Abstract
The inflammatory cytokine stem cell factor (SCF, ligand of c-kit receptor) has been implicated as a pro-oncogenic driver and an adverse prognosticator in several human cancers. Increased SCF levels have recently been reported in a small series of patients with chronic lymphocytic leukemia (CLL), however its precise role in CLL pathophysiology remains elusive. In this study, CLL cells were found to express predominantly the membrane isoform of SCF, which is known to elicit a more robust activation of the c-kit receptor. SCF was significantly overexpressed in CLL cells compared to healthy tonsillar B cells and it correlated with adverse prognostic biomarkers, shorter time-to-first treatment and shorter overall survival. Activation of immune receptors and long-term cell-cell interactions with the mesenchymal stroma led to an elevation of SCF primarily in CLL cases with an adverse prognosis. Contrariwise, suppression of oxidative stress and the BTK inhibitor ibrutinib lowered SCF levels. Interestingly, SCF significantly correlated with mitochondrial dynamics and hypoxia-inducible factor-1a which have previously been linked with clinical aggressiveness in CLL. SCF was able to elicit direct biological effects in CLL cells, affecting redox homeostasis and cell proliferation. Overall, the aberrantly expressed SCF in CLL cells emerges as a key response regulator to microenvironmental stimuli while correlating with poor prognosis. On these grounds, specific targeting of this inflammatory molecule could serve as a novel therapeutic approach in CLL
Introduction
Chronic inflammation facilitates the survival, proliferation and immune evasion of chronic lymphocytic leukemia (CLL) cells in their tissue microenvironments.1,2 CLL cells aberrantly express pro-inflammatory mediators such as cytokines and chemokines due to intrinsic abnormalities (e.g., mutations in MYD88, NOTCH1 or other genes) or extrinsic determinants (e.g., antigenic stimulation of B-cell receptors [BcR] and Toll-like receptors [TLR]).1,3 Moreover, CLL cells further exacerbate the inflammatory character of their respective tumor milieu by modulating their surrounding cells in a pro-oncogenic fashion.4
Inflammation in the CLL milieu is linked with the clinical heterogeneity of the disease since increased levels of cytokines/chemokines correlate with adverse prognostic markers such as high ZAP70 and CD38 expression, unmutated IGHV genes, aggressive stereotyped subsets (e.g., subset #1) and adverse prognostic chromosomal aberrations such as del(11q) and del(17p).5–7 Gradually, inflammation favors the expansion of more aggressive CLL clones that ultimately lead to chemorefractoriness.1,7 Hence, deciphering the complex biology of inflammation in the CLL microenvironment is paramount for the successful "bench-to-bedside" transition of novel therapeutics that will drastically improve the outcomes of CLL patients.
Recently, the mitogenic cytokine stem cell factor (SCF, KIT ligand, encoded by the KITL gene) was found to be increased in the serum of CLL patients and also expressed by CLL cells infiltrating the bone marrow and lymph nodes.7-9 SCF is a pro-inflammatory glycoprotein that, upon homo-dimerization, binds to the c-kit receptor (KIT gene, CD117), a class III receptor tyrosine kinase that is primarily localized on the cell surface but can also be found in the cytoplasm and the nucleus.10-12 There are two major alternative splice variants of the KITL gene, distinguished based on the presence or not of a specific exon (exon 6) that encodes a proteolytic cleavage site. The variant of the KITL gene carrying exon 6 encodes a transiently transmembrane isoform that is swiftly released after proteolytic cleavage from metalloproteases such as MMP-9 (soluble SCF, KL-1). The variant lacking exon 6 encodes a predominantly transmembrane isoform that can also be processed by metalloproteases albeit with less affinity (membrane SCF, KL-2). The KL-2 isoform elicits a more durable activation of c-kit whereas the KL-1 isoform elicits a more rapid but transient activation due to ensuing receptor internalization.13,14 Both isoforms induce cell proliferation and differentiation albeit with distinct efficacy depending on the tissue involved, the available synergistic growth factors and the developmental stage of the c-kit+ cells.12,15 Furthermore, the KL-1 isoform serves as a chemoattractant whereas the KL-2 isoform is involved in cellular adhesion.16,17 Both SCF isoforms increase in inflamed tissues due to reactive oxygen species (ROS) and potentiate the alleviation of excessive oxidative stress, thus functioning as compensatory survival factors.18,19
SCF is considered an adverse prognostic marker in cancer, correlating with shorter overall and relapse-free survival as well as increased angiogenesis.20 Furthermore, activation of c-kit by SCF binding promotes survival, proliferation, stemness, immune evasion and drug resistance of malignant cells while it potentiates their metastatic dissemination. 15,21-23 SCF overproduction by cancer cells leads to a plethora of interactions with surrounding c-kit+ cells which underpins the prevalence of the tumor microenvironment at the expense of the neighboring normal cellular niches.24
There is scant information regarding the role of SCF in the pathobiology of CLL. Realizing this knowledge gap, here we sought to investigate the expression profile of SCF in CLL and also explore how SCF could be modulated by the CLL microenvironment. We report that SCF overexpression in CLL is associated with adverse prognosis and a shorter time-to-first treatment. Moreover, we provide evidence that immune and redox signaling induce SCF expression in CLL cells, which affects mitochondrial dynamics and cell proliferation, highlighting a crucial role for this inflammatory growth factor in CLL cell homeostasis.
Methods
Chronic lymphocytic leukemia patients
Blood samples were collected with informed consent from 75 patients diagnosed with CLL according to the guidelines of the International Workshop on Chronic Lymphocytic Leukemia/National Cancer Institute (iwCLL/NCI).The patients were either untreated or off therapy for at least 6 months prior to sampling.25 The study was approved by the local Ethics Review Committee of the participating institutions. Demographic, clinical and biological data for the entire cohort of patients are given in Online Supplementary Table S1 and specifically for the ibrutinib cohort in Online Supplementary Table S2.
Cell purification, cultures and stimuli
CD19+ B cells were negatively selected from whole blood and cultured in the presence or absence of specific ligands for certain periods, depending on the assay. A detailed description of the cell culture conditions and stimuli is provided in the Online Supplementary Material.
Quantification of KITLG and KIT mRNA expression
Total cellular RNA was isolated from purified B cells with the Qiagen RNAeasy mini kit (QIAGEN), including a DNAse incubation step. KITLG and KIT mRNA levels were quantified by realtime quantitative polymerase chain reaction (RQ-PCR) (for more details please see the Online Supplementary Methods).
Western blotting and flow cytometry studies
Protein expression of SCF and hypoxia-inducible factor-1 alpha (HIF-1a) was assessed by western blotting and flow cytometry while cell viability, apoptosis, mitochondrial mass, membrane potential and intracellular expression of Ki-67 were analyzed by flow cytometry. A detailed description of these studies is provided in the Online Supplementary Material and Online Supplementary Table S3.
