Abstract
Next-generation sequencing (NGS) is used to investigate the presence of somatic mutations. The utility of incorporating routine sequencing to guide diagnosis and therapeutic decisions remains unclear. We report the findings of an observational, multicenter study that aimed to assess the impact of somatic mutation testing by NGS in a reallife setting of chronic myeloid malignancies. A total of 177 patients were enrolled, partitioned into two overlapping groups. In group A (n=94), the indication was to search for clonal hematopoiesis, in a context of suspected myelodysplastic syndrome or myeloproliferative neoplasia. In group B (n=95), the theranostic impact of somatic mutations was studied. A panel of 34 genes was used on DNA extracted from blood or bone marrow samples. Within group A, the detection of clonal hematopoiesis supported the diagnosis of chronic myeloid malignancies for 31 patients while the absence of clonal hematopoiesis ruled out the suspected diagnosis in 47 patients. Within group B, NGS identified prognostically relevant somatic mutations in 32 patients, which had a therapeutic impact in 18 cases. By determining the presence or absence of somatic mutations, the application of NGS in daily practice was found to be useful for an integrated final diagnosis in 83% of the patients. Moreover, the search for somatic mutations had a prognostic impact that led to treatment modification in 19% of the cases. This study outlines the fact that adequate implementation of new investigations may have a significant positive medico-economic impact by enabling appropriate management of patients.
Introduction
During the past decade, high-throughput sequencing has increased knowledge on the genomic landscape of hematologic malignancies. Somatic mutations have thus become new biomarkers that are useful in clinical practice to improve diagnosis, prognosis stratification and targeted treatment. In France, the implementation of next-generation sequencing (NGS) platforms by the Institut National du Cancer (INCa) has enabled physicians to investigate these markers since 2013. Non-acute myeloid malignancies, i.e., myelodysplastic syndromes (MDS) and myeloproliferative neoplasms (MPN) represent good indications for NGS because of their relatively high incidence of mutations and the ease with which they can be assessed in peripheral blood samples1-3 in time frames compatible with decision-making.
More than 40 genes are now known to be potentially recurrently mutated in MDS,1 mixed MDS/MPN syndromes2 and MPN.3 In a context of idiopathic cytopenia of undetermined significance (ICUS), the presence of somatic mutations can support the diagnosis of clonal cytopenia of undetermined significance (CCUS).4,5 In MDS, several mutations, including those of TP53, EZH2, ETV6, RUNX1 or ASXL1, have been shown to be strongly associated with decreased overall survival,6 independently of the risk group according to the Revised International Prognostic Scoring System (IPSS-R).7 Other studies have shown that the addition of molecular testing allows for a better prognostication of the outcome of patients with chronic myelomonocytic leukemia (CMML)8 or MPN.3 It has therefore been suggested that the IPSS-R and Dynamic International Prognostic Scoring System (DIPSS)9 scores could be improved by incorporating information on molecular abnormalities.10 This is possibly pertinent since targeted therapies are now emerging for the treatment of MDS and MPN, which was long limited to a few drugs of moderate efficacy, making the search for somatic mutations of relevance in order to be able to propose the best available treatment.11
Given the potential advantages of the broad screening capacity of NGS, this technique is now often used without proper published international guidelines. In order to explore the added value of somatic mutation testing by NGS, we conducted an observational multicenter study aimed at assessing the impact of somatic mutation testing by NGS in a real-life setting, focusing on the impact of implementation of this testing on diagnosis, prognosis and treatment in selected populations.
Methods
Patients
In our University Hospital, all NGS requests must be validated during regional multidisciplinary meetings. Indications for NGS analysis include challenging cases of suspected MDS or MPN in which the detection of somatic mutations could help the diagnosis, prognostic assessment or search for theranostic markers. This occurs after morphology/pathology, flow cytometry and cytogenetics have already been performed in order to apply World Health Organization (WHO) recommendations. All patients for whom NGS testing was prescribed between October 2014 and March 2019 were included in the present study. All provided their consent according to local ethical rules. In a first cohort of patients (group A), the indication for NGS was to confirm or rule out a suspected diagnosis. In a second cohort, (group B), therapeutic decisions were expected to be supported by the detection or not of prognostic somatic mutations.
