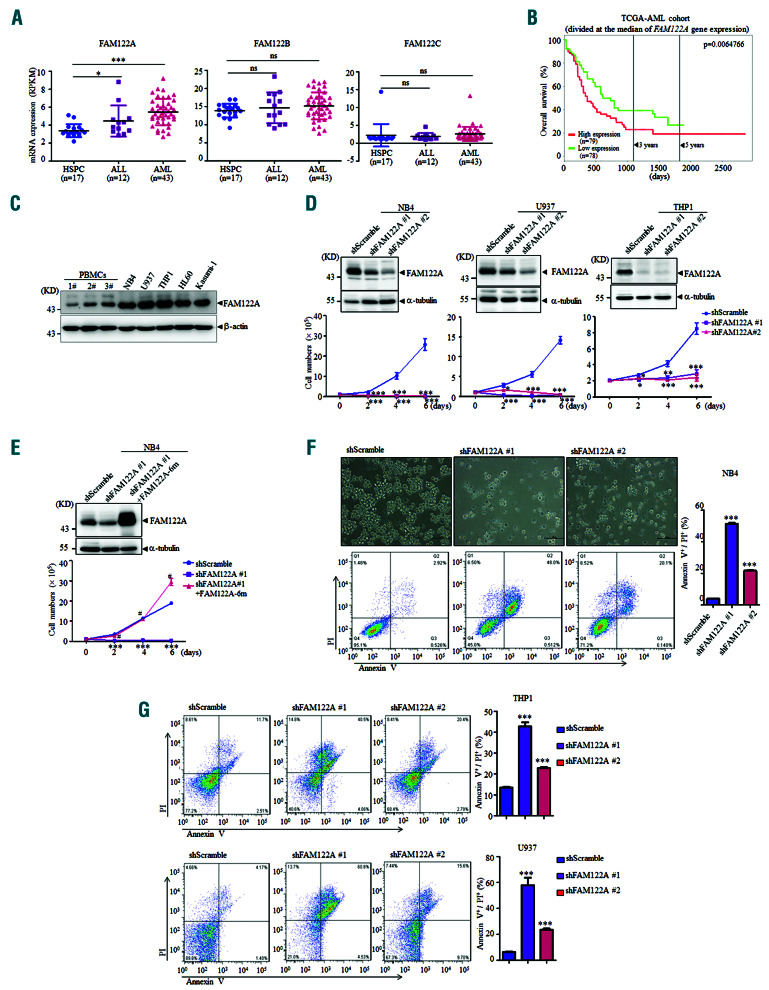
Acute myeloid leukemia (AML) is a heterogeneous clonal disorder of hematopoietic stem and progenitor cells (HSPC), characterized by uncontrolled proliferation, differentiation blockage and reduced apoptosis. Although progressive advances have been made in AML treatment over the last decades, the 5-year survival rates of AML patients are still low, especially for elderly patients.1,2 It is, therefore, necessary to understand the molecular pathological mechanisms and develop more effective treatment strategies for this disease. Given its frequent inactivation in AML cases, the importance of protein phosphatase 2A (PP2A) as a tumor suppressor and promising target for therapy has been highlighted recently.3 The loss of PP2A activation occurs at different levels in AML, either with the mutation and downregulation of PP2A subunits or overexpression of PP2A inhibitors SET, CIP2A and SET interacting protein.4,5 Pharmacological restoration of PP2A activity by its activator (FTY720) effectively antagonizes leukemogenesis.6 It was shown that FAM122A (family with sequence similarity 122a), a highly conserved protein among a variety of mammalian species, interacts with PP2A-Aa and -B55a (a scaffold and regulatory subunit of the PP2A complex) and promotes the polyubiquitination and degradation of its Ca*******subunit,7 and FAM122A sumoylation increases the degradation of PP2A-Ca protein together with the reduced phosphatase activity of PP2A.8 Recently, we demonstrated that FAM122A maintains the growth of hepatocellular carcinoma cells through promoting MAPK/AKT signaling.9 However, the normal function and pathophysiological significance of FAM122A are still largely unknown. Here we investigated whether FAM122A has a role in the growth of AML cells and AML development.