Detection of soluble stem cell factor in chronic lymphocytic leukemia cell cultures by enzyme-linked immunosorbent assay
Cell-free supernatants of primary CLL cells stimulated with CpG for 24 h or not (control) were harvested and analyzed using the Human Stem Cell Factor ELISA Kit (Abcam) following the manufacturer’s instructions.
Immunohistochemistry
Immunohistochemistry was performed on 5 mm thick sections of analyzed samples (for details see Online Supplementary Methods) by using SCF antibody (Abcam). Six lymph node samples, three bone marrow biopsies with CLL infiltration along with biopsy specimens of reactive lymph nodes were also stained with an anti- CD117 antibody (Dako). For immunostaining a Bond Max™ autostainer (Leica Microsystems, Wetzlar, Germany) was used.
Gene silencing by siRNA methodology
Purified CD19+ B cells from MEC1 CLL cells were transfected with 100 nM of FlexiTube siRNA directed against human SCF or 40 nM of AllStars Negative Control siRNA (Qiagen), using the Amaxa Human B Cell Nucleofector Kit (Lonza) as previously reported.26 MEC1 cells were collected and processed for flow cytometry analysis at the indicated time points.
Statistical analysis
The statistical significance of bivariate relationships between variables normally distributed was assessed with the use of the Student t-test. When the underlying distribution was not normal, the Mann–Whitney or Wilcoxon test was applied. For all comparisons, the level of statistical significance was set at P=0.05. Survival analysis was conducted to assess the impact of SCF on overall survival and time-to-first treatment (TTFT). All statistical analyses were performed with the use of GraphPad Prism 5 software (La Jolla, CA, USA), and R. Detailed information about the statistical analysis can be found in the Online Supplementary Methods.
Results
Chronic lymphocytic leukemia cells predominantly express the membrane isoform of stem cell factor
First, we investigated the expression profile of KITLG gene splice variants in CLL cells by RQ-PCR analysis. Using specific primers for each variant, we observed that CLL cells expressed transcripts that encode the membrane (KL-2) isoform only, albeit generally at lower levels than the control HL-60 promyelocytic leukemia cells (Online Supplementary Figure S1A-C).
Next, we investigated SCF protein expression in CLL cells by western blotting analysis, utilizing a monoclonal antibody that recognizes a common epitope in the common extracellular domain (N-terminus) of both SCF isoforms, and detected a single band at 37 kDa.27,28 We also used an antibody specific for the KL-1 isoform which recognized the protein domain encoded by the exon 6-7 junction and detected a significantly weaker single band at 32 kDa (Online Supplementary Figure S1D and E). On these grounds, we concluded that CLL cells predominantly express the membrane (KL-2) SCF isoform.
High stem cell factor expression is an adverse prognosticator in chronic lymphocytic leukemia
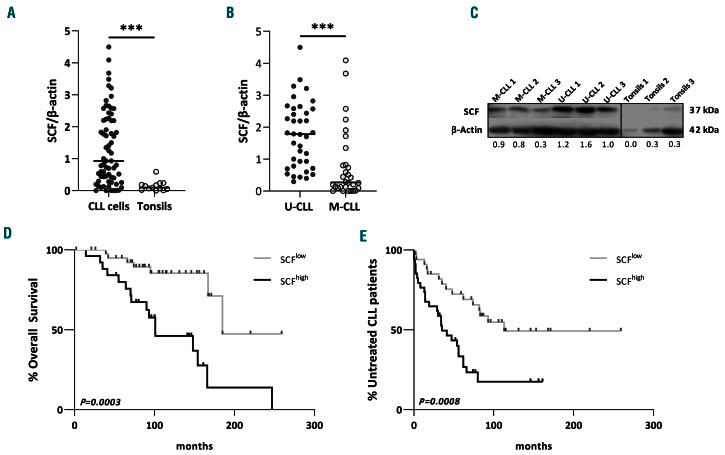
Western blot analysis with an anti-SCF antibody that recognized both SCF isoforms disclosed significantly (fold-difference [FD]: 9.8; P<0.001) higher overall SCF levels in negatively selected CD19+ tumor B cells from 69 CLL patients compared to CD19+ tonsillar B cells from 13 healthy donors (Figure 1A). Within CLL, increased SCF protein levels correlated significantly with CD38 positivity (nCD38+=17, nCD38-=51, FD:2.35; P=0.03), ZAP70 positivity (nZAP70+=20, nZAP70-=30, FD: 2.9; P=0.0167) and trisomy 12 (nTri12+=8, nTri12-=43, FD: 3; P=0.0488) (Online Supplementary Figure S2A-C). Interestingly, increased SCF protein levels correlated strongly with the mutational status of the clonotypic IGHV genes; indeed, CLL cases with unmutated IGHV genes (U-CLL) showed significantly higher SCF levels compared to those cases with mutated IGHV genes (M-CLL) (nU-CLL=38, nM-CLL=30; FD: 6.6; P<0.001) (Figure 1B and C). Flow cytometry analysis for the detection of SCF+ cells confirmed significantly higher SCF protein expression in U-CLL than in M-CLL (nUCLL =16, nM-CLL=9, FD:4.9; P=0.0086) (Online Supplementary Figure S2D and E).
Analysis of SCF protein levels with regard to overall survival (OS) and TTFT in CLL cases revealed that patients with high SCF protein levels (SCFhigh) had significantly shorter OS (P<0.001) and TTFT (P<0.001) compared to patients with low SCF levels (SCFlow) (Figure 1D and E, Online Supplementary Table S4).
Figure 1.
Stem cell factor is overexpressed in chronic lymphocytic leukemia cells and correlated with adverse prognosis. (A, B) Comparison of the expression of stem cell factor (SCF) protein in chronic lymphocytic leukemia (CLL) cells compared to healthy tonsillar B cells (A) and in IG-unmutated (U-CLL) compared to IG-mutated (M-CLL) cells, showing overexpression in U-CLL cases (B). The dot plots show SCF protein levels normalized to β-actin from western blot studies. Each dot represents one case. (C) Representative western blot for the distinct SCF probing in samples from healthy tonsils, U-CLL and M-CLL cases. (D, E) SCFhigh cases have shorter overall survival (OS) (D) and shorter time-to-first treatment (TTFT) (E) compared to SCFlow cases, as shown by Kaplan-Meier OS and TTFT curves for CLL patients with high SCF expression (normalized SCF protein levels to β-actin >1.7 for OS and >0.93 for TTFT) and low SCF expression (normalized SCF protein expression to β-actin ≤1.7 for OS and ≤0.93 for TTFT). Cut-off points were generated using time-dependent receiver operating characteristic curve analysis. The Mann–Whitney U and the log-rank test were applied as appropriate to evaluate statistical significance; *P<0.05, **P<0.01, ***P<0.001.
