Abstract
Background/Aims
Gut microbiota ferments indigestible food that rests in the colon to produce short-chain fatty acids (SCFAs) acetate, propionate, and butyrate. Colonic SCFA stimulate the synthesis of serotonin which is central in irritable bowel syndrome (IBS) pathophysiology. Reduced SCFA have been linked to specific IBS symptoms like colonic hyperalgesia and hypersensitivity. SCFA enter the colonocyte mainly via 2 energy-dependent monocarboxylate transporters, MCT1 (SLC16A1) and SMCT1 (SLC5A8). We investigated specific gut microbiota, SCFA concentrations, and monocarboxylate transporter mRNA expression in patients with IBS.
Material and Methods
A total of 30 IBS patients—15 constipation-predominant (C-IBS) and 15 diarrhoea-predominant (D-IBS)—and 15 healthy controls were recruited. Bacteroidetes and Bifidobacterium species were analyzed using quantitative polymerase chain reaction (qPCR) on stool samples. SCFA concentrations were determined by gas chromatography/mass spectroscopy of stool samples. Monocarboxylate transporter mRNA was quantified by qPCR on colon biopsy specimens.
Results
Bacteroides was significantly increased in the D-IBS group compared with the C-IBS group and healthy controls. Bifidobacterium was significantly reduced in both IBS groups. SCFA ratios were altered in both IBS groups with a reduction of all 3 measured SCFA in C-IBS and acetic acid in D-IBS. MCT1 and SMCT1 were significantly reduced in C-IBS and D-IBS.
Conclusion
In agreement with findings of previous studies, the microbiota assessed were significantly altered inferring dysbiosis in IBS. SCFA and their ratios were significantly altered in both IBS groups. SCFA transporters, MCT1 and SMCT1 were significantly reduced in both IBS groups, suggesting reduced colonocyte SCFA transfer. SCFA availability and transfer into the colonocytes may be important in IBS pathogenesis and should be prospectively studied.
Keywords: Gut microbiota, short chain fatty acids, monocarboxylate transporters, irritable bowel syndrome
INTRODUCTION
Irritable bowel syndrome (IBS) is one of the most common functional gastrointestinal disorders. Typical IBS phenotypes include constipation type (C-IBS), diarrhoea type (D-IBS), mixed type (IBS-M), and unspecific (IBS-U). In IBS-U, patients meet the diagnostic criteria for IBS, but their bowel habits cannot be accurately categorized. IBS pathophysiology remains largely unknown (1), and treatment is mostly symptomatic. It is believed to be a multifactorial disorder with contributing factors including visceral hypersensitivity, deranged gut motility, and a dysfunctional gut-brain axis among others (2, 3).
Increasingly, an association between gut microbiota derangement and IBS symptoms is being recognized (4). Although great variability exists in the gut microbiota quantities between different studies in IBS, it is clear that certain bacteria, especially Lactobacillus, Enterobacteriaceae, Bacteroidetes, and Bifidobacterium among others, are significantly altered (5).
Gut microbiota produce short-chain fatty acids (SCFAs) by fermentation of indigestible food debris, especially fibre (6). SCFAs have important immunological and regulatory functions and are the apparent interface between gut microbiota and host (7). In addition, they have important metabolic, protective, and trophic functions (8). As part of their metabolic role, they regulate proliferation and differentiation of colonic cells, are important in cancer prevention, produce vitamins, and regulate fat storage. SCFAs protect gut integrity against pathogenic microbes and toxins using various methods including regulating neutrophil trafficking. Protection also includes cancer prevention by inducing apoptosis in cancer cells. The major trophic functions involve the regulation of proliferation and differentiation of colon epithelial cells and maturation of the immune system (9, 10).
Data suggest that SCFAs may play a role in IBS pathophysiology (11). Acetate, propionate, and butyrate are the main SCFAs produced by gut microbiota. Colonic luminal SCFAs stimulate the synthesis of serotonin which is central in IBS pathophysiology (12). Propionate, mainly produced by Bacteroides, is responsible for the release of serotonin from the enterochromaffin cells to activate 5-HT4 receptors and stimulate the enteric peristaltic reflex (13, 14). Both propionate and butyrate induce tonic contractions of the colon (15).
Of the three main SCFAs, butyrate is important in the maintenance of colon health. It is the principal energy source for colonocytes and plays an important anti-inflammatory role (16). Butyrate modulates IBS with specific reference to colonic hyperalgesia and hypersensitivity (17). Data showed that butyrate-producing microbiota are reduced in IBS (18). Hence, reduced butyrate concentration can potentially influence IBS symptoms.