Next-generation sequencing analysis
In order to detect somatic mutations, a customized, targeted panel of 34 genes (145 kbp) was applied to DNA extracted from peripheral blood or bone marrow samples (Online Supplementary Table S1). The selection of these 34 genes followed INCa recommendations published in 2013 and updated in 201612 and was completed by a review of the literature. DNA libraries, built with the Haloplex® target enrichment protocol (Agilent Technologies, Santa Clara, CA, USA), were paired-end sequenced with a MiSeq® Instrument (Illumina, San Diego, CA, USA). Data were analyzed using an in-house pipeline including trimmed reads alignment to the GRCh34 human reference genome (February 2009 assembly) with BWA-MEM, tumor variant detection by three variant callers (GATK HaplotypeCaller, VarScan and SAMTools) and variant annotations with public databases (gnomAD, COSMIC, dbSNP, ClinVar) and in silico predictors in the case of unknown variants (CADD, SIFT, PolyPhen-2, MutationTaster), using updated versions whenever available. Mutations were considered significant if they reached a good quality score (DP4), a minimum variant allele frequency of at least 1% and a minimum of 20 reads supporting the variant for hotspots or 50 reads for non-hotspot variants. Variants of undetermined significance (VUS) with no clear association to a disease were defined according to Li et al.13 Final reports were delivered to clinicians in real time, without mentioning variants identified as polymorphisms.
Group A: the impact of next-generation sequencing on diagnosis
Clonal hematopoiesis was defined by the presence of at least one somatic mutation. The detection of clonal hematopoiesis was integrated with suspected diagnoses of MDS or MPN according to the criteria detailed below.
Idiopathic cytopenia of undetermined significance and clonal cytopenia of undetermined significance
In the case of ICUS with morphological evidence of dysplasia but without MDS-related cytogenetic abnormalities, the detection of clonal hematopoiesis was used to retain a diagnosis of MDS. In the context of ICUS14 without morphological evidence of dysplasia and without MDS-related cytogenetic abnormalities, the presence of somatic mutations defined a diagnosis of CCUS.5
Chronic myelomonocytic leukemia
In cases of suspected CMML with or without minimal evidence of dysplasia, WHO criteria include the presence of acquired clonal cytogenetic or molecular genetic abnormalities (i.e., somatic mutations) as supportive of the diagnosis.15
Aplastic anemia
In a context of bone marrow hypoplasia, the absence of clonal hematopoiesis was used to exclude a diagnosis of hypoplastic MDS (hMDS). In addition, the detection of PIGA somatic mutations helped to make a diagnosis of aplastic anemia (AA).15,16
Myeloproliferative neoplasms
In cases of suspected MPN without BCR-ABL fusion and without any of the three classic driver mutations (JAK2V617F, CALR and MPL) as detected by classical methods, the WHO classification recommends searching for the most frequent accompanying mutations (e.g., ASXL1, EZH2, TET2, IDH1, IDH2, SRSF2 and/or SF3B1) to help determine the clonal nature of the disease.15
Group B: the impact of next-generation sequencing on prognosis
In group B, the diagnoses of MDS, MPN/MDS - including CMML - and MPN were established according to the WHO classification. 15
Scores of the IPSS-R,7 CMML-specific Prognostic Scoring System (CPSS),17 DIPSS9 and the MYSEC prognostic model18 were used for standard prognosis assessment of MDS, MPN/MDS, CMML and primary or secondary myelofibrosis. Molecular markers considered to indicate a poor prognosis for MDS or MPN/MDS, excluding CMML, were TP53, EZH2, ETV6, RUNX1 and ASXL1 mutations, as reported by Bejar et al.6 The detection of an SF3B1 mutation was considered a good prognostic sign.19 The Itzykson8 and/or CPSS-Mol20 scores, which respectively integrate ASXL1 mutation only or RUNX1, NRAS, SETBP1 and ASXL1, were used for the prognostic assessment of CMML. SRSF2, ASXL1, IDH1, IDH2 and EZH2 mutations were indicative of a worse prognosis in the context of myelofibrosis.21 The ASXL1 mutation was also considered to be associated with poor prognosis in patients with AA.16,22,23
Results
Patients’ characteristics
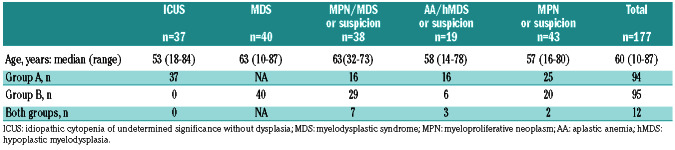
One hundred and seventy-seven patients (100 males and 77 females), originating from ten hospitals were included. Their median age was 60 years old (range, 10-87). These patients had ICUS or MDS (n=77), MPN/MDS (n=38), AA or a suspicion of hMDS (n=19) and MPN or suspected MPN (n=43) (Table 1).
