In order to identify novel prognostic markers and actionable targets, we performed whole-exome/-transcriptome sequencing of relapsed childhood B-cell precursor (BCP) acute lymphoblastic leukemia (ALL) diagnosed in the Czech Republic. In patients with “B-other ALL” (BCP-ALL negative for ETV6-RUNX1, BCR-ABL1, TCF3-PBX1, and KMT2A rearrangement, hyperdiploidy [51-67 chromosomes] and hypodiploidy [<44 chromosomes]), we found recurrent mutations of codons H1038 and Q1072 of the zinc finger E-box binding homeobox 2 (ZEB2) gene. The ZEB2 gene is located on 2q22.3 and encodes a transcriptional repressor implicated in the pathogenesis of early T-cell ALL, possibly via deregulation of IL7R-JAK/STAT signaling.1,2 It is known that codons H1038 and Q1072 are located in the C-terminal zinc finger coding-region (DNA binding region), but the impact of the mutations (ZEB2mut) on ZEB function is unknown. Although recurrence of ZEB2mut in BCP-ALL is being traced in a growing number of genomic studies,3-6 the biological and clinical roles of these mutations in BCP-ALL have not been described so far.
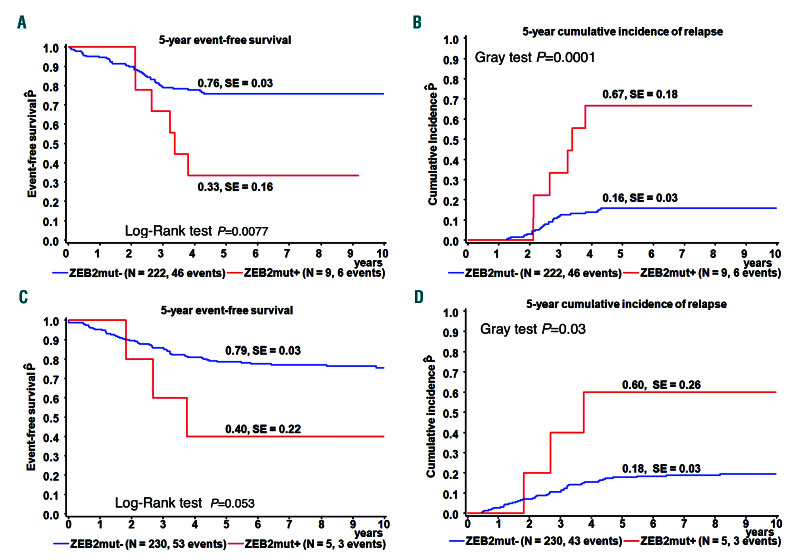
In this study, we aimed to determine the frequency and clinical relevance of ZEB2mut in childhood B-other ALL. Using whole-exome sequencing, RNA-sequencing and amplicon sequencing of DNA or complementary DNA (cDNA), we analyzed 231 and 36 Czech children with newly manifesting and relapsed B-other ALL, respectively (the so-called “discovery cohorts”), consecutively diagnosed between November 2002 and December 2017 and treated according to several successive Berlin-Frankfurt- Münster (BFM)-based protocols (see Online Supplementary Data). We detected ZEB2mut in 3.8% (9/231) of newly diagnosed B-other ALL. ZEB2mut was associated with significantly worse event-free survival and a higher relapse rate (Figure 1A and B). In accordance with the higher relapse rate, we found significant enrichment of ZEB2mut in the discovery relapse cohort (8/36, 29%; P=0.0005). Interestingly, two out of the total nine relapses were isolated extramedullary relapses and one was a combined extramedullary and bone marrow relapse.
To validate these findings, we analyzed 626 and 102 children with newly diagnosed and relapsed B-other ALL diagnosed and treated in Germany (the so-called “validation cohorts”). The frequency of ZEB2mut in the validation cohort of newly manifesting leukemias (enrolled into the Associazione Italiana di Ematologia e Oncologia Pediatrica [AIEOP]-BFM ALL 2000 and 2009 protocols) was 2.7% (17/626). While ZEB2mut was associated with a significantly higher relapse frequency in the patients enrolled into the AIEOP-BFM ALL 2000 study (Figure 1C and D), only 1/12 ZEB2mut patients enrolled in the AIEOP-BFM ALL 2009 study relapsed and two patients died without relapse. In the validation relapse cohort, ZEB2mut frequency was 4.9% (5/102), which was significantly lower than that in the discovery relapse cohort.
Figure 1.
Outcome analysis. (A-D) Event-free survival and cumulative incidence of relapse at 5 years according to ZEB2mut status in patients with newly diagnosed B-other acute lymphocytic leukemia in the discovery cohort (A, B) and those from the validation cohort who were treated on the AIEOP-BFM ALL 2000 protocol (C,D). SE: standard error.