Once produced, SCFAs are transported from the luminal aspect of the colon into the colonocyte via passive diffusion and/or carrier-mediated transport. However, as the physiological luminal pH ranges from 5.6 to 6.6, the major SCFAs are in an anionic form, not available for simple diffusion (19). Thus, active transport through monocarboxylate transporters (MCT) appears to be the main transport mode. The MCT system consists of (i) the electroneutral H+-coupled MCT1 of which SLC16A is the major one and (ii) the electrogenic Na+ - coupled SMCT1 of which SLC5A8 is the main transporter (20). MCT1 is expressed more than SMCT1. MCT1 transports all SCFAs, whereas SMCT1 transports butyrate more than acetate and propionate (21). The role of MCT in IBS pathogenesis has not been investigated so far.
The association between SCFAs and IBS symptomatology is inconclusive with conflicting reports (1). This study aims to investigate the relationship between SCFAs and their transportation across the colonic epithelium in the different IBS subtypes. We investigated the ratio of SCFA production and their transporters, MCT1 and SMCT1, in the same patient population. In addition, to corroborate alteration of gut microbiota, we investigated two specific gut microbiota, Bacteroides and Bifidobacterium, which have been previously identified with IBS.
MATERIALS AND METHODS
Ethical consideration
The study was approved by the Human Ethics Committee of the Nelson Mandela University (NMU) (H07SciBCM-001). Written informed consent to participate was obtained from each participant on enrollment.
Study population
A total of 30 patients were recruited, 15 in C-IBS and 15 in D-IBS subgroups and 15 healthy controls. The control group consisted of healthy patients scheduled for surveillance colonoscopy as part of cancer prevention. Inclusion criteria: age between 18 and 60, fulfilling Rome III criteria for diagnosis of IBS. Exclusion criteria: unable/unwilling to give informed consent or undergo colonoscopy, pregnancy, pre-, pro- or antibiotic therapy in last 3 months. Patients with abnormal colonoscopy finding and/or abnormal histology were also excluded.
Stool samples
All subjects gave a stool sample prior to colonoscopy. Patients were instructed to save the stool sample in a −20°C freezer after obtaining the specimen. Once brought to the clinic, the stool samples were stored at −80°C until analysis. SCFAs (acetate, propionate, and butyrate) were determined in the stool using gas chromatography/mass spectrometry (GC/MS). The phyla Bacteroides and Bifidobacterium were determined in the stool using qPCR.
Colon biopsies
All study participants underwent colonoscopy, and proximal (caecum and ascending colon) colon biopsies were taken. Biopsy samples were placed in RNA, and later, left at room temperature for 24 h and then stored at −20°C until analysis. Monocarboxylate transporter mRNA was quantified from colon biopsy specimens using qPCR.
Stool microbiota analysis
Fecal bacterial DNA was isolated using the QIAmp DNA Stool Mini Kit (Qiagen, Hilden, Germany): 200 mg of stool sample was vortexed in 500 μL of Buffer ASL for 1 min and centrifuged at 5000x g for 5 min. Supernatants were kept on ice, and pellets were resuspended in 500 μL of Buffer ASL and repeated 5 times. To maximize bacterial cell lysis, supernatants were pooled and treated with 10 mg/mL of lysozyme (Roche Diagnostics, Mannheim, Germany) for 30 min at 37°C. Samples were heated for 10 min at 95°C. The rest of the procedure followed protocol specifications. DNA extraction samples were quantified using fluorometric analysis via the Qubit fluorometer.
qPCR quantification of Bacteroides spp. and Bifidobacterium bifidum genomes in DNA extracted from stool: Bacteroides spp. genomes were quantified based on the presence of a specific 16S rRNA gene segment which is highly homologous to members of this family using the Bacteroides spp. quantification kit (Primer Design, Camberley, UK). The Bifidobacteria bifidum quantification kit (Primer Design, Camberley, UK) was used to determine the copy number of B.bifidum 16S – 23S intergenic spacer regions from the DNA extracted from stool DNA. A standard curve was set up using serial dilutions of the positive control template provided by the kits. Results are expressed as values that equate to log10 of total copy numbers detected.
SCFA analysis
Fecal dry weight was used in the SCFA analysis. To determine the dry weight of stool samples, three 200 mg samples were collected from different regions of the stool of all patients. All samples were centrifuged 13.4x g for 10 min to pellet stool matter and remove excess liquid. Miniscule insertions were made in the lids of the collection tubes, and the tubes were subsequently snap-frozen in liquid nitrogen. Samples were then placed in a vacuum jar and connected to a VirTis temperature-monitored vacuum system for 24 h to completely freeze-dry all stool samples. Samples were then re-weighed to determine the dry weight of stool matter in each sample.
