ABSTRACT
RNA-binding proteins are important regulators of RNA metabolism and are of critical importance in all steps of the gene expression cascade. The role of aberrantly expressed RBPs in human disease is an exciting research field and the potential application of RBPs as a therapeutic target or a diagnostic marker represents a fast-growing area of research.
Aberrant overexpression of the human RNA-binding protein La has been found in various cancer entities including lung, cervical, head and neck, and chronic myelogenous leukaemia. Cancer-associated La protein supports tumour-promoting processes such as proliferation, mobility, invasiveness and tumour growth. Moreover, the La protein maintains the survival of cancer cells by supporting an anti-apoptotic state that may cause resistance to chemotherapeutic therapy.
The human La protein represents a multifunctional post-translationally modified RNA-binding protein with RNA chaperone activity that promotes processing of non-coding precursor RNAs but also stimulates the translation of selective messenger RNAs encoding tumour-promoting and anti-apoptotic factors. In our model, La facilitates the expression of those factors and helps cancer cells to cope with cellular stress. In contrast to oncogenes, able to initiate tumorigenesis, we postulate that the aberrantly elevated expression of the human La protein contributes to the non-oncogenic addiction of cancer cells. In this review, we summarize the current understanding about the implications of the RNA-binding protein La in cancer progression and therapeutic resistance. The concept of exploiting the RBP La as a cancer drug target will be discussed.
KEYWORDS: La autoantigen, La protein, La/SSB, SUMO, LARP, RNA-binding, RBP, TOP mRNAs, La-related proteins, Laminin B1, CCND1, cyclin D1, Bcl2, XIAP, BiP, Mdm2, posttranslational modification, PTM, IRES, translation, proliferation, invasion, tumour-promoting, anti-apoptotic, non-oncogenic addiction, cancer
1. The RNA-binding protein La
RNA-binding proteins (RBPs) are important regulators of RNA metabolism and are of critical importance in all steps of the gene expression cascade. Over 2,000 RBPs are known and they interact with RNA via different binding surfaces[1], [2]. The role of aberrantly expressed RBPs [3–5] in human disease, such as cancer, but also viral infections, is an important and exciting research field and the potential application of RBPs as either a therapeutic target or a diagnostic marker represents a fast-growing research field.
The RBP La was first described in systemic autoimmune diseases such as Lupus-associated (La) or Sjögren’s syndrome B (SSB) auto-antigen [6,7]. The ubiquitously expressed La protein has three RNA-binding domains referred to as La motif, RNA Recognition Motif 1 (RRM1) and RNA Recognition Motif 2 (RRM2) (Fig. 1) [8–14]. The La motif together with the RRM1 comprise the so-called ‘La module’, a feature characterizing all family members of La-related proteins (LARPs) [9,10,15–18] of which some are cancer-associated [19–26]. Interestingly, in contrast to the La homolog found in yeast (Saccharomyces cerevisiae, referred to as Lhp1; and Schizosaccharomyces pombe La, referred to as Sla1p) the La homologs in mammalian, drosophila, amoeba (Dictyostelium discoideum), and flagellate protozoa (Trypanosoma brucei) contain, in addition to the La module, the RRM2 located in the C-terminal region [8,10,27]. Whereas the La module is mainly involved in binding to the uridinylated 3ʹ-terminus common to the 3ʹend of all RNA Pol III transcripts, the additional RNA-binding domain RRM2 allows binding of the La protein to internal RNA sequences/structural elements as found in viral and cellular target mRNAs. For binding of internal RNA sequences or structures both RNA-binding domains, RRM1 and RRM2, are required [16,28–30]. It is considered that the RRM2 confers cellular functions to La proteins, which cannot be fulfilled by yeast La homologs. Besides its RNA-binding domains, La has a nuclear localization signal (NLS) [31], a nucleolar localization signal (NoLS) [32,33], a nuclear retention element (NRE) [31,34,35], and amino acid residues in RRM1 implicated in the nuclear export of La [34] ensuring nuclear-cytoplasmic shuttling [31,36–41] of the otherwise mostly nuclear localized La protein. The La protein contains a dimerization domain between amino acids 293 to 348 in its C-terminal domain [42]. Although a minimal dimerization motif has not been established, the deletion of those amino acids prevents dimerization and addition of the recombinant expressed dimerization domain impairs in vitro translation. It is considered that the expression of this dimerization domain acts in a dominant-negative manner in cells [43–45].
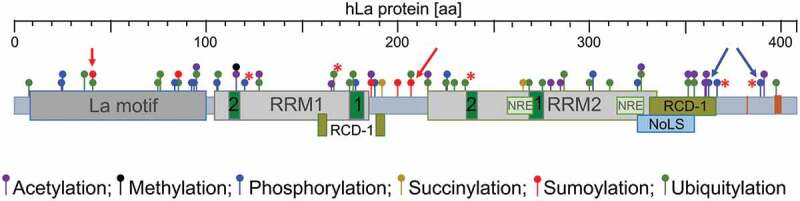
Figure 1.
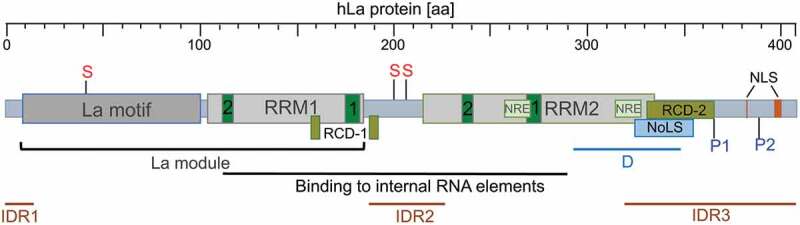
Domain structure and key PTMs of human La protein (hLa, 408 aa)
RRM = RNA Recognition Motif; green box = RiboNucleoProtein (RNP) motif; RCD = RNA chaperone domain (amino acids required for RCD-1 activity are located in the loop 3 and the 53 helix of RRM1) [29,52,53]; NRE = Nuclear Retention Element [31,34,35]; NoLS = Nucleolar Localization Signal [32,33]; NLS = Nuclear Localization Signal [31]; D = Dimerization domain [42]; IDR = Internal Disordered Region [54]; PTM = PostTranslational Modification; S = Sumoylation site [111,113]; P = Phosphorylation site [29,37,100,101].
It has been suggested earlier that the La protein possesses helicase activity [46,47]. Furthermore, it is well described that La is an RNA chaperone assisting structural changes in different RNA elements [48–51]. Two RNA chaperone domains (RCD-1) [52,53] and RCD-2 [29] have been identified previously. Furthermore, La contains three intrinsic disordered regions (IDRs) that are probably involved in RNA folding and the processing of bound RNA molecules [54]. Comparing structural information available for the La motif, RRM1, and RRM2 located adjacent to the IRDs, we find that the IDR boundaries are not sharply defined. The first IRD starts with the N-terminal end of hLa and ends approximately at aa 15 [54], however structural information available for the N-terminal end of the La motif suggests an overlap of a few amino acids with a helical fold of La [17,18,55]. The second IRD (approximately aa 187 to 228, [54]) located in the linker region between RRM1 and RRM2 has no obvious overlap with protein folds at the C-terminal end of RRM1 or the N-terminal end of RRM2. In contrast, the alpha helix (aa 308 to 325) located at the C-terminal end of RRM2 [27] overlaps with the predicted start of the third IRD (approximately aa 318 to 408, [54]). Hence, until a full structure of RNA-associated La is determined, the exact boundaries between the IRDs and the RNA-binding domains will remain elusive.
The multifunctional La protein has been described as promoting two critical cellular processes: (I) the processing of non-coding RNA precursors and (II) the translation of selective mRNAs. Via the La module, La binds to the uridinylated 3ʹ-terminus (UUU-3ʹOH) of RNA polymerase III precursor transcripts such as pre-tRNAs and protects them from exonucleolytic decay and ensures proper RNA folding [10,12,17,18,48,56,57]. The La protein also regulates the synthesis of specific tRNA-derived miRNAs [58,59] and is implicated in the biosynthesis of miRNAs by binding to pri- and pre-precursors [60,61]. The role and function of La in processing of RNA polymerase III precursor transcripts and the biogenesis of non-coding RNAs will not be reviewed herein.
The La protein has been identified as an internal ribosome entry site (IRES) trans-acting factor (ITAF) in viral RNA translation [42,62–66] and the translation of IRES-containing cellular mRNAs, as we will discuss below.
Moreover, recent studies showed that murine La (mLa) and human La (hLa) proteins are intensively altered by post-translational modifications (PTM) such as linkage of the small ubiquitin-like modifier (SUMO) to specific lysine residues and phosphorylation of specific serine and threonine residues, as will be reviewed below.
In this review, we will summarize the understanding of cancer-associated La including known La gene mutations and aberrant expression level as found in various cancer entities. The analysis of next-generation sequencing data sets shows a correlation between increased La expression and patient survival, as discussed below. Hence, clinical evidence is growing that elevated La expression contributes to cancer progression.
We will review PTMs of the La protein and their potential role in cancer. Finally, we will summarize the status quo about the role of La in cancer progression, drug resistance, and associated cellular pathways and molecular mechanisms regulated by La. The focus of this review is the role of murine and human La in cancer pathobiology and mRNA translation.
Experiments to directly test whether La is an oncogene, such as BALB/c-3T3 cell transformation assays, have – to our knowledge – not been studied yet. However, it has been shown that small hairpin (sh)RNA-mediated depletion of mLa in EMT-transformed hepatocytes reduces subcutaneous tumour growth in murine models [39]. Those experiments demonstrate that La is required for cancer growth in mice, although they do not allow concluding that La is an oncogene per se causing malignant transformation. Commonly, an oncogene arises by mutations changing the functionality of the encoded gene to promote cancer pathobiology. So-called ‘driver’ mutations [67] are causing a direct advantage for cancer cells, oftentimes expressed as a significant gain in survival and growth, resulting in chemotherapeutic resistance and unrestricted proliferation of cancer cells. In contrast, ‘passenger’ mutations [67] are considered as a byproduct of the transformation process and are not oncogenic per se. Often passenger mutations do not or only indirectly support cancer progression, whereas some of those mutations are considered as tremendously important since they support survival and growth leading to a non-oncogenic addiction of cancer cells. The concept of non-oncogenic addiction is well established [68,69] and refers to the function of a specific cellular factor, which is not an oncogene per se but is required for cancer cells to cope with cellular stress in order to be excessively viable, resistant, and aggressive. Cancer cells experience a vast amount of stress caused by misregulated cellular processes developed during the malignant transformation process such as increased proliferation, genomic instability, and metabolic challenges. To overcome this increased intrinsic stress and to be functional, cancer cells depend on non-oncogenic addiction genes. As we will argue in this review, the La gene encodes a non-oncogenic addiction factor and aberrant expression or post-translational modifications of La may modulate its non-oncogenic addiction functionality. In this line, targeting La represents a promising and novel therapeutic strategy in order to raise the cellular stress of cancer cells to an unendurable level inevitably resulting in cancer cell death but sparing normal cells.
2. Expression of La in cancer
Protein expression is regulated at the transcriptional and post-transcriptional level. The activity of a protein is frequently regulated by post-translational modifications. In cancer cells, gene mutations often change the protein expression level or protein function by altering the amino acid sequence. In this chapter, we will review the information about cancer-associated mutations in the human La (hLa) gene and summarize the knowledge about the mRNA and protein expression of hLa in cancer.
Almost thirty years ago, it was found that murine La protein expression is increased by twofold in spontaneously transformed fibroblasts BALB/3T12-3 (contact-insensitive and tumorigenic) relative to their non-transformed counterpart cell line BALB/3T3 (contact-sensitive and non-tumorigenic) as well as in murine sarcoma virus-transformed rat fibroblasts (KNRK) relative to the non-transformed counterpart cell line NRK [70]. Thereafter, it has also been shown that the protein expression of hLa is elevated in numerous immortal cell lines derived from solid tumours and blood cancer tissue relative to its expression in the corresponding primary cell type [71]. This study included CD3-enriched peripheral blood lymphocytes versus Jurkat T-lymphocytes, normal human bronchial epithelium (NHBE) versus A549 bronchogenic carcinoma cells, prostate epithelial cells (PrEC) versus PC-3 and LNCaP prostate cancer cells, CD14-enriched monocytes versus U937 myeloid leukaemia cells, buccal cavity epithelial cells versus oropharyngeal squamous cell carcinoma SSC-25 cells, and human mammary epithelial cells (HMEC) versus breast cancer MCF-7 and MDA-MB-231 cells.
Aberrant expression of hLa protein in patient-derived tumour samples was first reported in 2003. In chronic myelogenous leukaemia (CML) hLa protein is more abundant in patient-derived CML-BC (blast crisis) and CML-APcd34+ (accelerated phase) than CML-CP (chronic phase) mononuclear cells [43]. Interestingly, the authors presented data supporting the notion that the hLa protein level is increasing during CML progression [43]. Furthermore, it has been shown that hLa levels correlate with BCR/ABL oncoprotein levels and tyrosine kinase activity. This study gave the first hint about the molecular mechanism leading to elevated hLa protein levels in BCR/ABL-transformed cells. Treatment of BCR/ABL-transformed cells with tyrosine kinase inhibitor STI571 (Gleevec/Imatinib) revealed that BCR/ABL signalling leads to enhanced hLa protein level although hLa mRNA levels stayed unchanged in STI571-treated when compared to control-treated cells. In addition, the finding that hLa protein stability dropped from t1/2 = 19.8h to t1/2 = 11h upon STI571 treatment revealed that BCR/ABL signalling triggers the stabilization of the hLa protein [43]. This observation suggests that BCR/ABL-induced oncogenic signalling increases hLa protein levels by a post-transcriptional mechanism.
Subsequently, it has been shown that hLa is not only overexpressed in haematopoietic malignancies but also in solid tumours. Tissue microarray analysis revealed significant elevated expression of the hLa protein in cervical squamous cell carcinoma (SCC) tissue compared to normal tumour adjacent tissue [72]. The hLa protein level was also aberrantly elevated in head and neck tumour samples applying an oral cavity tissue microarray consisting of 9 normal tumour adjacent tissues and 42 SCC tissues, diagnosed as SCC tissue ranging from grades I to III [73]. Furthermore, in lung cancer >95% of tumour cells showed robust nuclear La staining, whereas in the normal lung tissue samples approximately 50% of cell nuclei had weak-to-moderate hLa staining [74]. Additionally, hLa protein and mRNA levels were elevated in primary myelofibrosis due to activation of the JAK2 signalling[75]. To define a potential correlation between hLa protein expression in various tumour entities, tumour grades, and tumour recurrence a comprehensive analysis of larger numbers of tumour tissue samples with matched normal tissue complemented with patients’ data are required to develop deeper concepts about the contribution of aberrant hLa expression in cancer pathogenesis. Moreover, those studies might reveal that hLa expression can be used as a prognostic marker in certain cancer entities.