Besides FAM122A, the FAM122 family has two other members, FAM122B and FAM122C, which share 60% and 30% amino acid sequence identity, respectively, with FAM122A. We examined the mRNA expression levels of these three FAM122 members in a panel of samples from patients with AML and acute lymphoblastic leukemia as well as human CD34+ HSPC by analyzing the public RNA-sequencing database (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE48173). As shown in Figure 1A, the levels of mRNA expression of FAM122A, but not of FAM122B and FAM122C, were significantly increased in patients with AML or acute lymphoblastic leukemia compared to the levels in normal HSPC. A similar pattern of higher expression of FAM122A was also found in bone marrow cells from patients with four subtypes of AML with different karyotypes, that is, t(8;21), t(15;17), inv(16)/t(16;16) and t(11q23)/MLL, compared with the expression in hematopoietic stem cells enriched for Lin−CD34+CD38−CD90+CD45RA− in normal bone marrow cells (Online Supplementary Figure S1A) (http://servers.binf.ku.dk/hemaexplorer/). The Cancer Genome Atlas datasets also showed that AML patients with high FAM122A mRNA expression had shorter overall survival (Figure 1B), suggesting a potentially oncogenic role of FAM122A in AML. Moreover, FAM122A protein expression was also higher in AML cell lines, compared to the level in peripheral blood mononuclear cells from three healthy individuals (Figure 1C).
To assess the function of FAM122A in AML, three human AML cell lines – (i) NB4, an acute promyelocytic leukemia cell line carrying the t(15;17), (ii) U937 and (iii) THP1 with the MLL-AF9 fusion gene – were transfected with lentiviruses expressing short hairpin RNA (shRNA) targeting two distinct regions of FAM122A mRNA (designated shFAM122A#1 and shFAM122A#2) together with a non-specific shRNA (shScramble). The results showed that FAM122A could be effectively knocked down with these two pairs of shRNA in all three AML cells (upper panels, Figure 1D), and knockdown of FAM122A caused a substantial decrease in cell growth compared to that of shScramble-expressing NB4 cells (bottom panels, Figure 1D), which could be significantly rescued by re-expression of FAM122A (Figure 1E). Meanwhile, FAM122A silencing also suppressed the growth of normal CD34+ cells to some degree (Online Supplementary Figure S1B-D), suggesting that there is a differential requirement of FAM122A for the growth of normal HSPC and AML cells.
We continued to investigate whether FAM122A knockdown induces cell death and/or altered cell cycle distribution. Examining NB4 cells, we found that fewer live cells were visible under light microscopy among cells transduced with shFAM122A-lentivirus for 4 days, compared with their control cells (upper panels, Figure 1F). NB4 cells transfected with shFAM122A#2 and especially shFAM122A#1 displayed significant increases in cells double-positive for annexin V and propidium iodide without significant alteration of the cell cycle distribution (bottom panels of Figure 1F and data not shown), indicating that FAM122A silencing led to cell apoptosis. Similar results were also observed in U937 and THP1 cells (Figure 1G), although these effects became obvious after transduction for 6 days. The role of FAM122A in leukemic cell growth in vivo was further investigated using a mouse model of AML driven by MLL-AF9, which is produced by t(9;11)(p22;q23), frequently found in infant AML and associated with the M5 subtype of AML.10 MLL-AF9 leukemic cells with a yellow fluorescent protein (YFP) tag from bone marrows of leukemic mice were engineered to express either green fluorescent protein (GFP)-tagged shControl or shRNA that target murine Fam122a (designated shFam122a#3 and shFam122a#4) with a lentivirus system (Figure 2A). GFP+/YFP+ cells in each condition purified by sorting were assessed for infection efficiency and colony formation, as well as AML development through injecting the sorted leukemic cells into lethally irradiated mice (all mice were used and cared for according