Microenvironmental triggering through immune receptors induces stem cell factor expression in chronic lymphocytic leukemia
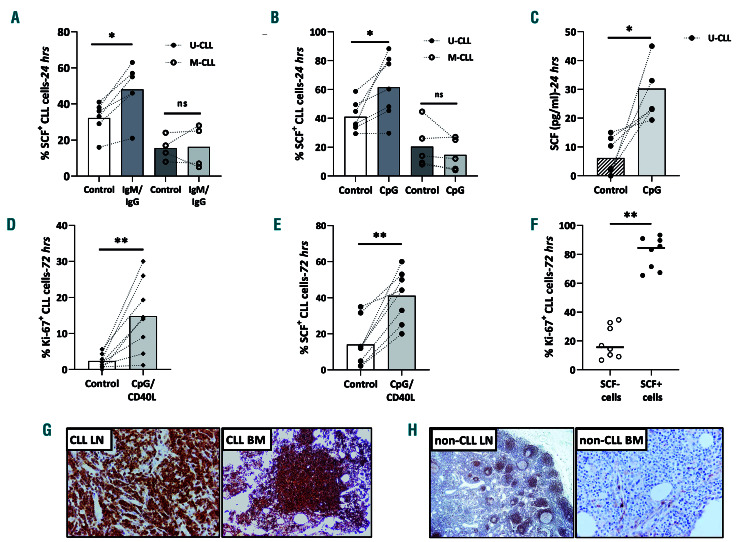
We investigated the impact of BcR activation on SCF expression after stimulating negatively selected CLL cells from U-CLL clones with anti-IgM or anti-IgG (depending on the heavy chain isotype expressed by the malignant cells) for 24 h. Western blotting and flow cytometry analysis revealed that BcR crosslinking significantly induced SCF expression in U-CLL (n=9, FD:1.6; P=0.0039 and n=6, FD:1.5; P=0.03 respectively), while, as expected, no significant change was detected in M-CLL (n=4), likely due to their overall weak responsiveness to BcR stimulation (Figure 2A, Online Supplementary Figure S3A-C).29
Subsequently, we studied the effect of TLR9 signaling on SCF expression by stimulating negatively selected malignant cells from U-CLL clones (n=7) and M-CLL clones (n=5) with CpG for 24 h. Flow cytometry analysis unveiled that, compared to untreated controls, TLR9 activation conferred a significant increase in SCF+ cells in U-CLL (FD: 1.7; P=0.0156) as opposed to a (non-significant) decrease in MCLL (FD: 1.65) (Figure 2B, Online Supplementary Figure S3D). Furthermore, enzyme-linked immunosorbent studies revealed that TLR9 activation by CpG also triggered higher secretion of soluble SCF into the supernatants of UCLL cell cultures compared to the amount secreted by untreated controls (FD: 9.3; P=0.0156) (Figure 2C).
We next investigated whether SCF expression is related to CLL proliferation. To this purpose, negatively selected malignant cells from eight CLL cases were stimulated with CpG/CD40L for 72 h and the percentages of SCF+ and proliferating (Ki-67+) CLL cells were determined. The anticipated increase in proliferating cells (FD: 7.1; P=0.0078) (Figure 2D) was accompanied by an increase in SCF+ cells (FD: 3.7; P=0.0078) (Figure 2E). Most Ki-67+ CLL cells were also SCF+ (85% Ki-67+/SCF+ CLL cells, 15% Ki-67+/SCFCLL cells; P=0.0078) (Figure 2F, Online Supplementary Figure S3E).
Prompted by this observed correlation between SCF and proliferation in CLL, we suppressed SCF expression in the proliferating MEC1 CLL cell line by short interfering (si)RNA and analyzed Ki-67 positivity in three independent biological replicates. In line with our data, SCF downmodulation by two of the four siRNA used (FD:1.2 for Si 9 and FD:2 for Si 8; P=0.05) was accompanied by decreases in the proliferative fraction of MEC1 cells (FD:1.3; P=0.05 for Si 9, FD:1.2; P=0.07 for Si 8) (Online Supplementary Figure S3F and G).
Figure 2.
Microenvironmental triggering induces stem cell factor in chronic lymphocytic leukemia cells. (A) Comparison of stem cell factor (SCF)+ cells (flow cytometry for viable/SCF+ chronic lymphocytic leukemia [CLL] cells) from IG-unmutated CLL (U-CLL) and IG-mutated (M-CLL) cases, untreated (Control) cases and cases treated with IgM/IgG. (B) Comparison of SCF+ cells (flow cytometry for viable/SCF+ CLL cells) from U-CLL and M-CLL cases, untreated (Control) cases and cases treated with CpG. (C) Comparison of secreted SCF (pg/mL; determined by enzyme-linked immunosorbent assay in cellular supernatants) from U-CLL cells, untreated (Control) and CpG-treated cells. (D, E) Comparison of proliferating cells (Ki-67+) (D) and SCF+ cells (%) (E) in untreated (Control) and CpG/CD40L-treated CLL cases, as determined by flow cytometry. (F) The majority of proliferating (Ki-67+) CLL cells were also SCF+ as determined by flow cytometry. (G) Lymph node (left) and the corresponding bone marrow biopsy (right) from the same CLL patient showing CLL invading cells with aberrant SCF expression. (H) Reactive, non-CLL lymph node (left) exhibiting SCF positivity mostly in mantle zones but not in germinal centers and reactive, non-CLL bone marrow (right) exhibiting scant SCF positivity in scarce immune and endothelial cells. Interconnected dots represent one case in two different conditions while bars represent median values. The Wilcoxon P test was applied to evaluate statistical significance; *P<0.05, **P<0.01, ***P<0.001. FD: fold difference.
To further corroborate our findings, we used immunohistochemistry to analyze the expression of SCF in biopsy samples from bone marrow (n=6) and lymph nodes (n=7) where the proliferative fraction of CLL cells reside; in two cases, concurrent, paired lymph node/bone marrow biopsy samples were available. In all cases, the vast majority of the CLL cells invading the lymph nodes and/or the bone marrow showed SCF positivity, presenting a granular cytoplasmic staining pattern; in some cases, membranous localization of the immunostain was also observed. There was no difference in staining pattern intensity between lymph nodes and bone marrow biopsies, including the two cases with matched biopsies from both sites. Endothelial cells and macrophages were also SCF immunopositive (Figure 2G, Online Supplementary Figure S4). As controls, we studied reactive lymph nodes, in which SCF positivity was observed in mantle zones of the follicles, and in some interfollicular lymphoid cells, whereas germinal centers were negative or showed moderate positivity. Moreover, in a non-CLL reactive bone marrow biopsy, sparse SCF immunopositivity was only traced in a small number of macrophages and endothelial cells (Figure 2H).
