Abstract
Abstract
Senecavirus A (SVA) is an emerging picornavirus that has been associated with vesicular disease and neonatal mortality in swine. The construction of SVA virus carrying foreign reporter gene provides a powerful tool in virus research. However, it is often fraught with rescuing a recombinant picornavirus harboring a foreign gene or maintaining the stability of foreign gene in the virus genome. Here, we successfully generated recombinant SVA GD05/2017 viruses (V-GD05-clone) expressing the green fluorescent protein (iLOV), red fluorescent protein (RFP), or NanoLuc luciferase (Nluc). These recombinant viruses have comparable growth kinetics to the parental virus. Genetic stability analysis indicated that V-GD05-iLOV was highly stable in retaining iLOV gene for more than 10 passages, while V-GD05-RFP and V-GD05-Nluc lost the foreign genes in five passages. In addition, high-intensity fluorescent signals were found in the V-GD05-RFP- and V-GD05-iLOV-infected cells by fluorescence observation and flow cytometry analysis, and the luciferase activity assay could quantitatively monitor the replication of V-GD05-Nluc. In order to identify the porcine cell receptor for SVA, anthrax toxin receptor 1 (ANTXR1) was knocked out or overexpressed in the ST-R cells. The ANTXR1 knock-out cells lost the ability for SVA infection, while overexpression of ANTXR1 significantly increased the cell permissivity. These results confirmed that ANTXR1 was the receptor for SVA to invade porcine cells as reported in the human cells. Overall, this study suggests that these SVA reporter viruses will be useful tools in elucidating virus pathogenesis and developing control measures.
Key points
• We successfully generated SVA viruses expressing the iLOV, RFP, or Nluc.
• The iLOV was genetically stable in the V-GD05-iLOV genome over ten passages.
• ANTXR1 was the receptor for SVA to invade porcine cells.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00253-021-11181-6.
Keywords: Senecavirus A, Reverse genetics, Reporter virus, Porcine ANTXR1
Introduction
Senecavirus A (SVA) is the only species in the genus Senecavirus of family Picornaviridae (Adams et al. 2015; Chen et al. 2019). Like all picornaviruses, SVA is a non-enveloped viruses approximately in size 25–30 nm with icosahedral symmetry (Adams et al. 2015; Venkataraman et al. 2008a; Wang et al. 2020a, 2020b). SVA has a single-stranded, positive-sense RNA genome with approximately 7300 nucleotides (nt) in length, which comprises 5’ and 3’ untranslated region (UTR) and encodes a single open reading frame (ORF) that is subsequently processed to 12 polypeptides in the standard picornavirus L-4-3-4 layout: leader protein (Lpro); the four structural proteins VP4 (1A), VP2 (1B), VP3 (1C), and VP1 (1D); the remaining seven non-structural proteins (nsp) 2A, 2B, 2C, 3A, 3B, 3Cpro, and 3Dpol (Chen et al. 2016; Hales et al. 2008). During translation of the SVA polyprotein, primary cleavage is mediated by the 3C protease, except that ribosomal “skipping” event happens at the C terminus of 2A, which results in separation of capsid proteins (P1) region from the non-structural protein coding region (Donnelly et al. 2001; Liu et al. 2019; Venkataraman et al. 2008b).
SVA was first discovered from a PER.C6 cell culture in the USA in 2002 (Venkataraman et al. 2008b). Since then, the virus has been developed as an oncolytic agent due to its selective tropism for human tumor cells. Thereafter, SVA has been associated with sporadic cases of vesicular disease in pigs in the USA and Canada (Corner 2012; Pasma et al. 2008). After 2014–2015, outbreaks of SVA-associated vesicular disease (SAVD) have been reported in a number of swine-producing countries around the world (Canning et al. 2016; Guo et al. 2016; Hause et al. 2016; Leme et al. 2016; Montiel et al. 2016; Saporiti et al. 2017; Vannucci et al. 2015; Wu et al. 2016). So far, many aspects of knowledge such as SVA transmission and pathogenesis remain highly lacking. Reverse genetics is a powerful tool to address the issues with developing recombinant reporter virus. Formerly, full-length cDNA infectious clones of SVV-001 (Poirier et al. 2012) and KS15-01 (Chen et al. 2016) harboring the coding sequence of green fluorescent protein (GFP) were constructed, respectively. Unfortunately, foreign genes were unstable in the virus genome through a few passages. Moreover, GFP-tagged SVA was not well suitable for accurate quantification in monitoring the virus replication. Therefore, we initiated the development of novel SVA reporter viruses to deeply characterize the virus life cycle. In the study, we aimed to construct recombinant SVA carrying green fluorescent protein (iLOV), red fluorescent protein (RFP), or Nanoluciferase (Nluc) genes inserted at the junction site between 2A and 2B. To retain the native coding sequences of virus proteins surrounding the foreign gene, we inserted the porcine teschovirus 2A-like (T2A), a stop-restart translational element, at the side of the 2B product within the SVA polyprotein. This study will provide insight into the stability of foreign genes of different sizes in the SVA genome by characterizing these novel reporter viruses and exploit one of their potentials in identifying the cell receptor for SVA infection of porcine cells.
Materials and methods
Cells and viruses
BHK-21, HEK293T cells, ST-R cells (IFN-α/β receptor-knockout ST cell line), and ST-R ANTXR1 KO cells (ANTXR1-knockout ST-R cell line) were cultured in the DMEM (Gibco, Carlsbad, CA) supplemented with 10% fetal bovine serum (Sigma-Aldrich, St. Louis, MO) at 37 °C and 5% CO2. The wild-type SVA GD05/2017 (GenBank MH316116) strain was isolated and kept at our laboratory (Wang et al. 2019).
Antibodies and reagents
The SVA-VP3 and -3C mAbs were made by our laboratory. The Myc and GAPDH mAbs were purchased from Sigma (Sigma-Aldrich, St. Louis, MO). The Dylight 488-labeled goat anti-mouse IgG, Dylight 549-labeled goat anti-rabbit IgG, HRP-labeled goat Anti-mouse IgG, and HRP-labeled goat anti-rabbit IgG were purchased from Abbkine (Wuhan, China). Lipofectamine® 3000 was purchased from Invitrogen (Shanghai, China). ANTXR1 mAb was purchased from Bioss (Beijing, China).