to Shanghai Jiao Tong University School of Medicine and national guidelines). The efficient silencing effects of FAM122A were confirmed in both shFam122a#3- and shFam122a#4-expressing cells and FAM122A inhibition significantly abrogated colony-forming capacity in methylcellulose (Figure 2B and C). The purified MLL-AF9 leukemic cells expressing two Fam122a-shRNA together with shControl in equal numbers (2,000 or 5,000 cells) were transplanted into lethally irradiated recipient mice. As shown in Figure 2D, the recipient mice transplanted with shFam122a#3 and shFam122a#4 cells had significantly longer disease latency and extended survival, as determined by mean survival (50 or 53 days in shFam122a#3- or shFam122a#4- expressing mice compared to 25 days in control mice with 2,000 cells; 43 or 50 days in shFam122a#3- or shFam122a#4-expressing mice compared to 22 days in controls with 5,000 cells). Flow assisted cell sorting analysis of hematopoietic organs and peripheral blood further showed that mice transplanted with both Fam122a-shRNA leukemic cells displayed significantly lower percentages of YFP+ cells in peripheral blood, bone marrow and spleen, compared with those of shControl counterparts (Figure 2E). The sizes and weights of livers and spleens from FAM122A-silenced recipients appeared much smaller than those of control recipients (Figure 2F and G), accompanied by less leukemic cell infiltration (Figure 2H). Collectively, these observations strongly suggest that FAM122A is a critical regulator of AML cell growth and development in vitro and in vivo.
Figure 1.
FAM122A is required for the growth and survival of acute myeloid leukemia cells. (A) The expression levels of FAM122A, FAM122B, and FAM122C mRNA were compared among hematopoietic stem and progenitor cells (HSPC), acute lymphoblastic leukemia and acute myeloid leukemia (AML) patients’ samples in RNA-sequencing data from the Gene Expression Omnibus dataset (GSE48173). Each point presents the related gene expression of individual patients with horizontal lines indicating the median expression level in each group. *P<0.05 and ***P<0.001, compared with HSPC. ns: not statistically significant. (B) Kaplan-Meier plot showing the correlation between FAM122A expression and overall survival of AML patients from The Cancer Genome Atlas database. The patients were stratified into a high- and low-expression groups according to the median value of FAM122A expression. A log-rank test was performed to compare the survival curves of these two groups. P<0.01 was considered to indicate a statistically significant difference. (C) FAM122A protein was compared by western blot between three normal human peripheral blood mononuclear cell specimens and the indicated AML cell lines. (D) The effects of FAM122A knockdown in AML cells were confirmed by western blot (upper panels). The growth rates of FAM122A knockdown cells (designated shFAM122A#1 and shFAM122A#2) and control cells (shScramble) were monitored and analyzed by a cell counter (lower panels). (E) Re-expression of FAM122A with a shRNA-resistant plasmid carrying six mutations in shFAM122A#1-expressing NB4 cells (designated shFAM122A#1/FAM122A-6m) was confirmed by western blot (upper panel). The growth of the indicated cells was also monitored by a cell counter (lower panel). (F) FAM122A-knockdown NB4 cells were observed under microscopy and photographed (upper panels, scale bar = 100 mm) after transduction with lentiviruses expressing shRNA for 4 days, and their degree of apoptosis was analyzed by calculating annexinV+/propidium iodide+ cells with flow cytometry (FACS) (bottom panels). (G) FAM122A knockdown U937 and THP1 cells, together with their corresponding control cells, were subjected to FACS analysis for apoptosis after infection with shRNA viruses for 6 days (left panels). (F, G) The analyzed quantification data are shown on the right of the FACS data. *P<0.05, **P<0.01 and ***P<0.001, compared with the corresponding shScramble cells. #P<0.01 compared with shFAM122A#1-expressing cells.
Figure 2.