NGS was performed for 94 patients in group A, within a median of 4 months of hematologic detection of anomalies. Group B consisted of 95 patients, with a median follow- up from molecular assessment to last news of 11 months. Twelve patients belonged to both groups.
Within group A, the most frequent blood count anomalies were cytopenia (68%: anemia [56%], thrombocytopenia [28%], neutropenia [30%] or pancytopenia [16%]) followed by thrombocytosis (16%), and monocytosis (13%). Rare patients had thrombosis with a suspicion of MPN but no blood count anomalies (3%). Bone marrow smears and/or biopsy evidenced no significant dysplastic anomalies in 53% of the cases. Cytogenetic abnormalities not specific for MDS, such as chromosome Y loss or chromosome 20 deletion, were observed in 8% of the cases. Karyotyping was normal in 72% or failed in 5% of the cases. No karyotyping was performed in seven cases of thrombocytosis, two cases of erythrocytosis, one cases of thrombosis with normal blood count and one case of hypereosinophilic syndrome. Before molecular assessment, the context was ICUS (with or without morphological evidence of dysplasia) (39%), suspicion of MPN/MDS (17%) or AA/hMDS (17%) (Table 1, Online Supplementary Table S2). All suspected cases of MPN (27%) were negative for the three classic driver mutations.
The majority of patients in group B were diagnosed with MDS (42%), followed by MPN/MDS (31%), MPN (21%) or hypoplasia (6%). Most patients with MDS, MPN/MDS and MPN were classified as low risk (60%) according to the respective scoring systems (Table 1).
Impact of mutations detected by next-generation sequencing on diagnosis (group A)
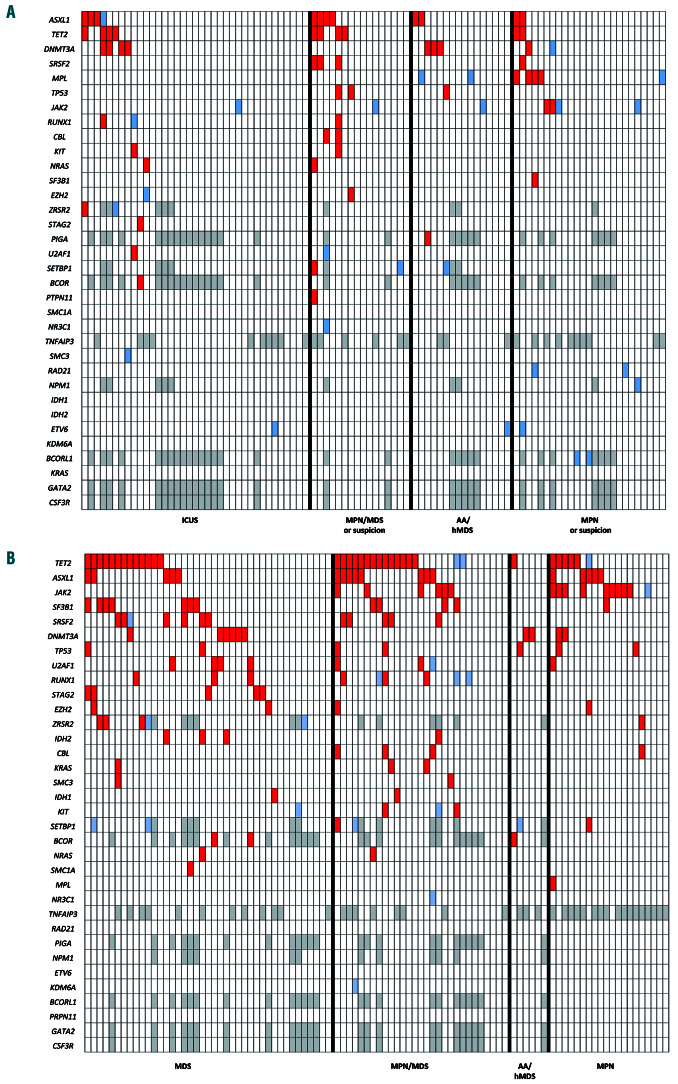
Thirty-three percent of the patients (31/94) had at least one mutation in favor of clonal hematopoiesis (range for the whole study cohort, 0-7). Twenty-six of them had a normal karyotype and five had a non-specific chromosomal abnormality (i.e., del(20q) or -Y). The most frequently mutated genes were ASXL1 (12%), TET2 (11%) and DNMT3A (9%) (Figure 1A, Online Supplementary Table S2). Patients with mutations were significantly older (median: 67 years vs. 48 years in those without mutations; P<0.001) but no difference in frequency of mutations was seen according to the context of suspected disease. Patients with clonal hematopoiesis also had significantly more myelemia (1.6% vs. 0.2%; P=0.005) and a higher red cell distribution index (16% vs. 14%; P=0.01). The signs of dysplasia observed on bone marrow smears and/or biopsy were not associated with the presence of a detected somatic mutation (P=not significant).