Construction of SVA full-length cDNA clones
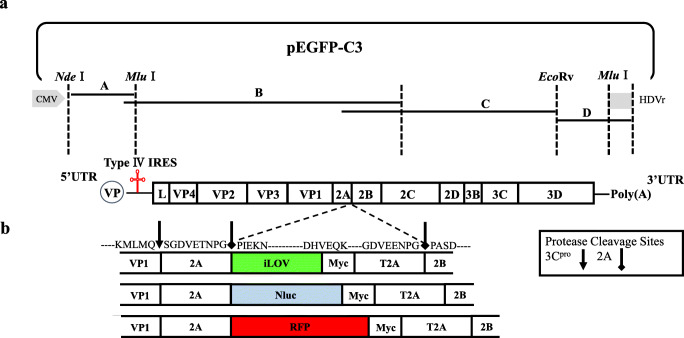
The full-length SVA cDNA clones were constructed as described previously (Wang et al. 2020a, 2020b). Four separate fragments (named A to D) were amplified from the SVA GD05/2017 cDNA and assembled into pEGFP-C3 as shown in the Fig. 1a. The resulting full-length cDNA clone was designated as pC3-SVA-GD05. To construct recombinant SVA expressing reporter proteins, the iLOV, RFP, and Nluc genes were amplified from the pUC57-Nluc, pTag-RFP, and pUC57-iLOV. The foreign genes fused with a T2A at its C terminus were inserted between the 2A and 2B of pC3-SVA-GD05.These recombinant plasmids were sequenced and designated as pC3-SVA-GD05-Nluc-T2A, pC3-SVA-GD05-iLOV-T2A, and pC3-SVA-GD05-RFP-T2A (Fig. 1b). Primers were designed based on the conserved genomic regions of multiple SVA strains (Tables S1 and S2).
Fig. 1.
The schematic diagram for the construction of SVA. a Schematic diagram of the full-length SVA GD05/2017 genome and construction of the full-length cDNA clone. b Schematic presentation of the insertion of iLOV, Nluc, and RFP into GD05/2017 genome. Cleavage of polypeptide by 3C was indicated by arrowhead, while ribosome skipping was indicated by diamond head
Recovery of recombinant viruses
BHK-21 cells were seeded in the 6-well plates and transfected with pC3-SVA-GD05 and its recombinants. Transfection was conducted using Lipofectamine® 3000 following the manufacturer’s instructions. At 72-h post-transfection, cell culture supernatants from the BHK-21 cells were transferred to the ST-R cells. Cytopathic effect (CPE) was monitored daily after infection. The recombinant virus recovered from pC3-SVA-GD05 was designated as V-GD05-clone. The same method was used to rescue the iLOV, RFP, and Nluc-reporter viruses (V-GD05-iLOV, V-GD05-RFP, and V-GD05-Nluc). The viral protein expression in the recombinant virus-infected cells was detected by fluorescence observation, western blot, luciferase activity assay, and flow cytometry.
Genetic stability of reporter viruses in the ST-R cells
Reporter viruses had been serially cultured from the first passage (P1) to the tenth passage (P10) in the ST-R cells. The viral RNAs of P3, P5, and P10 virus stocks were extracted and reverse transcribed into cDNA. The foreign gene regions were examined by PCR and sequence analysis.
The growth curve of recombinant viruses
ST-R cells were infected with P3 recombinant viruses at an MOI of 0.1. The culture supernatants were collected at 6, 12, 24, 36, 48, and 60 hpi, respectively. Virus titer was determined by CPE observation, and quantified as 50% tissue culture infective dose (TCID50) mL-1 according to the Reed-Muench method (Reed and Muench 1938).
Immunofluorescence assay
The ST-R cell monolayers were fixed with 4% paraformaldehyde in PBS (pH 7.4) for 10 min and then permeabilized with 0.1% TritonX-100 and 2% BSA in PBS for 30 min at room temperature. After 1-h incubation with primary antibodies, cell monolayers were washed with PBS for three times and further incubated for 1 h with Dylight 488 anti-rabbit IgG. The cell nuclei were stained with 4, 6-diamidino-2-phenylindole-dihydrochloride (DAPI) performed as suggested by the manufacturer (Molecular Probes). After three times washing by PBS, cells were analyzed under a fluorescent microscope, and pictures were taken with Leica Cell Imaging System (Leica, Germany).
Western blot analysis
The ST-R cells were treated with lysis buffer (Beyotime Biotech, Shanghai, China). The cell lysate supernatant was mixed with 5×loading buffer containing 5% β-mercaptoethanol and denatured at 100 °C for 10 min. Proteins were separated on a 12% SDS-PAGE gel and blotted onto nitrocellulose membrane. The membranes were blocked with 5% skim milk in the Tris-buffered saline (TBS) containing Tween 20 at room temperature for 2 h. The membranes were incubated with primary antibodies at 4 °C overnight. After three times washing with TBST, the membranes were incubated by HRP-conjugated secondary antibodies. At last, the target proteins were visualized using the chemiluminescence imaging system (Tanon, Shanghai, China).
Flow cytometry analysis
The SVA-infected ST-R cells were harvested by treatment of 0.25% trypsin and then washed twice with PBS at 1000 rpm for 5 min, following by suspending cells in the PBS. The specific fluorescence of iLOV and RFP was measured upon excitation at 488 and 561 nm, respectively. iLOV fluorescence was analyzed through FL1 channel and RFP fluorescence was analyzed through FL2 channel. Data from 10,000 events were recorded, and the data was analyzed using CytExpert Software 2.3 (Beckman Coulter, USA).
Luciferase activity assay
ST-R cells in the 6-well plates were infected with V-GD05-Nluc at an MOI of 0.1. At 12 h, 18 h, and 24 h post infection, Nano-Glo® Luciferase Assay System (Promega, USA) was used to detect the Nluc activity according to the manufacturer’s instructions.
Construction and analysis of ST-R ANTXR1 KO cell line
The sgRNA was designed based on the gene of porcine ANTXR1 through http://crispor.tefor.net/. The sequences of sgRNAs targeting the first exon of ANTXR1 were #1F: 5’-CACCGCTCATCTGCGCCGGGCAAG-3’; #1R: 5’-AAACCTTGCCCGGCGCAGATGAGC-3’; #2F: 5’-CACCGCAGGTCAAATCCCCCGTAGC-3’; #2R: 5’-AAACGCTACGGGGGATTTGACCTGC-3’. To generate lentivirus expressing sgRNA, HEK293T cells were co-transfected with Lenti-ANTXR1-sgRNA, packaging vector psPAX2, and envelop vector pMD2.G using Lipofectamine® 3000. Culture supernatants containing lentiviruses were harvested at 24 h and 48 h post-transfection and then passed through a 0.45 μM-pore-size filter. ST-R cells were infected by filtered lentiviruses expressing ANTXR1-sgRNAs to develop ST-R ANTXR1 KO stable cell line. The knockout of ANTXR1 gene was identified by DNA sequencing and western blot.
Overexpression of ANTXR1 in the ST-R cells
Porcine ANTXR1 gene was amplified from the cDNA of ST-R cells and inserted into pCAGGS-Myc vector. ST-R cells were seeded in the 6-well plates and transfected with pCAGGS-Myc-pig-ANTXR1 (2.5 μg/well). At 24 h, 36 h, and 48 h post-transfection, the cells were infected with V-GD05-RFP, V-GD05-iLOV, and V-GD05-Nluc at an MOI of 0.1 for 24 h. The expression of porcine ANTXR1 was detected by western blot.