Mitochondrial reactive oxygen species, mesenchymal stroma and hypoxia regulate stem cell factor within the chronic lymphocytic leukemia microenvironment
Since SCF induction can be driven by oxidative stress, we investigated whether the observed induction of SCF expression after microenvironmental triggering in CLL cells might be related to changes in redox homeostasis. To this purpose, we quantified the mitochondrial mass as well as the membrane potential of activated CLL cells because aberrant ROS in CLL cells derive from the mitochondria.3
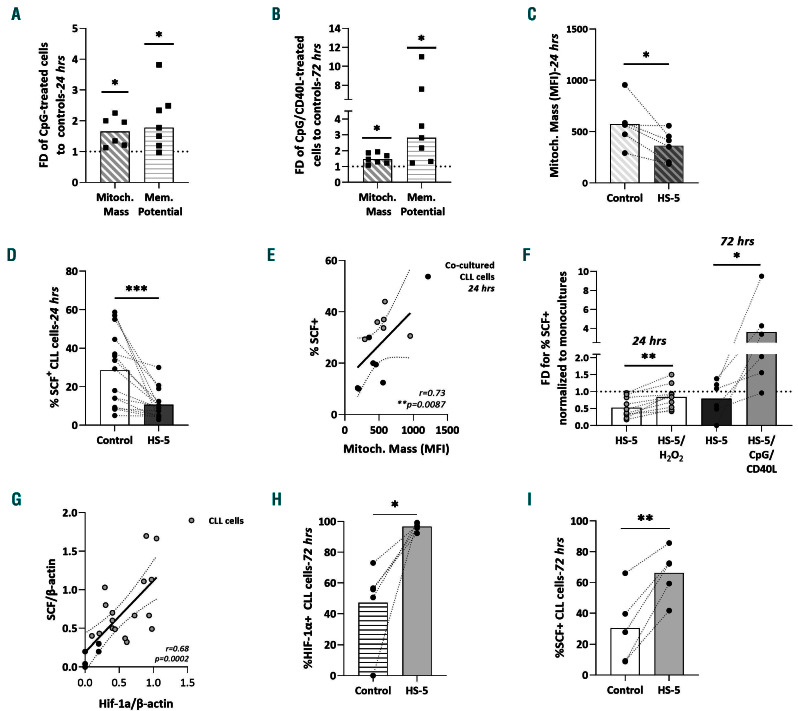
Of note, in tandem with augmented SCF expression, significant increases were found in both mitochondrial mass (n=6, FD: 1.7, P=0.01) and membrane potential (n=7, FD: 1.8; P=0.01) in TLR9-stimulated versus unstimulated CLL cells (Figure 3A, Online Supplementary Figure S5A). Moreover, in parallel to augmented proliferation/SCF levels, a significant increase in mitochondrial mass was also found in TLR9/CD40-stimulated versus unstimulated CLL cells (n=7, FD: 1.5; P=0.01). Interestingly, a similar but more pronounced effect was detected for the membrane potential of the activated CLL cells (n=7, FD: 2.8; P=0.01) (Figure 3B, Online Supplementary Figure S5B).
To further explore the possibility of a causal relationship between mitochondrial ROS and SCF regulation in CLL, we suppressed the oxidative stress of CLL cells in vitro and measured SCF levels. To this end, we used short-term (24 h) co-cultures of CLL cells with fully confluent HS-5 mesenchymal cells which are a known source of precursors for the anti-oxidant glutathione.31,32 As expected, HS-5 cells conferred significant anti-apoptotic protection to co-cultured CLL cells (n=8, FD: 1.6; P=0.0156) (Online Supplementary Figure S5C) and also decreased their mitochondrial mass, in keeping with the literature (n=6, FD: 1.5; P=0.03) (Figure 3C, Online Supplementary Figure S5D).31,32 Of note, short-term co-culture with HS-5 cells triggered a significant reduction in the number of SCF+ CLL cells (n=15, FD: 2.9; P<0.001) (Figure 3D, Online Supplementary Figure S5E) which significantly correlated with the suppression of mitochondrial mass (n=6, r=0.73; P=0.0087) (Figure 3E). Moreover, we incubated co-cultures of CLL cells from ten cases with HS-5 cells with an oxidative agent (H2O2) in order to induce ROS production and determined the number of SCF+ cells by flow cytometry. As expected, this stimulation significantly increased the number SCF+ cells (FD=2; P=0.002); an even more pronounced effect was observed when HS-5/CLL cell co-cultures were stimulated in vitro by CpG/CD40L for 72 h which, as described above, induces mitochondrial ROS (n=6, FD: 3.2; P=0.03) (Figure 3F).
Next, we explored the association of HIF-1a with SCF protein expression. HIF-1a is a major hypoxia regulator that is abnormally expressed in CLL cells even under normoxia, critically relying on both mitochondrial ROS to avoid proteasomal degradation by the PHD/pVHL proteins but also the mesenchymal stroma.33-35 Western blotting analysis of healthy tonsillar B cells (n=5) and CLL cells (n=18) unveiled significant overexpression of HIF-1a protein levels in the latter (FD: 5; P<0.001), in line with the literature (Online Supplementary Figure S5F and G).36 Interestingly, in CLL a significant correlation emerged between HIF-1a and SCF protein levels (n=23, r=0.68; P<0.001) (Figure 3G). To further strengthen the correlation between SCF and HIF-1a, we also studied long-term cocultures of SCFhigh U-CLL cells with HS-5 cells for 72 h, a setting known to simulate the bone marrow CLL milieu more closely, augmenting critical survival pathways such as hypoxia and glycolysis, and identified a significant increase in HIF-1 levels in CLL cells (n=5, FD:1.76; P=0.0192) (Figure 3H, Online Supplementary Figure S5H).35,37 The co-cultured CLL cells also manifested higher SCF positivity than untreated controls (n=5, FD: 2.67; P=0.0026), in agreement with our immunohistochemical data (Figure 3I, Online Supplementary Figure S5I).