Available next-generation sequencing data combined with patients’ information allows establishing associations between transcript levels of the gene of interest and clinical outcome. We used the R2 website (‘R2: Genomics Analysis and Visualization Platform (http://r2.amc.nl) or PROGgeneV2 – Pan Cancer Prognostics Database (http://genomics.jefferson.edu/proggene/) to retrieve analysed high-throughput sequencing data sets from a variety of cancer entities. Interestingly, the provided data analysis revealed that high La mRNA expression correlates significantly with poor survival in some cancer entities – namely neuroblastoma, lung adenocarcinoma, and acute myeloid leukaemia (Fig. 2), however high La mRNA levels are not in all cancer entities associated with poor survival as found for bladder cancer or ovarian cancer (not shown). In summary, accumulating data demonstrate that the mRNA and/or protein level of hLa are aberrantly elevated in various cancer entities and that high hLa expression often correlates with poor survival. So far, only little is known about the transcriptional regulation of the La gene [76–78], the translation of La mRNA by a potential IRES element located in the 5ʹ-untranslated region (5ʹUTR) [78], the regulation of La protein stability, and cleavage of La protein by specific proteases [43,79–82]. Hence, the molecular mechanism regulating La transcription and protein abundance in normal versus cancer cells and under various cellular conditions should be studied in the near future.
Figure 2.
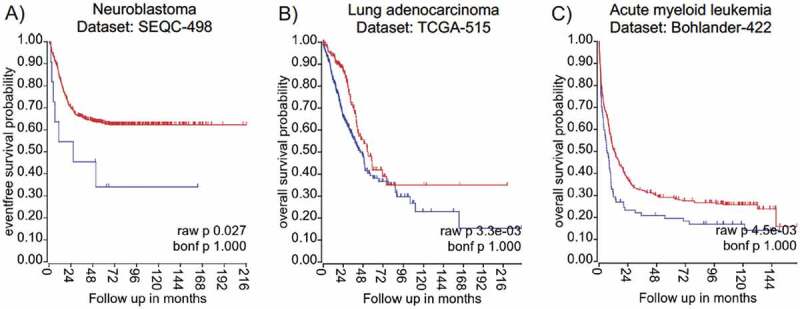
Kaplan Meyer survival curves suggest elevated expression of hLa mRNA as indicator for poor cancer prognosis
The conclusion that high hLa mRNA expression correlates with low survival is drawn from: A) The neuroblastoma data set SEQC-498 consists of 498 cases including 487 patient samples with low (red) La and 11 with high (blue) La mRNA expression levels, leading to a significant difference (p = 0,027). B) The lung adenocarcinoma data set TCGA-515 consists of 515 cases including 187 patient sample with low (red) La and 328 with high (blue) La mRNA expression levels, leading to a significant difference (p = 0,0033). C) The acute myeloid leukaemia data set Bohlander-422 consists of 422 cases including 335 patient sample with low (red) La and 87 with high (blue) La mRNA expression levels, leading to a significant difference (p = 0,0045). Datasets and Kaplan Meier curves were derived from the R2Platform (https://r2.amc.nl). Red lines = low La mRNA expression; blue lines = high La mRNA expression.
Not only should the level of protein expression be under consideration but also the occurrence of cancer-associated mutations in the gene of interest. To address this question it is beneficial to use the NCI’s Genomic Data Commons (GDC) data portal of the National Institute of Health (https://portal.gdc.cancer.gov) and the cBioPortal for Cancer Genomics developed at Memorial Sloan Kettering Cancer Centre (www.cbioportal.org). The retrieved analysed data of 44,366 patients included in 176 studies revealed a total of 100 mutations in the hLa gene. By calculating the frequency of mutations in the hLa gene only 10 out of 35 different cancer entities displayed an alteration frequency of the hLa gene of more than 1%, with a maximal frequency of about 4% in bladder/urinary tract cancer due to gene amplification (Fig. 3). Furthermore, no significant correlation between the frequency of hLa gene mutations and the survival of patients has been found when combining data of 32 of The Cancer Genome Atlas (TCGA) PANCancer studies. Even when studying 25 different cancer entities only in liver cancer and myeloid cancer was hLa gene amplification and deep deletion significantly associated with poor survival (data not shown). In addition, hLa gene alterations were not found in cancer entities of the peripheral nervous system, the testis or bone (data not shown). Interestingly, a few of the 100 alterations within the hLa gene (Fig. 4) may affect post-translational modifications (PTM) of the hLa protein (Fig. 5). For example, in colon cancer phosphorylation at threonine 389 (T389) might be impaired due to the T389R mutation found in this type of cancer. Furthermore, some mutations are occurring in close proximity to sites of amino acid modifications, which could affect the consensus sequence for a specific PTM. Whether genetic alterations leading to changes in PTM can cause a cancer-relevant phenotype must be studied. In summary, mutations in the La gene only occurred in one-third of the different cancer entities tested and only rarely was a correlation between La mutations and patients’ survival observed. However, whether one or the other mutation acts as ‘driver’ by actually changing the functionality of La fostering cancer progression remains to be experimentally addressed. By applying the CRISPR-Cas9 gene-editing system it is possible to introduce specific mutations in the La gene and to study the impact of a specific La mutation on cell proliferation, chemoresistance and tumour growth in murine models.
Figure 3.
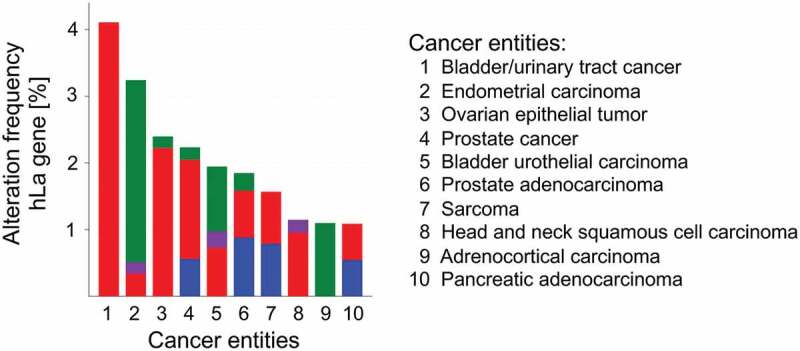
Alteration frequency of the human La in different cancer entities
Human La gene alteration frequency found in different cancer entities. Top 10 cancer types are shown. Bars indicate type of gene alteration: Green = Point mutation; Red = Amplification; Purple = Fusion; Blue = Deep Deletion; Gray = Multiple Alterations. The Top 10 cancer types shown; A and B are adapted graphs derived from cBioPortal (www.cbioportal.org). Data were retrieved from: https://www.cbioportal.org. using the gene symbol SSB.
Figure 4.
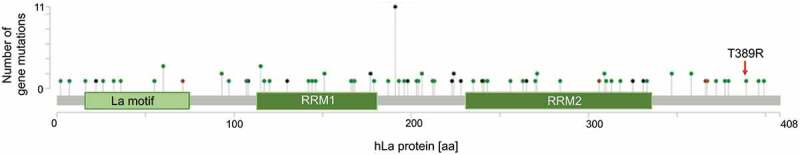
Human La gene alterations
Lollipop diagram indicating the distribution of common gene alterations in the open reading frame of the hLa gene. The phosphorylation site T389 is changed to R389 in colon cancer (red arrow). The diagram is based on data obtained from 44,366 patients included in 176 studies. The somatic mutation frequency is 0.2%. Sixty-one missense mutation (green dots), 29 truncating mutations (black dots, including nonsense, non-stop, frameshift deletion, frameshift insertion, splice site mutations), 4 inframe deletions or insertions mutations (brown dots) and 6 mutations of other types (purple dots) are shown. The y-axis shows how often the mutation was found in this patient cohort. Data were retrieved from: https://www.cbioportal.org. using the gene symbol SSB.
Figure 5.
Posttranslational modifications (PTM) in the human La protein
Published PTMs are indicated such as sumoylation site K41 and K208 (red arrows) and phosphorylation site S366 and T389 (blue arrows). Most of those PTM sites have been identified by high-throughput mass spectrometry. (Adapted from www.phosphosite.org). Red stars indicated identified gene alterations leading to changes in the indicated PTM sites.
Taken together, hLa mRNA levels are significantly elevated in certain cancer entities and often correlate with poor clinical prognosis. Elevated hLa protein levels have been documented for liquid as well as solid tumour entities, and for some even without increased hLa mRNA expression, indicating post-transcriptional regulation of hLa expression in some cancer entities e.g. CML [43]. The frequency of hLa gene mutations found in cancer is overall very low and in only two cancer entities a significant association with poor patients’ survival has been documented in the databases, however this conclusion does not rule out that a specific mutation within the hLa gene has a – so far unknown – major impact on cancer progression.
The data suggest further that hLa is not a standalone, causative oncogene, which – when overexpressed or mutated – is sufficient to promote cancer. It appears more convincing that hLa requires cancer-type specific cellular settings, changing its expression and posttranslational modifications, to facilitate cancer. Hence, in-depth correlation studies might illuminate tumour-promoting gene expression circuits requiring elevated hLa expression for manifestation and functionality and suggests that the hLa protein is rather important for non-oncogenic addiction helping cancer cells to adapt to cellular stress. The concept of non-oncogenic addiction [68,69] is based on the finding that cancer cells have a higher stress burden than normal cells and that cellular factors critical for stress response pathways are eminently required for cancer cells to cope with stress. Targeting such a cellular factor (also referred to as synthetic lethal factor) in combination with low dosed conventional chemotherapeutic drugs may predominantly weaken the ability of cancer cells to handle cellular stress [68,69]. As we will see below, hLa supports a tumour-promoting and an anti-apoptotic state of cancer cells suggesting that targeting the hLa protein impairs the ability of cancer cells to cope with stress and, consequently, favourably sensitizes cancer cells for chemotherapeutic treatment.
3. Cancer-associated La and the expression of tRNA and tRFs
As mentioned in the introduction, La is required for proper maturation of RNA polymerase III (Pol III) transcripts, many of which are critical components of the translation machinery during protein synthesis such as transfer RNAs (tRNAs) or the 5S ribosomal RNA (rRNA). Hence, in the context of this review, it is important to summarize the available information about the potential impact of an aberrantly elevated La protein level in cancer cells on the expression of RNA Pol III transcripts and the occurrence of tRNA-derived fragments (tRFs). As an example, it is well established that tRNA levels are not constant and are significantly increased in cancer cells [83–85]. Furthermore, fluctuation of the tRNA level has been reported for different metabolic stages and can have a strong effect on the translation of certain mRNAs containing rare codons or upstream open reading frames (uORFs) [86–89]. Additionally, it has been found that tRFs [90] are associated with cancer progression [91,92]. To which extent elevated La protein levels in cancer cells are modulating mature tRNA level and/or the generation of tRFs has to be determined. However, studies using La knockout (KO) or conditional La KO mice suggest no major differences in mature RNA Pol III transcripts such as mouse Y3 RNA and most tRNAs. As an example, only a small reduction in mature tRNATyr has been found although the maturation of the intron-containing pre-tRNATyr was altered in mLa KO cells of the fore brain [93]. Interestingly, in the mouse cortex of conditional mLa KO mice, the processing of the 5.8S rRNA, an RNA Pol I transcript, was disturbed but it is not clear whether the level of mature 5.8S rRNA was reduced, as would be expected [94].
As long as we do not have a comprehensive set of data on mature RNA Pol III transcript level [95] in La-depleted or KO cells, or a correlation between elevated La and tRNA levels in different cancer entities, we assume that the reported differences in tRNA levels in cancer cells are due to the metabolic stage and/or the misregulation of transcription factors regulating RNA Pol III transcription, some of which are actually oncogenes such as Myc [96]. Alike, we are not aware of studies showing that tRFs are generated in a La-dependent manner. Interestingly, experimental depletion of hLa induces the generation of tRNA-derived miRNAs [58,59]. Hence, experimental or therapeutic approaches that change elevated La levels in cancer cells are not likely to impact the level of mature RNA Pol III transcript but may add alternative processing pathways of RNA Pol III precursor transcripts due to lower La levels resulting in small non-coding RNAs or tRFs with regulatory functions in cancer pathobiology. Because of the large number of different RNA Pol III precursor transcripts such as tRNA, 5S rRNA, U6 snRNA, 7SL RNA, which are all bound by the La protein during maturation, future studies may reveal a novel La-dependent regulatory Pol III transcript pathway in cancer pathobiology.
4. Post-translational modifications of the La protein
Post-translational modifications (PTM) regulate cellular localization, stability, expression level, and functionality of proteins. PTMs appear in response to extracellular and/or intracellular cues, often exist in a reversible manner, and multiple PTMs can occur in a single protein. Genome-wide proteomic studies revealed phosphorylation, sumoylation, ubiquitylation, and acetylation sites in hLa (Fig. 5, entry name: LA_HUMAN; gene name: SSB; protein ID: 05455; https://www.uniprot.org and https://www.phosphosite.org). Phosphorylation of hLa is well established and has been reported previously [97–100]. In summary, recombinant hLa can be phosphorylated by protein kinase A (PKA), protein kinase C (PKC), and casein kinase 2 (CK2) in vitro [100]. Whether PKA and PKC also phosphorylate endogenous hLa remains to be demonstrated. In contrast, phosphorylation of hLa by CK2 and AKT at serine 366 (S366) and threonine 389 (T389), respectively, has been characterized in more detail. S366 is phosphorylated by CK2 in vitro as well as in cells [100,101]. Phosphorylation of S366 regulates recycling of RNA Pol III transcription complexes, 5ʹ-end processing of pre-tRNA molecules, subcellular localization, and translation of 5′-terminal oligopyrimidine tracts (TOP) mRNAs [36,101–104]. Initial studies did not support the concept that hLa phosphorylation affects its subcellular localization [100], however later studies using an S366 phosphorylation (pS366)-specific antibody suggests that hLapS366 localizes to the nucleus whereas non-phosphorylated hLa is found in the nucleolus and cytoplasm [33]. Moreover, mLa and hLa can be phosphorylated by AKT at T301 or T389, respectively [29,37]. Platelet-derived growth factor (PDGF) triggers the activation of the phosphoinositide 3-kinase (PI3K) pathway and leads to AKT-mediated phosphorylation of mLa at T301. Phosphorylation of mLa at T301 stimulates the nuclear export and the association of mLa with polyribosomes and a selective set of translationally active mRNAs. Hence, AKT-mediated phosphorylation of mLa promotes protein synthesis of mRNAs encoding many cancer-associated factors as RNA sequencing analysis revealed [37]. In contrast in hLa, AKT-mediated phosphorylation of recombinant hLa occurs at T389 and impairs La’s RNA chaperone activity in vitro. Furthermore, expression of a green fluorescent (gfp)-tagged phosphorylation-deficient mutant hLa (gfpLaT389A) in cells suggests that T389 phosphorylation contrarily regulates Cap-dependent and IRES-dependent translation [29].
Importantly, the aberrant expression/activity of CK2 and AKT are implicated in cancer by supporting pro-proliferative and pro-survival pathways and regulating cell plasticity during tumour spread [105–110]. Hence, the phosphorylation of hLa by these two cancer-associated protein kinases implies a potential important regulation of the stability and functionality of hLa in cancer cells. As mentioned earlier constitutive active JAK2 has been reported to elevate hLa mRNA levels [75], however it remains to be studied whether this signalling pathway, which is also implicated in facilitating tumour-promoting processes such as epithelial-mesenchymal transition (EMT), leads to hLa phosphorylation.