Identification of ANTXR1 as cell receptor using SVA reporter viruses
ST-R ANTXR1 KO stable cell lines and porcine ANTXR1-overexpressed ST-R cells were used to identify the cell receptor for SVA infection. The ST-R ANTXR1 KO cells were infected with V-GD05-iLOV, V-GD05-RFP, or V-GD05-Nluc (MOI=0.1). At 36 hpi, the fluorescence in the V-GD05-iLOV and V-GD05-RFP infected cells was observed by fluorescence microscope. The luciferase activity of V-GD05-Nluc infected cells was measured at 18 hpi by Nano-Glo® Luciferase Assay. Additionally, after 24, 36, and 48 h post-transfection, porcine ANTXR1 overexpressed ST-R cells were infected with V-GD05-RFP, V-GD05-iLOV, or V-GD05-Nluc (MOI=0.1). At 24 hpi, fluorescence and luciferase activities of the infected cells were detected.
Statistical analysis
Statistical analyses were performed by using one-way analysis of variance (ANOVA) using GraphPad InStat Prism software (version5.0). Statistical significance was expressed as a P value less than 0.05 or 0.01.
Results
Construction of SVA infectious clone
As shown in Fig. 1a, a panel of fragments that span the entire SVA GD05/2017 genome was assembled into pEGFP-C3 to yield pC3-SVA-GD05. In the virus infectious clone, a CMV promoter was placed at the 5’ terminus of the virus genome, while a polyA tail of 22 residues was inserted at the 3’ end. To construct the reporter-tagged SVA viruses, the junction site between the 2A and 2B was selected for insertion of foreign gene. The iLOV, RFP, and Nluc fused with a T2A peptide were inserted into pEGFP-C3-GD05 (Fig. 1b), and the resulting plasmids were named as pC3-SVA-GD05-iLOV-T2A, pC3-SVA-GD05-RFP-T2A, and pC3-SVA-GD05-Nluc-T2A. During the translation of virus polypeptide, the ribosome skips at TNPG↓P of the SVA 2A sequence and continues in frame to produce a reporter-T2A fusion protein and then skips a second time at the T2A SNPG↓P sequence and continues in frame to translate the remainder of the GD05/2017 polyprotein. The advantage of this strategy is capable of retaining the native N or C-terminuses of virus proteins surrounding the foreign genes.
The recovery and characterization of recombinant viruses
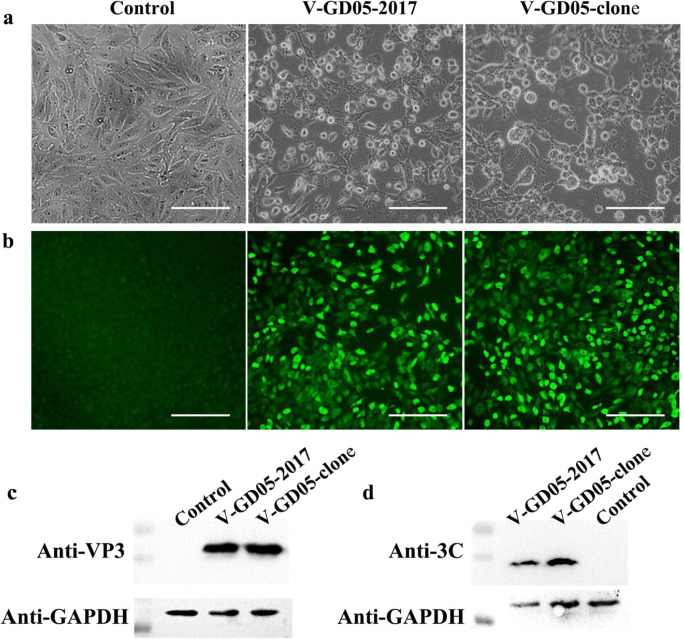
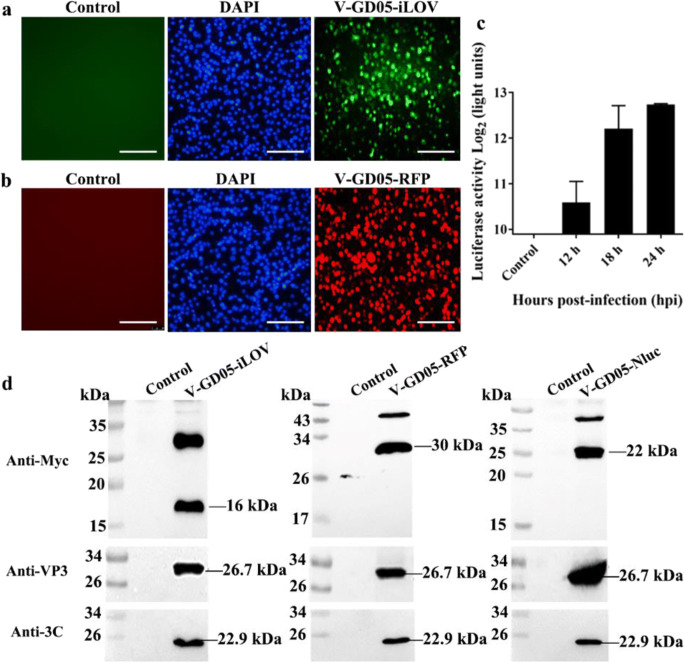
To rescue the recombinant viruses, pC3-SVA-GD05, pC3-SVA-GD05-iLOV-T2A, pC3-SVA-GD05-RFP-T2A, or pC3-SVA-GD05-Nluc-T2A were transfected into BHK-21 cells. Cell culture supernatants from BHK21 cells were further passaged onto ST-R cells to generate V-GD05-clone, V-GD05-iLOV, V-GD05-RFP, and V-GD05-Nluc. In comparison with the parental virus, the V-GD05-clone displayed a similar cytopathic effect (Fig. 2a), and the expression of VP3 could be detected by immunofluorescence (Fig. 2b). Meanwhile, the expression of VP3 and 3C could also be detected by western blot (Fig. 2c and 2d). For characterization of reporter viruses, the iLOV (green) (Fig. 3a) and RFP (red) (Fig. 3b) fluorescence signals were observed in the V-GD05-iLOV- and V-GD05-RFP-infected ST-R cells at 36 hpi, while the Nluc luciferase activities were also detectable in the V-GD05-Nluc-infected ST-R cells at 12-24 hpi (Fig. 3c). In the western blot assays (Fig. 3d), the bands of iLOV-T2A (16 kDa), RFP-T2A (30 kDa), and Nluc-T2A (22 kDa) were clearly identified, suggesting that the fusion proteins have been processed as expected. However, a larger band was always present in each assay, which may be a partially processed polypeptide containing the reporter proteins. The finding is not unusual in that 2A peptide-mediated cleavage efficiency is not 100%.
