Abstract
More than 30% of the world population suffers from allergy. Allergic individuals are characterized by the production of immunoglobulin E (IgE) antibodies against innocuous environmental allergens. Upon allergen recognition IgE mediates allergen-specific immediate and late-phase allergic inflammation in different organs. The identification of the disease-causing allergens by demonstrating the presence of allergen-specific IgE is the key to precision medicine in allergy because it allows tailoring different forms of prevention and treatment according to the sensitization profiles of individual allergic patients. More than 30 years ago molecular cloning started to accelerate the identification of the disease-causing allergen molecules and enabled their production as recombinant molecules. Based on recombinant allergen molecules, molecular allergy diagnosis was introduced into clinical practice and allowed dissecting the molecular sensitization profiles of allergic patients. In 2002 it was demonstrated that microarray technology allows assembling large numbers of allergen molecules on chips for the rapid serological testing of IgE sensitizations with small volumes of serum. Since then microarrayed allergens have revolutionized research and diagnosis in allergy, but several unmet needs remain. Here we show that detection of IgE- and IgG-reactivity to a panel of respiratory allergens microarrayed onto silicon elements is more sensitive than glass-based chips. We discuss the advantages of silicon-based allergen microarrays and how this technology will allow addressing hitherto unmet needs in microarray-based allergy diagnosis. Importantly, it described how the assembly of silicon microarray elements may create different microarray formats for suiting different diagnostic applications such as quick testing of single patients, medium scale testing and fully automated large scale testing.
Keywords: allergy, allergen, IgE, molecular diagnosis, microarrayed allergens, allergen chip, precision medicine
Background
The major difference between allergic patients and healthy, non-allergic subjects is that allergic patients produce IgE antibodies against certain environmental antigens, termed allergens, whereas non-allergic subjects produce IgG antibodies (1, 2). IgE antibodies bind specifically to high (FcεRI) and low affinity receptors (FcεRII) for IgE present on inflammatory cells which become activated by IgE-allergen immune complexes to release inflammatory mediators, cytokines, and proteases and/or to activate allergen-specific T cells (3, 4). Therefore, allergen contact induces in allergic patients containing specific IgE, allergic inflammation in different tissues and organs leading to a variety of allergic symptoms comprising rhinoconjunctivitis (hay fever), asthma, skin inflammation, gastrointestinal symptoms, and systemic symptoms such as anaphylactic shock (5). By contrast, IgG recognition of allergens does not trigger allergic inflammation because allergen-IgG immune complexes cannot bind to Fcε receptors and thus fail to trigger allergic inflammation. IgE antibodies occur in very small concentrations in the blood and therefore were identified only in 1966 (6). Due to their importance for triggering allergic reactions already in 1967 the first serological test for measuring allergen-specific IgE in the blood of allergic patients was developed and termed radioallergosorbent test (RAST) (7). Before the discovery of IgE antibodies, allergic sensitization was diagnosed by exposing subjects with suspected allergic sensitization to extracts made from the disease-causing allergen sources in order to study if this would induce immediate allergic inflammation. One of the first descriptions of controlled allergen provocation dates back to a study performed by Charles Blackley in 1873 (8). Since the induction of allergic inflammation resulting from the activation of mast cells by IgE-allergen immune complexes occurs within few minutes, IgE-associated allergy was also termed immediate type hypersensitivity in the classical description of the four types of immunological hypersensitivity of the immune system published by Coombs & Gell (9). Accordingly the diagnosis of allergy has been based on three elements, one is the case history trying to relate the occurrence of allergic symptoms in a patient to exposure to certain allergen sources; the second element is trying to induce allergic reactions in the patient by exposing the person to the allergen source and recording of subsequent allergic symptoms; and the third by confirming IgE sensitization by demonstrating the presence of IgE antibodies specific for the allergen source in the blood or tissue fluids of the patient (10). Traditionally, testing is performed exactly in the described order by starting with the anamnesis followed by provocation testing and final confirmation of sensitization by measuring specific IgE antibodies.