Ibrutinib downregulates stem cell factor expression in chronic lymphocytic leukemia
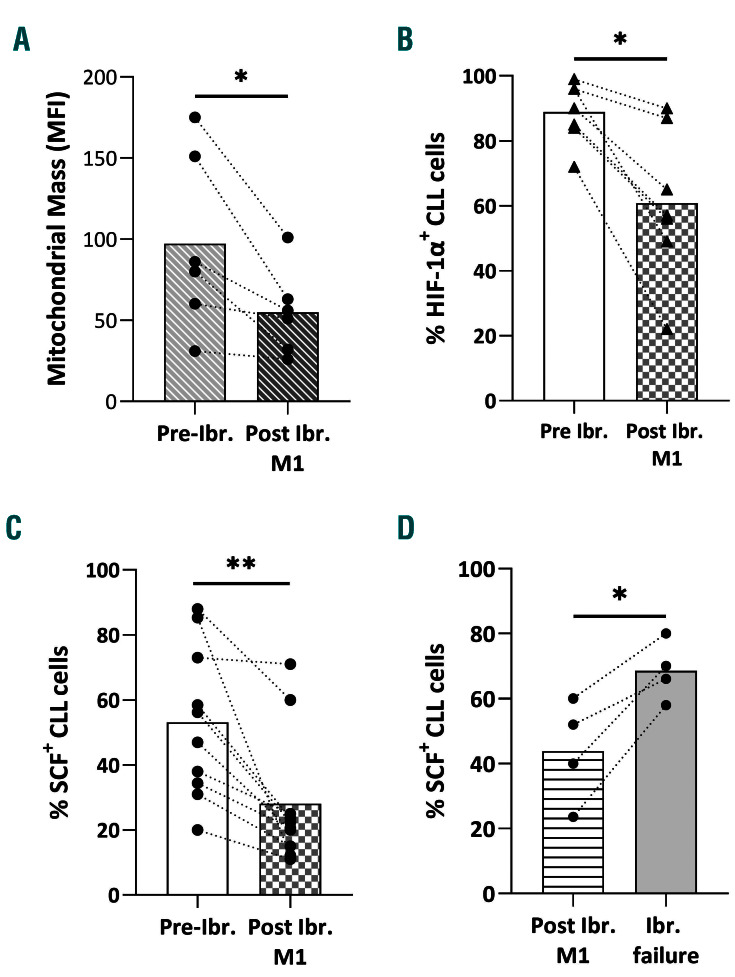
Recent literature suggests that the pharmacological mechanism of action of ibrutinib involves curtailment of inflammatory cytokines/chemokines as well as perturbations of mitochondrial dynamics.38,39 With this in mind, we assessed mitochondrial dynamics, hypoxia and SCF expression in ten CLL patients under ibrutinib therapy by profiling samples taken at two time points, namely before treatment initiation and at 1 month under treatment, and found that ibrutinib led to: (i) significant suppression of mitochondrial mass (n=6, FD: 1.6; P=0.0313) (Figure 4A, Online Supplementary Figure S6A), (ii) significant downregulation of HIF-1a expression (n=7, FD: 1.6; P=0.0156) (Figure 4B, Online Supplementary Figure S6B); and (iii) a significant decrease in SCF levels (n=10, FD: 2.4; P=0.002) (Figure 4C, Online Supplementary Figure S6C). Moreover, we analyzed longitudinal samples from four CLL patients who lost their response to ibrutinib therapy and observed that SCF expression showed a statistically significant increase at ibrutinib failure (FD:1.5, P=0.01) (Figure 4D, Online Supplementary Figure S6D).
The stem cell factor/c-kit signaling axis is active in chronic lymphocytic leukemia
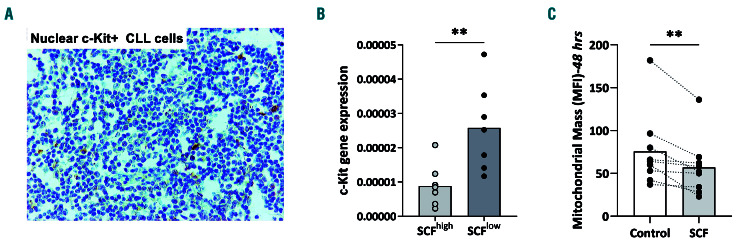
To further explore the role of SCF in the CLL microenvironment, we first analyzed the expression profile of ckit, the receptor for SCF. Immunohistochemical analysis of c-kit expression in CLL biopsies disclosed immunopositivity in tryptase-positive mast cells that were invading the lymph nodes and, mainly, the bone marrow of CLL patients in large numbers, in keeping with the literature (Online Supplementary Figure S7A and B).40 CLL cells were negative for typical surface c-kit expression. However, in some cases, a few neoplastic lymphoid cells exhibited weak c-kit nuclear immunopositivity (Figure 5A). A similar pattern emerged in RQ-PCR studies in negatively selected CLL cells in which KIT transcripts were detected at low abundance, albeit at significantly higher (FD:3.26; P=0.0039) levels in SCFlow cases (n=7) than in SCFhigh cases (n=8) (Figure 5B, Online Supplementary Figure S7C). It is also relevant to mention that KIT transcripts were detected in our previous transcriptome analysis of adverse prognosis CLL (more particularly, stereotyped subsets #6 and #8),41 hence strengthening the present observations.
Next we stimulated CLL cells with soluble SCF and measured mitochondrial dynamics. We found that SCF significantly suppressed mitochondrial mass (n=9, FD:1.2; P=0.0039) (Figure 5C, Online Supplementary Figure S7D), indicating functional SCF/c-kit signaling in CLL cells. There was no difference in the anti-oxidant effect of SCF between SCFhigh and SCFlow cases.
Figure 3.
Mitochondrial redox homeostasis modulates stem cell factor in chronic lymphocytic leukemia. (A) Mitochondrial mass (determined by mean fluorescent intensity, MFI) and membrane potential (% +) of CpG-treated IG-unmutated chronic lymphocytic leukemia (U-CLL) cells: flow cytometry analysis of viable, CpG-treated U-CLL cells normalized to respective untreated controls. (B) Mitochondrial mass (MFI) and membrane potential (% +) of U-CLL cells treated with CpG/CD40L: flow cytometry analysis of viable CpG/CD40L-treated U-CLL cells normalized to respective untreated controls. (C) Mitochondrial mass (MFI) of CLL cells co-cultured with the mesenchymal HS-5 cells. Interconnected dots represent one case in two different conditions and bars represent median values. (D) Stem cell factor (SCF) protein expression in CLL cells from short-term co-cultures with HS-5 cells: flow cytometry analysis for viable CD19+/SCF+ CLL cells. Interconnected dots represent one case in two different conditions while bars represent median values. (E) Spearman correlation of SCF protein expression and mitochondrial mass in CLL cells cultured alone (gray dots) and co-cultured with HS-5 cells (black dots). The x axis represents the MFI of MitoGreen probing for mitochondrial mass while the y axis the percentage viable CD19+/SCF+ CLL cells. (F) SCF protein expression in CLL cells co-cultured with HS-5 cells ± H2O2 and ± CpG/CD40: flow cytometry analysis of viable CD19+/SCF+ co-cultured CLL cells normalized to viable CD19+/SCF+ monocultured CLL cells. (G) Spearman correlation of SCF protein expression and HIF-1α protein expression in healthy tonsillar B cells (black dots) and CLL B cells (gray dots). The x axis represents HIF-1a protein levels while the y axis represents SCF protein levels, both normalized to β-actin; data are derived from western blot studies. (H, I) Protein expression of hypoxia-inducible factor-1α (HIF-1a) (H) and SCF (I) in SCFhigh U-CLL cells from long-term co-cultures (72 h) with HS-5 cells: flow cytometry analysis for viable CD19+/SCF+ CLL cells. Interconnected dots represent one case in two different conditions while bars represent median values. The Wilcoxon P test was applied to evaluate statistical significance. *P<0.05, **P<0.01, ***P<0.001. FD: fold difference.