Fig. 2.
Recovery and identification of recombinant virus V-GD05-clone. a ST-R cells were infected with V-GD05/2017 and V-GD05-clone. After 24 h, CPE was observed under microscopy. b Cells were fixed and treated with VP3 mAb followed by addition of Dylight 488 anti-rabbit IgG as secondary antibody. The bar = 200 μm. c V-GD05/2017 and V-GD05-clone infected cells at an MOI of 0.1 were analyzed by western blot with the VP3 mAb. d V-GD05/2017 and V-GD05-clone infected cells at an MOI of 0.1 were analyzed by western blot with 3C mAb
Fig. 3.
Characterization of SVA reporter viruses. BHK-21 cells were transfected by pC3-SVA-GD05-iLOV-T2A, pC3-SVA-GD05-RFP-T2A. At 3 dpi, cell culture supernatant from the transfected cells was passaged onto ST-R cells. a and b The fluorescence were observed in the V-GD05-iLOV- or V-GD05-RFP-infected cells at 36 hpi. The bar = 200 μm. c The luciferase activities were detected in the V-GD05-Nluc-infected ST-R cells at 12-24 hpi. d Cell lysates from the reporter viruses infected ST-R cells infected with V-GD05-iLOV, V-GD05-RFP, V-GD05-Nluc, or mock-infected cells were harvested at 36 hpi. The expression of reporter and viral proteins were analyzed by western blot using myc, VP3, and 3C mAbs
Genetic stability of reporter viruses
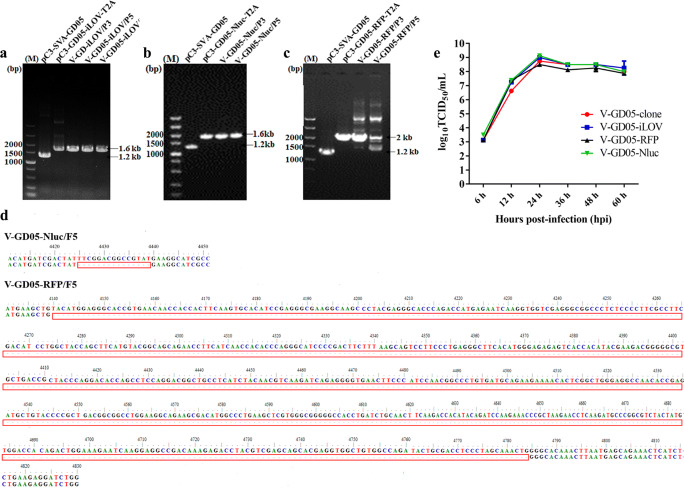
Picornaviruses are generally limited by genetic instability as an expression vector, and it is therefore necessary to investigate the genetic stability of reporter genes in the SVA genome. Following the initial recovery of rescued viruses in BHK-21, viruses were serially passaged 10 times in the ST-R cells to yield P1 to P10 virus stocks. RT-PCR and sequencing analysis of the P3, P5, and P10 stocks revealed that exogenous RFP and Nluc genes were relatively stable which were retained within the SVA genome over five passages (P1 to P5) (Figs. 4b–d). In comparison, the iLOV gene was consistently intact with no mutations or deletions in the SVA genome through 10 passages (Fig. 4a). Although the results showed that SVA tolerated the iLOV, RFP, and Nluc genes at different degrees, it remained to investigate whether the insertion of additional genes of 300–700 nt into the genome would affect the efficiency of virus replication. Accordingly, virus growth comparisons were performed among V-GD05-clone, V-GD05-iLOV, V-GD05-RFP, and V-GD05-Nluc. After infection with recombinant viruses, the ST-R cell culture supernatants were collected at 6, 12, 24, 36, 48, and 60 hpi to measure the virus titer. As shown in Fig. 4e, replication kinetics of the reporter viruses were similar to that of the V-GD05-clone, where all of viruses reached the peak titers at 24 hpi. The above results showed the virus replications of V-GD05-iLOV, V-GD05-RFP, and V-GD05-Nluc were not drastically affected in vitro.
Fig. 4.
Genetic stability and growth curve of SVA reporter viruses. a Identification of iLOV gene in the V-GD05-iLOV of P3, P5, and P10 virus stocks with the pC3-SVA-GD05 and pC3-SVA-GD05-iLOV-T2A as controls. b Identification of Nluc gene in the V-GD05-Nluc of P3 and P5 virus stocks with the pC3-SVA-GD05 and pC3-SVA-GD05-Nluc-T2A as controls. c Identification of RFP gene in the V-GD05-RFP of P3 and P5 virus stocks with the pC3-SVA-GD05 and pC3-SVA-GD05-RFP-T2A as controls. d Sequencing analysis of the V-GD05-RFP and V-GD05-Nluc deletion variants. e ST-R cells were infected with SVA reporter viruses at the MOI of 0.1 and the virus titers in the cell culture supernatants were analyzed at 6, 12, 24, 36, 48, and 60 hpi
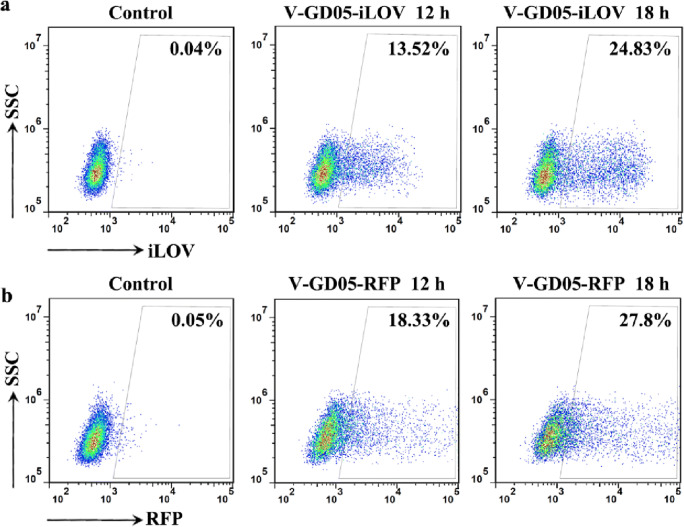
The construction of fluorescent or luminescent SVA offers alternative systems to detect SVA-infected cells in vitro and in vivo studies. We next explored flow cytometry by which V-GD05-iLOV and V-GD05-RFP replication can be detected and quantified in cells. As shown in Fig. 5, V-GD05-iLOV and V-GD05-RFP-infected ST-R cells were capable of being detected and quantified using flow cytometry. The numbers of positive ST-R cells expressing fluorescent proteins increased from 13.52 to 24.83% (V-GD05-iLOV) and from 18.33 to 27.8% (V-GD05-RFP) at 12 hpi and 18 hpi. Moreover, luciferase activities could be detected in the V-GD05-Nluc-infected ST-R cells as high as close to 213 comparing to 20 in the control cells at 24 hpi (Fig. 3c).