Traditional Forms of Allergy Diagnosis
Traditional allergy diagnosis always starts with a detailed anamnesis trying to identify the presence or absence of allergic symptoms. The next step is to try associating the occurrence of symptoms with contact to certain allergen sources and to verify that controlled exposure to allergen extracts prepared from the allergen source will elicit an allergic reaction. For this purpose, allergen extracts are prepared from the natural allergen sources. These allergen extracts represent mixtures of allergenic and non-allergenic, potentially also irritating substances which may elicit an inflammatory reaction without underlying IgE sensitization. Some examples are the presence of histamine in fish or adverse reactions to milk due to lactose intolerance (11). Accordingly the next step for confirming the condition of an IgE-associated allergy is to verify that the patient serum contains IgE antibodies which react specifically with the allergen extract. However, the demonstration of the presence of allergen-specific IgE with allergen extracts is problematic. First of all, the disease-causing allergen molecules cannot be identified with allergen extracts because they represent mixtures of different allergen molecules and non-allergenic materials. Furthermore, the quality of allergen extracts may strongly vary and depend on various factors which are out of the control of the manufacturer. For example, certain allergens may be lacking in certain extracts (12) and there may be contaminations with allergens from other sources just to name a few problems. It also seems that allergen extracts for in vivo provocation testing are becoming less available because they do not meet current standards for medical products (13). Accordingly the use of allergen extracts has several disadvantages which are reviewed in (14). In order to address the urgent needs and bottlenecks of diagnostic allergens the European Academy of Allergy and Clinical Immunology (EAACI) has founded a task force which has set up an action plan to maintain the supply of diagnostic allergen extracts (15). This action plan comprises i.) a simplification of authorization, ii.) specific regulations for special types of extracts, iii.) new models beyond the current model of homologous allergens, iv.) simplification of pharmacovigilance reporting, v.) reduction of regulation fees and vi.) reimbursement for diagnostic allergen extracts. Nevertheless, the practice of traditional allergy diagnosis becomes now challenged with the appearance of molecular allergy diagnosis.
Molecular Allergy Diagnosis
Shortly after the first allergen-encoding DNAs had been isolated (16–18), the first two studies were published showing that one can replace complex allergen extracts such as birch pollen or grass pollen extract with a few defined recombinant allergen molecules for IgE-based serological diagnosis without losing sensitivity or specificity (19, 20). These results were quite surprising because at that time it was thought that allergen molecules may exist in different isoforms with variable IgE reactivity and that it may be impossible to find one isoform which would be suitable for the diagnosis in all patients (21, 22). Furthermore, it was not clear if a few allergen molecules would be sufficient to cover the IgE epitopes of whole allergen extracts. The early studies performed with recombinant allergens for diagnosis also indicated that patients who are sensitized to a certain allergen source may react with different molecules in this source and accordingly show different clinical phenotypes that need “patient-tailored treatment concepts” (19). Subsequently, an increasing number of allergen molecules from different sources were produced and recombinant allergens became available for the first time in the most commonly used fully automated in vitro allergy diagnosis system, the ImmunoCAP system (23). However, the principle of ImmunoCAP testing was that one test provided only one information so that for each allergen source several allergen molecule-based tests would be needed to cover the spectrum of the allergens of the allergen source. Thus molecular testing with single allergen molecules would have increased the costs for testing considerably. It was therefore, clear that using this test one would always start testing for allergen-specific IgE with allergen extracts and only if deemed necessary and affordable, one would continue with molecular testing.