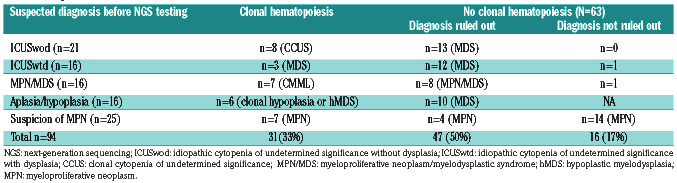
The detection of clonal hematopoiesis by NGS allowed us to retain the diagnosis of myeloid malignancy for 17 patients (18%) (3 MDS, 7 MPN/MDS and 7 MPN), eight cases of CCUS and six cases of bone marrow hypoplasia with clonal hematopoiesis (Table 2). In most patients with AA/hMDS (n=10/16), the absence of clonal hematopoiesis favored a diagnosis of idiopathic AA, while clonal hematopoiesis detected in six cases supported a diagnosis of hMDS. Thirteen mutations were observed in seven patients with suspected MPN. Among those, four had MPL mutations, respectively two subclonal hotspot p.Trp515Leu (not previously detected by Sanger sequencing) and three non-canonical mutations (p.Trp515Ser, p.His499Valfs*46, p.Tyr591Asp).
Considering the patients without detected clonal hematopoiesis (n=63), the initial suspected diagnosis of myeloid malignancy was ruled out in 47/63 (75%) of the cases including 25/26 suspected MDS (96%), 8/9 suspected MPN/MDS (89%), all suspected hMDS (10/10) and 4/18 suspected MPN (22%). In these patients, cytopenias were ultimately mainly considered as of peripheral origin or ICUS and monocytosis as reactive. In the absence of clonal hematopoiesis, no diagnosis could be ruled out for most suspected cases of MPN (14/18) (Table 2, Online Supplementary Table S2) because of morphological anomalies on bone marrow biopsy (data not shown).
VUS were found in 26 cases (28%) (Figure 1A, Online Supplementary Table S2). The most frequent ones were observed in JAK2 (p.Asn1108Ser, p.Arg1063His, p.Ile35Thr), MPL (c.1565+5C>T, p.Pro70Leu, p.Gln433Arg, p.Ala622Pro), ETV6 (p.Ala329Thr, p.Pro223Leu, p.Ala329Thr) and BCORL1 (p.Arg21His, p.Ala612Thr, p.Ile1022Thr, p.Arg1183Gln). Among patients with VUS, 14 had no clear somatic mutations, no proof of clonal hematopoiesis and no significant association with a given suspected context. Of note, identification of a poor prognostic mutation led to intensification of therapy in 3/12 patients in group A.
Table 1.
Partition of the patients according to diagnosis or suspected diagnosis.
Figure 1.
Profile and frequency of mutations in the two study groups. (A) Profile of mutations according to the suspected context in diagnosis group A. Red: somatic mutations; blue: variant of undetermined significance (VUS); gray: not evaluated. (B) Profile of mutations according to the proven pathology in prognosis group B. Red: somatic mutations; blue: VUS, gray: not evaluated. ICUS: idiopathic cytopenia of undetermined significance without dysplasia; MDS: myelodysplastic syndrome; MPN: myeloproliferative neoplasm; AA: aplastic anemia; hMDS: hypoplastic myelodysplasia.
Table 2.
Diagnostic assessment.
Table 3.
Prognostic evaluation according to the pathology.
Table 4.