Fig. 5.
Detection of SVA reporter virus-infected cells by flow cytometry. ST-R cells were infected with V-GD05-iLOV (a) and V-GD05-RFP (b) at an MOI of 0.1. After 12 or 18 h, cells were analyzed by flow cytometry
Using reporter viruses to identify the cell receptor for SVA infection
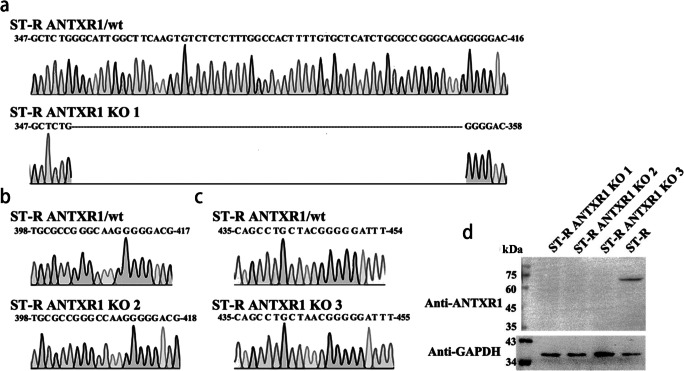
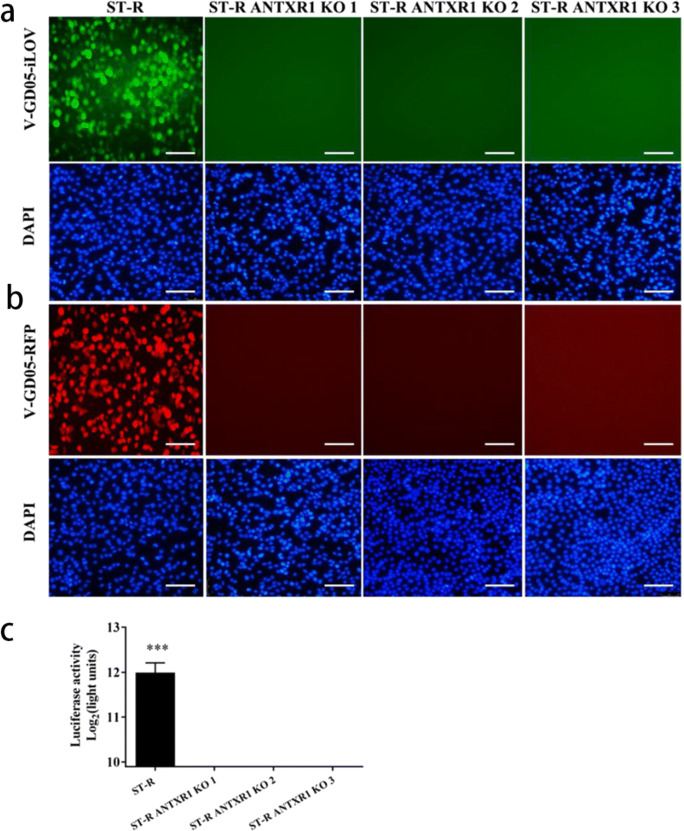
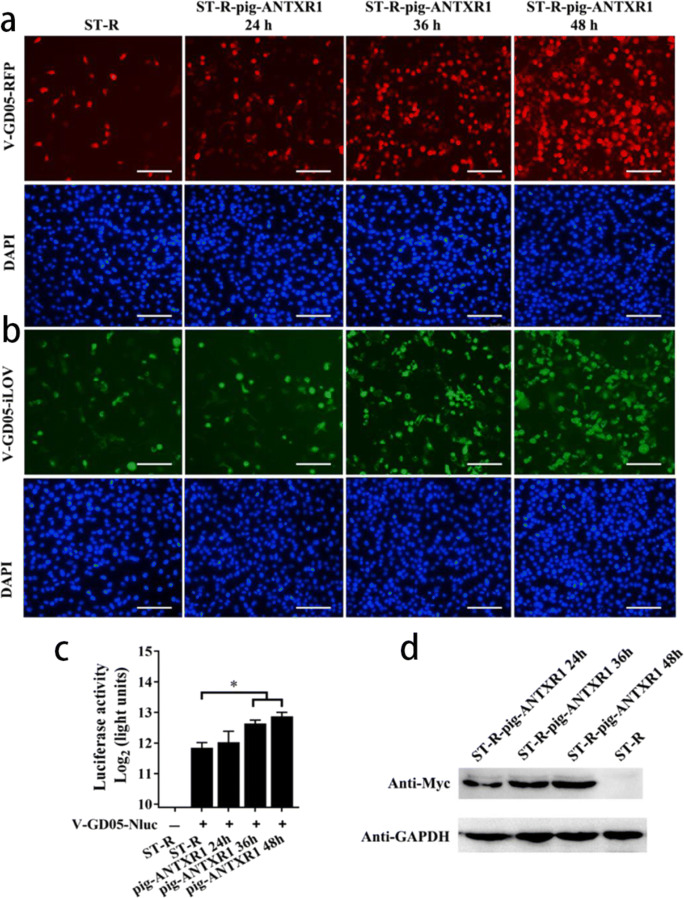
It was reported that ANTXR1 was the cellular receptor for entry of SVV-001 into human cells. To investigate the effect of porcine ANTXR1 for SVA infection, we initially knocked out (KO) the ANTXR1 in the ST-R cells and developed three KO cell lines (ST-R ANTXR1 KO1, KO2, KO3). We sequenced the target region of the genomic DNA extracted from ST-R ANTXR1 KO cell lines to analyze the mutation effect. The sequencing results showed that there was a deletion of 58 nucleotides in the target region of ST-R ANTXR1 KO1 cells (Fig. 6a). We also identified the ST-R ANTXR1 KO2 (Fig. 6b) and ANTXR1 KO3 cells (Fig. 6c), both of which harbored frameshift mutations within the first exon of ANTXR1. Then, the expression of ANTXR1 protein in the ST-R ANTXR1 KO cells was investigated. As expected, porcine ANTXR1 in the ST-R ANTXR1 KO cell lines were unable to detect by western blot using the ANTXR1-specific mAb (Fig. 6d). The ST-R ANTXR1 KO cells were infected with reporter viruses at the MOI of 0.1. After 36 h, there was no fluorescent signal observed in the ST-R ANTXR1 KO cells infected by V-GD05-iLOV (Fig. 7a) and V-GD05-RFP (Fig. 7b), and luciferase activities were also undetectable in the V-GD05-Nluc-infected ST-R ANTXR1 KO cells (Fig. 7c). Furthermore, the porcine ANTXR1 was overexpressed in the ST-R cells transfected by pCAGGS-Myc-pig-ANTXR1. As shown in the Fig. 8d, the expression of Myc-tagged ANTXR1 was identified by western blot. At 24, 36, and 48 h post-transfection, SVA reporter viruses were then added to infect the ANTXR1-overexpressed cells. Compared with ST-R cells, RFP (Fig. 8a), iLOV (Fig. 8b) fluorescence signal and Nluc activities (Fig. 8c) were significantly increased in the ANTXR1-overexpressed ST-R cells. The above results indicate that ANTXR1 is the cell receptor for SVA infection of porcine cells.