A modified method by Yap (22) was used to extract and quantify SCFAs from stool samples. Fecal samples (1g) were added to 2 ml of GC-quality Methanol (Merck Chemicals, Darmstadt, Germany). Samples were homogenized by vortexing them for 5 min, followed by centrifugation at 5000x g for 30 min at 4°C. Approximately 1 μl of supernatant was injected by an autosampler into an Agilent Technologies gas chromatograph system and mass spectrum detector (Agilent Technologies) based on the method described by Scheppach et al. (23). A polar BP21 capillary column (SGE), 30 m in length with an internal diameter of 0.25 mm and inner walls coated with bonded polyethylene glycol was used. The oven temperature gradient was programmed for 60°C to 110°C 10°C/min, 110°C to 150°C 5°C/min and from 150°C to 230°C 120°C/min. The temperatures of both the injector and detector were 230°C. The inlet was operated in a splitless mode. Helium gas was used at a flow rate of 1.22 mL/min (30 cm/s). All data were analyzed using the software system MSD Chemstation G1701EA Version E.02.01.1177. 5 mM valeric acid was used as internal standard.
Monocarboxylate transporter analysis
Quantitative real-time polymerase chain reaction (qPCR) was used to quantify mRNA of butyrate transporters. Total RNA was extracted from the colon biopsy samples using Bio-Rad Aurum™ RNA extraction kit (Bio-Rad Laboratories Inc, Hercules, California, USA) following the manufacturer’s instructions. cDNA was synthesized from 1 μg of RNA, using the reverse transcription BioRad iScript® cDNA synthesis kit (Bio-Rad Laboratories Inc, Hercules, California, USA.), in a 20 μL reaction mixture, according to the manufacturer’s instructions. The samples were then frozen at −80°C until the qPCR experiment.
Real-time qPCR reactions were run on an iCycler IQ® system (Bio-Rad Laboratories Inc, Hercules, California, USA). Each well of a 96 well PCR plate contained 20 μL consisting of 1 μL of the above cDNA, 10 μL SYBR®Green supermix (Bio-Rad Laboratories Inc, Hercules, California, USA), and 500 nM forward and reverse primers and nuclease-free PCR grade water (Ambion Inc, Austin, Texas, USA). For no template controls (NTC), the cDNA sample was replaced with nuclease-free water (Ambion, Austin, Texas, USA). Each sample and NTC was run in duplicate.
The protocol for all qPCR runs comprised of 3-min Taq polymerase activation at 95°C and 40 cycles of denaturing at 95°C for 30 s, 30 s at appropriate annealing temperature (Table 1), and extension at 72°C for 30 s.
Table 1.
Forward and reverse sequences for all genes and their annealing temperatures.
| Gene of interest | Forward sequence | Reverse sequence | Annealing temp (°C) |
|---|---|---|---|
| SLC16A1 | 5′-AGTAGTTATGGGAAGAGTCAGCA-3′ | 5′-GTCGGGCTACCATGTCAACA-3′ | 59 |
| SLA5A8 | 5′-GCCTATGATGGTGGAAGAT-3′ | 5′-GTTGACACCGTAGATGCT-3′ | 59 |
|
| |||
| Reference genes | Forward sequence | Reverse sequence | Annealing temp (°C) |
|
| |||
| SF3A1 | Commercial primer, obtained from GeNorm™ Normalization kit. (Primer Design, UK) | 60 | |
| ALUsq | 5′-CATGGTGAAACCCCGTCTCTA-3 | 5′-GCCTCAGCCTCCCGAGTAG-3′ | 60 |
| ALUsx | 5′-TGGTGAAACCCCGTCTCTACTAA-3′ | 5′-CCTCAGCCTCCCGAGTAGCT-3′ | 60 |
The amplification reaction was followed by a melt curve to verify the specificity of the reaction. The plate was heated to 95°C, and then cooled to 1°C below the annealing temperature for each primer pair to ensure that all the DNA was double stranded. The temperature was increased in increments of 0.5°C for 30 s up to 95°C to melt the double-stranded DNA.
The Cq values for the reference genes were transformed into relative quantification data using the ΔCq method, and they were analyzed using the GeNorm software (Primer Design, Camberley, UK) to determine the optimal set of reference genes. Genes of interest were analyzed following the above qPCR protocol, and Cq values were analyzed on the qBase version 2 software (BioGazelle, Ghent, Belgium). The GeNorm™ Normalization (Primer Design, Camberley, UK) protocol was used to identify the most suitable reference genes for this study. One reference gene, SF3A1, and 2 Alu repeats, ALUsq and ALUsx(24), were the most stable.
All the relevant genes used in the analyses in this study are tabulated in Table 1, with their respective forward and reverse sequences as well as melting temperature.
Statistical Analysis
Statistical comparisons were examined with one-way analysis of variance (ANOVA), followed by the Mann-Whitney U test. Demographic characteristics were summarized with descriptive statistics. Bar graphs were drawn using Microsoft Excel. All statistical parameters were calculated as mean ± standard deviation. The results were considered significantly different at p<0.05.
The results are displayed in graphs – first as an illustration of numerical differences and second as fold changes. The final value divided by the initial value. If this number was less than one the (negative) reciprocal is listed).
RESULTS
The demographic details for all 30 patients and 15 controls are shown in Table 2.
Table 2.