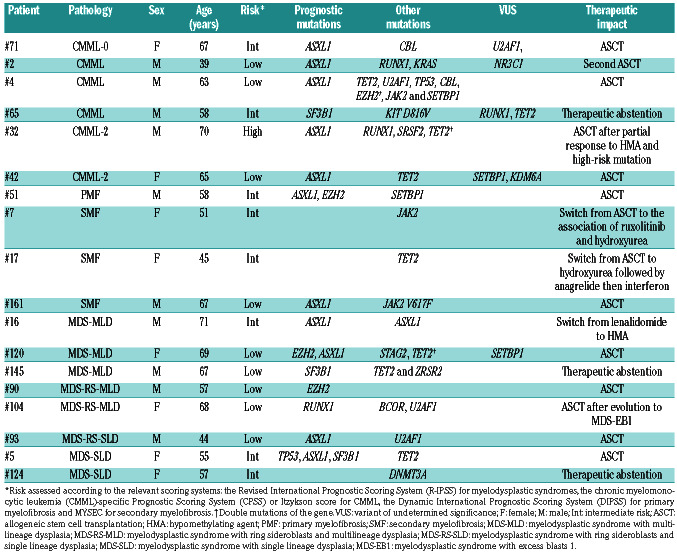
Patients in whom therapy was changed in the light of the next-generation sequencing data.
Prognostic/theranostic impact of mutations detected by next-generation sequencing (group B)
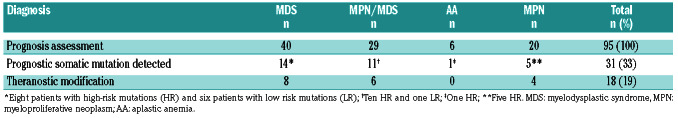
The median number of mutations in group B was two (range for the whole cohort, 0-9) and 75% of these patients had at least one mutation (Figure 1B, Online Supplementary Table S2). NGS identified prognostic mutations in 31/95 (33%) patients at different proportions according to the pathology, i.e., 14/40 (35%) in MDS, 11/29 (38%) in MPN/MDS, 5/17 (29%) in myelofibrosis and 1/6 (17%) in AA. Among the 24 patients with poor prognostics mutations, the conclusion was to proceed to an allogeneic stem cell transplantation for 12 of them and to start a hypomethylating agent in another one (Table 4). Among these patients, ASXL1 was the most frequent poor prognostic marker with therapeutic impact (n=11). Conversely, SF3B1, a mutation associated with a good prognosis, was identified in six cases of MDS and one of MPN/MDS. In five patients, this information led to a deescalation of treatment intensity, i.e., allogeneic stem cell transplantation was postponed because of the presence of a good prognostic mutation (SF3B1) or absence of poor prognostic mutations.
Of note, VUS were observed in 18/95 cases, three being detected in RUNX1, thereby potentially conveying a poor prognosis.
Discussion
The interest of high-throughput NGS has been widely demonstrated in onco-hematology, mostly in clinical trials, but evaluation of these new molecular data outside clinical research is lacking. In the same way as for the usage of a drug, it appears essential to evaluate the medico-economic impact on clinical decisions of the use of these new tools, which are modifying the workload and costs in diagnostic laboratories.24 Nevertheless, the use of high-throughput sequencing to aid diagnosis and treatment decisions in chronic myeloid malignancies has seldom been evaluated in “real life”. Here, we retrospectively examined the impact of NGS assays on a series of 177 patients from ten centers. The impact on diagnosis and therapeutic decisions were assessed separately by dividing the patients into two groups.
The main objective of NGS for diagnostic purposes is the search for somatic mutations to provide or not proof of clonal hematopoiesis in favor of myeloid malignancies when standard diagnostic criteria for the disease have not been reached with classical tests (cytology, cytogenetics, pathology). ASXL1, DNMT3A and TET2mutations are the most frequent but can be age-related25 and observed in the hematopoietic cells of apparently healthy older individuals without MDS i.e., clonal hematopoiesis of indeterminate potential (CHIP).26 Consequently, the interpretation of such molecular abnormalities in subjects over 70 years of age requires a multidisciplinary discussion. Clonal hematopoiesis was identified here in 33% of the patients and a firm diagnosis of myeloid malignancy was retained in 18%. The diagnosis of CCUS was reached in 9% of the patients and had an impact on the continuation of clinical follow-up because there was a higher risk that these patients would develop a myeloid neoplasm.5,27 Conversely, because more than 85% patients with MDS have one or more somatic mutations that can be detected using a minimal panel of recurrently mutated genes,1 the absence of mutation provided significant help to exclude a diagnosis of MDS in the vast majority of cases. In the context of aplasia, the absence of clonal hematopoiesis and the presence of a PIGA mutation (mostly associated with AA) helped to exclude a diagnosis of hMDS.16,23
Overall, in this series, the search for clonal hematopoiesis by NGS was useful for 83% of the patients, allowing a diagnosis to be confirmed (33%) or excluded (50%). Nevertheless, it is essential to integrate these results with clear diagnostic criteria.28 The absence of detection of clonal hematopoiesis could interestingly help to exclude MDS, hMDS or mixed MPN/MDS but not MPN.