Fig. 6.
Identification of porcine ANTXR1 knocked-out cell lines. a Sequencing chromatogram of partial ANTXR1 gene in the ST-R cells and ST-R ANTXR1 KO1 cell line. b Sequencing chromatogram of partial ANTXR1 gene in the ST-R cells and ST-R ANTXR1 KO2 cell line. c Sequencing chromatogram of partial ANTXR1 gene in the ST-R cells and ST-R ANTXR1 KO3 cell line. d The expression of ANTXR1 protein in the ST-R and three KO cell lines were detected by western blot using the ANTXR1-specific mAb
Fig. 7.
Identification of ANTXR1 as cell receptor using SVA reporter viruses. ST-R and ST-R ANTXR1 KO cells were infected with V-GD05-iLOV(a) and V-GD05-RFP (b) at an MOI of 0.1, and the fluorescence was observed after 36 h infection. Nuclei were stained blue (DAPI). The bar = 200 μm. c ST-R and ST-R ANTXR1 KO cells were infected with V-GD05-Nluc at an MOI of 0.1. At 18 hpi, the luciferase activities were detected
Fig. 8.
Overexpression of ANTXR1 increases the cell permissivity for SVA infection. ST-R cells were transfected with pCAGGS-Myc-pig-ANTXR1. At 24 h, 36 h, and 48 h post-transfection, the cells were infected with V-GD05-RFP (a), V-GD05-iLOV (b), and V-GD05-Nluc (c) at an MOI of 0.1 for 24 h. (d) The expression of myc-tagged porcine ANTXR1 protein were detected through western blot using the myc-specific mAb
Discussion
Reporter-expressing viruses are useful tools in vitro and in vivo studies, such as quantitative analysis of viral replication, monitoring virus transport, screening antiviral agents, and identification of virus cell receptor, which do not require specific immunostaining of viral proteins (Li et al. 2016). In this report, we described the development and characterization of three SVA reporter recombinants encoding Nluc, iLOV, and RFP. The reporter viruses showed growth kinetics comparable to the V-GD05-clone (Fig. 4e). These results indicated that an efficient reverse genetic system has been established for rescuing SVA reporter viruses. For construction of reporter viruses, the foreign genes were inserted between the 2A and 2B genes with the addition of a Teschovirus 2A protease cleavage site at its C-terminus. Upon the cleavage by SVA 2A and Teschovirus 2A, the reporter proteins (iLOV, RFP, and Nluc) were released and detected (Fig. 3).
The SVA reporter virus expressing EGFP has been constructed using SVV-001 and KS15-01 strains in previous studies, but it was unstable in continuous three to five passages. In order to determine the size of exogenous gene that could be stably inserted into the SVA genome between 2A and 2B, we constructed three recombinant SVA expressing reporter proteins of different sizes. The flavoprotein improved LOV (iLOV) is a novel green fluorescent protein derived from the blue light receptor phototropin (Chapman et al. 2008). For certain applications such as fluorescent-tagged recombinant virus, the iLOV has been used as an alternative to the traditional GFP (~ 700 nt) for its smaller size (~ 300 nt) (Seago et al. 2013; van den Wollenberg et al. 2015). Recently, this method had been applied to screen antivirals in Rhinovirus-iLOV-infected HeLa cells (Han et al. 2018), where the number of green fluorescence-positive-infected cells decreased when they were treated with known flavivirus inhibitor (such as bafilomycin). As a novel engineered product, Nanoluciferase (Nluc) has been developed from a deep-sea shrimp luciferase by directed evolution. Not only is the Nluc smaller ( ~500 nt) than either firefly or Renilla luciferase, it is also more stable to environmental conditions as well as produces brighter and more sustained luminescence (Hall et al. 2012). In the cases of Nluc-expressing flaviviruses, the enzyme activity was quantified in a simple manner or a high-throughput assay to assess the antiviral activity of potential inhibitors in vitro (Cao et al. 2018; Pierson et al. 2017). The brilliantly red fluorescent protein TagRFP (RFP) is derived from Entacmaea quadricolor with a size of ~ 700 nt comparable to the one of GFP (Subach et al. 2008). It is characterized with long time fluorescence and high pH resistance, making it an attractive tag for protein studies. When these fluorescent and Nluc reporters with distinct properties are tagged into SVA genome, they offer particular benefits in the virus research.
After continuous passage of SVA reporter viruses, the results showed that the RFP and Nluc gene fragments were lost after three to five passages, while the iLOV was genetically stable over ten passages through cell culture using the same strategy. Comparing to the previous reports on the EGFP-tagged SVA, the RFP and Nluc have the similar issue of unstability in the SVA genome under a few passage. In the FMDV study, a foreign gene of 300–400 nt was considered as the maximum size for stable insertion into virus genome (Seago et al. 2013), which coincides with the current finding that the iLOV of ~ 300 nt was capable of being stably maintained in the SVA. The reasons for loss of foreign gene quickly during viral passage may involve more than one factor, such as the packaging limit of capsid, genome recombination, codon usage bias, and RNA structure (Song et al. 2012). Interestingly, a recent study show that the RNA-dependent RNA polymerase (RdRp) played a critical role in SVA recombination, where specific nonsynonymous mutations in the RdRp were capable of reducing recombination to occur, thereby increasing the stability of foreign gene in the virus genome (Li et al. 2019). In the future study, we are interested to investigate if it works on our SVA reporter viruses.
ANTXR1, also known as tumor endothelial marker 8 (TEM8), was first identified as the cell surface receptor of anthrax toxin (Miles et al. 2017). Unlike ANTXR2, another anthrax toxin receptor with a wide distribution in the adult tissues, ANTXR1, is abundant in the tumor cells and the vasculature of developing embryos (Cao et al. 2018; Chaudhary et al. 2012). Among 1037 cell lines in the Cancer Cell Line Encyclopedia, over 63% of cell lines exceed the expression cutoff of ANTXR1 (Miles et al. 2017). ANTXR1 has recently been identified as the cellular receptor for entry of SVV-001 into human cells. Although porcine ANTXR1 and human ANTXR1 share a close sequence homology of 97%, there were R88Q and D156E mutations on the porcine ANTXR1. The two residues on the N terminal of ANTXR1 play an indispensable role on interacting with SVA VP proteins. Therefore, the question remains whether it is the case for porcine SVA infection. Here, we knocked out or overexpressed the porcine ANTXR1 on the ST-R cells. After SVA reporter virus infection, there was no specific fluorescence and luciferase activity detectable in the ST-R ANTXR1 KO cells infected with V-GD05-iLOV, V-GD05-RFP, and V-GD05-Nluc, while overexpression of porcine ANTXR1 could increase the cellular permissivity for SVA infection. The results confirmed that ANTXR1 was the receptor for SVA GD05/2017 to invade porcine cells.