Discussion
In the present study, we assessed SCF expression in a well-characterized CLL cohort and also explored how the microenvironment might modulate SCF protein levels in CLL cells. We found that SCF is overexpressed in CLL B cells compared to healthy B cells and that the membrane isoform (KL-2) predominates. Interestingly, a striking discrepancy was noted between elevated SCF protein levels versus low-abundant KITLG transcripts, indicative of posttranslational modifications or distinct decay kinetics for KITLG transcripts and SCF protein levels which merit further molecular investigation.
By correlating SCF expression with the clinicobiological characteristics of the CLL patients included in the study, we report for the first time a strong association between SCF overexpression and adverse prognosis in CLL since increased SCF protein levels were associated with several adverse prognostic biological features (e.g., CD38 and ZAP70 positivity, unmutated IGHV genes) as well as shorter overall survival and TTFT. These findings are in line with a growing body of evidence regarding the negative prognostic significance of SCF in solid tumors e.g., hepatocellular carcinoma, colorectal carcinoma, prostate cancer and pancreatic ductal adenocarcinoma.42-45 Moreover, a significant association was identified between SCF overexpression and trisomy 12, which might be attributed to a gene dosage effect considering that the KITLG gene locus is located on chromosome 12.12
Stimulation of immune receptors (BcR, TLR9, CD40) was found to modulate the levels of both membrane and soluble SCF, thus highlighting the dynamic nature of SCF expression in CLL. It is worth mentioning that strong SCF protein expression characterized the rapidly dividing, Ki- 67+ CLL cells, while SCF downmodulation using siRNA decreased the proliferation of MEC1 CLL cells. These findings are in keeping with the herein presented immunohistochemical evidence of intense expression of SCF in the bone marrow and lymph nodes of CLL patients, where the proliferating fraction of CLL cells resides.9
Our work also suggests that SCF protein levels are affected by mitochondrial redox homeostasis of CLL cells within the tumor microenvironment. Such modulation of SCF expression by oxidative stress has been previously reported in astrocytes and endothelial cells, prompting the hypothesis that in the short-term SCF contributes to compensatory cellular repair mechanisms while in the longterm it can further incite the inflammatory milieu by attracting immune cells.18,19 Recent studies in healthy B cells, T cells and macrophages showed that inflammatory responses to immune signals are wired by mitochondrial dynamics.46 In the case of CLL cells, mitochondrial ROS levels are significantly higher than in healthy B cells due to intensified mitochondrial biogenesis. This aberrant oxidative stress affects CLL cells and their surrounding cells by establishing inflammation and immune evasion in the CLL milieu.30 Our data point towards a similar trend for SCF regulation in CLL by showing for the first time that proliferating CLL cells significantly augment mitochondrial ROS in tandem with SCF protein levels.
A possible causal relationship between mitochondrial ROS and SCF was suggested by our finding that both were significantly suppressed in short-term co-culture with HS-5 mesenchymal stromal cells. In this context, the mesenchymal stroma was recently found to supply CLL cells with cysteine, thus leading to a marked increase of glutathione, alluding to the anti-oxidant capacity of mesenchymal stromal cells in the CLL microenvironment.31 Unsurprisingly, therefore, the suppression of SCF in CLL cells by HS-5 mesenchymal stromal cells was reversed once the oxidative stimuli of H2O2 and CpG/CD40L were introduced in the co-cultures.
Our study also revealed a significant correlation between the protein levels of SCF and HIF-1a in CLL cells. Recent experiments with the mitochondrial complex I inhibitor BAY 87-2243 in CLL cells have shown that, as in several other solid tumors, the stability of HIF-1a is tightly controlled by mitochondrial ROS.35 Additionally, SCF is amplified in CLL cells in parallel with HIF-1a during long term co-cultures with the mesenchymal stroma, likely as a reflection of the upregulation of glycolytic markers (e.g., c-myc).35,36,47 Moreover, proliferation centers, known to be hypoxic in CLL, were also found to be highly SCF-positive. 48 For these reasons, the herein identified association of HIF-1a with SCF cannot be viewed as paradoxical and recalls findings in certain solid malignancies such as, pancreatic ductal adenocarcinoma, in which HIF-1a can function as a transcription factor for the KITLG gene.44
Figure 4.
Ibrutinib leads to downregulation of mitochondrial mass, hypoxiainducible factor-1a and stem cell factor. (A) Mitochondrial mass in chronic lymphocytic leukemia (CLL) samples before and after 1 month of ibrutinib therapy determined by flow cytometry analysis for viable CLL cells. (B) Hypoxia-inducible factor (HIF)-1a protein expression in CLL samples before and after 1 month of ibrutinib therapy determined by flow cytometry analysis for viable HIF-1a+ CLL cells. (C) Stem cell factor (SCF) protein expression in CLL samples before and after 1 month of ibrutinib therapy determined by flow cytometry for viable SCF+ CLL cells. (D) SCF protein expression (determined by flow cytometry for viable SCF+ CLL cells) in samples from CLL patients after 1 month of ibrutinib therapy and loss of response to treatmment. Interconnected dots represent one case in two different conditions and bars represent median values. The Wilcoxon P test was applied to evaluate statistical significance. *P<0.05, **P<0.01. MFI: mean fluorescence intensity; ibr: ibrutinib
Figure 5.
Functional implications of stem cell factor in chronic lymphocytic leukemia cells. (A) Immunohistochemical staining showing weak nuclear immunopositivity for c-Kit of the scarce chronic lymphocytic leukemia (CLL) cells residing in the lymph nodes. (B) Real-time quantitative polymerase chain reaction analysis of KIT transcripts in stem cell factor (SCF)high CLL cases and SCFlow CLL cases. (C) Mitochondrial mass in CLL cells treated or not (control) with 100 ng/mL SCF for 48 h. Interconnected dots represent one case in two different conditions and bars represent median values. The Wilcoxon P test was applied to evaluate statistical significance; *P<0.05, **P<0.01, ***P<0.001. FD: fold difference.
The convergence of immunoproliferative and redox signaling for the regulation of SCF in CLL is further supported by the results of our studies with ibrutinib, a BTK inhibitor that: (i) impinges on proliferative signals conveyed to CLL cells by their neighboring cells via several receptors, such as BcR, TLR and CD40; (ii) suppresses the expression of several chemoattractants and inflammatory cytokines from CLL cells; and, (iii) dishevels the metabolism of CLL cells, inciting perturbations in their mitochondrial dynamics.38,39 Indeed, longitudinal profiling revealed significant mitochondrial changes in CLL cells after treatment, along with significant suppression of SCF and HIF-1a, thus underlining the importance of immune signaling, hypoxia and mitochondrial ROS for the overexpression of SCF in CLL.