Table 1.
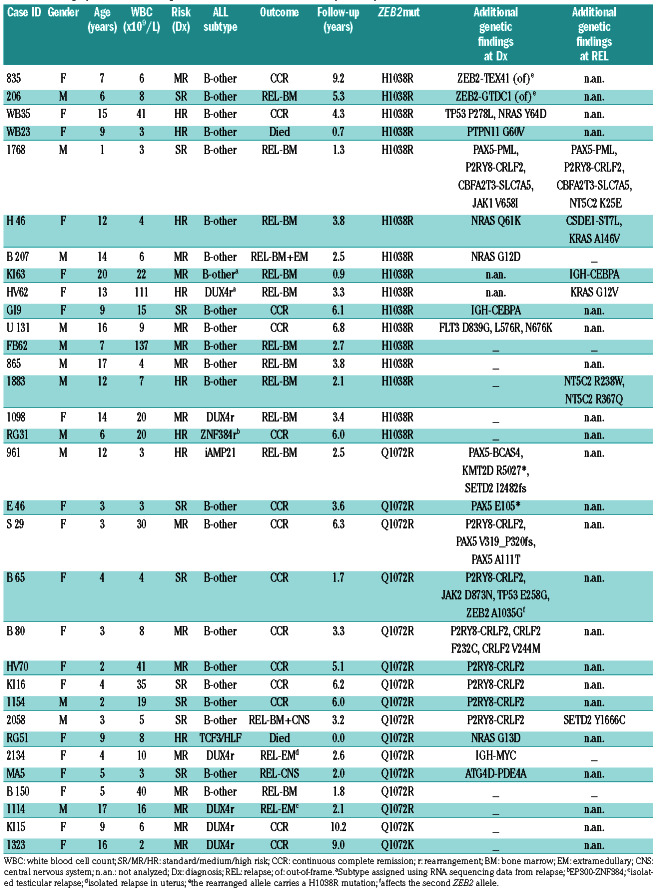
Demographic, clinical and genetic characteristics of ZEB2mut-positive patients.
Figure 2.
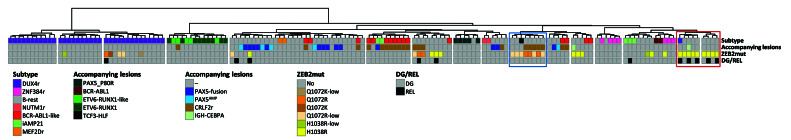
Unsupervised hierarchical clustering. Unsupervised hierarchical clustering based on the expression of 800 most differentially expressed genes (across the whole sample set, n =156) was performed using the ward.D method and Euclidean distance linkage. The figure shows the resultant dendrogram. The sample set includes samples from the patients with ZEB2-mutation-positive B-other acute lymphocytic leukemia (ALL) in the present study and samples from the patients with B-other ALL, BCR-ABL1-positive ALL and ETV6-RUNX1-positive ALL from our previous study.15 The BCR-ABL1-positive and ETV6-RUNX1-positive samples were included in the sample set to demonstrate the strength of co-clustering of ETV6-RUNX1-like and BCR-ABL1-like samples with their fusion-positive counterparts. The ETV6-RUNX1-like and BCR-ABL1-like samples were identified by supervised hierarchical clustering analyses (Online Supplementary Figure S1). The left-to-right order of samples can be found in Online Supplementary Table S4. ZEB2 H1038 and Q1072 mutation clusters are highlighted by red and blue boxes, respectively. DG: diagnosis; REL: relapse; r: rearrangement; low: variant allele frequency at the DNA level (or complementary DNA level if DNA not analyzed) <20%.
The median time to relapse in all ZEB2mut-positive patients was 2.6 years from original diagnosis (range, 0.9-5.3); the median follow-up time of those who remained in first continuous complete remission was 6.1 years (range, 1.7-10.2). Among 24 ZEB2mut-positive patients treated on frontline protocols utilizing minimal residual disease-based risk stratification, 7, 11 and 6 patients were stratified to standard, medium and high risk arms, respectively.
Of the total 32 ZEB2mut patients identified within our study (for patients’ characteristics including distribution into cohorts, see Table 1 and Online Supplementary Table S1), the ZEB2mut was already detected at the initial manifestation of leukemia in 29. Among these patients, we found a male to female ratio of 0.53, a median age at diagnosis of 8 years (10/29 patients were ≥10 years old) and a median initial white blood cell count of 8.2x109/L (only 1 patient had a white blood cell count ≥50x109/L). The H1038R, Q1072R and Q1072K mutations were found in 14, 13 and 2 patients, respectively. At the DNA level, the diagnostic variant allele frequencies of ZEB2mut ranged from 2% to 79% (variant allele frequency ≤20% in 12/25 cases), pointing to a frequent subclonality (and thus an unlikely primary origin of ZEB2mut). Variant allele frequencies of ZEB2mut alleles at the transcript level were mostly higher or comparable to those at the DNA level. Among 14 analyzed patients with paired diagnostic/relapse samples, the ZEB2mut present at diagnosis was preserved in ten and lost in one patient, while in three patients the ZEB2mut from relapse was undetectable at diagnosis and thus potentially newly gained.