FAM122A promotes acute myeloid leukemia development in vivo. (A) Schematics of the experimental process of the bone marrow transplantation assay. MLL-AF9-expressing mouse leukemic cells were infected with GFP-tagged shControl or shFam122a lentiviruses. Flow-sorted GFP+ cells were assayed to verify the knockdown effect and colony forming ability. Subsequently, 2,000 or 5,000 GFP+ cells, mixed with 2x105 normal bone marrow cells, were transplanted into lethally irradiated recipient mice for survival calculation. (B, C) GFP+ MLL-AF9 leukemia cells from each shRNA condition were assessed for FAM122A knockdown effects (B) and colony formation (scale bar = 100 mm) (C). (D) Kaplan-Meier survival curve analysis of mice transplanted with 2,000 or 5,000 purified MLLAF9 cells expressing GFP-tagged shControl or shFam122a (n=5; log-rank test). (E-H) Animals were transplanted with 2,000 acute myeloid leukemia (AML) cells and after 21 days the percentages of GFP+/YFP+ cells in peripheral blood, bone marrow and spleen from indicated recipient mice were analyzed by flow cytometry (E). The spleens (F) and livers (G) of indicated mice were photographed and weighed. Images of hematoxylin & eosin-stained specimens of liver and spleen from indicated mice (H) were observed by microscopy (scale bar = 100 mm). **P<0.01 and ***P<0.001 compared with shControl-expressing AML mice. CFU: colony-forming unit; GFP: green fluorescent protein; YFP: yellow fluorescent protein; BM: bone marrow; PB: peripheral blood; SP: spleen.
Figure 3.
The effect of FAM122A silencing on phosphorylatios of PP2A substrates and MYC expression. (A) Acute myeloid leukemia (AML) cells with FAM122A silencing and reexpression (shFAM122A#1-FAM122A-6m) were subjected to western blot for detection of the indicated proteins. (B) The indicated cells were treated with or without 50 nM okadaic acid (OA) for 6 h (NB4 cells), or 20 nM OA for 12 h (U937 cells), and then western blotting was performed to examine protein levels. (C) The MYC mRNA expression level was examined in FAM122A knockdown AML cells. Data are means with the bar indicating the standard deviation in an independent experiment. ***P<0.001 compared with shScramble cells. (D, E) FAM122A knockdown NB4 cells, together with shScramble cells, were treated with or without 20 mΜ MG132 for 4 h (D), or cyclohexamide at a dose of 100 mg/mL at different time points (E), followed by western blot to examine the indicated proteins. CHX: cyclohexamide.
Many key substrates of PP2A are involved in the regulation of cell growth and apoptosis, including pro-proliferation regulators AKT, ERK, MYC11 and antiapoptotic factors Bcl-212 and BAD.13 Considering that FAM122A was previously identified as a PP2A inhibitor, we further examined the phosphorylation states of a series of PP2A substrates. FAM122A silencing significantly decreased the phosphorylation of several PP2A substrates, including MYC at Ser62, ERK at Thr202/Tyr204, BAD at Ser112 and Bcl-2 at Ser70, but not the substrate of tyrosine phosphatase SHP-1 such as JAK2 (Figure 3A). The effect of decreased phosphorylation could be abolished when FAM122A was re-expressed in shFAM122A#1 cells, indicating the altered phosphorylation events are specifically regulated by FAM122A, although we did not find a change in AKT phosphorylation. FAM122A knockdown also reduced total MYC and Bcl-2 protein levels (Figure 3A). Treatment with okadaic acid, a PP2A inhibitor, significantly abrogated FAM122A silencingdecreased phosphorylation of ERK and BAD, but not that of MYC, suggesting that FAM122A maintains AML cell growth and survival possibly by modulating PP2A activity (Figure 3B). Indeed, PP2A-Cα silencing partially rescued FAM122A knockdown-triggered apoptosis (Online Supplementary Figure S2).
MYC is essential for the occurrence and progression of AML,14 and there is mounting evidence that Ser62 phosphorylation is critical for maintaining the stability of MYC protein.15 However, we found that treatment withokadaic acid or PP2A-Ca knockdown (Online Supplementary Figure S2B) did not affect phosphorylated MYC in FAM122A knockdown cells, suggesting that the reduction in phosphorylated MYC was not due directly to PP2A. FAM122A knockdown resulted in decreased MYC mRNA levels (Figure 3C), but did not affect the stability or degradation of the MYC protein (Figure 3D and E). AF9-induced leukemic cells with FAM122A silencing also demonstrated a decrease in MYC protein (Figure 2B). These results strongly imply that FAM122A promotes AML cell growth also by maintaining MYC transcriptional expression.