Molecular studies are also interesting to guide treatment decisions, especially through prognostic evaluation. In group B, 33% of the patients had a molecular anomaly with a reported prognostic impact. Treatment was modified for 19%, in most cases with a reinforcement of therapy and/or a decision to perform allografting.
Integration of a search for molecular abnormalities by NGS is thus shown here to have a clear impact on patients’ therapeutic management provided that it complements a thorough diagnostic algorithm and multidisciplinary indication. Currently, decisions on therapeutic intensification are rarely based on the integration of molecular data given the lack of international consensus.
These new tools also reveal the presence of VUS for which the somatic or constitutional origin as well as the pathogenicity are still unknown. This may be problematic when such VUS are observed in genes whose mutations carry a prognostic impact. Our data show that in the cases of VUS in RUNX1, patients were not ultimately considered as being at high risk. Non-hematopoietic DNA testing and updating of international databases for the interpretation of these VUS are required. Among patients for whom treatment has not been modified, a minority was found to harbor a high-risk mutation and a majority of cases lacked risk-conferring mutations. Again, NGS results cannot be considered individually but should be integrated with the usual IPSS-R markers.29
In summary, we report a benefit in terms of diagnosis and therapeutic impact for, respectively, 83% and 19% of the patients in a large real-life cohort. This outcome is interesting given that decisions to run these tests were made in a critically explored context and not in a systematic fashion. The medico-economic implications of such integrative and multidisciplinary prescription and analysis deserve specific, thorough investigations. The latter would involve a calculation of the savings generated by promptly stopping investigations and alleviating treatment after ruling out a neoplasm, including the reduced costs generated by the decision of not performing an allograft. This should be balanced by the cost of adapted therapy resulting from the positive identification of a neoplastic disease. Such a large scale microcosting evaluation was not the aim of this study. It can however already be suspected, from the results of this work, that a proper use of relatively expensive NGS assays is largely balanced by an improved and more reasoned therapeutic management of patients.
Supplementary Material
Acknowledgments
The authors would like to thank Laetitia Aubert, Marie- Christine Boursier, Emilie Brangoulo, Cécile Lagarde, Veronique Chenais and Amandine Sebie for their excellent technical help with molecular analyses. We would also like to thank all the members of the HUGO group involved in this work.
References
- 1.Papaemmanuil E, Gerstung M, Malcovati L, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122(22):3616-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patel BJ, Przychodzen B, Thota S, et al. Genomic determinants of chronic myelomonocytic leukemia. Leukemia. 2017;31(12):2815-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grinfeld J, Nangalia J, Baxter EJ, et al. Classification and personalized prognosis in myeloproliferative neoplasms. N Engl J Med. 2018;379(15):1416-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Valent P, Orazi A, Steensma DP, et al. Proposed minimal diagnostic criteria for myelodysplastic syndromes (MDS) and potential pre-MDS conditions. Oncotarget. 2017;8(43):73483-73500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malcovati L, Gallì A, Travaglino E, et al. Clinical significance of somatic mutation in unexplained blood cytopenia. Blood. 2017;129(25):3371-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bejar R, Stevenson K, Abdel-Wahab O, et al. Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med. 2011;364(26):2496-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greenberg PL, Tuechler H, Schanz J, et al. Revised International Prognostic Scoring System for myelodysplastic syndromes. Blood. 2012;120(12):2454-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Itzykson R, Kosmider O, Renneville A, et al. Prognostic score including gene mutations in chronic myelomonocytic leukemia. J Clin Oncol. 2013;31(19):2428-2436. [DOI] [PubMed] [Google Scholar]