Modification of mLa and hLa by the small ubiquitin-like modifier (SUMO) has been first shown in rat neurons and this modification has been linked to the anterograde transport in rat neurons and the association with sumoylated mLa with dynein [111]. Continuation of the study revealed that sumoylation of recombinant hLa increases hLa’s RNA-binding activity [112] and RNA immunoprecipitation followed by RNA sequencing showed that sumoylated hLa is differently associated with about 3,000 mRNAs compared to sumoylation-deficient (∆SUMO) hLa [113]. Interestingly, the proliferation of cells expressing gfp-tagged sumoylation-deficient La (gfpLa∆SUMO) was impaired when compared to wildtype La (gfpLaWT) expressing cells. Investigation of the underlying mechanism showed that proliferation was hampered due to proteasome-dependent destabilization of signal transducer and activator of transcription 3 (STAT3) in gfpLa∆SUMO expressing cells [113]. A growing body of data suggests that aberrant activation of the sumoylation pathway, especially under cellular stress, contributes to cancer [114,115]. Hence, these data suggest that an increase in hLa sumoylation has a pro-proliferative effect in cancer cells.
Taken together, extra- and intra-cellular signalling pathways including stress pathways often activated in cancer cells are regulating a myriad of effector proteins by PTMs. The hLa protein is modified by phosphorylation and sumoylation (Fig. 5) and the modifying kinases and modification cascades are activated in cancer cells. Thus, efforts to illuminate a role of hLa in cancer has to consider that the degree of PTMs varies from one cell type to another and changes quickly in response to extra- and intra-cellular stimuli, such as stress signals caused by chemotherapeutic drug treatment.
5. The role of the La protein in cancer pathobiology
So far, we have summarized information about the expression of hLa in cancer, cancer-associated alterations within the hLa gene, and regulation of hLa by post-translational modifications. Now, we will focus on the role of La in cancer pathobiology.
5.1. Proliferation, migration and invasiveness of cancer cells
As mentioned above, hLa expression is elevated in many cancer-derived cell lines. Small interference RNA (siRNA)-mediated depletion of hLa in cancer cell lines is a useful tool to study the role and function of hLa in cancer-associated cellular processes such as cell proliferation, migration, and invasion in vitro.
A first report applying La-specific siRNAs revealed that La depletion reduces cell growth of cervical carcinoma cell line HeLa without the induction of apoptosis [116]. In a later study and by using again La-specific siRNAs a profound defect in HeLa cell proliferation was determined [72]. Analysis of the underlying molecular mechanism revealed that hLa promotes translation of the proto-oncogene CCND1 (cyclin D1), a key factor controlling the cell cycle transition from G1 to S phase. Importantly, upon overexpression of gfp-tagged siRNA-resistant hLa wildtype protein (gfpLasiR_WT) in La-depleted HeLa cells, CCND1 expression and cell proliferation was restored. In the same study, it has been shown that La depletion in prostate cancer cell lines LNCaP and PC3 also caused a defect in cell proliferation, which was again accompanied by reduced CCND1 expression [72].
Extending those studies to head and neck cancer revealed very similar data. Cell proliferation of squamous cell carcinomas (SCC) cells was impaired upon hLa depletion, however no profound effect on CCND1 expression was observed [73]. Moreover, hLa depletion led to significantly reduced SCC cell motility and invasiveness. Overexpression of siRNA-resistant hLa (gfpLasiR_WT) in hLa-depleted cancer cells restored cell motility. Besides cell proliferation, motility and invasiveness, which are important cancer- and metastasis-promoting processes, the expression of β-catenin and matrix metalloproteinase 2 (MMP-2) was significantly reduced in hLa-depleted cancer cells [73]. It is well known that β-catenin and MMPs are strongly associated with cancer progression [117–119] and it remains to be investigated by which mechanism hLa regulates the expression of those factors.
5.2. Tumour growth in murine models
Although the La protein supports cancer-promoting processes it does not necessarily mean that La is required for tumour growth. By using murine Ras-transformed hepatocellular cancer (HCC) cells (MIM-R), which underwent TGFβ-induced EMT (MIM-RT), the Mikulits laboratory showed in mouse experiments that depletion of mLa in mesenchymal (MIM-RT) but not epithelial (MIM-R) cells impairs subcutaneous tumour growth [39]. The role of hLa in tumour growth is further supported by our unpublished data demonstrating that prostate cancer cells stably overexpressing gfp-tagged mutant hLa defective in dimerization, post-translational modifications, or RNA-binding are forming fewer and smaller tumours in xenograft experiments compared to wildtype hLa (TH, GS unpublished data).
5.3. Survival of cancer cells
One of the hallmarks of cancer cells is a pro-survival phenotype established by either reduced or non-functional expression of pro-apoptotic factors and/or the overexpression of anti-apoptotic proteins. The X-linked inhibitor of apoptosis (XIAP) acts as an anti-apoptotic protein inhibiting caspase 3 and 9 activities. The first hint that hLa supports an anti-apoptotic state came from studies in HeLa cells [120,121]. In this study, the authors reported that hLa interacts with an RNA element located in the 5ʹUTR of the mRNA encoding XIAP. By using a biscistronic reporter system they showed that the 5ʹUTR contains an internal ribosome entry site (IRES) element bound by hLa.
An increase in XIAP expression is expected to correlate with reduced apoptosis and to counteract the efficiency of drugs intended to trigger apoptosis in cancer cells. Interestingly, it has been shown that overexpression of a trans-dominant negative hLa protein [42] downregulated Mdm2 level, an oncoprotein and negative regulator of tumour suppressor p53, and increased sensitivity towards Adriamycin-induced apoptosis in murine 32D-BCR/ABL myeloid precursor cells [43]. Hence, those data suggest that a hLa-dependent increase of the Mdm2 level augments survival of cancer cells expressing constitutively active tyrosine kinases and contributes to the progression of CML into blast crisis [122]. Interestingly, treatment of BCR/ABL-positive cells with the tyrosine kinase inhibitor STI-571 (Imatinib) not only reduced hLa [43] but also XIAP expression [123] suggesting that hLa regulates XIAP expression in BCR/ABL-positive cells and thereby counteracts Adriamycin-induced apoptosis.
The B-cell lymphoma 2 (Bcl2) protein, a mitochondrial membrane-associated anti-apoptotic factor, is likewise encoded by an mRNA containing an IRES element in the 5ʹUTR [124,125]. Remarkably, studies using head and neck squamous cell carcinoma (HNSCC) cell lines revealed that hLa depletion is associated with increased sensitivity towards the chemotherapeutic drug cisplatin. Analysing this observation exposed that hLa depletion caused a decline in Bcl2 protein expression and that exogenous Bcl2 expression increased cisplatin resistance, demonstrating that hLa-dependent Bcl2 expression contributes to an anti-apoptotic state of HNSCC cells [45].
Importantly, La depletion did not induce apoptosis as shown previously [72,73,116]. However, the role of cancer-associated hLa in counteracting apoptosis supports cancer cells surviving cellular stress induced by chemotherapeutic drugs, such as cisplatin or Adriamycin, and accentuates the view that La acts as a non-oncogenic addiction factor. Moreover, the expression of XIAP – an anti-apoptotic factor encoded by an IRES-containing mRNA – was also reduced in La-depleted HNSCC cells [45]. As we will review below, La regulates the translation of XIAP and Bcl2 mRNA.
6. The La protein facilitates the translation of selective mRNAs encoding tumour-promoting and anti-apoptotic factors
At the beginning of this chapter, we summarize information about a potential role of the La protein in protein synthesis. Here we will focus on reviewing examples in which La fosters the translation of selective mRNAs encoding tumour-promoting and anti-apoptotic factors in cancer cells.
Messenger RNAs carry at the 5ʹ-end a 7-methylguanosine, known as a Cap structure, ensuring protection, nuclear export and translation of mRNAs [126]. Under normal circumstances, most cellular mRNAs are capped and translated in a Cap-dependent manner. Translational initiation at the Cap structure is mediated by a number of translation initiation factors and is considered being rate-limiting [126,127].
Cap-dependent translation is often impaired in cells responding to specific environmental stimuli and cellular stress conditions. Cancer cells, especially, have to cope with a variety of stress conditions such as hypoxia or reactive oxygen species (ROS), and induction of apoptosis by chemotherapeutic treatment [128]. Under those conditions most Cap-dependent translated cellular mRNAs are translated at a lower rate, however mRNAs containing an IRES in their 5ʹUTR can be translated by a Cap-independent, IRES-mediated mechanism [121,129–132]. Whereas viral IRESs are well studied and classified, based on structural and sequence similarities, cellular IRES are too divergent for a systematic classification [129]. Initially, the hLa protein was discovered as an IRES trans-acting factor (ITAF), supporting IRES-mediated translation of viral mRNAs, such as poliovirus and hepatitis C virus [44,62,63,65,66,133,134]. In the viral model, hLa binds to RNA elements located at the 5ʹUTR and contributes to start site selection, most likely by assisting re-arrangements of structural features due to its RNA chaperone activity [63]. Although accumulating data suggest that hLa plays a role in translation of selective mRNAs encoding tumour-promoting and anti-apoptotic factors in cancer cells the molecular mechanism is still not clear. Since the discovery of hLa as ITAF, a role of La in mRNA translation has been studied using in vitro translation assays, biscistronic reporter assays, and polyribosomal gradient centrifugations.
In vitro translation assays exploit, most often, rabbit reticulocyte lysates or translation-active cell extracts prepared form cancer cell lines. In an effort to understand the internal initiation of ribosomes on model mRNAs observed in in vitro translation assays, the contribution of general RBPs, which are present at low concentrations in rabbit reticulocyte lysates, has been addressed [135]. Ribosomes can initiate translation on uncapped mRNAs by binding to sequence elements present in the 5ʹUTR and it has been found that addition of recombinant RBPs (e.g. hLa) to rabbit reticulocyte lysate impairs the translation of uncapped mRNA, most likely because ribosome binding sites are covered by RBPs. In those experiments, native recombinant La was used and post-translational modifications such as phosphorylation and sumoylation pathways often activated in cancer cells were not considered.
Cells respond to various stress signals with the activation of specific protein kinases phosphorylating the eukaryotic translation initiation factor 2 alpha (eIF2alpha) at serine 51. This phosphorylation event inhibits protein synthesis [136,137], however translation of specific mRNAs with uORFs is still translated and encoding proteins facilitating the adaption to cellular stress [138]. One of those kinases is protein kinase RNA-activated (PKR, also referred to as interferon-induced, double-stranded RNA (dsRNA)-dependent activated protein kinase or EIF2AK2), which can be activated by double-stranded RNA (dsRNA) in cells [139,140]. Upon binding of dsRNA, for example originated from viral RNA, autophosphorylation of PKR occurs and activated PKR phosphorylates eIF2alpha. Phosphorylated eIF2alpha inhibits the formation of the pre-initiation complex and consequently impairs Cap-dependent translation [141,142]. Due to its RNA chaperone activity, it has been postulated that hLa inhibits PKR activation by unwinding dsRNA [50,143] and thereby maintains Cap-dependent translation. Interestingly, not only does viral-derived dsRNA regulate PKR activity but also small non-coding RNAs such as small nucleolar (sno)RNAs [144], nc886, also referred to as pre-miR886 [145,146], Alu RNAs [147], and the binding of PKR to dsRNA-structured inverted Alu repeats (IRAlu’s) located in the 3ʹUTR of some mRNAs [148,149]. Since La is implicated in the processing of U3 snoRNAs [150] and binds the 3ʹ-end of RNA Pol III transcripts such as nc886 and B1-Alu RNAs [151,152], elevated La expression in cancer cells may foster the expression of those RNAs and thereby upregulate the activity of PKR. On the other hand, due to its RNA chaperone activity La might unwind those dsRNA structures and thereby downregulate the activity of PKR, which could explain the dual function of PKR as tumour suppressor or tumour promoting factor depending on the cancer type [153,154].
Another important aspect comes from studies aiming to test whether hLa protein is associated with ribosomal subunits and translating ribosomes. hLa has been found to bind to the 18S rRNA and co-fractionates with the 40S ribosomal subunit [155]. Studying the distribution of the mLa or hLa protein in polyribosomal gradients demonstrated that La is present in the monoribosomal fractions, the 80S peak and in polyribosomal fractions [37,156,157 and TH unpublished data], strongly supporting the view that La is not only associated with free small ribosomal subunits but is also part of the translating ribosome. As shown for mLa, AKT-mediated phosphorylation changes the distribution of mLa in the sucrose gradient in that more mLa is present in the polyribosomal fractions correlating with an increase in actively translated mRNAs (see below, [37]).
The RNA chaperone hLa might actually execute important functions within the pre-initiation complex while scanning the 5ʹUTR for the authentic translational start site. During this process, the mRNA moves through the channel of the 40S subunit until the translational start site has been found [126]. While scanning the 5ʹUTR, the 43S subunit has to be able to remove secondary structures possibly impairing efficient proceeding otherwise. Besides the eIFA [158,159], a helicase associated with the pre-initiation complex, the helicase DHX29 [160,161] facilitates the unfolding of hindering RNA structural elements in the 5ʹUTR. DHX29 is associated with the 40S ribosomal subunit and might interact with the 18S RNA [162]. Upon recognition of the translational start site, the 48S complex assembles. Interestingly, in cell-based assays, it has been shown that siRNA-mediated depletion of hLa or the expression of a dominant-negative La mutant (LaDN) prevents the formation of the 48S complex during hepatitis C virus and poliovirus translation initiation [44], suggesting inefficient recognition of the correct start site in absence of functional hLa protein [134].
A comprehensive sequence analysis of translation start sites revealed a consensus sequence, referred to as a Kozak sequence, surrounding the AUG start codon that predicts the strength of a translational start site [163,164]. Interestingly, it has been shown that hLa preferentially binds RNA oligonucleotides with an AUG as well as a strong Kozak sequence [45,165]. The findings imply that hLa binds near or at a strong translational start site of selective mRNAs. Considering the RNA chaperone activity of La, it is reasonable to assume that La associates with the 43S subunit via binding to the 18S ribosomal RNA [155] and thereby facilitates the unfolding of hairpin structures located in close proximity of translational start sites resulting in efficient mRNA translation (Fig. 6A). In this view, depletion of hLa or expression of hLa dominant-negative mutants will impair translation initiation of selective mRNAs sharing structural elements located near to or harbouring the translational start site [29,45].
Figure 6.