We utilized RNA sequencing to investigate the presence of subtype-defining genetic lesions, subtype-defining gene-expression signatures4,6-14 and other known genetic lesions in ZEB2mut-positive ALL.
Of 32 ZEB2mut-positive ALL cases, six were classified as DUX4-rearranged; the TCF3-HLF fusion and ZNF384 rearrangement were found in one case each, while no subtype-defining lesion was detected by RNA sequencing in 24 cases. Among these, iAMP21 was found by routine cytogenetic investigation in one case, while it was not consistently investigated in all patients. ETV6- RUNX1-like and BCR-ABL1-like gene expression signatures were not detected in any patient (Online Supplementary Figure S1). Thus, the majority of ZEB2mutpositive cases (23/32; 72%) did not belong to any established ALL subtype.9
Variant and fusion analyses of RNA sequencing data revealed that additional genes/pathways were recurrently affected in ZEB2mut-positive cases (Table 1). Of the 28 ZEB2mut cases analyzed by RNA sequencing at initial leukemia manifestation, seven had the P2RY8-CRLF2 fusion. Of note, this fusion was significantly enriched in cases with the Q1072 mutations compared to those with the H1038 mutation (47% vs. 0%, P=0.007). On the other hand, Ras pathway-activating mutations (mutations in NRAS, PTPN11 and FLT3) tended to be more frequent in cases with the H1038 mutation (38% vs. 7%, P=0.07). Interestingly, in addition to genetic differences, patients with the two ZEB2mut types also differed by age. Patients with an H1038 ZEB2mut were significantly older than those with a Q1072 ZEB2mut (median age 12.4 vs. 3.8 years; P=0.02). There was no significant difference in white blood cell count and a modest trend towards a higher frequency of males in H1038 ZEB2mutpositive patients than in Q1072 ZEB2mut-positive ones (P=0.13).
Other recurrently somatically affected genes were PAX5 (n=2) and TP53 (n=2). One patient harbored the IGH-CEBPA fusion, which was also found in an additional patient at relapse (unfortunately, in this patient we could not investigate its presence at initial diagnosis). Of note, a recent transcriptomic study described a rare subgroup of BCP-ALL defined by gene expression signature, which was enriched for the ZEB2 H1038 mutation and the IGH-CEBPE fusion (likely functionally similar to IGH-CEBPA).3 However, unlike in our patients, these two genetic lesions did not co-occur in individual patients with the respective gene expression signature in the cited study. Last but not least, three of 32 patients harbored additional aberrations of ZEB2 itself: a missense mutation affecting the second allele was found in one patient and out-of-frame fusions predicted to result in C-terminal truncation and modification of the ZEB2 protein were found in two patients (Table 1, Online Supplementary Figure S2); in both, the H1038 mutation was located on the rearranged allele.
We further utilized RNA sequencing data to analyze the potential presence of gene expression signatures associated with ZEB2mut. Unsupervised hierarchical clustering analyses revealed clusters that were enriched for either Q1072 or H1038 ZEB2mut (Figure 2). None of these clusters involved all patients with the respective ZEB2mut, and, moreover, the “H1038 ZEB2mut cluster” involved additional patients without ZEB2mut. Thus, ZEB2mut seems to impact the gene expression signature, albeit less specifically and prominently compared to (at least some) subtype-defining genetic lesions (e.g., DUX4r, ETV6-RUNX1).9,15 Whether the potential difference in gene expression signatures associated with H1038 versus Q1072 ZEB2mut truly derives from the ZEB2mut type or whether it is (also) attributable to differences in the spectra of accompanying genetic lesions remains unclear.