In order to utilize the increasing numbers of allergen molecules which were developed by time for diagnostic testing, new test platforms were needed. It was a co-incidence that microarray technology became available for printing nucleic acids onto chips leading at that time. The first DNA chips were manufactured by the company Affymetrix which was based in Santa Clara, California, and a similar technology was applied by the Vienna start-up company VBC Genomics headed by Manfred Müller from Vienna, Austria which had the instruments for printing microarrays. In a collaboration between Manfred Müller and Rudolf Valenta, which was the first to develop chips containing microarrayed allergen molecules were then developed (24). which were not only one of the first protein arrays for diagnostic purposes but also represented the first microarrays for in vitro allergy diagnosis (24). These micro-arrays contained more than 90 different allergen molecules from different allergen sources provided by researchers from all over the world. The exciting thing with allergen microarrays was that one could test IgE reactivity simultaneously to a large number of different allergen molecules with a few microliters of serum or other body fluids. Compared to other existing allergy test systems the allergen chip thus represented a breakthrough which may be also considered a “disruptive technology” because it had the potential to change allergy diagnosis completely. One year after the appearance of the study describing the first allergen chip, the concept of microarray-aided allergy diagnosis was considered the first time (25).
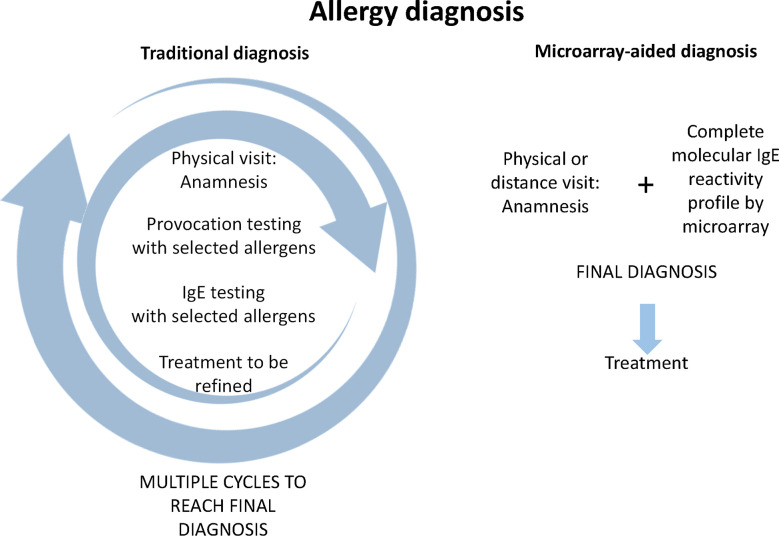
Figure 1 provides a comparison of traditional allergy diagnosis, which in principle is an approach driven by the hypothesis developed by the physician based on the anamnesis. According to the information collected by the anamnesis the physician selects certain allergen sources for a first round of in vivo provocation testing which is usually conducted by skin testing, and eventually the confirmation of IgE sensitization by serology is performed. According to the in vivo provocation results which can be evaluated during the first visit, the physician may already make first preliminary treatment recommendations. Results from serology are usually not available on the same day, and often it may turn out that the original hypothesis, what the culprit allergens might have been, may need refinement through additional anamnesis and repeated testing (Figure 1, left). Therefore traditional allergy diagnosis requires usually multiple visits until a more or less complete picture of the sensitization profile of the patient has been obtained. It is needless to say that the traditional allergy testing path may become challenging because it may require repeated consultations and thus long time to reach a final diagnosis, and this may reduce patients’ compliance (Figure 1).
Figure 1.
Comparison of traditional allergy diagnosis and microarray-aided allergy diagnosis. In traditional allergy diagnosis (left part) an anamnesis of allergic symptoms is recorded which serves as the basis for targeted provocation testing, usually skin testing with a limited number of allergen extracts selected according to the anamnesis and eventually collection of a serum sample for measuring IgE specific for the suspected allergen sources. In the best case, the patient receives first treatment suggestions according to skin test results. Usually at least one, but often several additional visits are necessary to adjust the treatment to the IgE test results and/or to conduct further targeted testing to determine more precisely the patients sensitization profile and to further adjust treatment. Regarding microarray-aided allergy diagnosis (right part), it can be envisioned that the first visit can be conducted even in a virtual, telemedicine-like form because no provocation testing is required. The anamnesis and complete molecular IgE reactivity profile would be available to the specialist online who could then prescribe precise treatment taking clinical information and the complete sensitization profile into consideration.