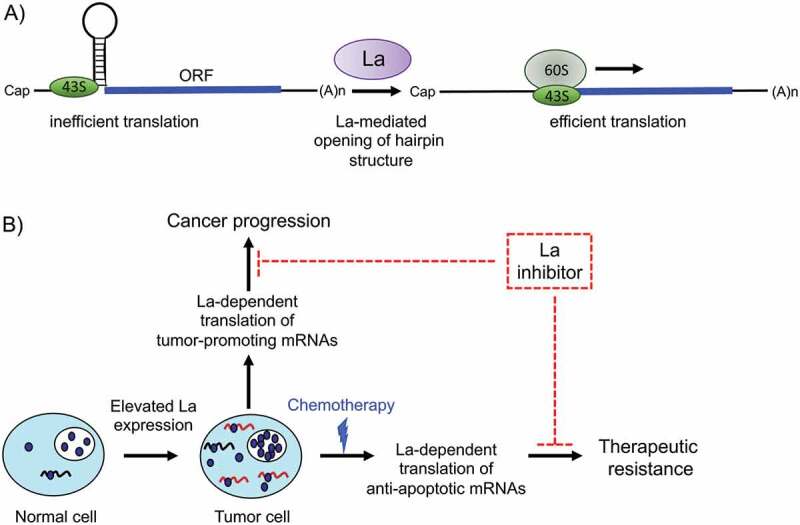
A model for the role of human RNA-binding protein La in cancer progression and therapeutic resistance
A) The RNA chaperone La facilitates translation of mRNAs harbouring hairpin structures in close proximity to the translational start site. An RNA hairpin acts as a road block for the scanning ribosomal 43S subunit. Upon La-assisted helix destabilization of the RNA hairpin, the 43S subunit reaches the translational start site, formation of the 80S ribosome occurs and mRNA translation starts. B) The model is based on the assumption that La facilitates the translation of mRNA regulons encoding factors implicated in cancer pathobiology (tumour-promoting mRNAs) and survival (anti-apoptotic mRNAs). Blocking the binding of La to selective mRNAs may impair cancer progression and the ability of cancer cells to cope with cellular stress under chemotherapeutic treatment. ORF: Open Reading Frame.
A more recent study revealed an interesting new role of hLa in protein synthesis. The Bayfield laboratory found that hLa binds to the poly(A) tail of mRNAs. In mRNA transfection experiments they showed that amino acid residues which are important for poly(A) binding are also required to enhance mRNA translation [157]. Hence, not only the binding of hLa to RNA elements located in the 5ʹUTR but also the binding to poly(A) tails appears to promote efficient mRNA translation. Future work in the field is expected to answer the question of how hLa regulates the translation of selective mRNAs and how cancer-promoting pathways influence La’s role in protein synthesis.
As mentioned above, murine La can be phosphorylated by AKT at T301 [37]. Interestingly, polyribosomal gradient analysis of extracts prepared from cell lines expressing gfp-tagged wildtype mLaWT or mutant mLaT301A revealed that only phosphorylated mLa co-fractionates with translating ribosomes and that a phosphorylation-deficient mutant of mLa stays in monoribomal fractions [37]. If native mLa is associated with the 43S subunit and phosphorylated mLa with translating ribosomes, mLaT301 phosphorylation represents an important regulatory posttranslational modification. Hence, cancer-promoting signalling cascades leading to AKT-mediated phosphorylation of mLa are expected to change the distribution of mRNAs between actively translated and translationally inactive mRNAs in a La-dependent manner – a malignant process contributing to the onco-proteome of cancer cells.
Additionally, human La is phosphorylated by CK2 at hLaS366 and polyribosomal gradient analysis of the TOP mRNA encoding ribosomal protein L37 in cells overexpressing wildtype hLaWT or the mutant hLaS366A revealed that more L37 mRNA is associated with translating ribosomes in hLaWT and less in hLaS366A expressing cells, suggesting that phosphorylation of hLa at serine 366 promotes L37 mRNA translation [104]. Since L37 mRNA is associated preferentially with native, non-phosphorylated hLa [36] and not with S366 phosphorylated hLa (LapS366), it suggests that phosphorylation at hLaS366 enhances the association of hLa with the ribosomes rather than with the L37 mRNA. This observation is puzzling because LapS366 has been reported to preferentially localize to the nucleoplasm [36] and suggests that LapS366 is already associated with the 40S ribosomal subunit in the nucleus. Of note, the role of La in TOP mRNA translation is controversially discussed [166] and it is important to consider that the 5ʹUTR of TOP mRNAs is very short and not folded in stable secondary structures when compared to other mRNAs reported to be regulated by La. Still, it is well established that CK2 phosphorylates La and that CK2 activity is associated with cancer progression [167,168] suggesting that the fraction of phosphorylated LaS366 increases in cancer cells and regulates the translation of a subset of mRNAs in a similar way as observed for mLa [37].
Nevertheless, these findings demonstrate that phosphorylation of residues in the C-terminal domain of mLa (threonine 301) and hLa (serine 366, threonine 389) are critical and correlate with increased association of selective mRNAs with translating ribosomes. Those PTMs should be considered when using recombinant La in in vitro translation assays. The above-reviewed studies clearly suggest that La can modulate translation and that it depends on cellular conditions such as impaired Cap-dependent translation during cellular stress, viral infection, the presence of dsRNA, signalling pathways triggering PTMs of the La protein, La’s RNA chaperone activity, the presence of Cap-independent translation initiation elements, and the Poly(A) tail.
According to the IRESite database (IRESsite.org), more than 68 viral and 115 cellular IRES elements have been reported as off 2009. Whereas IRES-dependent translation of viral RNAs is well studied and of significant importance for the viral life cycle, IRES-dependent translation of cellular mRNAs is often controversially discussed and critical controls are required to demonstrate IRES-mediated translation of a given cellular mRNA [169,170]. Furthermore, it is also very important to consider whether the amount of protein produced in an IRES-dependent manner is of biological significance [170,171]. Most of the mRNAs translationally regulated by La contain IRES elements in their 5ʹUTR such as the tumour-promoting factors BiP, Laminin B1, CCND1, and the anti-apoptotic factors Bcl2 and XIAP.
The concept of Cap-independent, IRES-mediated translation initiation might not be the only way by which La supports translation. We have also to consider that selective mRNAs with certain characteristics surrounding the start site are translated in a La-dependent manner. Below we will review studies investigating the translational control of mRNAs encoding factors implicated in tumour-promoting or anti-apoptotic processes.
6.1. Laminin B1
Epithelial to mesenchymal transition (EMT) plays a key role in establishing a malignant cancer phenotype. Transforming growth factor β (TGFβ) can induce EMT in a number of epithelial cell types such as in hepatocytes [172]. EMT is associated with increased cell motility and survival, and often a cancer stem-like cell (CSC) phenotype resulting in chemoresistance, metastasis and poor clinical outcome [173–175]. Although EMT is primarily regulated on the transcriptional level [176], recent studies suggest that RBP-dependent translation of selective mRNAs is of importance as well [177]. The overexpression of RBP YB1 in non-invasive breast cancer cells induces EMT and activates IRES-dependent translation of SNAIL1, a transcriptional master regulator of EMT [178]. Furthermore, binding of RBP hnRNPE1 and translation elongation factor eEF1A1 to the TGFβ-activated translational (BAT) RNA element blocks the translational elongation of selective mRNAs and this block is released during TGFβ-induced EMT [179,180].
Laminin B1 is part of the extracellular matrix and is elevated in hepatocellular carcinoma (HCC) [181]. Laminin B1 is bound by the Laminin receptor, which is highly expressed in metastatic cancer and promotes survival, angiogenesis and cell invasion [182,183]. Laminin B1 protein has been shown to be translationally upregulated during EMT [184] suggesting a potential regulation of Laminin B1 mRNA translation by an TGFβ-dependent mechanism. Interestingly, it has been found that the 5ʹUTR of Laminin B1 mRNA contains an IRES element [184]. This Laminin B1 IRES has been tested in different mouse cell lines: immortalized mouse hepatocytes (MIM-1-4), MIM-1-4 cells transformed by oncogenic Ras (MIM-R) and the cell line MIM-RT derived from MIM-R cells that have undergone TGFβ-induced EMT. The Laminin B1 IRES is more active in invasive MIM-RT cells than in MIM-R or MIM-1-4 cells suggesting that the MIM-RT cell line gained some cancerous features during the TGFβ-induced EMT [39,185]. The 335nucleotides long 5ʹUTR of Laminin B1 mRNA contains an IRES as demonstrated by two different bicistronic vector systems. Furthermore, the Laminin B1 IRES is bound by mLa and recombinant hLa promotes translation of a Laminin B1 5ʹUTR reporter when using rabbit reticulocytes extracts, which were cleared for the La protein before in vitro translation [185]. Of note, recombinant hLa, which is not modified by PTMs, are sufficient to promote Laminin B1 IRES activity in this in vitro system. About 30% of Laminin B1 IRES activity is mediated by an 5ʹUTR element spanning from nucleotide −82 to 0, representing a region just upstream of the translational start site. However, it remains to be determined to which sequence/structural element the mLa and hLa protein binds within the Laminin B1 5ʹUTR. Interestingly, an increase in cytoplasmic La has been reported for MIM-RT cells as well as TGFβ-treated MIM-R cells. As shown previously, platelet-derived growth factor (PDGF) treatment of glia cells induced a cytoplasmic localization of mLa, which was associated with AKT-mediated phosphorylation of mLa [37]. Moreover, Gotzmann et al. found that TGFβ treatment induced the PDGF receptor and activated PDGF signalling in the murine hepatocytes cell line MMH-R [186], raising the question of whether PDGF regulates cytoplasmic accumulation of mLa. Indeed, recently it has been shown that TGFβ treatment-induced cytoplasmic accumulation of mLa and activated the Laminin B1 IRES. Interestingly, in the murine hepatocyte model, the cytoplasmic accumulation of La and the increase of Laminin B1 IRES activity was dependent on PDGF and MAPK/ERK signalling and only to a lower extend on PI3K/AKT signalling[39]. Furthermore, in sequential histological staining of human hepatocellular carcinoma (HCC) tissue, activation PDGFRα signalling correlated with strong cytoplasmic La staining and higher Laminin B1 expression within the invasive front suggesting also an active PDGFRα/La/Laminin B1 pathway during cancer progression in HCC patients [187].
6.2. CCND1
CCDN1 (cyclin D1) is a cooperative oncogene often overexpressed in cancer tissue and is suggested to support tumorigenesis [188]. CCND1 expression is induced by mitogens and upon its association with cyclin-dependent kinase (CDK), CCND1 contributes to entry of the cell cycle into the S-phase and cell proliferation [189–191]. Hence, CCND1 is a key player in the regulation of cell proliferation. However, in addition, CCND1 might affect cell proliferation via an alternative route, since it is established that CCND1 has CDK-independent functions [192].
Interestingly, it has been shown that the 5ʹUTR of CCND1 mRNA contains an IRES element [193]. The CCND1 IRES is basically inactive in PTEN negative cells suggesting that AKT signalling regulates its activity [193–195]. The 5ʹUTR is 209 nucleotides long and the minimal IRES is located between nucleotides −209 and −45. The hnRNPA1 protein was described to be a positive regulator upon phosphorylation [196]. Like hnRNPA1, hLa is a regulator of CCND1 IRES-mediated translation. The hLa protein is associated with CCND1 mRNA and stimulates a bicistronic CCND1 IRES reporter as well as in vitro transcribed and transfected bicistronic CCND1 IRES reporter mRNA, suggesting strongly that hLa stimulates IRES-dependent CCND1 mRNA translation [72]. In conclusion, these studies indicate that cancer-associated hLa stimulates Cap-independent translation of CCND1 mRNA, however future work is needed to address in detail the molecular mechanism.
6.3. XIAP
The X-linked inhibitor of apoptosis (XIAP) belongs to a class of proteins known as inhibitors of apoptosis [197]. XIAP is able to protect stressed cells against apoptosis by direct binding of caspase 3, 7, or 9 via its baculoviral IAP repeat (BIR) domains [198]. It has been observed that XIAP levels are elevated in a number of different cell types after induction of stress such as ionizing radiation or serum starvation [199]. Interestingly, the XIAP mRNA level remained nearly unchanged, suggesting a posttranscriptional regulation of XIAP under those conditions. Cloning of several parts of the XIAP 5ʹUTR into bicistronic expression vectors was employed to define a functional IRES element between position nucleotides −162 and −1, which allows XIAP expression under cellular stress [199]. Several different XIAP IRES constructs were created and used to demonstrate that alternative splicing, which has been reported to occur with specific bicistronic XIAP IRES constructs [200,201], did not account for IRES-mediated expression [202]. Furthermore, transfection of RNA transcripts synthesized from a variety of different XIAP IRES reporter constructs in vitro confirmed the presence of an IRES in the XIAP 5ʹUTR [202].
Searching for cellular factors affecting XIAP expression led to the finding that the hLa protein facilitates IRES-mediated translation of XIAP mRNA. Furthermore, in HeLa cells, hLa is associated with the XIAP IRES RNA element expressed from a bicistronic XIAP IRES reporter [120]. The hLa binding site has been mapped within the IRES element between nucleotide −62 and −32 upstream of the authentic translational start site [120]. The authors also demonstrated that a dominant-negative mutant of La impairs IRES-mediated translation in vitro as well as in HeLa cells [120]. Two XIAP mRNA isoforms are known. The isoform with the short 5ʹUTR is translated in a Cap-dependent manner and does not contain an IRES element, whereas the longer much less abundant XIAP mRNA isoform contains the IRES element [202]. The IRES-containing isoform might be important to assure XIAP expression under conditions of cellular stress when Cap-dependent translation is impaired due to the treatment of cancer cells with chemotherapeutic drugs.
The hLa protein is overexpressed in various cancer entities such as head and neck cancer squamous cell carcinoma, lung and breast cancer in which XIAP protein has also been described to be overexpressed [45,203,204], suggesting that XIAP protein is elevated in those cancer entities due to high La protein expression. Accordingly, studies showed that expression of a dominant-negative hLa mutant reduced XIAP protein expression [120], and depletion of hLa shifted XIAP mRNA from translationally active to inactive fractions in sucrose gradient analyses [45]. In the future, it will be interesting to study whether the protein expression of XIAP and hLa correlates in patients’ samples and to determine in detail the molecular mechanism by which hLa supports XIAP IRES-mediated mRNA translation in cancer cells.
6.4. Bcl2
The finding that hLa regulates mRNA translation of the anti-apoptotic protein XIAP and the observation that hLa-depleted cells are more sensitive towards cisplatin treatment, proposed that additional anti-apoptotic factors are regulated by hLa in cancer cells. Indeed, it has been found that B-cell lymphoma 2 (Bcl2) protein expression is reduced in hLa-depleted cells of the HNSCC cell line [45]. Analysis of the underlying mechanism led to the finding that hLa binds in close proximity to the translational start site of Bcl2 mRNA. This region can fold, as predicted by the mfold program [205], into a stable hairpin structure, which can be destabilized by the RNA chaperone hLa in an in vitro RNA chaperone assay [45]. Furthermore, luciferase reporter assays revealed that the RNA chaperone domain of hLa is required to support Bcl2 5ʹUTR-dependent reporter protein expression. Also applying sucrose gradient analyses, Bcl2 mRNA as well as XIAP mRNA shifted from translationally active to inactive fractions in hLa-depleted HNSCC cells [45]. Whether hLa promotes Bcl2 translation via stimulating the recruitment of ribosomes to the Bcl2 IRES element [124,125] or by remodelling structural elements located in close proximity to the translational start site remains to be addressed experimentally. So far, RNA immunoprecipitations, RNA pull-down experiments, and in vitro RNA chaperone assays imply that the hLa protein binds to the Bcl2 mRNA and assists structural changes of the Bcl2 translational start site [45].