Demographic details of the study population.
| Control group | C-IBS | D-IBS | |
|---|---|---|---|
| Age | 41.3 | 41.0 | 42.1 |
| Sex (F/M) | 11/4 | 11/4 | 11/4 |
| BMI | 29.25 | 27.83 | 28.46 |
| Smoking | 1 (M) | 1 (M) | 0 |
| Duration of disease (years) | 10±4 | 9±7 |
BMI: Body Mass Index; C-IBS: constipation-predominant; D-IBS: diarrhoea-predominant.
Microbiota analysis
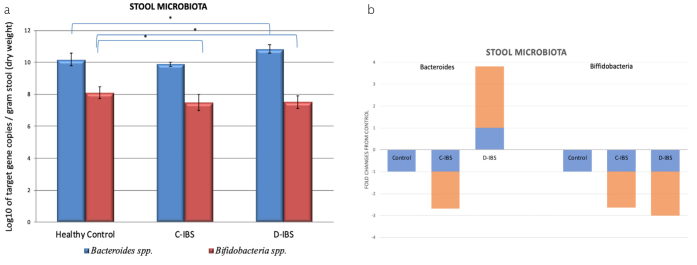
The number of Bacteroides 16S rRNA copies in stool samples of patients with D-IBS was significantly higher than those in stool samples of healthy controls and C-IBS patients (p<0.05). The number of Bifidobacteria 16S–23S intergenic spacer region copies were significantly lower in patients with C-IBS and D-IBS compared to those in the healthy control group (p<0.05). Results are displayed in Figure 1.
Figure 1. a, b.
Results of the 2 gut microbiota, Bacteroides and Bfidobacteria analysed. (a) Quantitative real time PCR analysis of Bacteroides spp. and Bifidobacteria spp. in the stool of healthy, C-IBS and D-IBS patient. n = 15 in each group. (b) * denotes p<0.05. Significance level compared against healthy control subjects. (b) Fold change of qPCR analysis of Bacteroides spp. and Bifidobacteria spp. in stool samples of C-IBS and D-IBS patients compared with healthy control subjects. Fold change for healthy control subjects is +1 or −1, represented by blue graphs, respective fold changes for C-IBS and D-IBS patients are represented by orange graph.
Results of stool microbiota analysis confirmed a significant quantitative change in abundance for both Bacteroides spp. and Bifidobacteria spp. in both IBS groups compared to the control group, suggesting the presence of dysbiosis.
SCFA
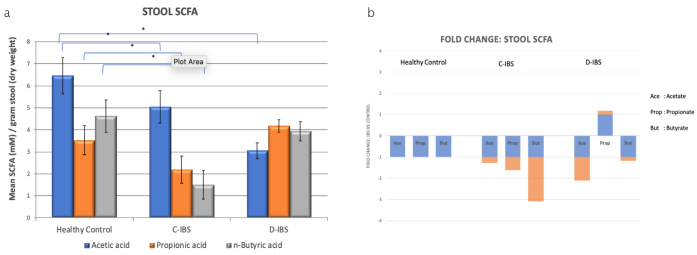
The expression ratio of the main SCFAs: acetate, propionate, and butyrate in the healthy control group was 6.5: 3.5: 4.5. (Figure 2). For C-IBS the expression ratio was 5.0: 2.0: 1.5, whereas for D-IBS it was 3.0: 4.1: 4.0 in D-IBS. There was significant reduction in acetate levels in both C-IBS and D-IBS compared to the control group. The ratio of the three SCFAs are different in both C-IBS and D-IBS compared to the control group. Most noticeable is the decrease of propionic acid in C-IBS, whereas it is increased in D-IBS when compared to the control group. This result is statistically significant. For butyrate specifically, there was statistically significant reduction in C-IBS compared to healthy control group, whereas it remained unchanged in D-IBS (although trending lower).
Figure 2. a, b.
Short chain fatty acids results analysed by gas chromatography/mass spectroscopy on stool samples. (a) Concentration of acetic acid, propionic acid and n-butyric acid in stool samples from healthy controls, C-IBS and D-IBS patients. n=15 in each group. * denotes p<0.05. Significance level compared against Healthy control patients. (b) Fold change of acetic acid, propionic acid and n-butyric acid concentrations in stool samples of C-IBS and D-IBS patients compared to healthy control subjects. Fold change for healthy control subjects is +1 or −1, represented by blue graphs, respective fold changes for C-IBS and D-IBS patients are represented by orange graph.
Monocarboxylate transporters
MCT1
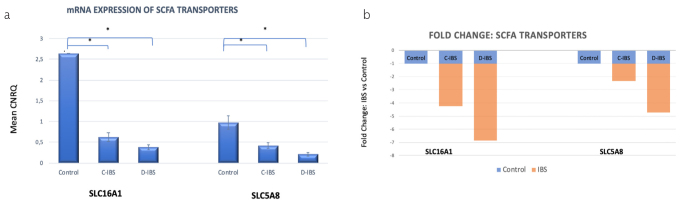
The relative expression of SLC16A1 mRNA was high in normal colonic mucosa, compared to the IBS groups. For the IBS groups, SLC16A1 mRNA was significantly reduced for both C-IBS (p=0.000004) and D-IBS (p=0.0001) compared to the healthy control group (Figure 3).