By contrast, microarray-aided allergy diagnosis has the potential to reduce the time to reach the final diagnosis and to select the correct treatment albeit it may be initially more expensive because it would place the assessment of the complete sensitization profile of the patient already at the beginning of the first consultation. One could imagine that based on validated allergy questionnaires such as the ISAAC questionnaire (26) already the general practitioner (GP) initiates microarray-based allergy screening in subjects with suspected allergy. The patient could then have a first consultation with a specialist presenting already the anamnesis and the complete molecular IgE reactivity profile so that the specialist can immediately determine the best treatment practice. In contrast to the traditional “hypothesis-driven approach”, this pathway is reminiscent of a “discovery approach” because the comprehensive IgE test result might stimulate the specialist to refine the anamnesis according to the IgE sensitization profile. Obviously the microarray-aided diagnosis would need in vivo provocation testing only in certain cases for confirmatory purposes, if at all and hence the consultation could be performed also in a telemedicine-based format. The advantage of such a telemedicine-based approach is that it reduces the number of visits, avoids eventual long travelling, and remote areas in a country can easily benefit from specialist knowledge (Figure 1, right). In this context the possibilities of mobile health (mHealth) technology using mobile communication devices to support and improve health-related services, data flow, patient management, surveillance, and disease management should be mentioned. A task force of EAACI has reviewed all these possibilities, discussed advantages, limitations, and risks such as data protection, and provided recommendations (27). In the context of precision medicine, the potential of changing the practice from clinician- to patient-centered health care is highlighted.
Some recent reports underline the potential of molecular diagnosis in the field of health economics and suggest that it can help to save costs for the diagnosis of respiratory allergies (28) and food allergies (29).
Currently, allergy diagnosis is still dominated by the traditional approach of diagnosis as indicated in Figure 1, but more and more specialists in allergy start to use molecular allergy diagnosis, and already quite a few prefer microarray-aided diagnosis as shown also in Figure 1. There are many different reasons for the different preferences. For example, traditional allergy diagnosis is often preferred because it is currently reimbursed by the health care system, whereas serological diagnosis and, in particular microarray diagnosis, requires laboratory facilities and a different mode of reimbursement shared between specialists and diagnostic laboratories. Factors limiting the increased use of microarray-guided diagnosis are that practitioners must be skilled in interpreting complex molecular test results and/or have well-trained algorithms for clinical decision making (i.e., clinical decision support systems) available. Information on sensitizations not linked to symptoms require appropriate and time-consuming information of patients, and certain allergen molecules for obtaining complete results may still need to be discovered and included in the micro-arrays. Finally, it will be necessary to reduce the costs of micro-arrays, which are often driven not only by costs of goods for manufacturing but also by costs due to quality control, validation, and registration. However, once the latter issues are resolved one may expect that microarray-based allergy diagnosis will become a highly cost-effective diagnostic tool. One must consider that microarray-based allergy diagnostic approaches can provide more than hundred individual test results from one sample and present the comprehensive picture of allergen sensitization, which can be used for precision medicine treatment. In fact, unrecognized and untreated allergy is a major cost factor for the management of allergic diseases. With the adherence to proper treatment and the precise diagnosis and proper management of allergic diseases, it has been estimated that high costs can be saved (30, 31).