In summary, we have successfully constructed the iLOV, RFP, or Nluc tagged-SVA reporter viruses using a Chinese strain in this study. It was found that the iLOV-tagged SVA could maintain a high level of stable passage in cells. Furthermore, we examined the feasibility of SVA reporter virus as a research tool by fluorescence observation, flow cytometry, and luciferase activity. Importantly, our studies on the ANTXR1-knockout or -overexpressed ST-R cell suggested that ANTXR1 was the cellular receptor for SVA invasion of pig-derived cells. We believe that these reporter virus tools are useful for studying SVA pathogenesis and vaccine development as well as for screening novel anti-SVA agents.
Supplementary Information
(PDF 225 kb)
Acknowledgments
We thank all of members at Dr. Chen’s lab for their suggestions and excellent technical assistance
Authors’ contributions
MW carried out most of the experiments. CM and MC carried out complementary experiments. All authors analyzed the data. ZC and CM conceived and supervised the study. ZC, MW and CM designed the experiments and wrote the manuscript.
Funding
This work was funded by the National Natural Science Foundation of China (grant no. 31772748), the National Key Research and Development Program of China (2017YFD0500104), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD) to Z.C.
Data availability
The recombinant SVA viruses generated in this study can be obtained from the corresponding author upon reasonable request.
Declarations
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Minmin Wang and Chunxiao Mou contributed equally to this work.
References
- Adams MJ, Lefkowitz EJ, King AM, Bamford DH, Breitbart M, Davison AJ, Ghabrial SA, Gorbalenya AE, Knowles NJ, Krell P, Lavigne R, Prangishvili D, Sanfacon H, Siddell SG, Simmonds P, Carstens EB. Ratification vote on taxonomic proposals to the international committee on taxonomy of viruses (2015) Arch Virol. 2015;160(7):1837–1850. doi: 10.1007/s00705-015-2425-z. [DOI] [PubMed] [Google Scholar]
- Canning P, Canon A, Bates JL, Gerardy K, Linhares DC, Piñeyro PE, Schwartz KJ, Yoon KJ, Rademacher CJ, Holtkamp D, Karriker L. Neonatal mortality, vesicular lesions and lameness associated with senecavirus A in a U.S. Sow Farm. Transbound Emerg Dis. 2016;63(4):373–378. doi: 10.1111/tbed.12516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Zhang R, Liu T, Sun Z, Hu M, Sun Y, Cheng L, Guo Y, Fu S, Hu J, Li X, Yu C, Wang H, Chen H, Li X, Fry EE, Stuart DI, Qian P, Lou Z, Rao Z. Seneca valley virus attachment and uncoating mediated by its receptor anthrax toxin receptor 1. Proc Natl Acad Sci U S A. 2018;115(51):13087–13092. doi: 10.1073/pnas.1814309115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman S, Faulkner C, Kaiserli E, Garcia-Mata C, Savenkov EI, Roberts AG, Oparka KJ, Christie JM. The photoreversible fluorescent protein iLOV outperforms GFP as a reporter of plant virus infection. Proc Natl Acad Sci U S A. 2008;105(50):20038–20043. doi: 10.1073/pnas.0807551105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary A, Hilton Mary B, Seaman S, Haines Diana C, Stevenson S, Lemotte Peter K, Tschantz William R, Zhang Xiaoyan M, Saha S, Fleming T, St. Croix B. TEM8/ANTXR1 blockade inhibits pathological angiogenesis and potentiates tumoricidal responses against multiple cancer types. Cancer Cell. 2012;21(2):212–226. doi: 10.1016/j.ccr.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Yuan F, Li Y, Shang P, Schroeder R, Lechtenberg K, Henningson J, Hause B, Bai J, Rowland RRR, Clavijo A, Fang Y. Construction and characterization of a full-length cDNA infectious clone of emerging porcine senecavirus A. Virology. 2016;497:111–124. doi: 10.1016/j.virol.2016.07.003. [DOI] [PubMed] [Google Scholar]
- Chen L, Zhang J, Wang M, Pan S, Mou C, Chen Z. Pathogenicity of two chinese seneca valley virus (SVV) strains in pigs. Microb Pathog. 2019;136:103695. doi: 10.1016/j.micpath.2019.103695. [DOI] [PubMed] [Google Scholar]
- Corner SSK. Seneca valley virus and vesicular lesions in a pig with idiopathic vesicular disease. J Vet Sci Technol. 2012;3(6):1–3. doi: 10.4172/2157-7579.1000123. [DOI] [Google Scholar]
- Donnelly MLL, Luke G, Mehrotra A, Li X, Hughes LE, Gani D, Ryan MD. Analysis of the aphthovirus 2A/2B polyprotein ‘cleavage’ mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal ‘skip’. J Gen Virol. 2001;82(Pt 5):1013–1025. doi: 10.1099/0022-1317-82-5-1013. [DOI] [PubMed] [Google Scholar]
- Guo B, Piñeyro PE, Rademacher CJ, Zheng Y, Li G, Yuan J, Hoang H, Gauger PC, Madson DM, Schwartz KJ, Canning PE, Arruda BL, Cooper VL, Baum DH, Linhares DC, Main RG, Yoon KJ. Novel senecavirus A in swine with vesicular disease, United States, July 2015. Emerg Infect Dis. 2016;22(7):1325–1327. doi: 10.3201/eid2207.151758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hales LM, Knowles NJ, Reddy PS, Xu L, Hay C, Hallenbeck PL. Complete genome sequence analysis of seneca valley virus-001, a novel oncolytic picornavirus. J Gen Virol. 2008;89(Pt 5):1265–1275. doi: 10.1099/vir.0.83570-0. [DOI] [PubMed] [Google Scholar]