Figure 3. a, b.
SCFA transporters analysed in the colonic mucosa by quantitative real-time PCR. (a) Monocarboxylate transporters SLC16A1 and SLC5A8 mRNA expression by qPCR. CNRQ = Calibrated Normalized Relative Quantities, normalized to mean of all samples. n=15 in each group. * denotes p<0.05. Significance level compared to Healthy control. (b) Fold change of The mean CNRQ for qPCR values of SLC16A1 and SLC5A8 transporters in the mucosal biopsies of C-IBS and D-IBS patients compared with healthy control subjects. Fold change for healthy control subjects is +1 or −1, represented by blue graphs, respective fold changes for C-IBS and D-IBS patients are represented by orange graph.
SMCT1
The relative expression of SLC5A8 mRNA was significantly reduced for both C-IBS (p=0.041) and D-IBS (p=0.02) compared to the healthy control group.
SLC16A1 and SLC5A8 were significantly down-regulated in C-IBS with low butyrate concentration levels, suggesting that reduced amounts of SCFAs were transported into the colonocyte. Similarly, SLC16A1 and SLC5A8 were also significantly down-regulated in the presence of ostensibly normal butyrate concentration levels in D-IBS. The monocarboxylate transporters, MCT1 and SMCT1 were significantly reduced in both IBS groups.
DISCUSSION
This study evaluated SCFAs in IBS in combination with their transporters. The results of this study confirmed significant derangement in the SCFA concentrations and their relative ratios of expression. The results further showed significant reduction in the 2 monocarboxylate transporters tested, suggesting that the SCFAs are not transported into the colonocyte where they are active.
Dysbiosis or altered gut microbiota diversity has been documented in many IBS studies, and results from this study confirmed significant differences in the two microbiota quantified, supporting dysbiosis previously described (25, 26). Our results are consistent with those of Parker et al. (27) who found a significant increase in Bacteroides and a significant decrease in Bifidobacteria when rectal biopsies of IBS patients were compared with those of healthy controls.
Fecal Bifidobacterium was significantly reduced in both C-IBS and D-IBS in this study and various other studies (28, 29). As a modulator of abdominal pain and bloating in IBS, one can speculate that the reduced concentration of Bifidobacteria may explain, to some extent, these symptoms in this patient population. For Bacteroides, results from various studies have been mixed with unchanged or reduced levels in C-IBS and increased levels in D-IBS (28–30). Results from our study confirmed the same trend. It is apparent that a complex interaction exists between gut microbiota, diet, and SCFAs in health and disease. Although we have not included diet as a measure in our study, it has a profound effect on gut microbiota composition and SCFA production. This association should be prospectively studied. Currently, however, on a larger scale, the results of studies on the intestinal microbiota of IBS subjects are inconsistent and occasionally contradictory (26).
SCFA ratios were significantly altered in both C-IBS and D-IBS compared to healthy controls. As SCFAs are a direct result of gut microbiota fermentation, one can reliably deduce that this altered SCFA ratio resulted from dysbiosis. As previously shown, altered SCFA concentrations affect serotonin expression which is known to impact IBS.
Results from this study showed significant reduction in all three major SCFAs, i.e., acetate, propionate, and butyrate in patients with C-IBS. However, a significant decrease was only observed for acetate in D-IBS patients compared with that in healthy controls. An increase in propionate was also noted in our patients with D-IBS, but not C-IBS compared with healthy controls. This result is not surprising as propionate is known to stimulate peristalsis. It is interesting to note that Bacteroides, one of the key synthesizers of propionate, was also found to be increased in D-IBS. Butyrate, a key promoter of colonic health and the main energy provider for the colonocyte, was significantly reduced in C-IBS compared to the healthy control group, but its concentration remained unchanged in D-IBS. These results are similar to those of Ringel-Kulka et al. and others (31–34).
Normal gut function is influenced not only by the amount and relative abundance of various SCFAs but also by the ratio of one SCFA to the other. From various studies, it appears that the propionate: butyrate ratio is an important biomarker for IBS (16, 34). In our study, the propionate: butyrate ratio in healthy controls was 0.75, whereas in C-IBS and D-IBS, they were 1.45 and 1.07, respectively. It appears these ratios are important in disease states and may as well be useful adjuncts to current investigations in IBS and other disease states like CRC and IBD.
