaired hematopoiesis in primary myelofibrosis. J Clin Invest. 2017;127(4):1316-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khajuria RK, Munschauer M, Ulirsch JC, et al. Ribosome levels selectively regulate translation and lineage commitment in human hematopoiesis. Cell. 2018; 173(1):90-103.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gautier E-F, Ducamp S, Leduc M, et al. Comprehensive proteomic analysis of human erythropoiesis. Cell Rep. 2016; 16(5):1470-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiśniewski JR, Hein MY, Cox J, Mann M. A “proteomic ruler” for protein copy number and concentration estimation without spikein standards. Mol Cell Proteomics. 2014;13(12):3497-3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gautier E-F, Leduc M, Cochet S, et al. Absolute proteome quantification of highly purified populations of circulating reticulocytes and mature erythrocytes. Blood Adv. 2018;2(20):2646-2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Puigbò P, Bravo IG, Garcia-Vallve S. CAIcal: a combined set of tools to assess codon usage adaptation. Biol Direct. 2008;3:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mills KI, Kohlmann A, Williams PM, et al. Microarray-based classifiers and prognosis models identify subgroups with distinct clinical outcomes and high risk of AML transformation of myelodysplastic syndrome. Blood. 2009;114(5):1063-1072. [DOI] [PubMed] [Google Scholar]
- 18.Tebaldi T, Re A, Viero G, et al. Widespread uncoupling between transcriptome and translatome variations after a stimulus in mammalian cells. BMC Genomics. 2012; 13:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leppek K, Das R, Barna M. Functional 5’ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat Rev Mol Cell Biol. 2018;19(3):158-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanson G, Coller J. Codon optimality, bias and usage in translation and mRNA decay. Nat Rev Mol Cell Biol. 2018;19(1):20-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miettinen TP, Björklund M. Modified ribosome profiling reveals high abundance of ribosome protected mRNA fragments derived from 3’ untranslated regions. Nucleic Acids Res. 2015;43(2):1019-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Floor SN, Doudna JA. Tunable protein synthesis by transcript isoforms in human cells. Elife. 2016;5:e10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrett LW, Fletcher S, Wilton SD. Regulation of eukaryotic gene expression by the untranslated gene regions and other noncoding elements. Cell Mol Life Sci. 2012; 69(21):3613-3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delli Ponti R, Marti S, Armaos A, Tartaglia GG. A high-throughput approach to profile RNA structure. Nucleic Acids Res. 2017;45(5):e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009; 324(5924):218-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pechmann S, Frydman J. Evolutionary conservation of codon optimality reveals hidden signatures of cotranslational folding. Nat Struct Mol Biol. 2013;20(2):237-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gardin J, Yeasmin R, Yurovsky A, Cai Y, Skiena S, Futcher B. Measurement of average decoding rates of the 61 sense codons in vivo. eLife. 2014;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Presnyak V, Alhusaini N, Chen Y-H, et al. Codon optimality is a major determinant of mRNA stability. Cell. 2015;160(6):1111-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu C-H, Dang Y, Zhou Z, et al. Codon usage influences the local rate of translation elongation to regulate co-translational protein folding. Mol Cell. 2015;59(5):744-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu Q, Medina SG, Kushawah G, et al. Translation affects mRNA stability in a codon-dependent manner in human cells. Elife. 2019;8:e45396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bazzini AA, Del Viso F, Moreno-Mateos MA, et al. Codon identity regulates mRNA stability and translation efficiency during the maternal-to-zygotic transition. EMBO J. 2016;35(19):2087-2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grimson A, Farh KK-H, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27(1):91-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rissland OS, Subtelny AO, Wang M, et al. The influence of microRNAs and poly(A) tail length on endogenous mRNA-protein complexes. Genome Biol. 2017;18(1):211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bretones G, Álvarez MG, Arango JR, et al. Altered patterns of global protein synthesis and translational fidelity in RPS15-mutated chronic lymphocytic leukemia. Blood. 2018; 132(22):2375-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mills EW, Wangen J, Green R, Ingolia NT. Dynamic regulation of a ribosome rescue pathway in erythroid cells and platelets. Cell Rep. 2016;17(1):1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farrar JE, Vlachos A, Atsidaftos E, et al. Ribosomal protein gene deletions in Diamond-Blackfan anemia. Blood. 2011; 118(26):6943-6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tuller T, Waldman YY, Kupiec M, Ruppin E. Translation efficiency is determined by both codon bias and folding energy. Proc Natl Acad Sci U S A. 2010;107(8):3645-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horos R, Ijspeert H, Pospisilova D, et al. Ribosomal deficiencies in Diamond- Blackfan anemia impair translation of transcripts essential for differentiation of murine and human erythroblasts. Blood. 2012; 119(1):262-272. [DOI] [PubMed] [Google Scholar]
- 39.Pereboom TC, Bondt A, Pallaki P, et al. Translation of branched-chain aminotransferase- 1 transcripts is impaired in cells haploinsufficient for ribosomal protein genes. Exp Hematol. 2014;42(5):394-403.e4. [DOI] [PubMed] [Google Scholar]
- 40.Archer SK, Shirokikh NE, Beilharz TH, Preiss T. Dynamics of ribosome scanning and recycling revealed by translation complex profiling. Nature. 2016;535(7613):570-574. [DOI] [PubMed] [Google Scholar]
- 41.Tanguay RL, Gallie DR. Translational efficiency is regulated by the length of the 3’ untranslated region. Mol Cell Biol. 1996;16(1):146-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vicens Q, Kieft JS, Rissland OS. Revisiting the closed-loop model and the nature of mRNA 5’-3’ communication. Mol Cell. 2018;72(5):805-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brogna S, Wen J. Nonsense-mediated mRNA decay (NMD) mechanisms. Nat Struct Mol Biol. 2009;16(2):107-113. [DOI] [PubMed] [Google Scholar]
- 44.Fernandes LD, de Moura APS, Ciandrini L. Gene length as a regulator for ribosome recruitment and protein synthesis: theoretical insights. Sci Rep. 2017;7(1):17409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sudmant PH, Lee H, Dominguez D, Heiman M, Burge CB. Widespread accumulation of ribosome-associated isolated 3’ UTRs in neuronal cell populations of the aging brain. Cell Rep. 2018;25(9):2447-2456.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Young DJ, Guydosh NR, Zhang F, Hinnebusch AG, Green R. Rli1/ABCE1 recycles terminating ribosomes and controls translation reinitiation in 3’UTRs in vivo. Cell. 2015;162(4):872-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson MK, Gilbert WV. mRNA lengthsensing in eukaryotic translation: reconsidering the “closed loop” and its implications for translational control. Curr Genet. 2017; 63(4):613-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arava Y, Wang Y, Storey JD, Liu CL, Brown PO, Herschlag D. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2003;100(7):3889-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weinberg DE, Shah P, Eichhorn SW, Hussmann JA, Plotkin JB, Bartel DP. Improved ribosome-footprint and mRNA measurements provide insights into dynamics and regulation of yeast translation. Cell Rep. 2016;14(7):1787-1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rogers DW, Böttcher MA, Traulsen A, Greig D. Ribosome reinitiation can explain lengthdependent translation of messenger RNA. PLoS Comput Biol. 2017;13(6):e1005592. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw and preprocessed HTA 2.0 microarray data are publicly available at the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database (GEO; GSE108822). The raw and preprocessed proteomic data are available via ProteomeXchange with identifiers PXD008650 and PXD009258. Several published datasets were used: (GEO GSE126523; GSE15061; GSE89183; GSE85864; GSE95854).