- Hall MP, Unch J, Binkowski BF, Valley MP, Butler BL, Wood MG, Otto P, Zimmerman K, Vidugiris G, Machleidt T, Robers MB, Benink HA, Eggers CT, Slater MR, Meisenheimer PL, Klaubert DH, Fan F, Encell LP, Wood KV. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem Biol. 2012;7(11):1848–1857. doi: 10.1021/cb3002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M, Rajput C, Hinde JL, Wu Q, Lei J, Ishikawa T, Bentley JK, Hershenson MB. Construction of a recombinant rhinovirus accommodating fluorescent marker expression. Influenza Other Resp. 2018;12(6):717–727. doi: 10.1111/irv.12602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hause BM, Myers O, Duff J, Hesse RA. Senecavirus A in Pigs, United States, 2015. Emerg Infect Dis. 2016;22(7):1323–1325. doi: 10.3201/eid2207.151591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leme RA, Oliveira TE, Alcântara BK, Headley SA, Alfieri AF, Yang M, Alfieri AA. Clinical manifestations of senecavirus A infection in neonatal pigs, brazil, 2015. Emerg Infect Dis. 2016;22(7):1238–1241. doi: 10.3201/eid2207.151583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Li L-F, Yu S, Wang X, Zhang L, Yu J, Xie L, Li W, Ali R, Qiu H-J. Applications of replicating-competent reporter-expressing viruses in diagnostic and molecular virology. Viruses. 2016;8(5):127. doi: 10.3390/v8050127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wang H, Shi J, Yang D, Zhou G, Chang J, Cameron CE, Woodman A, Yu L. Senecavirus-specific recombination assays reveal the intimate link between polymerase fidelity and RNA recombination. J Virol. 2019;93(13):e00576–e00519. doi: 10.1128/JVI.00576-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Li X, Wu M, Qin L, Chen H, Qian P. Seneca valley virus 2C and 3C(pro) induce apoptosis via mitochondrion-mediated intrinsic pathway. Front Microbiol. 2019;10:1202. doi: 10.3389/fmicb.2019.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles LA, Burga LN, Gardner EE, Bostina M, Poirier JT, Rudin CM. Anthrax toxin receptor 1 is the cellular receptor for seneca valley virus. J Clin Invest. 2017;127(8):2957–2967. doi: 10.1172/JCI93472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montiel N, Buckley A, Guo B, Kulshreshtha V, VanGeelen A, Hoang H, Rademacher C, Yoon KJ, Lager K. Vesicular disease in 9-week-old pigs experimentally infected with senecavirus A. Emerg Infect Dis. 2016;22(7):1246–1248. doi: 10.3201/eid2207.151863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasma T, Davidson S, Shaw SL. Idiopathic vesicular disease in swine in Manitoba. Can Vet J. 2008;49(1):84–85. [PMC free article] [PubMed] [Google Scholar]
- Pierson TC, Brecher M, Li Z, Liu B, Zhang J, Koetzner CA, Alifarag A, Jones SA, Lin Q, Kramer LD, Li H. A conformational switch high-throughput screening assay and allosteric inhibition of the flavivirus NS2B-NS3 protease. PLoS Pathog. 2017;13(5):e1006411. doi: 10.1371/journal.ppat.1006411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier JT, Reddy PS, Idamakanti N, Li SS, Stump KL, Burroughs KD, Hallenbeck PL, Rudin CM. Characterization of a full-length infectious cDNA clone and a GFP reporter derivative of the oncolytic picornavirus SVV-001. J Gen Virol. 2012;93(Pt 12):2606–2613. doi: 10.1099/vir.0.046011-0. [DOI] [PubMed] [Google Scholar]
- Reed LJ, Muench H (1938) A simple method of estimating fifty percent endpoints. Am J Hygiene 27:493–497
- Saporiti V, Fritzen JTT, Feronato C, Leme RA, Lobato ZIP, Alfieri AF, Alfieri AA. A ten years (2007–2016) retrospective serological survey for seneca valley virus infection in major pig producing states of brazil. Vet Res Commun. 2017;41(4):317–321. doi: 10.1007/s11259-017-9697-6. [DOI] [PubMed] [Google Scholar]
- Seago J, Juleff N, Moffat K, Berryman S, Christie JM, Charleston B, Jackson T. An infectious recombinant foot-and-mouth disease virus expressing a fluorescent marker protein. J Gen Virol. 2013;94(Pt 7):1517–1527. doi: 10.1099/vir.0.052308-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Liu L, Edwards SV, Wu S. Resolving conflict in eutherian mammal phylogeny using phylogenomics and the multispecies coalescent model. Proc Natl Acad Sci U S A. 2012;109(37):14942–14947. doi: 10.1073/pnas.1211733109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subach OM, Gundorov IS, Yoshimura M, Subach FV, Zhang JH, Gruenwald D, Souslova EA, Chudakov DM, Verkhusha VV. Conversion of red fluorescent protein into a bright blue probe. Chem Biol. 2008;15(10):1116–1124. doi: 10.1016/j.chembiol.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Wollenberg DJM, Dautzenberg IJC, Ros W, Lipińska AD, van den Hengel SK, Hoeben RC. Replicating reoviruses with a transgene replacing the codons for the head domain of the viral spike. Gene Ther. 2015;22(3):267–279. doi: 10.1038/gt.2014.126. [DOI] [PubMed] [Google Scholar]
- Vannucci FA, Linhares DC, Barcellos DE, Lam HC, Collins J, Marthaler D. Identification and complete genome of seneca valley virus in vesicular fluid and sera of pigs affected with idiopathic vesicular disease, brazil. Transbound Emerg Dis. 2015;62(6):589–593. doi: 10.1111/tbed.12410. [DOI] [PubMed] [Google Scholar]
- Venkataraman S, Reddy SP, Loo J, Idamakanti N, Hallenbeck PL, Reddy VS. Crystallization and preliminary X-ray diffraction studies of seneca valley virus-001, a new member of the picornaviridae family. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008;64(Pt 4):293–296. doi: 10.1107/S1744309108006921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman S, Reddy SP, Loo J, Idamakanti N, Hallenbeck PL, Reddy VS. Structure of seneca valley virus-001: an oncolytic picornavirus representing a new genus. Structure. 2008;16(10):1555–1561. doi: 10.1016/j.str.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Chen L, Pan S, Mou C, Shi K, Chen Z. Molecular evolution and characterization of novel seneca valley virus (SVV) strains in South China. Infect Genet Evol. 2019;69:1–7. doi: 10.1016/j.meegid.2019.01.004. [DOI] [PubMed] [Google Scholar]
- Wang M, Jin S, Chen L, Chen Z. Complete genome sequencing and construction of an infectious clone of a SVA GD05/2017strain. J Yangzhou Univ. 2020;41(2):51–56. [Google Scholar]
- Wang J, Mou C, Wang M, Pan S, Chen Z. Transcriptome analysis of senecavirus A-infected cells: Type I interferon is a critical anti-viral factor. Microb Pathog. 2020;147:104432. doi: 10.1016/j.micpath.2020.104432. [DOI] [PubMed] [Google Scholar]
- Wu Q, Zhao X, Chen Y, He X, Zhang G, Ma J. Complete genome sequence of seneca valley virus CH-01-2015 identified in China. Genome Announc. 2016;4(1):e01509–e01515. doi: 10.1128/genomeA.01509-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 225 kb)
Data Availability Statement
The recombinant SVA viruses generated in this study can be obtained from the corresponding author upon reasonable request.