Our study documented the presence of low-abundant KIT transcripts in CLL cells while also showing that their stimulation with soluble SCF suppressed mitochondrial mass, indicative of functional SCF/c-kit signaling. Besides a possible SCF/c-kit autocrine interaction within CLL cells, SCF may also function as a receptor with signaling capacity, as was recently revealed in thymic cells and could arguably apply to CLL cells as well.49 The detection of large numbers of c-kit+ mast cells, especially in CLL bone marrow biopsies, highlights paracrine stimulation of c-kit reactive microenvironment by SCF+ CLL cells as an alternative, not mutually exclusive mode for SCF-induced modulation of CLL cell physiology, which merits thorough investigation in future studies.
In conclusion, the present study offers novel perspectives on microenvironmental interactions in the CLL microenvironment by focusing on the previously unexplored role of SCF. On the basis of our findings, SCF emerges as an important player in the regulation of redox and proliferative homeostasis in CLL cells.40 Considering our finding that SCF is associated with adverse prognosis and modulated by the CLL microenvironment, selective targeting of SCF may represent a novel therapeutic paradigm for CLL.
Supplementary Material
Funding Statement
Funding: This work was supported in part by Greek Government Departmental funds for graduate student research distributed to the Laboratory of Pharmacology (to AST) of the Department of Pharmacy of Aristotle University of Thessaloniki, Greece; the GCH-CLL project funded by ERA NET TRANSCAN-2 Joint Translational Call for Proposals 2014 (JTC 2014); the ODYSSEUS Program, implemented under the “Action for the Strategic Development on the Research and Technological Sector”, funded by the Operational Program "Competitiveness, Entrepreneurship and Innovation" (NSRF 2014-2020) and cofinanced by Greece and the European Union (European Regional Development Fund); the NEoteRIC, H2020-ICT-2018-20/H2020-ICT-2019-2 (grant agreement number: 871330). Part of this work will be submitted to the faculty of the Department of Pharmacy of Aristotle University of Thessaloniki, Greece as part of the requirements for awarding a doctorate degree in Pharmacology to GIG.
References
- 1.Rozovski U, Keating MJ, Estrov Z. Targeting inflammatory pathways in chronic lymphocytic leukemia. Crit Rev Oncol Hematol. 2013;88(3):655-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fabbri G, Dalla-Favera R. The molecular pathogenesis of chronic lymphocytic leukaemia. Nat Rev Cancer. 2016;16(3):145-162. [DOI] [PubMed] [Google Scholar]
- 3.Schulz A, Toedt G, Zenz T, Stilgenbauer S, Lichter P, Seiffert M. Inflammatory cytokines and signaling pathways are associated with survival of primary chronic lymphocytic leukemia cells in vitro: a dominant role of CCL2. Haematologica. 2011;96(3):408-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farinello D, Wozińska M, Lenti E, et al. A retinoic acid-dependent stroma-leukemia crosstalk promotes chronic lymphocytic leukemia progression. Nat Commun. 2018;9(1):1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan X-J, Dozmorov I, Li W, et al. Identification of outcome-correlated cytokine clusters in chronic lymphocytic leukemia. Blood. 2011;118(19):5201-5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Del Giudice I, Chiaretti S, Santangelo S, et al. Stereotyped subset #1 chronic lymphocytic leukemia: a direct link between B-cell receptor structure, function, and patients’ prognosis. Am J Hematol. 2014;89(1):74-82. [DOI] [PubMed] [Google Scholar]
- 7.Karmali R, Paganessi LA, Frank RR, et al. Aggressive disease defined by cytogenetics is associated with cytokine dysregulation in CLL/SLL patients. J Leukoc Biol. 2013;93(1): 161-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aydin R, Herold T, Holdenrieder S, et al. Alterations of the chemokine microenvironment in chronic lymphocytic leukemia. Blood. 2013;122(21):1619. [Google Scholar]
- 9.Fox MF, Pontier A, Gurbuxani S, Sipkins DA. Stem cell factor expression in B cell malignancies is influenced by the niche. Leuk Lymphoma. 2013;54(10):2274-2280. [DOI] [PubMed] [Google Scholar]
- 10.Hsueh Y-S, Chang HH, Shan Y-S, et al. Nuclear KIT induces a NFKBIB-RELA-KIT autoregulatory loop in imatinib-resistant gastrointestinal stromal tumors. Oncogene. 2019;38(38):6550-6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu H, Chen X, Focia PJ, He X. Structural basis for stem cell factor-KIT signaling and activation of class III receptor tyrosine kinases. EMBO J. 2007;26(3):891-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lennartsson J, Rönnstrand L. Stem cell factor receptor/c-Kit: from basic science to clinical implications. Physiol Rev. 2012;92(4):1619-1649. [DOI] [PubMed] [Google Scholar]
- 13.Tabone-Eglinger S, Wehrle-Haller M, Aebischer N, Jacquier M-C, Wehrle-Haller B. Membrane-bound Kit ligand regulates melanocyte adhesion and survival, providing physical interaction with an intraepithelial niche. FASEB J. 2012;26(9):3738-3753. [DOI] [PubMed] [Google Scholar]
- 14.Oriss TB, Krishnamoorthy N, Raundhal M, et al. Cutting edge: MMP-9 inhibits IL-23p19 expression in dendritic cells by targeting membrane stem cell factor affecting lung IL- 17 response. J Immunol. 2014;192(12):5471-5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foster B, Zaidi D, Young T, Mobley M, Kerr B. CD117/c-kit in cancer stem cell-mediated progression and therapeutic resistance. Biomedicines. 2018;6(1):31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang B, Lei Z, Zhang G, et al. SCF-mediated mast cell infiltration and activation exacerbate the inflammation and immunosuppression in tumor microenvironment. Blood. 2018;112(4):1269-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tabone-Eglinger S, Calderin-Sollet Z, Pinon P, et al. Niche anchorage and signaling through membrane-bound Kit-ligand/c-kit receptor are kinase independent and imatinib insensitive. FASEB J. 2014;28(10):4441-4456. [DOI] [PubMed] [Google Scholar]
- 18.Genis L, Dávila D, Fernandez S, Pozo-Rodrigálvarez A, Martínez-Murillo R, Torres-Aleman I. Astrocytes require insulin-like growth factor I to protect neurons against oxidative injury. F1000Res. 2014(2):1-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Afonyushkin T, Oskolkova O V, Bochkov VN. Oxidized phospholipids stimulate production of stem cell factor via NRF2 dependent mechanisms. Angiogenesis. 2018;21(2): 229-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang M, Ma Q, Hu H, et al. Stem cell factor/ c-kit signaling enhances invasion of pancreatic cancer cells via HIF-1a under normoxic condition. Cancer Lett. 2011;303(2):108-117. [DOI] [PubMed] [Google Scholar]