Individual SCFAs have varied functions in maintaining colon health and function. The transport of the SCFAs from the lumen to the colonocyte is therefore important. During homeostasis, SCFAs are transported into the colonocyte mainly via MCT1 (SLC 16A1) or SMCT1 (SLC 5A8) transporters (35–37). Therefore, as previously shown, reduced MCT1 and MCT1 result in a reduced butyrate uptake by the colonocyte (38). However, a recent study in mice, found that MCT1 is up-regulated in the presence of butyrate (39). There is a significant increase in the expression of these transporters in various cancer types, including colorectal cancer, but no studies on these transporters have been performed in IBS (37). In our study, the mRNA expression of both of these transporters was significantly reduced in both IBS groups, in the presence of reduced SCFA, especially in C-IBS. It is not clear whether this is a cause or a consequence of the underlying disease process. Logically, reduced SCFA levels combined with a reduction in the monocarboxylate transporters will result in reduced SCFA uptake into the colonocyte. Although the reduced SCFA concentration resulted from microbiota dysbiosis, the reason for the reduced transporter expression is not clear and needs further investigation. Goncalves et al. showed that butyrate uptake was reduced by inhibition of MCT1 and SMCT by primary bile salts, especially chenodeoxycholic acid, which is abundant in the terminal ileum and proximal colon (20). Another study showed an increase in fecal primary bile salts in patients with D-IBS (29). The effect of primary bile salts on SCFA transporters and SCFA uptake and a possible link to IBS symptoms needs further exploration.
This study focused on SCFA concentrations and their transporters in patients with IBS. Conclusive remarks about the above were hampered by two limitations in the study. First, there was no diet intervention or patient-kept dietary record in this study. Diet has a direct influence on gut microbiota and hence on SCFA production. However, evidence showed that unless drastic, short-term dietary modifications only have a modest effect on the microbiome (40). However, causal associations between the parameters studied cannot be inferred. Second, we did not investigate an exhaustive list of gut microbiota. Current IBS literature states that dysbiosis is established in IBS. Therefore, only two microbiota were analyzed quantitatively to corroborate previous findings of these microbiota in IBS.
To conclude, results from this study confirmed gut microbiota derangement in patients with IBS. Furthermore, we showed a reduction and disequilibrium of SCFAs in both IBS groups. Monocarboxylate transporters also significantly reduced in the presence of low SCFA levels, suggesting reduced SCFA uptake into the colonocyte. These factors may be important in IBS pathophysiology and require prospective investigation. Such studies should include a bigger study population, dietary analysis, a comprehensive and targeted microbiota analysis, and primary bile acids in the context of SCFA and monocarboxylate transporters in IBS.
MAIN POINTS.
The microbiota studied in this IBS population showed significant derangement inferring dysbiosis as a possible aetiogenic factor.
SCFA’s, a product of microbiota fermentation, and their ratios were significantly altered in both C- and D-IBS and an association with symptoms can be readily deduced.
Reduction in the SCFA transporters as showed in this study implies that SCFA do not reach their target in IBS and may have causal links.
Acknowledgements
We want to thank Dr Sharlene Govender from the Nelson Mandela University for the technical assistance with microbiota extraction and analysis
Footnotes
Presented in: This study was presented in the SAGES (South African Gastroenterology Association) Annual Congress, August, 2015. (Best Oral Presentation).
Ethics Committee Approval: This study was approved by the Human Ethics Committee of the Nelson Mandela University (NMU) (H07SciBCM-001) in 2012.
Informed Consent: Written informed consent was obtained from the patients who participated in this study.
Peer-review: Externally peer-reviewed.
Author Contributions: Concept – E.S., S.R.; Design – E.S., R.T., S.R.; Supervision – S.R.; Resource – E.S., S.R.; Materials – E.S., R.T., S.R.; Data Collection and/or Processing – E.S., R.T.; Analysis and/or Interpretation – E.S., R.T., S.R.; Literature Search – E.S., S.R.; Writing – E.S., S.R.; Critical Reviews – S.R.
Conflict of Interest: The authors have no conflict of interest to declare.
Financial Disclosure: National Research Foundation South Africa; Grant number: 2075266.