Measurement of Allergen-Specific IgE and IgG Responses With Microarrayed Allergens
Microarrayed allergens can be used for measuring simultaneously IgE reactivity to a large number of different allergen molecules with very small volumes of serum. In this context, it should be mentioned that it has been shown that allergen-specific IgE can be also measured in plasma and other body fluids such as nasal secretions and in milk samples (32–35). Moreover, it has been shown that dried blood spots on paper can be recovered for specific IgE and IgG measurements which allows sending serum samples as paper dried blood spots in simple envelops without requiring expensive packaging, cooling, and sophisticated transportation (36). Several recent studies have confirmed the importance of allergen-specific IgG antibodies for the protective effects of AIT, and the measurement of allergen-specific IgG antibodies which block allergic patients IgE binding to allergens is therefore considered as an important biomarker for the clinical efficacy of AIT (37). Accordingly, certain commercial allergen arrays allow measuring specific IgE and IgG antibodies (e.g. Thermo Fisher ImmunoCAP ISAC Immuno-solid-phase Allergen Chip, which contains 112 allergens from 51 allergen sources) (38); however, this has not been shown for all available allergen arrays (e.g., MADx Allergen Explorer ALEX; containing 282 allergens: 156 extracts and 126 components) (39). The measurement of allergen-specific IgG antibodies in cohorts has provided new insights in beneficial functions of IgG. For example it has been suggested that allergen-specific IgG transmitted from the mother to the child via the placenta during pregnancy may protect the off-spring from allergic sensitization (40). Likewise, evidence has been provided that the production of allergen-specific IgG antibodies follows different pathways and mechanisms than those involved in the production of allergen-specific IgE antibodies (2, 41–43).
Examples for the Use of Microarrayed Allergens in Allergy Research
Microarrayed allergens have been used in research to address several questions. For example, it has been shown that adult allergic patients do not change their IgE sensitization profiles for a decade demonstrating that there is no acquisition or loss of IgE sensitizations in adult allergic patients (44). Furthermore, information was obtained about the characteristics of IgE sensitization profiles in different populations. For example, it was found that sensitization to clinically relevant grass pollen allergens is rare in tropical climates and that most of the grass pollen-specific IgE is directed to non-allergenic carbohydrate epitopes (45). In another study it was observed that man-made changes of the environment as for example obtained by replanting of certain plants can alter the allergic sensitization profiles towards plant-derived allergens in populations within two generations (46).
However, without any doubt, the most exciting results were obtained when microarrayed allergen molecules were used to study the development of the allergic sensitization profiles in longitudinal birth cohorts allowing to analyze the evolution of IgE sensitization profiles from birth to early adolescence (47, 48). Importantly it was found that allergy frequently starts with early asymptomatic IgE sensitization and that early assessment of IgE sensitization profiles and IgE-levels allow predicting the development of allergy later in life (49–52). This finding suggests that for allergy similar as for other diseases such as cancer, cardiovascular diseases, and metabolic disease, early screening in the form of a preventive medical examination by determination of IgE sensitization profiles early in life might allow initiating preventive measures (e.g., allergen avoidance, early allergen-specific immunotherapy) (53–55) to prevent the development of allergic symptoms (i.e., secondary prevention) later in life.
Precision Medicine by Chip-Based Allergy Diagnosis
In allergy, like in other important diseases, it has become clear that it is necessary to transform healthcare towards the principles of “P4 Medicine” for predictive, preventive, personalized (precision), and participatory medicine by developing new diagnostic and predictive tests as well as therapies and preventive strategies which affect the course of disease or prevent the development of disease (56). Allergy is ideally suitable for a precision medicine approach because patients are sensitized to different allergens and allergen combinations and suffer from a wide variety of symptoms which may change during the course of disease. Furthermore, there are several different strategies for the treatment of allergy available which require the identification of the disease allergens. It is also clear that early preventive measures should be more effective than late mending of severe disease (5, 57). Accordingly, it has been suggested that molecular allergy diagnosis improves treatment especially in pediatric allergy (58). In this context, examples of how molecular diagnosis helped in the diagnosis of children suffering from complex allergic sensitizations and tailoring the treatment according to the needs of the children should be mentioned (59). Furthermore, evidence accumulates that allergic phenotypes and symptoms are associated with certain patterns and/or levels of allergen-specific IgE in children and adult allergic patients suggesting that serological surrogate parameters for diagnosis can be developed (60–63).