- 21.Pan PY, Wang GX, Yin B, et al. Reversion of immune tolerance in advanced malignancy: modulation of myeloid derived suppressor cell development by blockade of SCF function. Blood. 2007;111(1):219-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Belloc F, Airiau K, Jeanneteau M, et al. The stem cell factor-c-KIT pathway must be inhibited to enable apoptosis induced by BCR-ABL inhibitors in chronic myelogenous leukemia cells. Leukemia. 2009;23(4):679-685. [DOI] [PubMed] [Google Scholar]
- 23.Roy L Das, Curry JM, Sahraei M, et al. Arthritis augments breast cancer metastasis: Role of mast cells and SCF/c-Kit signaling. Breast Cancer Res. 2013;15(2):R32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colmone A, Amorim M, Pontier AL, Wang S, Jablonski E, Sipkins DA. Leukemic cells create bone marrow niches that disrupt the behavior of normal hematopoietic progenitor cells. Science. 2008;322(5909):1861-1865. [DOI] [PubMed] [Google Scholar]
- 25.Hallek M, Cheson BD, Catovsky D, et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute–Working Group 1996 guidelines. Blood. 2008;111(12):5446-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Papakonstantinou N, Ntoufa S, Chartomatsidou E, et al. The histone methyltransferase EZH2 as a novel prosurvival factor in clinically aggressive chronic lymphocytic leukemia. Oncotarget. 2016;7(24):35946-35959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu HS, Clogston CL, Wypych J, et al. Posttranslational processing of membrane-associated recombinant human stem cell factor expressed in Chinese hamster ovary cells. Arch Biochem Biophys. 1992;298(1):150-158. [DOI] [PubMed] [Google Scholar]
- 28.Zazo Seco C, Serrão De Castro L, Van Nierop JW, et al. Allelic mutations of KITLG, encoding KIT ligand, cause asymmetric and unilateral hearing loss and Waardenburg syndrome type 2. Am J Hum Genet. 2015;97(5):647-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Packham G, Krysov S, Allen A, et al. The outcome of B-cell receptor signaling in chronic lymphocytic leukemia: proliferation or anergy. Haematologica. 2014;99(7):1138-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jitschin R, Hofmann AD, Bruns H, et al. Mitochondrial metabolism contributes to oxidative stress and reveals therapeutic targets in chronic lymphocytic leukemia. Blood. 2014;123(17):2663-2672. [DOI] [PubMed] [Google Scholar]
- 31.Zhang W, Trachootham D, Liu J, et al. Stromal control of cystine metabolism promotes cancer cell survival in chronic lymphocytic leukaemia. Nat Cell Biol. 2012;14(3): 276-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang W, Pelicano H, Yin R, et al. Effective elimination of chronic lymphocytic leukemia cells in the stromal microenvironment by a novel drug combination strategy using redoxmediated mechanisms. Mol Med Rep. 2015;12(5):7374-7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mills EL, Kelly B, Logan A, et al. Succinate dehydrogenase supports metabolic repurposing of mitochondria to drive inflammatory macrophages. Cell. 2016;167(2):457-470.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fuhrmann DC, Brüne B. Mitochondrial composition and function under the control of hypoxia. Redox Biol. 2017;12:208-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Griggio V, Vitale C, Todaro M, et al. HIF-1a is overexpressed in leukemic cells from TP53- disrupted patients and is a promising therapeutic target in chronic lymphocytic leukemia. Haematologica. 2020;105(4):1042-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghosh AK, Shanafelt TD, Cimmino A, et al. Aberrant regulation of pVHL levels by microRNA promotes the HIF/VEGF axis in CLL B cells. Blood. 2009;113(22):5568-5574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jitschin R, Braun M, Qorraj M, Saul D, Blanc K Le, Zenz T. Stromal cell-mediated glycolytic switch in CLL cells involves. Blood. 2015;125(22):3432-3437. [DOI] [PubMed] [Google Scholar]
- 38.Niemann CU, Herman SEM, Maric I, et al. Disruption of in vivo chronic lymphocytic leukemia tumor–microenvironment interactions by ibrutinib – findings from an investigator- initiated phase II study. Clin Cancer Res. 2016;22(7):1572-1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pelicano H, Feng L, Wierda WG, Keating MJ, Huang P. Metabolic alterations induced by ibrutinib in CLL cells as a basis for drug combinations to enhance ibrutinib therapeutic activity. Cancer Res. 2017;77(13 Suppl):4408. [Google Scholar]
- 40.Ribatti D. Mast cells in lymphomas. Crit Rev Oncol Hematol. 2016;101:207-212. [DOI] [PubMed] [Google Scholar]
- 41.Papakonstantinou N, Ntoufa S, Tsagiopoulou M, et al. Integrated epigenomic and transcriptomic analysis reveals TP63 as a novel player in clinically aggressive chronic lymphocytic leukemia. Int J Cancer. 2019;144(11):2695-2706. [DOI] [PubMed] [Google Scholar]
- 42.Bellone G, Smirne C, Carbone A, et al. KIT/stem cell factor expression in premalignant and malignant lesions of the colon mucosa in relationship to disease progression and outcomes. Int J Oncol. 2006;29(4):851-859. [PubMed] [Google Scholar]
- 43.Wang X, Ren H, Zhao T, et al. Stem cell factor is a novel independent prognostic biomarker for hepatocellular carcinoma after curative resection. Carcinogenesis. 2014;35(10):2283-2290. [DOI] [PubMed] [Google Scholar]
- 44.Gao C, Li S, Zhao T, et al. SCF, regulated by HIF-1a, promotes pancreatic ductal adenocarcinoma cell progression. PLoS One. 2015;10(3):1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alabiad MA, Harb OA, Taha HF, El Shafaay BS, Gertallah LM, Salama N. Prognostic and clinic-pathological significances of SCF and COX-2 expression in inflammatory and malignant prostatic lesions. Pathol Oncol Res. 2019;25(2):611-624. [DOI] [PubMed] [Google Scholar]
- 46.Weinberg SE, Sena LA, Chandel NS. Mitochondria in the regulation of innate and adaptive immunity. Immunity. 2015;42(3): 406-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rigoni M, Riganti C, Vitale C, et al. Simvastatin and downstream inhibitors circumvent constitutive and stromal cellinduced resistance to doxorubicin in IGHV unmutated CLL cells. Oncotarget. 2015;6(30): 1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Serra S, Brusa D, Vaisitti T, et al. Adenosine signaling mediates hypoxic responses in the chronic lymphocytic leukemia microenvironment. Blood. 2015;126(23):47-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Buono M, Thézénas ML, Ceroni A, Fischer R, Nerlov C. Bi-directional signaling by membrane- bound KitL induces proliferation and coordinates thymic endothelial cell and thymocyte expansion. Nat Commun. 2018;9(1): 1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.