6.5. BiP
The mRNA encoding the molecular chaperone binding immunoglobulin protein (BiP), also referred to as 78-kDa glucose-regulated protein (GRP78) or heat shock 70 kDa protein 5 (HSPA5), contains an IRES element within the 5ʹUTR [206]. BiP belongs to the family of Hsp70 proteins and is mainly localized to the endoplasmic reticulum (ER). Elevated BiP expression is associated with cancer progression, patient survival and drug resistance probably facilitated by cell membrane standing BiP and PI3K/AKT oncogenic pathway activation [207,208].
The BiP IRES was first reported by the Sarnow’s laboratory [206]. They showed that the IRES element consists of two parts (nucleotides −1 to −53 and −162 to −220) and is bound by two proteins referred to as p60 and p90. Subsequent studies found that hLa binds to a region spanning nucleotides −115 to −207 within the BiP IRES [209]. Using a bicistronic BiP IRES reporter, they showed that gfp-tagged hLa stimulates the BiP IRES-dependent luciferase expression in COS-7 cells [209]. The BiP 5ʹUTR contains a conserved structured region just upstream of the translational start site from nucleotides −129 to −222, which contains the region (−115 to −207) bound by hLa [209,210]. Hence, the model in which hLa facilitates translation by unwinding structural elements located in close proximity of the start site would be in agreement with those data.
In summary, reports accumulate supporting the notion that La stimulates the mRNA translation of factors contributing to chemoresistance such as XIAP, Bcl2, and BiP as well as Survivin, BclxL, and Bag1 (GS unpublished data). Notably, BclxL and Bag1 are encoded by IRES-containing mRNAs [211–213] and stem-loop structures within the 5ʹUTR that potentially could impede mRNA translation was proposed recently for the Survivin mRNA [214]. The theory of RNA regulons is based on the finding that a specific RBP interacts with different mRNAs and jointly controls an expression network modulating a particular cellular process [215,216]. In this case, the data suggest that the La protein controls a pro-survival RNA regulon contributing to chemoresistance and strongly support the notion that RBP La acts as non-oncogenic addiction factor by establishing a pro-survival state.
6.6. Mdm2
The mouse double minute 2 homolog (Mdm2) is expressed in a variety of isoforms oftentimes originating from alternative splicing [217]. Mdm2 is an E3 ubiquitin-protein ligase triggering the ubiquitinylation and consequently the degradation of tumour suppressor p53 [218]. Hence, elevated Mdm2 expression often correlates with low p53 protein level in cancer cells.
The study by Trotta et al. revealed a correlation between the very low expression of p53 and the high expression of Mdm2 in BCR/ABL-expressing murine 32D cells [43]. Additionally, it has been shown that Mdm2 mRNA is translationally regulated in other cells [219–223], raising the question of whether translation of Mdm2 mRNA is up-regulated in BCR/ABL-expressing 32D cells.
Mdm2 contains two upstream open reading frames (uORFs), which contribute to translational regulation of Mdm2 [222–224]. Sucrose gradient analyses revealed that inhibition of BCR/ABL by STI571 shifted Mdm2 mRNA from translationally active to inactive fractions, demonstrating a BCL/ABL-dependent regulation of Mdm2 mRNA translation [43]. Further, the data showed that uORFs are not involved in this translational regulation of Mdm2 mRNA but rather a small sequence element located between nucleotides −62 and −36 upstream of the translational start site [43]. The authors further found that the La protein binds directly to this small sequence element located upstream of the Mdm2 translational start site and that expression of a dominant-negative hLa (LaDN) mutant impaired Mdm2 expression in cells and in in vitro translation assays [43].
A set of protein kinase inhibitors was used to study the impact of those kinases on La protein levels. The study by Trotta et al. found that STI571, PKC, and PI3K inhibitors decreased hLa levels suggesting that BCR/ABL-signalling stabilizes hLa protein in a PKC- and/or PI3K-dependent manner. It has been reported that PKC can phosphorylate recombinant hLa [100], however this aspect was not addressed in this study [43].
Taken together, mRNAs encoding tumour-promoting and anti-apoptotic factors, which are regulated by the hLa protein have in common that hLa binds most often in close proximity to the translational start site. Those binding regions are predicted to fold into secondary structures. Hence, hLa’s RNA chaperone activity might destabilize those structural elements to promote start site selection and 48S complex formation during translation initiation. In addition, for mLa it has been shown that signalling pathways trigger the cytoplasmic localization of mLa.
7. La protein as a novel cancer drug target
The overexpression of La protein in various cancer entities promotes translation of mRNAs encoding tumour-promoting and anti-apoptotic factors, and contributes to proliferation, migration, invasiveness, tumour growth, and chemo-resistance of cancer cells as detailed above. To support mRNA translation, the La protein must bind to RNA elements in those mRNAs. Hence, a La inhibitor blocking the binding of La to selective mRNAs, specifically during Cap-independent translation in cancer cells treated with chemotherapeutics provides an opportunity for a novel drug intervention point (Fig. 6B).
The feasibility of inhibiting RBP:RNA interactions by compounds has been demonstrated in a few cases, however only little is known about drug discovery approaches targeting the interaction between cellular RBPs and their target RNAs [225–228]. In previous studies, it has been shown that mLa protects hepatitis B virus (HBV) RNA against cytokine-induced degradation [81,229,230] and that the phosphorylation of La at serine 366 increases during HBV replication and promotes the HBV life cycle [231]. Hence, to test whether compounds can target hLa, a virtual screening approach has been used to identify active molecules and to show that they impair the HBV life cycle [232]. The hLa protein is also implicated in the translation of the hepatitis C virus (HCV) and efforts were reported to target the La:HCV RNA interaction by a small La-derived peptide [233].
Based on the finding that cancer-associated hLa contributes also to cancer progression and chemotherapeutic resistance, studies were initiated to identify compounds able to inhibit the binding of hLa to CCND1 and Bcl2 mRNA. Using a high-throughput fluorescence polarization assay, compounds were identified and tested in orthogonal assays to demonstrate that they impair binding of hLa to CCND1- or Bcl2-derived synthetic RNAs [234]. Most interestingly, those active compounds diminished the viability of cancer cells, but not normal cells. They specifically impaired the binding of hLa to CCND1 and Bcl2 mRNA in cells, reduced Bcl2 protein level and sensitized cancer cells for cisplatin-induced apoptotic cell death [234].
Taken together, various efforts have been reported aiming to target the hLa protein during the viral life cycle and cancer progression. Although the chemical space of those identified compounds and the required concentrations are suboptimal, those studies demonstrate the proof-of-concept, that the interaction between hLa and selective RNAs might represent a novel drug intervention point.
8. Outlook and open questions
In recent years experimental data have accumulated demonstrating that the La protein plays a significant role in cancer pathobiology. According to the e!Ensembl database and as reported previously [78,235] several La mRNA isoforms are expressed. Two mRNA isoforms, which differ in length but are expressing the the full-length hLa protein, next to three isoforms expressing shorter versions of the hLa protein are known. Interestingly, different promoters with or without an NF-kappa B binding site exist and alternative splicing can lead to hLa mRNA isoforms containing an IRES or not [78,235]. The potential role of those La isoforms in normal and in cancer cells remains to be tested.
Although it is not well understood which mechanism causes the overexpression of La in haematologic malignancies and in various solid tumour entities, it is clear that La is required to secure cell proliferation, migration, invasiveness, and tumour growth of cancer cells (Fig. 6B). However, many interesting questions remain to be answered. For example, although it is well documented that La modulates translation of various mRNAs encoding tumour-promoting and anti-apoptotic factors, the molecular mechanism is only poorly understood. Does La remodel RNA elements located in the 5ʹUTR of mRNAs and promote translation by easing the scanning of the 43 ribosomal subunit? The La protein associates with ribosomal subunits and is present in polyribosmal fractions containing translationally active mRNAs. Do those findings suggest a function of La during translational initiation and/or elongation? Since La is implicated in many different aspects of the RNA metabolism such as processing of non-coding RNAs, like tRNA and U6 snRNA [56], and is likely to be involved in miRNA biogenesis, it is challenging to differentiate between indirect and direct effects in studies applying depletion of La. The subcellular localization of the La protein and its functionality is modulated by PTMs. Hence, studying the impact of signalling pathways and cell plasticity on posttranslational modifications of La causing changes in its cellular functions is expected to reveal exciting new information about this protein during cancer progression. Moreover, the La protein interacts with a broad variety of RNA molecules. The contribution of the different RNA-binding surfaces in the binding of those different RNA sequence/structural elements remains an important area of research. Deeper understanding of the RNA-binding modes would allow the structure-based drug design of biologically active compounds able to block the binding of La to selective mRNAs encoding factors involved in cancer progression or viral replication.
In this review, we favour the view that La is a non-oncogenic addiction factor. As reported, La depletion sensitizes cancer cells for chemotherapeutics [43,45]. These kinds of experiments and comparing the response of La-depleted cancer cells to other stressors such as ER stress, serum deprivation or oxidative stress are ideal experimental settings to test the role of La in non-oncogenic addiction. In this experimental model reduction of La protein expression or functionality should preferentially harm cancer cells compared to normal cells, as has been shown for molecular compounds inhibiting La:mRNA complex formation [234].
Lastly, it is an open question which mechanisms are regulating the La protein expression level. So far, only marginal information is available on the promoter control of the La gene, the signals regulating La transcription, or the posttranscriptional regulation of La protein stability as observed in cancer cells. In this context it is important to note that only a few protein interaction partners of hLa have been described so far, namely nucleolin [33,236,237] and DEAH-box RNA helicase DDX15/hPrp43 [238]. Furthermore, about 150 proteins are listed in the BioGRID database (https://thebiogrid.org) as potential La interaction partners. The limited information on hLa interacting partners, hLa gene regulators, signalling pathways regulating hLa transcription, and posttranscriptional control represents a very exciting area of research.
La protein belongs to the family of La-related proteins (LARPs) and to our knowledge it is currently not known whether other LARPs interact with hLa or to which extend other LARPs are able to compensate for the function of hLa in depleted cells or knock-out models. The growing number of important cellular functions members of the LARP family are implicated in [10] suggests that future studies will reveal more critical cancer-associated functions of this protein family.
Acknowledgments
We thank the LARP Society for inspiration and wonderful discussions. We further thank our former lab members: Julia Dittmann, Alena Fedarovich, Sven Horke, Alexander Brock, Venkatesh Kota, Julia Kuehnert, Reycel Rodriguez, Avery Zierk, and Carly Farrell for experimental and technical support. We further thank the Verein zur Förderung krebskranker und körperbehinderter Kinder Ostbayern e.V. (VKKK) for the support.
Funding Statement
This work was supported by the German Research Foundation [SFB 960, B14 to T.H. and G.S.].