Microarrayed Allergens in Food and Respiratory Allergy
The diagnosis of food allergy is often challenging because the frequency of IgE-associated food allergy is often considered higher than it is in reality. For example, adverse reactions to cow’s milk due to lactose intolerance are much more common than IgE-mediated allergy (11). Although many clinicians consider only the double-blind, placebo-controlled food challenge as gold standard for the diagnosis of food allergy, this test is cumbersome, and there may be severe and even life-threatening side effects. Accordingly, alternative diagnostic tests are needed. So far a considerable number of food allergen molecules have been identified which are associated with severe, mild, or even no reactions allowing for serological testing of food allergy also by microarray-based IgE testing (64).
In this context a study demonstrating different IgE sensitization profiles in children suffering from severe peanut allergy and in peanut-sensitized but asymptomatic children should be mentioned (65). Screening for IgE sensitizations using a large panel of food allergen molecules is useful for several reasons. First, it allows testing simultaneously for IgE sensitization to a large panel of allergen molecules with high anaphylactic capacity to predict the risk of food allergy, and thus it may help to reduce hazardous food challenge testing (29, 66, 67). Second, and importantly, negative test results to a large panel of food allergen molecules are helpful in searching for other reasons of food intolerance beyond IgE-mediated allergy. Besides IgE testing to food allergen molecules, it has turned out that measuring IgE sensitizations to food allergen-derived peptides may be useful to discriminate patients with no or mild symptoms from those suffering from severe symptoms (68, 69). Microarrayed peanut allergen molecules were also used to investigate the course of peanut sensitization in childhood and to predict symptomatic peanut allergy in a birth cohort (70). Very recently it was found that oral allergy syndrome (OAS) to Bet v 1-related food allergen molecules of the pathogen-related protein 10 (PR10) family was associated with the levels of Bet v 1-specific IgE and the numbers of recognized PR10 molecules (61).
Microarrayed allergen molecules are not only useful for the diagnosis of IgE-mediated food allergies but are also considered for the diagnosis of asthma triggered by allergens in sensitized allergic patients (71). The two major trigger factors for asthma are allergens for patients with IgE sensitizations and infections with respiratory viruses, in particular with rhinoviruses (RVs) (72). For example, it has been shown for house dust mite allergy that children suffering from allergic asthma differ regarding their IgE reactivity profiles and ability to produce allergen-specific IgG antibodies (60). Children with asthma showed higher IgE levels to certain allergens, reacted with a larger number of molecules, and produced less allergen-specific IgG as compared to children suffering only from rhinitis (60). Furthermore, machine learning approaches have been suggested to identify pairwise interactions of IgE antibodies and their association with asthma based on microarray results (73). Allergen molecules from cat and dog, which are important for the development of respiratory allergy in childhood, have been identified in the Swedish BAMSE birth cohort (74).
For the diagnosis of RV-induced asthma, a chip containing peptides derived from the N-terminus of VP1 proteins from a representative number of RV strains covering RV-A, RV-B and RV-C species has been produced (75). This chip allowed measuring species-specific increases of RV-specific IgG antibodies in children who had experienced RV-induced asthma exacerbations, and cumulative IgG responses were higher in children with RV-induced exacerbations of respiratory illnesses (76, 77). Accordingly, it has been proposed to use microarrayed allergens and respiratory virus-derived peptides for diagnosis of allergen and/or RV-induced asthma and personalized treatment according to the test results (77).