In summary, our study shows that FAM122A is critical for maintaining AML cell growth and survival by modulating PP2A activity and sustaining MYC expression, findings which warrant further investigation into its potential role in the treatment of AML.
Supplementary Material
Acknowledgments
We thank Professors Cheng-Cheng Zhang (University of Texas Southwestern Medical Center, USA) and Jun-ke Zheng (Shanghai Jiao Tong University School of Medicine, China) for kindly providing the MLL-AF9 plasmid and Professor Stephen J. Elledge (Howard Hughes Medical Institute, USA) for the pINDUCER20.
Funding Statement
Funding: this work was supported by a grant from the National Natural Science Foundation (81572290, HY).
References
- 1.Ling Y, Zhang Z, Zhang H, Huang Z. Protein kinase inhibitors as therapeutic drugs in AML: advances and challenges. Curr Pharm Des. 2017;23(29):4303-4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Kouchkovsky I, Abdul-Hay M. Acute myeloid leukemia: a comprehensive review and 2016 update. Blood Cancer J. 2016; 6(7):e441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arriazu E, Pippa R, Odero MD. Protein phosphatase 2A as a therapeutic target in acute myeloid leukemia. Front Oncol. 2016; 6:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pippa R, Dominguez A, Malumbres R, et al. MYC-dependent recruitment of RUNX1 and GATA2 on the SET oncogene promoter enhances PP2A inactivation in acute myeloid leukemia. Oncotarget. 2017;8(33):53989-54003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lucas CM, Scott LJ, Carmell N, et al. CIP2A- and SETBP1-mediated PP2A inhibition reveals AKT S473 phosphorylation to be a new biomarker in AML. Blood Adv. 2018;2(9):964-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang Y, Huang Q, Lu Y, Li X, Huang S. Reactivating PP2A by FTY720 as a novel therapy for AML with C-KIT tyrosine kinase domain mutation. J Cell Biochem. 2012;113(4):1314-1322. [DOI] [PubMed] [Google Scholar]
- 7.Fan L, Liu MH, Guo M, et al. FAM122A, a new endogenous inhibitor of protein phosphatase 2A. Oncotarget. 2016;7(39):63887-63900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fan F, Zhao J, Liu Y, et al. Identifying the SUMO1 modification of FAM122A leading to the degradation of PP2A-Calpha by ubiquitinproteasome system. Biochem Biophys Res Commun. 2018;500(3):676-681. [DOI] [PubMed] [Google Scholar]
- 9.Zhou Y, Shi WY, He W, et al. FAM122A supports the growth of hepatocellular carcinoma cells and its deletion enhances doxorubicin- induced cytotoxicity. Exp Cell Res. 2020; 387(1):111714. [DOI] [PubMed] [Google Scholar]
- 10.Krivtsov AV, Twomey D, Feng Z, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442(7104):818-822. [DOI] [PubMed] [Google Scholar]
- 11.Dang CV. MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harb Perspect Med. 2013;3(8):a014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin SS, Bassik MC, Suh H, et al. PP2A regulates BCL-2 phosphorylation and proteasome-mediated degradation at the endoplasmic reticulum. J Biol Chem. 2006;281(32):23003-23012. [DOI] [PubMed] [Google Scholar]
- 13.Shamas-Din A, Brahmbhatt H, Leber B, Andrews DW. BH3-only proteins: orchestrators of apoptosis. Biochim Biophys Acta. 2011; 1813(4):508-520. [DOI] [PubMed] [Google Scholar]
- 14.Hoffman B, Amanullah A, Shafarenko M, Liebermann DA. The proto-oncogene c-myc in hematopoietic development and leukemogenesis. Oncogene. 2002;21(21):3414-3421. [DOI] [PubMed] [Google Scholar]
- 15.Noguchi K, Kitanaka C, Yamana H, Kokubu A, Mochizuki T, Kuchino Y. Regulation of c-Myc through phosphorylation at Ser-62 and Ser-71 by c-Jun N-terminal kinase. J Biol Chem. 1999; 274(46):32580-32587. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.