In summary, in this study we determined the frequency of H1038/Q1072 ZEB2mut in newly diagnosed and relapsed cases of pediatric B-other ALL and the impact of these mutations on outcome. While the discovery part of the study suggested that ZEB2mut is a potential risk factor, and the overall data from the discovery plus validation cohorts also confirmed significant enrichment of ZEB2mut in relapsed cases (26/857 at diagnosis vs. 13/138 at relapse; P=0.0013), the original finding was only partly validated in independent cohorts. We may only speculate whether differences in treatment between the AIEOP-BFM ALL 2000 and 2009 protocols, potential selection biases in validation cohorts (that were non-consecutive and the validation relapse cohort did not include isolated extramedullary relapses) and/or small numbers of ZEB2mut-positive patients could have contributed to these partially discordant findings. Although our study included more than 800 cases of newly diagnosed B-other ALL, the limited number of ZEB2mut-positive patients did not allow us to analyze the potential association of all biological and clinical features on relapse risk in deep detail. Nevertheless, we found no significant association between relapse occurrence and ZEB2mut type, ZEB2mut variant allele frequency, ALL subtype, concomitant genetic lesions, sex (although males tended to relapse more frequently than females: 70% vs. 32% relapse incidence, P=0.06), age, white blood cell count or risk. Given the relatively low frequency of ZEB2mut, and potential dependence of its prognostic impact on additional genetic/clinical factors, other studies on well-characterized larger cohorts would be needed to further elucidate the clinical relevance of the mutations, as well as biological studies addressing their functional consequences.
We found genetic and demographic differences between patients with distinct ZEB2 mutations. There are currently no experimental data supporting an assumption that distinct ZEB2mut types have the same (yet unknown) functional impact and biological consequences; moreover, ZB2 function can be further altered in patients with ZEB2 fusions. Thus, although we did not observe differences in relapse rate with respect to mutation type in our study, sub-analyses considering mutation types as separate entities should be emphasized in future studies.
Supplementary Material
Funding Statement
Funding: this study was supported by grants from the Czech Health Research Council (NV15-30626A) and Charles University (Primus/MED/28, UNCE 204012) and by the project (Ministry of Health, Czech Republic) for conceptual development of research organization 00064203 (University Hospital Motol, Prague, Czech Republic). The research infrastructure was supported by the Ministry of Education, Youth and Sports (NPU I n. LO1604 and LM2015091).
References
- 1.Goossens S, Peirs S, Van Loocke W, et al. Oncogenic ZEB2 activation drives sensitivity toward KDM1A inhibition in T-cell acute lymphoblastic leukemia. Blood. 2017;129(8):981-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goossens S, Radaelli E, Blanchet O, et al. ZEB2 drives immature Tcell lymphoblastic leukaemia development via enhanced tumourinitiating potential and IL-7 receptor signalling. Nat Commun. 2015; 6:5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li JF, Dai YT, Lilljebjorn H, et al. Transcriptional landscape of B cell precursor acute lymphoblastic leukemia based on an international study of 1,223 cases. Proc Natl Acad Sci U S A. 2018; 115(50):E11711-E11720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu YF, Wang BY, Zhang WN, et al. Genomic profiling of adult and pediatric B-cell acute lymphoblastic leukemia. EBioMedicine. 2016; 8:173-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schroeder MP, Bastian L, Eckert C, et al. Integrated analysis of relapsed B-cell precursor acute lymphoblastic leukemia identifies subtype-specific cytokine and metabolic signatures. Sci Rep. 2019; 9(1):4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang J, McCastlain K, Yoshihara H, et al. Deregulation of DUX4 and ERG in acute lymphoblastic leukemia. Nat Genet. 2016; 48(12):1481-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirabayashi S, Ohki K, Nakabayashi K, et al. ZNF384-related fusion genes define a subgroup of childhood B-cell precursor acute lymphoblastic leukemia with a characteristic immunotype. Haematologica. 2017;102(1):118-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu Z, Churchman M, Roberts K, et al. Genomic analyses identify recurrent MEF2D fusions in acute lymphoblastic leukaemia. Nat Commun. 2016;7:13331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gu Z, Churchman ML, Roberts KG, et al. PAX5-driven subtypes of B-progenitor acute lymphoblastic leukemia. Nat Genet. 2019; 51(2):296-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lilljebjorn H, Henningsson R, Hyrenius-Wittsten A, et al. Identification of ETV6-RUNX1-like and DUX4-rearranged subtypes in paediatric B-cell precursor acute lymphoblastic leukaemia. Nat Commun. 2016;7:11790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roberts KG, Li Y, Payne-Turner D, et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N Engl J Med. 2014; 371(11):1005-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Den Boer ML, van Slegtenhorst M, De Menezes RX, et al. A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: a genome-wide classification study. Lancet Oncol. 2009; 10(2):125-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mullighan CG, Su X, Zhang J, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009;360(5):470-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zaliova M, Kotrova M, Bresolin S, et al. ETV6/RUNX1-like acute lymphoblastic leukemia: a novel B-cell precursor leukemia subtype associated with the CD27/CD44 immunophenotype. Genes Chromosomes Cancer. 2017;56(8):608-616. [DOI] [PubMed] [Google Scholar]
- 15.Zaliova M, Stuchly J, Winkowska L, et al. Genomic landscape of pediatric B-other acute lymphoblastic leukemia in a consecutive European cohort. Haematologica. 2019;104(7):1396-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.