REFERENCES
- 1.James SC, Fraser K, Young W, McNabb WC, Roy NC. Gut Microbial Metabolites and Biochemical Pathways Involved in Irritable Bowel Syndrome: Effects of Diet and Nutrition on the Microbiome. J Nutr. 2020;150:1012–21. doi: 10.1093/jn/nxz302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Enck P, Aziz Q, Barbara G, et al. Irritable bowel syndrome. Nat Rev Dis Primers. 2016;2:16014. doi: 10.1038/nrdp.2016.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soares RL. Irritable bowel syndrome: a clinical review. World J Gastroenterol. 2014;20:12144–60. doi: 10.3748/wjg.v20.i34.12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Canakis A, Haroon M, Weber HC. Irritable bowel syndrome and gut microbiota. Curr Opin Endocrinol Diabetes Obes. 2020;27:28–35. doi: 10.1097/MED.0000000000000523. [DOI] [PubMed] [Google Scholar]
- 5.Rodino-Janeiro BK, Vicario M, Alonso-Cotoner C, Pascua-García R, Santos J. A Review of Microbiota and Irritable Bowel Syndrome: Future in Therapies. Adv Ther. 2018;35:289–310. doi: 10.1007/s12325-018-0673-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lopetuso LR, Petito V, Graziani C, et al. Gut Microbiota in Health, Diverticular Disease, Irritable Bowel Syndrome, and Inflammatory Bowel Diseases: Time for Microbial Marker of Gastrointestinal Disorders. Dig Dis. 2018;36:56–65. doi: 10.1159/000477205. [DOI] [PubMed] [Google Scholar]
- 7.Tan J, McKenzie C, Potamitis M, Thorburn AN, Mackay CR, Macia L. The role of short-chain fatty acids in health and disease. Adv Immunol. 2014;121:91–119. doi: 10.1016/B978-0-12-800100-4.00003-9. [DOI] [PubMed] [Google Scholar]
- 8.Laing B, Barnett MPG, Marlow G, Nasef NA, Ferguson LR. An update on the role of gut microbiota in chronic inflammatory diseases, and potential therapeutic targets. Expert Rev Gastroenterol Hepatol. 2018;12:969–83. doi: 10.1080/17474124.2018.1505497. [DOI] [PubMed] [Google Scholar]
- 9.Kasubuchi M, Hasegawa S, Hiramatsu T, Ichimura A, Kimura I. Dietary gut microbial metabolites, short-chain fatty acids, and host metabolic regulation. Nutrients. 2015;7:2839–49. doi: 10.3390/nu7042839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bultman SJ. Molecular pathways: gene-environment interactions regulating dietary fiber induction of proliferation and apoptosis via butyrate for cancer prevention. Clin Cancer Res. 2014;20:799–803. doi: 10.1158/1078-0432.CCR-13-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun Q, Jia Q, Song L, Duan L. Alterations in fecal short-chain fatty acids in patients with irritable bowel syndrome: A systematic review and meta-analysis. Medicine (Baltimore) 2019;98:e14513. doi: 10.1097/MD.0000000000014513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reigstad CS, Salmonson CE, Rainey JF, 3rd, et al. Gut microbes promote colonic serotonin production through an effect of short-chain fatty acids on enterochromaffin cells. FASEB J. 2015;29:1395–403. doi: 10.1096/fj.14-259598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grider JR, Piland BE. The peristaltic reflex induced by short-chain fatty acids is mediated by sequential release of 5-HT and neuronal CGRP but not BDNF. Am J Physiol Gastrointest Liver Physiol. 2007;292:G429–37. doi: 10.1152/ajpgi.00376.2006. [DOI] [PubMed] [Google Scholar]
- 14.Macfarlane S, Macfarlane GT. Regulation of short-chain fatty acid production. Proc Nutr Soc. 2003;62:67–72. doi: 10.1079/PNS2002207. [DOI] [PubMed] [Google Scholar]
- 15.Mitsui R, Ono S, Karaki S, Kuwahara A. Neural and non-neural mediation of propionate-induced contractile responses in the rat distal colon. Neurogastroenterol Motil. 2005;17:585–94. doi: 10.1111/j.1365-2982.2005.00669.x. [DOI] [PubMed] [Google Scholar]
- 16.Farup PG, Rudi K, Hestad K. Faecal short-chain fatty acids - a diagnostic biomarker for irritable bowel syndrome? BMC Gastroenterol. 2016;16:51. doi: 10.1186/s12876-016-0446-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kannampalli P, Shaker R, Sengupta JN. Colonic butyrate-algesic or analgesic? Neurogastroenterol Motil. 2011;23:975–9. doi: 10.1111/j.1365-2982.2011.01775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pozuelo M, Panda S, Santiago A, et al. Reduction of butyrate- and methane-producing microorganisms in patients with Irritable Bowel Syndrome. Sci Rep. 2015;5:12693. doi: 10.1038/srep12693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thibault R, Blachier F, Darcy-Vrillon B, de Coppet P, Bourreille A, Segain JP. Butyrate utilization by the colonic mucosa in inflammatory bowel diseases: a transport deficiency. Inflamm Bowel Dis. 2010;16:684–95. doi: 10.1002/ibd.21108. [DOI] [PubMed] [Google Scholar]
- 20.Goncalves P, Catarino T, Gregorio I, Martel F. Inhibition of butyrate uptake by the primary bile salt chenodeoxycholic acid in intestinal epithelial cells. J Cell Biochem. 2012;113:2937–47. doi: 10.1002/jcb.24172. [DOI] [PubMed] [Google Scholar]