- 9.Passamonti F, Cervantes F, Vannucchi AM, et al. A Dynamic International Prognostic Scoring System (DIPSS) predicts progression to acute myeloid leukemia in primary myelofibrosis. Blood. 2010;116(15):2857-2858. [DOI] [PubMed] [Google Scholar]
- 10.Guglielmelli P, Lasho TL, Rotunno G, et al. MIPSS70: mutation-enhanced international prognostic score system for transplantation- age patients with primary myelofibrosis. J Clin Oncol. 2018;36(4):310-318. [DOI] [PubMed] [Google Scholar]
- 11.Bejar R, Steensma DP. Recent developments in myelodysplastic syndromes. Blood. 2014;124(18):2793-2803. [DOI] [PubMed] [Google Scholar]
- 12.https://www.ecancer.fr/content/download/64146/575855/file/Listes_genes_minimales_analyse_usage_visee_diagnostique_NGS_fevrier_2016.pdf Last accessed February 2, 2020. [Google Scholar]
- 13.Li MM, Datto M, Duncavage EJ, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 2017;19(1):4-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valent P, Horny HP, Bennett JM, et al. Definitions and standards in the prognosis and treatment of the myelodysplastic syndromes: consensus statements and report from a working conference. Leuk Res. 2007;31(6):727-736. [DOI] [PubMed] [Google Scholar]
- 15.Swerdlow SH. International Agency for Research on Cancer. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Revised 4th edition. Lyon, 2017. [Google Scholar]
- 16.Stanley N, Olson TS, Babushok DV. Recent advances in understanding clonal haematopoiesis in aplastic anaemia. Br J Haematol. 2017;177(4):509-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Such E, Germing U, Malcovati L, et al. Development and validation of a prognostic scoring system for patients with chronic myelomonocytic leukemia. Blood. 2013; 121(15):3005-3015. [DOI] [PubMed] [Google Scholar]
- 18.Passamonti F, Giorgino T, Mora B, et al. A clinical-molecular prognostic model to predict survival in patients with post-polycythemia vera and post essential thrombocythemia myelofibrosis. Leukemia. 2017; 31(12):2726-2731. [DOI] [PubMed] [Google Scholar]
- 19.Malcovati L, Papaemmanuil E, Bowen DT, et al. for the Chronic Myeloid Disorders Working Group of the International Cancer Genome Consortium and of the Associazione Italiana per la Ricerca sul Cancro Gruppo Italiano Malattie Mieloproliferative. Clinical significance of SF3B1 mutations in myelodysplastic syndromes and myelodysplastic/myeloproliferative neoplasms. Blood. 2011;118(24): 6239-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elena C, Gallì A, Such E, et al. Integrating clinical features and genetic lesions in the risk assessment of patients with chronic myelomonocytic leukemia. Blood. 2016;128(10):1408-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vannucchi AM, Lasho TL, Guglielmelli P, et al. Mutations and prognosis in primary myelofibrosis. Leukemia. 2013;27(9):1861-1869. [DOI] [PubMed] [Google Scholar]
- 22.Kulasekararaj AG, Jiang J, Smith AE, et al. Somatic mutations identify a subgroup of aplastic anemia patients who progress to myelodysplastic syndrome. Blood. 2014;124(17):2698-2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshizato T, Dumitriu B, Hosokawa K, et al. Somatic mutations and clonal hematopoiesis in aplastic anemia. N Engl J Med. 2015;373(1):35-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garrison LP, Neumann PJ, Erickson P, Marshall D, Mullins CD. Using real-world data for coverage and payment decisions: the ISPOR Real-World Data Task Force report. Value Health. 2007;10(5):326-335. [DOI] [PubMed] [Google Scholar]
- 25.Genovese G, Kähler AK, Handsaker RE, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014; 371(26):2477-2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steensma DP, Bejar R, Jaiswal S, et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015; 126(1):9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cargo CA, Rowbotham N, Evans PA, et al. Targeted sequencing identifies patients with preclinical MDS at high risk of disease progression. Blood. 2015;126(21):2362-2365. [DOI] [PubMed] [Google Scholar]
- 28.Malcovati L, Papaemmanuil E, Ambaglio I, et al. Driver somatic mutations identify distinct disease entities within myeloid neoplasms with myelodysplasia. Blood. 2014;124(9):1513-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nazha A, Narkhede MS, Radivoyevitch T, et al. The Revised International Prognostic Scoring System “Molecular” (IPSS-Rm), a validated and dynamic model in treated patients with myelodysplastic syndromes (MDS). Blood. 2015;126(23):607. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.