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Corley M, Burns MC, Yeo GW.. How RNA-binding proteins interact with RNA: molecules and mechanisms. Mol Cell. 2020;78:9–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Hentze MW, Castello A, Schwarzl T, et al. A brave new world of RNA-binding proteins. Nat Rev Mol Cell Biol. 2018;19:327–341. [DOI] [PubMed] [Google Scholar]
- [3].Kechavarzi B, Janga SC. Dissecting the expression landscape of RNA-binding proteins in human cancers. Genome Biol. 2014;15:R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Galante PA, Sandhu D, de Sousa Abreu R, et al. A comprehensive in silico expression analysis of RNA binding proteins in normal and tumor tissue: identification of potential players in tumor formation. RNA Biol. 2009;6:426–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Gerstberger S, Hafner M, Ascano M, et al. Evolutionary conservation and expression of human RNA-binding proteins and their role in human genetic disease. Ad Exp Med Biol. 2014;825:1–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Mattioli M, Reichlin M. Heterogeneity of RNA protein antigens reactive with sera of patients with systemic lupus erythematosus. Description of a cytoplasmic nonribosomal antigen. Arthritis Rheum. 1974;17:421–429. [DOI] [PubMed] [Google Scholar]
- [7].Alspaugh MA, Talal N, Tan EM. Differentiation and characterization of autoantibodies and their antigens in Sjogren’s syndrome. Arthritis Rheum. 1976;19:216–222. [DOI] [PubMed] [Google Scholar]
- [8].Wolin SL, Cedervall T. The la protein. Annu Rev Biochem. 2002;71:375–403. [DOI] [PubMed] [Google Scholar]
- [9].Bousquet-Antonelli C, Deragon JM. A comprehensive analysis of the La-motif protein superfamily. RNA. 2009;15:750–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Maraia RJ, Mattijssen S, Cruz-Gallardo I, et al. The La and related RNA-binding proteins (LARPs): structures, functions, and evolving perspectives. Wiley Interdiscip Rev RNA. 2017;8(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Bayfield MA, Vinayak J, Kerkhofs K, et al. La proteins couple use of sequence-specific and non-specific binding modes to engage RNA substrates. RNA Biol. 2019;1–10. DOI: 10.1080/15476286.2019.1582955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kenan DJ, Keene JD. La gets its wings. Nat Struct Mol Biol. 2004;11:303–305. [DOI] [PubMed] [Google Scholar]
- [13].Chambers JC, Keene JD. Isolation and analysis of cDNA clones expressing human lupus La antigen. Proc Natl Acad Sci U S A. 1985;82:2115–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chambers JC, Kenan D, Martin BJ, et al. Genomic structure and amino acid sequence domains of the human La autoantigen. J Biol Chem. 1988;263:18043–18051. [PubMed] [Google Scholar]
- [15].Bayfield MA, Yang R, Maraia RJ. Conserved and divergent features of the structure and function of La and La-related proteins (LARPs). Biochim Biophys Acta. 1799(5–6);365–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Dock Bregeon AC, Lewis KA, Conte MR. The La-related proteins: structures and interactions of a versatile superfamily of RNA binding proteins. RNA Biol. 2019;1–16. DOI: 10.1080/15476286.2019.1695712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Dong G, Chakshusmathi G, Wolin SL, et al. Structure of the La motif: a winged helix domain mediates RNA binding via a conserved aromatic patch. Embo J. 2004;23:1000–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Teplova M, Yuan YR, Phan AT, et al. Structural basis for recognition and sequestration of UUU(OH) 3ʹ temini of nascent RNA polymerase III transcripts by La, a rheumatic disease autoantigen. Mol Cell. 2006;21:75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Stavraka C, Blagden S. The La-related proteins, a family with connections to cancer. Biomolecules. 2015;5:2701–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Koso H, Yi H, Sheridan P, et al. Identification of RNA-binding protein LARP4B as a tumor suppressor in glioma. Cancer Res. 2016;76:2254–2264. [DOI] [PubMed] [Google Scholar]
- [21].Ye L, Lin ST, Mi YS, et al. Overexpression of LARP1 predicts poor prognosis of colorectal cancer and is expected to be a potential therapeutic target. Tumour Biol. 2016;37:14585–14594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Seetharaman S, Flemyng E, Shen J, et al. The RNA-binding protein LARP4 regulates cancer cell migration and invasion. Cytoskeleton (Hoboken). 2016;73:680–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Burrows C, Abd Latip N, Lam SJ, et al. The RNA binding protein Larp1 regulates cell division, apoptosis and cell migration. Nucleic Acids Res. 2010;38:5542–5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hopkins TG, Mura M, Al-Ashtal HA, et al. The RNA-binding protein LARP1 is a post-transcriptional regulator of survival and tumorigenesis in ovarian cancer. Nucleic Acids Res. 2016;44:1227–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Mura M, Hopkins TG, Michael T, et al. LARP1 post-transcriptionally regulates mTOR and contributes to cancer progression. Oncogene. 2015;34:5025–5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ji X, Lu H, Zhou Q, et al. LARP7 suppresses P-TEFb activity to inhibit breast cancer progression and metastasis. eLife. 2014;3:e02907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Jacks A, Babon J, Kelly G, et al. Structure of the C-terminal domain of human La protein reveals a novel RNA recognition motif coupled to a helical nuclear retention element. Structure. 2003;11:833–843. [DOI] [PubMed] [Google Scholar]
- [28].Horke S, Reumann K, Schulze C, et al. The La motif and the RNA recognition motifs of human La autoantigen contribute individually to RNA recognition and subcellular localization. J Biol Chem. 2004;279:50302–50309. [DOI] [PubMed] [Google Scholar]
- [29].Kuehnert J, Sommer G, Zierk AW, et al. Novel RNA chaperone domain of RNA-binding protein La is regulated by AKT phosphorylation. Nucleic Acids Res. 2015;43:581–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Martino L, Pennell S, Kelly G, et al. Analysis of the interaction with the hepatitis C virus mRNA reveals an alternative mode of RNA recognition by the human La protein. Nucleic Acids Res. 2012;40:1381–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Simons FH, Broers FJ, Van Venrooij WJ, et al. Characterization of cis-acting signals for nuclear import and retention of the La (SS-B) autoantigen. Exp Cell Res. 1996;224:224–236. [DOI] [PubMed] [Google Scholar]
- [32].Horke S, Reumann K, Schweizer M, et al. Nuclear trafficking of La protein depends on a newly identified nucleolar localization signal and the ability to bind RNA. J Biol Chem. 2004;279:26563–26570. Epub 2004 Apr 1. [DOI] [PubMed] [Google Scholar]
- [33].Intine RV, Dundr M, Vassilev A, et al. Nonphosphorylated human La antigen interacts with nucleolin at nucleolar sites involved in rRNA biogenesis. Mol Cell Biol. 2004;24:10894–10904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Bayfield MA, Kaiser TE, Intine RV, et al. Conservation of a masked nuclear export activity of La proteins and its effects on tRNA maturation. Mol Cell Biol. 2007;27:3303–3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Intine RV, Dundr M, Misteli T, et al. Aberrant nuclear trafficking of La protein leads to disordered processing of associated precursor tRNAs. Mol Cell. 2002;9:1113–1123. [DOI] [PubMed] [Google Scholar]
- [36].Intine RV, Tenenbaum SA, Sakulich AL, et al. Differential phosphorylation and subcellular localization of La RNPs associated with precursor tRNAs and translation-related mRNAs. Mol Cell. 2003;12:1301–1307. [DOI] [PubMed] [Google Scholar]
- [37].Brenet F, Socci ND, Sonenberg N, Holland EC . Akt phosphorylation of La regulates specific mRNA translation in glial progenitors. Oncogene. 2009;28:128–139. [DOI] [PubMed] [Google Scholar]
- [38].Bachmann M, Pfeifer K, Schroder HC, et al. The La antigen shuttles between the nucleus and the cytoplasm in CV-1 cells. Mol Cell Biochem. 1989;85:103–114. [DOI] [PubMed] [Google Scholar]
- [39].Petz M, Them NC, Huber H, et al. PDGF enhances IRES-mediated translation of Laminin B1 by cytoplasmic accumulation of La during epithelial to mesenchymal transition. Nucleic Acids Res. 2012;40:9738–9749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Bachmann M, Chang S, Slor H, et al. Shuttling of the autoantigen La between nucleus and cell surface after UV irradiation of human keratinocytes. Exp Cell Res. 1990;191:171–180. [DOI] [PubMed] [Google Scholar]
- [41].Fok V, Friend K, Steitz JA. Epstein-Barr virus noncoding RNAs are confined to the nucleus, whereas their partner, the human La protein, undergoes nucleocytoplasmic shuttling. J Cell Biol. 2006;173:319–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Craig A, Svitkin Y, Lee H, et al. The La autoantigen contains a dimerization domain that is essential for enhancing translation. Mol Cell Biol. 1997;17:163–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Trotta R, Vignudelli T, Candini O, et al. BCR/ABL activates mdm2 mRNA translation via the La antigen. Cancer Cell. 2003;3:145–160. [DOI] [PubMed] [Google Scholar]
- [44].Costa-Mattioli M, Svitkin Y, Sonenberg N. La autoantigen is necessary for optimal function of the poliovirus and hepatitis C virus internal ribosome entry site in vivo and in vitro. Mol Cell Biol. 2004;24:6861–6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Heise T, Kota V, Brock A, et al. The La protein counteracts cisplatin-induced cell death by stimulating protein synthesis of anti-apoptotic factor Bcl2. Oncotarget. 2016;7:29664–29676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Bachmann M, Pfeifer K, Schroder HC, et al. Characterization of the autoantigen La as a nucleic acid-dependent ATPase/dATPase with melting properties. Cell. 1990;60:85–93. [DOI] [PubMed] [Google Scholar]
- [47].Huhn P, Pruijn GJ, van Venrooij WJ, et al. Characterization of the autoantigen La (SS-B) as a dsRNA unwinding enzyme. Nucleic Acids Res. 1997;25:410–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Chakshusmathi G, Kim SD, Rubinson DA, et al. A La protein requirement for efficient pre-tRNA folding. Embo J. 2003;22:6562–6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Wolin SL, Wurtmann EJ. Molecular chaperones and quality control in noncoding RNA biogenesis. Cold Spring Harb Symp Quant Biol. 2006;71:505–511. [DOI] [PubMed] [Google Scholar]
- [50].Xiao Q, Sharp TV, Jeffrey IW, et al. The La antigen inhibits the activation of the interferon-inducible protein kinase PKR by sequestering and unwinding double-stranded RNA. Nucleic Acids Res. 1994;22:2512–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Belisova A, Semrad K, Mayer O, et al. RNA chaperone activity of protein components of human Ro RNPs. RNA. 2005;11:1084–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Hussain RH, Zawawi M, Bayfield MA. Conservation of RNA chaperone activity of the human La-related proteins 4, 6 and 7. Nucleic Acids Res. 2013;41:8715–8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Naeeni AR, Conte MR, Bayfield MA. RNA chaperone activity of human La protein is mediated by variant RNA recognition motif. J Biol Chem. 2012;287:5472–5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Kucera NJ, Hodsdon ME, Wolin SL. An intrinsically disordered C terminus allows the La protein to assist the biogenesis of diverse noncoding RNA precursors. Proc Natl Acad Sci U S A. 2011;108:1308–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Alfano C, Sanfelice D, Babon J, et al. Structural analysis of cooperative RNA binding by the La motif and central RRM domain of human La protein. Nat Struct Mol Biol. 2004;11:323–329. [DOI] [PubMed] [Google Scholar]
- [56].Pannone BK, Xue D, Wolin SL. A role for the yeast La protein in U6 snRNP assembly: evidence that the La protein is a molecular chaperone for RNA polymerase III transcripts. Embo J. 1998;17:7442–7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Yoo CJ, Wolin SL. The yeast La protein is required for the 3ʹ endonucleolytic cleavage that matures tRNA precursors. Cell. 1997;89:393–402. [DOI] [PubMed] [Google Scholar]
- [58].Hasler D, Meister G. From tRNA to miRNA: RNA-folding contributes to correct entry into noncoding RNA pathways. FEBS Lett. 2016;590:2354–2363. [DOI] [PubMed] [Google Scholar]
- [59].Hasler D, Lehmann G, Murakawa Y, et al. The lupus autoantigen La prevents mis-channeling of tRNA fragments into the human microRNA pathway. Mol Cell. 2016;63:110–124. [DOI] [PubMed] [Google Scholar]
- [60].Liu Y, Tan H, Tian H, et al. Autoantigen La promotes efficient RNAi, antiviral response, and transposon silencing by facilitating multiple-turnover RISC catalysis. Mol Cell. 2011;44:502–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Liang C, Xiong K, Szulwach KE, et al. Sjogren syndrome antigen B (SSB)/La promotes global microRNA expression by binding microRNA precursors through stem-loop recognition. J Biol Chem. 2013;288:723–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Belsham GJ, Sonenberg N, Svitkin YV. The role of the La autoantigen in internal initiation. Curr Top Microbiol Immunol. 1995;203:85–98. [DOI] [PubMed] [Google Scholar]
- [63].Svitkin YV, Meerovitch K, Lee HS, et al. Internal translation initiation on poliovirus RNA: further characterization of La function in poliovirus translation in vitro. J Virol. 1994;68:1544–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Svitkin YV, Pause A, Sonenberg N. La autoantigen alleviates translational repression by the 5ʹ leader sequence of the human immunodeficiency virus type 1 mRNA. J Virol. 1994;68:7001–7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Ali N, Pruijn GJ, Kenan DJ, et al. Human La antigen is required for the hepatitis C virus internal ribosome entry site-mediated translation. J Biol Chem. 2000;275:27531–27540. [DOI] [PubMed] [Google Scholar]
- [66].Ali N, Siddiqui A. The La antigen binds 5ʹ noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site-mediated translation. Proc Natl Acad Sci U S A. 1997;94:2249–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Pon JR, Marra MA. Driver and passenger mutations in cancer. Annu Rev Pathol. 2015;10:25–50. [DOI] [PubMed] [Google Scholar]
- [68].Nagel R, Semenova EA, Berns A. Drugging the addict: non-oncogene addiction as a target for cancer therapy. EMBO Rep. 2016;17:1516–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Solimini NL, Luo J, Elledge SJ. Non-oncogene addiction and the stress phenotype of cancer cells. Cell. 2007;130:986–988. [DOI] [PubMed] [Google Scholar]
- [70].Rother RP, Thomas PS. La/SSB ribonucleoprotein levels increased in transformed cells. Clin Exp Immunol. 1991;83:369–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Al-Ejeh F, Darby JM, Brown MP. The La autoantigen is a malignancy-associated cell death target that is induced by DNA-damaging drugs. Clin Cancer Res. 2007;13:5509s–18s. [DOI] [PubMed] [Google Scholar]
- [72].Sommer G, Dittmann J, Kuehnert J, et al. The RNA-binding protein La contributes to cell proliferation and CCND1 expression. Oncogene. 2011;30:434–444. [DOI] [PubMed] [Google Scholar]
- [73].Sommer G, Rossa C, Chi AC, et al. Implication of RNA-binding protein La in proliferation, migration and invasion of lymph node-metastasized hypopharyngeal SCC cells. PLoS One. 2011;6:e25402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Staudacher AH, Al-Ejeh F, Fraser CK, et al. The La antigen is over-expressed in lung cancer and is a selective dead cancer cell target for radioimmunotherapy using the La-specific antibody APOMAB(R). EJNMMI Res. 2014;4:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Nakatake M, Monte-Mor B, Debili N, et al. JAK2(V617F) negatively regulates p53 stabilization by enhancing MDM2 via La expression in myeloproliferative neoplasms. Oncogene. 2012;31:1323–1333. [DOI] [PubMed] [Google Scholar]
- [76].Bachmann M, Hilker M, Grolz D, et al. Different La/SS-B mRNA isoforms are expressed in salivary gland tissue of patients with primary Sjogren’s syndrome. J Autoimmun. 1996;9:757–766. [DOI] [PubMed] [Google Scholar]