- 21.den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud DJ, Bakker BM. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res. 2013;54:2325–40. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yap KW, Mohamed S, Abd Manap YM, Maznah I, Meyer D. Dose-response effects of inulin on the faecal short-chain fatty acids content and mineral absorption of formula-fed infants. Nutr Food Sci. 2005;35:208–19. doi: 10.1108/00346650510605603. [DOI] [Google Scholar]
- 23.Scheppach WM, Fabian CE, Kasper HW. Fecal short-chain fatty acid (SCFA) analysis by capillary gas-liquid chromatography. Am J Clin Nutr. 1987;46:641–6. doi: 10.1093/ajcn/46.4.641. [DOI] [PubMed] [Google Scholar]
- 24.Vossaert L, O’Leary T, van Neste C, et al. Reference loci for RT-qPCR analysis of differentiating human embryonic stem cells. BMC Molecular Biology. 2013;14:1–7. doi: 10.1186/1471-2199-14-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bennet SM, Ohman L, Simren M. Gut microbiota as potential orchestrators of irritable bowel syndrome. Gut Liver. 2015;9:318–31. doi: 10.5009/gnl14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong SN, Rhee PL. Unraveling the ties between irritable bowel syndrome and intestinal microbiota. World J Gastroenterol. 2014;20:2470–81. doi: 10.3748/wjg.v20.i10.2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parkes GC, Rayment NB, Hudspith BN, et al. Distinct microbial populations exist in the mucosa-associated microbiota of sub-groups of irritable bowel syndrome. Neurogastroenterol Motil. 2012;24:31–9. doi: 10.1111/j.1365-2982.2011.01803.x. [DOI] [PubMed] [Google Scholar]
- 28.Gobert AP, Sagrestani G, Delmas E, et al. The human intestinal microbiota of constipated-predominant irritable bowel syndrome patients exhibits anti-inflammatory properties. Sci Rep. 2016;6:39399. doi: 10.1038/srep39399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duboc H, Rainreau D, Rajca S, et al. Increase in fecal primary bile acids and dysbiosis in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol Motil. 2012;24:513–20. e246–7. doi: 10.1111/j.1365-2982.2012.01893.x. [DOI] [PubMed] [Google Scholar]
- 30.Chassard C, Dapoigny M, Scott KP, et al. Functional dysbiosis within the gut microbiota of patients with constipated-irritable bowel syndrome. Aliment Pharmacol Ther. 2012;35:828–38. doi: 10.1111/j.1365-2036.2012.05007.x. [DOI] [PubMed] [Google Scholar]
- 31.Rangel I, Sundin J, Fuentes S, Repsilber D, de Vos WM, Brummer RJ. The relationship between faecal-associated and mucosal-associated microbiota in irritable bowel syndrome patients and healthy subjects. Aliment Pharmacol Ther. 2015;42:1211–21. doi: 10.1111/apt.13399. [DOI] [PubMed] [Google Scholar]
- 32.Beards E, Tuohy K, Gibson G. Bacterial, SCFA and gas profiles of a range of food ingredients following in vitro fermentation by human colonic microbiota. Anaerobe. 2010;16:420–5. doi: 10.1016/j.anaerobe.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 33.Casen C, Vebø HC, Sekelja M, et al. Deviations in human gut microbiota: a novel diagnostic test for determining dysbiosis in patients with IBS or IBD. Aliment Pharmacol Ther. 2015;42:71–83. doi: 10.1111/apt.13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ringel-Kulka T, Choi CH, Temas D, et al. Altered Colonic Bacterial Fermentation as a Potential Pathophysiological Factor in Irritable Bowel Syndrome. Am J Gastroenterol. 2015;110:1339–46. doi: 10.1038/ajg.2015.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lecona E, Olmo N, Turnay J, et al. Kinetic analysis of butyrate transport in human colon adenocarcinoma cells reveals two different carrier-mediated mechanisms. Biochem J. 2008;409:311–20. doi: 10.1042/BJ20070374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iwanaga T, Kishimoto A. Cellular distributions of monocarboxylate transporters: a review. Biomed Res. 2015;36:279–301. doi: 10.2220/biomedres.36.279. [DOI] [PubMed] [Google Scholar]
- 37.Ferro S, Azevedo-Silva J, Casal M, Corte-Real M, Baltazar F, Preto A. Characterization of acetate transport in colorectal cancer cells and potential therapeutic implications. Oncotarget. 2016;7:70639–53. doi: 10.18632/oncotarget.12156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goncalves P, Martel F. Butyrate and colorectal cancer: the role of butyrate transport. Curr Drug Metab. 2013;14:994–1008. doi: 10.2174/1389200211314090006. [DOI] [PubMed] [Google Scholar]
- 39.Borthakur A, Priyamvada S, Kumar A, et al. A novel nutrient sensing mechanism underlies substrate-induced regulation of monocarboxylate transporter-1. Am J Physiol Gastrointest Liver Physiol. 2012;303:G1126–33. doi: 10.1152/ajpgi.00308.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rajilic-Stojanovic M, Jonkers DM, Salonen A, et al. Intestinal microbiota and diet in IBS: causes, consequences, or epiphenomena? Am J Gastroenterol. 2015;110:278–87. doi: 10.1038/ajg.2014.427. [DOI] [PMC free article] [PubMed] [Google Scholar]