- [77].Hilker M, Troster H, Grolz D, et al. The autoantigen La/SS-B: analysis of the expression of alternatively spliced La mRNA isoforms. Cell Tiss Res. 1996;284: 383–389. [DOI] [PubMed] [Google Scholar]
- [78].Carter MS, Sarnow P. Distinct mRNAs that encode La autoantigen are differentially expressed and contain internal ribosome entry sites. J Biol Chem. 2000;275:28301–28307. [DOI] [PubMed] [Google Scholar]
- [79].Huang M, Ida H, Arima K, et al. La autoantigen translocates to cytoplasm after cleavage during granzyme B-mediated cytotoxicity. Life Sci. 2007;81:1461–1466. [DOI] [PubMed] [Google Scholar]
- [80].Romero V, Fellows E, Jenne DE, et al. Cleavage of La protein by granzyme H induces cytoplasmic translocation and interferes with La-mediated HCV-IRES translational activity. Cell Death Differ. 2009;16:340–348. [DOI] [PubMed] [Google Scholar]
- [81].Heise T, Guidotti LG, Chisari FV. La autoantigen specifically recognizes a predicted stem-loop in hepatitis B virus RNA. J Virol. 1999;73:5767–5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Ayukawa K, Taniguchi S, Masumoto J, et al. La autoantigen is cleaved in the COOH terminus and loses the nuclear localization signal during apoptosis. J Biol Chem. 2000;275:34465–34470. [DOI] [PubMed] [Google Scholar]
- [83].Pavon-Eternod M, Gomes S, Geslain R, et al. tRNA over-expression in breast cancer and functional consequences. Nucleic Acids Res. 2009;37:7268–7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Mahlab S, Tuller T, Linial M. Conservation of the relative tRNA composition in healthy and cancerous tissues. RNA. 2012;18:640–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Pavon-Eternod M, Gomes S, Rosner MR, et al. Overexpression of initiator methionine tRNA leads to global reprogramming of tRNA expression and increased proliferation in human epithelial cells. RNA. 2013;19:461–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407–450. [DOI] [PubMed] [Google Scholar]
- [87].Wan Makhtar WR, Browne G, Karountzos A, et al. Short stretches of rare codons regulate translation of the transcription factor ZEB2 in cancer cells. Oncogene. 2017;36:6640–6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Laplante M, Sabatini DM. Regulation of mTORC1 and its impact on gene expression at a glance. J Cell Sci. 2013;126:1713–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Mattijssen S, Arimbasseri AG, Iben JR, et al. LARP4 mRNA codon-tRNA match contributes to LARP4 activity for ribosomal protein mRNA poly(A) tail length protection. eLife. 2017;12:6. pii: e28889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Sobala A, Hutvagner G. Transfer RNA-derived fragments: origins, processing, and functions. Wiley Interdiscip Rev RNA. 2011;2:853–862. [DOI] [PubMed] [Google Scholar]
- [91].Grafanaki K, Anastasakis D, Kyriakopoulos G, et al. Translation regulation in skin cancer from a tRNA point of view. Epigenomics. 2019;11:215–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Soares AR, Santos M. Discovery and function of transfer RNA-derived fragments and their role in disease. Wiley Interdiscip Rev RNA. 2017;8(5). [DOI] [PubMed] [Google Scholar]
- [93].Gaidamakov S, Maximova OA, Chon H, et al. Targeted deletion of the gene encoding the La autoantigen (Sjogren’s syndrome antigen B) in B cells or the frontal brain causes extensive tissue loss. Mol Cell Biol. 2014;34:123–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Blewett NH, Iben JR, Gaidamakov S, et al. La deletion from mouse brain alters pre-tRNA metabolism and accumulation of pre-5.8S rRNA, with neuron death and reactive astrocytosis. Mol Cell Biol. 2017;37(10):e00588-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Orioli A. tRNA biology in the omics era: stress signalling dynamics and cancer progression. Bioessays. 2017;39(3). [DOI] [PubMed] [Google Scholar]
- [96].Grewal SS. Why should cancer biologists care about tRNAs? tRNA synthesis, mRNA translation and the control of growth. Biochim Biophys Acta. 2015;1849:898–907. [DOI] [PubMed] [Google Scholar]
- [97].Francoeur AM, Chan EK, Garrels JI, et al. Characterization and purification of lupus antigen La, and RNA-binding protein. Mol Cell Biol. 1985;5:586–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Pfeifle J, Anderer FA, Franke M. Multiple phosphorylation of human SS-B/LA autoantigen and its effect on poly(U) and autoantibody binding. Biochim Biophys Acta. 1987;928:217–226. [DOI] [PubMed] [Google Scholar]
- [99].Bachmann M, Schroder HC, Wagner KG, et al. Purification and characterization of the Ro and La antigens. Modulation of their binding affinities to poly(U) by phosphorylation and the presence of ATP. Biol Chem Hoppe-Seyler. 1986;367:671–680. [DOI] [PubMed] [Google Scholar]
- [100].Broekhuis CH, Neubauer G, van Der Heijden A, et al. Detailed analysis of the phosphorylation of the human La (SS-B) autoantigen. (De)phosphorylation does not affect its subcellular distribution. Biochemistry. 2000;39:3023–3033. [DOI] [PubMed] [Google Scholar]
- [101].Fan H, Sakulich AL, Goodier JL, et al. Phosphorylation of the human La antigen on serine 366 can regulate recycling of RNA polymerase III transcription complexes. Cell. 1997;88:707–715. [DOI] [PubMed] [Google Scholar]
- [102].Fan H, Goodier JL, Chamberlain JR, et al. 5ʹ processing of tRNA precursors can be modulated by the human La antigen phosphoprotein. Mol Cell Biol. 1998;18:3201–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Intine RV, Sakulich AL, Koduru SB, et al. Control of transfer RNA maturation by phosphorylation of the human La antigen on serine 366. Mol Cell. 2000;6:339–348. [DOI] [PubMed] [Google Scholar]
- [104].Schwartz EI, Intine RV, Maraia RJ. CK2 is responsible for phosphorylation of human La protein serine-366 and can modulate rpL37 5ʹ-terminal oligopyrimidine mRNA metabolism. Mol Cell Biol. 2004;24:9580–9591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Filhol O, Giacosa S, Wallez Y, et al. Protein kinase CK2 in breast cancer: the CK2beta regulatory subunit takes center stage in epithelial plasticity. Cell Mol Life Sci. 2015;72:3305–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Tang H, Massi D, Hemmings BA, et al. AKT-ions with a TWIST between EMT and MET. Oncotarget. 2016;7:62767–62777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Ruzzene M, Pinna LA. Addiction to protein kinase CK2: a common denominator of diverse cancer cells? Biochim Biophys Acta. 2010;1804:499–504. [DOI] [PubMed] [Google Scholar]
- [108].Ruzzene M, Bertacchini J, Toker A, et al. Cross-talk between the CK2 and AKT signaling pathways in cancer. Adv Biol Regul. 2017;64:1–8. [DOI] [PubMed] [Google Scholar]
- [109].Vasudevan KM, Garraway LA. AKT signaling in physiology and disease. Curr Top Microbiol Immunol. 2010;347:105–133. [DOI] [PubMed] [Google Scholar]
- [110].Aoki M, Fujishita T. Oncogenic Roles of the PI3K/AKT/mTOR Axis. Curr Top Microbiol Immunol. 2017;407:153–189. [DOI] [PubMed] [Google Scholar]
- [111].van Niekerk EA, Willis DE, Chang JH, et al. Sumoylation in axons triggers retrograde transport of the RNA-binding protein La. Proc Natl Acad Sci U S A. 2007;104:12913–12918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Kota V, Sommer G, Durette C, et al. SUMO-modification of the La protein facilitates binding to mRNA in vitro and in cells. PLoS One. 2016;11:e0156365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Kota V, Sommer G, Hazard ES, et al. SUMO modification of the RNA-binding protein La regulates cell proliferation and STAT3 protein stability. Mol Cell Biol. 2018;38(2):e00129-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Seeler JS, Dejean A. SUMO and the robustness of cancer. Nat Rev Cancer. 2017;17:184–197. [DOI] [PubMed] [Google Scholar]
- [115].Eifler K, Vertegaal ACO. SUMOylation-mediated regulation of cell cycle progression and cancer. Trends Biochem Sci. 2015;40:779–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Watkins NJ, Lemm I, Ingelfinger D, et al. Assembly and maturation of the U3 snoRNP in the nucleoplasm in a large dynamic multiprotein complex. Mol Cell. 2004;16:789–798. [DOI] [PubMed] [Google Scholar]
- [117].Valenta T, Hausmann G, Basler K. The many faces and functions of beta-catenin. Embo J. 2012;31:2714–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Shang S, Hua F, Hu ZW. The regulation of beta-catenin activity and function in cancer: therapeutic opportunities. Oncotarget. 2017;8:33972–33989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Rosenthal EL, Matrisian LM. Matrix metalloproteases in head and neck cancer. Head Neck. 2006;28:639–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Holcik M, Korneluk RG. Functional characterization of the X-linked inhibitor of apoptosis (XIAP) internal ribosome entry site element: role of La autoantigen in XIAP translation. Mol Cell Biol. 2000;20:4648–4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Holcik M, Sonenberg N. TRANSLATIONAL CONTROL IN STRESS AND APOPTOSIS. Nat Rev Mol Cell Biol. 2005;6:318–327. [DOI] [PubMed] [Google Scholar]
- [122].Perrotti D, Calabretta B. Translational regulation by the p210 BCR/ABL oncoprotein. Oncogene. 2004;23:3222–3229. [DOI] [PubMed] [Google Scholar]
- [123].Fang G, Kim CN, Perkins CL, et al. CGP57148B (STI-571) induces differentiation and apoptosis and sensitizes Bcr-Abl-positive human leukemia cells to apoptosis due to antileukemic drugs. Blood. 2000;96:2246–2253. [PubMed] [Google Scholar]
- [124].Sherrill KW, Byrd MP, Van Eden ME, et al. BCL-2 translation is mediated via internal ribosome entry during cell stress. J Biol Chem. 2004;279:29066–29074. [DOI] [PubMed] [Google Scholar]
- [125].Kampen KR, Sulima SO, Verbelen B, et al. The ribosomal RPL10 R98S mutation drives IRES-dependent BCL-2 translation in T-ALL. Leukemia. 2019;33:319–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Hinnebusch AG, Ivanov IP, Sonenberg N. Translational control by 5ʹ-untranslated regions of eukaryotic mRNAs. Science. 2016;352:1413–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5:827–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Robichaud N, Sonenberg N. Translational control and the cancer cell response to stress. Curr Opin Cell Biol. 2017;45:102–109. [DOI] [PubMed] [Google Scholar]
- [129].Godet AC, David F, Hantelys F, et al. IRES trans-acting factors, key actors of the stress response. Int J Mol Sci. 2019;20(4):924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Kondrashov AV, Spriggs KA, Bushell M, et al. Co-ordinated regulation of translation following DNA damage. Cell Cycle. 2009;8:3067–3068. [DOI] [PubMed] [Google Scholar]
- [131].Spriggs KA, Bushell M, Willis AE. Translational regulation of gene expression during conditions of cell stress. Mol Cell. 2010;40:228–237. [DOI] [PubMed] [Google Scholar]
- [132].Spriggs KA, Stoneley M, Bushell M, et al. Re-programming of translation following cell stress allows IRES-mediated translation to predominate. Biol Cell. 2008;100:27–38. [DOI] [PubMed] [Google Scholar]
- [133].Meerovitch K, Pelletier J, Sonenberg N. A cellular protein that binds to the 5ʹ-noncoding region of poliovirus RNA: implications for internal translation initiation. Genes Dev. 1989;3:1026–1034. [DOI] [PubMed] [Google Scholar]
- [134].Pudi R, Srinivasan P, Das S. La protein binding at the GCAC site near the initiator AUG facilitates the ribosomal assembly on the hepatitis C virus RNA to influence internal ribosome entry site-mediated translation. J Biol Chem. 2004;279:29879–29888. [DOI] [PubMed] [Google Scholar]
- [135].Svitkin YV, Ovchinnikov LP, Dreyfuss G, et al. General RNA binding proteins render translation cap dependent [published erratum appears in EMBO J 1997 Feb 17;16(4):896]. Embo J. 1996;15:7147–7155. [PMC free article] [PubMed] [Google Scholar]
- [136].Koromilas AE. Roles of the translation initiation factor eIF2alpha serine 51 phosphorylation in cancer formation and treatment. Biochim Biophys Acta. 2015;1849:871–880. [DOI] [PubMed] [Google Scholar]
- [137].Donnelly N, Gorman AM, Gupta S, et al. The eIF2alpha kinases: their structures and functions. Cell Mol Life Sci. 2013;70:3493–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Wek RC. Role of eIF2alpha kinases in translational control and adaptation to cellular stress. Cold Spring Harbor Perspect Biol. 2018;10(7):a032870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Garcia MA, Gil J, Ventoso I, et al. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol Mol Biol Rev. 2006;70:1032–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Sadler AJ, Williams BR. Structure and function of the protein kinase R. Curr Top Microbiol Immunol. 2007;316:253–292. [DOI] [PubMed] [Google Scholar]
- [141].Clemens MJ, Elia A. The double-stranded RNA-dependent protein kinase PKR: structure and function. J Interferon Cytokine Res. 1997;17:503–524. [DOI] [PubMed] [Google Scholar]
- [142].Boye E, Grallert B. eIF2alpha phosphorylation and the regulation of translation. Curr Genet. 2019;66(2):293–297. [DOI] [PubMed] [Google Scholar]
- [143].James MC, Jeffrey IW, Pruijn GJ, et al. Translational control by the La antigen. Structure requirements for rescue of the double-stranded RNA-mediated inhibition of protein synthesis. Eur J Biochem. 1999;266:151–162. [DOI] [PubMed] [Google Scholar]
- [144].Youssef OA, Safran SA, Nakamura T, et al. Potential role for snoRNAs in PKR activation during metabolic stress. Proc Natl Acad Sci U S A. 2015;112:5023–5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Lee K, Kunkeaw N, Jeon SH, et al. Precursor miR-886, a novel noncoding RNA repressed in cancer, associates with PKR and modulates its activity. RNA. 2011;17:1076–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Lee YS, Kunkeaw N, Lee YS. Protein kinase R and its cellular regulators in cancer: an active player or a surveillant? Wiley Interdiscip Rev RNA. 2020;11:e1558. [DOI] [PubMed] [Google Scholar]
- [147].Chu WM, Ballard R, Carpick BW, et al. Potential Alu function: regulation of the activity of double-stranded RNA-activated kinase PKR. Mol Cell Biol. 1998;18:58–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Kim Y, Lee JH, Park JE, et al. PKR is activated by cellular dsRNAs during mitosis and acts as a mitotic regulator. Genes Dev. 2014;28:1310–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Elbarbary RA, Li W, Tian B, et al. STAU1 binding 3ʹ UTR IRAlus complements nuclear retention to protect cells from PKR-mediated translational shutdown. Genes Dev. 2013;27:1495–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Kufel J, Allmang C, Chanfreau G, et al. Precursors to the U3 small nucleolar RNA lack small nucleolar RNP proteins but are stabilized by La binding. Mol Cell Biol. 2000;20:5415–5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Maraia RJ, Kenan DJ, Keene JD. Eukaryotic transcription termination factor La mediates transcript release and facilitates reinitiation by RNA polymerase III. Mol Cell Biol. 1994;14:2147–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Goodier JL, Fan H, Maraia RJ. A carboxy-terminal basic region controls RNA polymerase III transcription factor activity of human La protein. Mol Cell Biol. 1997;17:5823–5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Marchal JA, Lopez GJ, Peran M, et al. The impact of PKR activation: from neurodegeneration to cancer. Faseb J. 2014;28:1965–1974. [DOI] [PubMed] [Google Scholar]
- [154].Kim SH, Gunnery S, Choe JK, et al. Neoplastic progression in melanoma and colon cancer is associated with increased expression and activity of the interferon-inducible protein kinase, PKR. Oncogene. 2002;21:8741–8748. [DOI] [PubMed] [Google Scholar]
- [155].Peek R, Pruijn GJ, Van Venrooij WJ. Interaction of the La (SS-B) autoantigen with small ribosomal subunits. Eur J Biochem. 1996;236:649–655. [DOI] [PubMed] [Google Scholar]
- [156].Cardinali B, Carissimi C, Gravina P, et al. La protein is associated with terminal oligopyrimidine mRNAs in actively translating polysomes. J Biol Chem. 2003;278:35145–35151. [DOI] [PubMed] [Google Scholar]
- [157].Vinayak J, Marrella SA, Hussain RH, et al. Human La binds mRNAs through contacts to the poly(A) tail. Nucleic Acids Res. 2018;46:4228–4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Lu WT, Wilczynska A, Smith E, et al. The diverse roles of the eIF4A family: you are the company you keep. Biochem Soc Trans. 2014;42:166–172. [DOI] [PubMed] [Google Scholar]
- [159].Chu J, Pelletier J. Targeting the eIF4A RNA helicase as an anti-neoplastic approach. Biochim Biophys Acta. 2015;1849:781–791. [DOI] [PubMed] [Google Scholar]
- [160].Pisareva VP, Pisarev AV, Komar AA, et al. Translation initiation on mammalian mRNAs with structured 5ʹUTRs requires DExH-box protein DHX29. Cell. 2008;135:1237–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Pisareva VP, Pisarev AV. DHX29 and eIF3 cooperate in ribosomal scanning on structured mRNAs during translation initiation. RNA. 2016;22:1859–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Dhote V, Sweeney TR, Kim N, et al. Roles of individual domains in the function of DHX29, an essential factor required for translation of structured mammalian mRNAs. Proc Natl Acad Sci U S A. 2012;109:E3150–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984;12:857–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. [DOI] [PubMed] [Google Scholar]
- [165].McBratney S, Sarnow P. Evidence for involvement of trans-acting factors in selection of the AUG start codon during eukaryotic translational initiation. Mol Cell Biol. 1996;16:3523–3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Meyuhas O, Kahan T. The race to decipher the top secrets of TOP mRNAs. Biochim Biophys Acta. 2015;1849:801–811. [DOI] [PubMed] [Google Scholar]
- [167].Mandato E, Manni S, Zaffino F, et al. Targeting CK2-driven non-oncogene addiction in B-cell tumors. Oncogene. 2016;35:6045–6052. [DOI] [PubMed] [Google Scholar]
- [168].Trembley JH, Wang G, Unger G, et al. Protein kinase CK2 in health and disease: CK2: a key player in cancer biology. Cell Mol Life Sci. 2009;66:1858–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Terenin IM, Smirnova VV, Andreev DE, et al. A researcher’s guide to the galaxy of IRESs. Cell Mol Life Sci. 2017;74:1431–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Gilbert WV. Alternative ways to think about cellular internal ribosome entry. J Biol Chem. 2010;285:29033–29038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Jackson RJ. The current status of vertebrate cellular mRNA IRESs. Cold Spring Harbor Perspect Biol. 2013;5(2):a011569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Fabregat I, Caballero-Diaz D. Transforming growth factor-beta-induced cell plasticity in liver fibrosis and hepatocarcinogenesis. Front Oncol. 2018;8:357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Mitra A, Mishra L, Li S. EMT, CTCs and CSCs in tumor relapse and drug-resistance. Oncotarget. 2015;6:10697–10711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Papageorgis P. TGFbeta signaling in tumor initiation, epithelial-to-mesenchymal transition, and metastasis. J Oncol. 2015;2015:587193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Saitoh M. Epithelial-mesenchymal transition is regulated at post-transcriptional levels by transforming growth factor-beta signaling during tumor progression. Cancer Sci. 2015;106:481–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Evdokimova V, Tognon CE, Sorensen PH. On translational regulation and EMT. Semin Cancer Biol. 2012;22:437–445. [DOI] [PubMed] [Google Scholar]
- [178].Evdokimova V, Tognon C, Ng T, et al. Translational activation of snail1 and other developmentally regulated transcription factors by YB-1 promotes an epithelial-mesenchymal transition. Cancer Cell. 2009;15:402–415. [DOI] [PubMed] [Google Scholar]
- [179].Chaudhury A, Hussey GS, Ray PS, et al. TGF-beta-mediated phosphorylation of hnRNP E1 induces EMT via transcript-selective translational induction of Dab2 and ILEI. Nat Cell Biol. 2010;12:286–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Hussey GS, Chaudhury A, Dawson AE, et al. Identification of an mRNP complex regulating tumorigenesis at the translational elongation step. Mol Cell. 2011;41:419–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Lim SO, Park SJ, Kim W, et al. Proteome analysis of hepatocellular carcinoma. Biochem Biophys Res Commun. 2002;291:1031–1037. [DOI] [PubMed] [Google Scholar]
- [182].Givant-Horwitz V, Davidson B, Reich R. Laminin-induced signaling in tumor cells: the role of the M(r) 67,000 laminin receptor. Cancer Res. 2004;64:3572–3579. [DOI] [PubMed] [Google Scholar]
- [183].Patarroyo M, Tryggvason K, Virtanen I. Laminin isoforms in tumor invasion, angiogenesis and metastasis. Semin Cancer Biol. 2002;12:197–207. [DOI] [PubMed] [Google Scholar]
- [184].Petz M, Kozina D, Huber H, et al. The leader region of Laminin B1 mRNA confers cap-independent translation. Nucleic Acids Res. 2007;35:2473–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Petz M, Them N, Huber H, et al. La enhances IRES-mediated translation of laminin B1 during malignant epithelial to mesenchymal transition. Nucleic Acids Res. 2012;40:290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Gotzmann J, Fischer AN, Zojer M, et al. A crucial function of PDGF in TGF-beta-mediated cancer progression of hepatocytes. Oncogene. 2006;25:3170–3185. [DOI] [PubMed] [Google Scholar]
- [187].Govaere O, Petz M, Wouters J, et al. The PDGFRalpha-laminin B1-keratin 19 cascade drives tumor progression at the invasive front of human hepatocellular carcinoma. Oncogene. 2017;36:6605–6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Kim JK, Diehl JA. Nuclear cyclin D1: an oncogenic driver in human cancer. J Cell Physiol. 2009;220:292–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Arnold A, Motokura T, Bloom T, et al. PRAD1 (cyclin D1): a parathyroid neoplasia gene on 11q13. Henry Ford Hosp Med J. 1992;40:177–180. [PubMed] [Google Scholar]
- [190].Diehl JA. Cycling to cancer with cyclin D1. Cancer Biol Ther. 2002;1:226–231. [DOI] [PubMed] [Google Scholar]
- [191].Ewen ME, Lamb J. The activities of cyclin D1 that drive tumorigenesis. Trends Mol Med. 2004;10:158–162. [DOI] [PubMed] [Google Scholar]
- [192].Fu M, Wang C, Li Z, et al. Minireview: cyclin D1: normal and abnormal functions. Endocrinology. 2004;145:5439–5447. [DOI] [PubMed] [Google Scholar]
- [193].Shi Y, Sharma A, Wu H, et al. Cyclin D1 and c-Myc internal ribosome entry site (IRES)-dependent translation is regulated by AKT activity and enhanced by rapamycin through a p38 MAPK and ERK-dependent pathway. J Biol Chem. 2005;280(12):10964-73. [DOI] [PubMed] [Google Scholar]
- [194].Frost P, Shi Y, Hoang B, et al. Regulation of D-cyclin translation inhibition in myeloma cells treated with mammalian target of rapamycin inhibitors: rationale for combined treatment with extracellular signal-regulated kinase inhibitors and rapamycin. Mol Cancer Ther. 2009;8:83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Gera JF, Mellinghoff IK, Shi Y, et al. AKT activity determines sensitivity to mammalian target of rapamycin (mTOR) inhibitors by regulating cyclin D1 and c-myc expression. J Biol Chem. 2004;279:2737–2746. [DOI] [PubMed] [Google Scholar]
- [196].Jo OD, Martin J, Bernath A, et al. Heterogeneous nuclear ribonucleoprotein A1 regulates cyclin D1 and c-myc internal ribosome entry site function through Akt signaling. J Biol Chem. 2008;283:23274–23287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Silke J, Vucic D. IAP family of cell death and signaling regulators. Methods Enzymol. 2014;545:35–65. [DOI] [PubMed] [Google Scholar]
- [198].Dubrez-Daloz L, Dupoux A, Cartier J. IAPs: more than just inhibitors of apoptosis proteins. Cell Cycle. 2008;7:1036–1046. [DOI] [PubMed] [Google Scholar]
- [199].Holcik M, Lefebvre C, Yeh C, et al. A new internal-ribosome-entry-site motif potentiates XIAP-mediated cytoprotection. Nat Cell Biol. 1999;1:190–192. [DOI] [PubMed] [Google Scholar]
- [200].Van Eden ME, Byrd MP, Sherrill KW, et al. Translation of cellular inhibitor of apoptosis protein 1 (c-IAP1) mRNA is IRES mediated and regulated during cell stress. RNA. 2004;10:469–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Baranick BT, Lemp NA, Nagashima J, et al. Splicing mediates the activity of four putative cellular internal ribosome entry sites. Proc Natl Acad Sci U S A. 2008;105:4733–4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Riley A, Jordan LE, Holcik M. Distinct 5ʹ UTRs regulate XIAP expression under normal growth conditions and during cellular stress. Nucleic Acids Res. 2010;38:4665–4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Aird KM, Ghanayem RB, Peplinski S, et al. X-linked inhibitor of apoptosis protein inhibits apoptosis in inflammatory breast cancer cells with acquired resistance to an ErbB1/2 tyrosine kinase inhibitor. Mol Cancer Ther. 2010;9:1432–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Krepela E, Dankova P, Moravcikova E, et al. Increased expression of inhibitor of apoptosis proteins, survivin and XIAP, in non-small cell lung carcinoma. Int J Onc. 2009;35:1449–1462. [DOI] [PubMed] [Google Scholar]
- [205].Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Macejak DG, Sarnow P. Internal initiation of translation mediated by the 5ʹ leader of a cellular mRNA. Nature. 1991;353:90–94. [DOI] [PubMed] [Google Scholar]
- [207].Roller C, Maddalo D. The molecular chaperone GRP78/BiP in the development of chemoresistance: mechanism and possible treatment. Front Pharmacol. 2013;4:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Casas C. GRP78 at the centre of the stage in cancer and neuroprotection. Front Neurosci. 2017;11:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Kim YK, Back SH, Rho J, et al. La autoantigen enhances translation of BiP mRNA. Nucleic Acids Res. 2001;29:5009–5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Le SY, Maizel JV Jr.. A common RNA structural motif involved in the internal initiation of translation of cellular mRNAs. Nucleic Acids Res. 1997;25:362–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Yoon A, Peng G, Brandenburger Y, et al. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science. 2006;312:902–906. [DOI] [PubMed] [Google Scholar]
- [212].Coldwell MJ, deSchoolmeester ML, Fraser GA, et al. The p36 isoform of BAG-1 is translated by internal ribosome entry following heat shock. Oncogene. 2001;20:4095–4100. [DOI] [PubMed] [Google Scholar]
- [213].Cobbold LC, Spriggs KA, Haines SJ, et al. Identification of internal ribosome entry segment (IRES)-trans-acting factors for the Myc family of IRESs. Mol Cell Biol. 2008;28:40–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Badura M, Braunstein S, Zavadil J, et al. DNA damage and eIF4G1 in breast cancer cells reprogram translation for survival and DNA repair mRNAs. Proc Natl Acad Sci U S A. 2012;109:18767–18772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–543. [DOI] [PubMed] [Google Scholar]
- [216].Bisogno LS, Keene JD. RNA regulons in cancer and inflammation. Curr Opin Genet Dev. 2018;48:97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Bartel F, Harris LC, Wurl P, et al. MDM2 and its splice variant messenger RNAs: expression in tumors and down-regulation using antisense oligonucleotides. Mol Cancer Res. 2004;2(1):29–35. [PubMed] [Google Scholar]
- [218].Brooks CL, Gu W. p53 ubiquitination: mdm2 and beyond. Mol Cell. 2006;21:307–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Landers JE, Cassel SL, George DL. Translational enhancement of mdm2 oncogene expression in human tumor cells containing a stabilized wild-type p53 protein. Cancer Res. 1997;57:3562–3568. [PubMed] [Google Scholar]
- [220].Landers JE, Haines DS, Strauss JF 3rd, et al. Enhanced translation: a novel mechanism of mdm2 oncogene overexpression identified in human tumor cells. Oncogene. 1994;9(9):2745–2750. [PubMed] [Google Scholar]
- [221].Capoulade C, Bressac-de Paillerets B, Lefrere I, et al. Overexpression of MDM2, due to enhanced translation, results in inactivation of wild-type p53 in Burkitt’s lymphoma cells. Oncogene. 1998;16:1603–1610. [DOI] [PubMed] [Google Scholar]
- [222].Brown CY, Mize GJ, Pineda M, et al. Role of two upstream open reading frames in the translational control of oncogene mdm2. Oncogene. 1999;18:5631–5637. [DOI] [PubMed] [Google Scholar]
- [223].Jin X, Turcott E, Englehardt S, et al. The two upstream open reading frames of oncogene mdm2 have different translational regulatory properties. J Biol Chem. 2003;278:25716–25721. [DOI] [PubMed] [Google Scholar]
- [224].Akulich KA, Sinitcyn PG, Makeeva DS, et al. A novel uORF-based regulatory mechanism controls translation of the human MDM2 and eIF2D mRNAs during stress. Biochimie. 2019;157:92–101. [DOI] [PubMed] [Google Scholar]
- [225].D’Agostino VG, Sighel D, Zucal C, et al. Screening approaches for targeting ribonucleoprotein complexes: a new dimension for drug discovery. SLAS Discov. 2019;24:314–331. [DOI] [PubMed] [Google Scholar]
- [226].Roos M, Pradere U, Ngondo RP, et al. A small-molecule inhibitor of Lin28. ACS Chem Biol. 2016;11:2773–2781. [DOI] [PubMed] [Google Scholar]
- [227].Muralidharan R, Mehta M, Ahmed R, et al. HuR-targeted small molecule inhibitor exhibits cytotoxicity towards human lung cancer cells. Sci Rep. 2017;7:9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [228].Wang Z, Bhattacharya A, Ivanov DN. Identification of small-molecule inhibitors of the HuR/RNA interaction using a fluorescence polarization screening assay followed by NMR validation. PLoS One. 2015;10:e0138780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229].Heise T, Guidotti LG, Cavanaugh VJ, et al. Hepatitis B virus RNA-binding proteins associated with cytokine-induced clearance of viral RNA from the liver of transgenic mice. J Virol. 1999;73:474–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [230].Heise T, Guidotti LG, Chisari FV. Characterization of nuclear RNases that cleave hepatitis B virus RNA near the La protein binding site. J Virol. 2001;75:6874–6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [231].Tang J, Zhang ZH, Huang M, et al. Phosphorylation of human La protein at Ser 366 by casein kinase II contributes to hepatitis B virus replication and expression in vitro. J Viral Hepatitis. 2013;20:24–33. [DOI] [PubMed] [Google Scholar]
- [232].Tang J, Huang ZM, Chen YY, et al. A novel inhibitor of human La protein with anti-HBV activity discovered by structure-based virtual screening and in vitro evaluation. PLoS One. 2012;7:e36363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [233].Izumi RE, Das S, Barat B, et al. A peptide from autoantigen La blocks poliovirus and hepatitis C virus cap-independent translation and reveals a single tyrosine critical for La RNA binding and translation stimulation. J Virol. 2004;78:3763–3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [234].Sommer G, Fedarovich A, Kota V, et al. Applying a high-throughput fluorescence polarization assay for the discovery of chemical probes blocking La: RNA interactions in vitro and in cells. PLoS One. 2017;12:e0173246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [235].Troster H, Metzger TE, Semsei I, et al. One gene, two transcripts: isolation of an alternative transcript encoding for the autoantigen La/SS-B from a cDNA library of a patient with primary Sjogrens’ syndrome. J Exp Med. 1994;180:2059–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [236].Padilla PI, Uhart M, Pacheco-Rodriguez G, et al. Association of guanine nucleotide-exchange protein BIG1 in HepG2 cell nuclei with nucleolin, U3 snoRNA, and fibrillarin. Proc Natl Acad Sci U S A. 2008;105:3357–3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [237].Fouraux MA, Bouvet P, Verkaart S, et al. Nucleolin associates with a subset of the human Ro ribonucleoprotein complexes. J Mol Biol. 2002;320:475–488. [DOI] [PubMed] [Google Scholar]
- [238].Fouraux MA, Kolkman MJ, Van der Heijden A, et al. The human La (SS-B) autoantigen interacts with DDX15/hPrp43, a putative DEAH-box RNA helicase. RNA. 2002;8:1428–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]