Microarrayed Allergens for Prescription and Monitoring of Allergen-Specific AIT
AIT is an allergen-specific form of therapeutic vaccination which is based on the administration of the disease-causing allergens or modifications thereof with the goal to induce allergen-specific protective IgG antibodies and alterations of the cellular immune response to reduce symptoms of allergy upon allergen contact. Accordingly, the accurate prescription of AIT requires that the culprit allergens are identified. Since allergen sources often contain cross-reactive allergens the identification of the culprit allergen source can be challenging. It has therefore been suggested to use marker allergen molecules which are specific for given allergen sources as diagnostic marker allergens for improving the prescription of AIT (78). The marker allergen concept can be applied to almost all allergen sources and accordingly has been suggested for several common respiratory allergen sources (79–81). The use of marker allergen molecules was suggested not only for refining the prescription of AIT but also for the monitoring of treatment response by measuring the development of allergen-specific IgG antibodies which are considered as biomarkers for the success of AIT (37, 82). In this context an interesting discovery was made which suggested that the ImmunoCAP ISAC microarray platform which utilizes small amounts of immobilized allergens is very useful for AIT monitoring. In fact, when small amounts of allergens are immobilized on the solid phase of immunological tests for detecting allergen-specific IgE, IgG antibodies can compete with IgE for allergen binding when they block epitopes recognized by patients IgE (83, 84).
When patients develop such blocking IgG antibodies in the course of AIT, these IgG antibodies will then compete with IgE antibodies which then causes a reduction of IgE binding similar as it is observed by skin testing when allergen-specific IgG antibodies block IgE-mediated mast cell activation thus leading to a reduction of skin responses. The reduction of allergen-specific IgE binding by blocking IgG antibodies can only be measured with IgE binding assays containing small amounts of immobilized allergens such as the ImmunoCAP ISAC chip but not by the traditional ImmunoCAP test which contains larger amounts of immobilized allergens (83–85). No other diagnostic platform with similar properties has been identified so far. In fact, two independent studies have shown the reduction of IgE binding to allergens by AIT-induced IgG antibodies on the ImmunoCAP ISAC platform and suggested it as a possible biomarker for AIT (86, 87).
Several studies support the concept of using molecular testing for the refined prescription of AIT (88), and the cost-effectiveness of molecular diagnosis as compared to traditional allergy diagnosis has been shown (89). Two more recent studies should be mentioned which have shown that it may be possible to enhance the success of AIT by selection of patients whose IgE reactivity profiles match the immunogenic components present in the AIT vaccines (90, 91). Accordingly, microarray-based molecular diagnosis seems to be well suited as companion diagnostic tool for the selection of patients for AIT and for the monitoring of treatment success.
Unmet Needs in MicroArray Technology
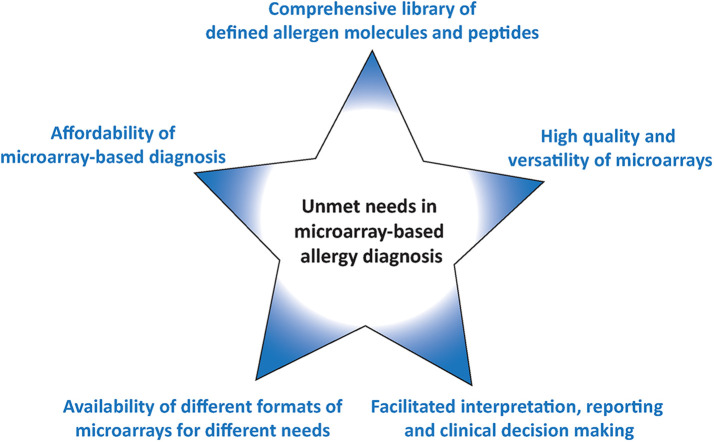
Since the first description of the use of microarrayed allergens in 2002 for allergy diagnosis the technology has become available world-wide and has been used extensively in research. However, several unmet needs remain which are summarized in Figure 2 and discussed below.
Figure 2.
Unmet needs in microarray-based allergy diagnosis.
The Library of Allergen Molecules and Peptides
The allergen molecules and allergen derived peptides and their quality can be considered the heart of any allergen microarray because it determines what application can be addressed with the allergen chip. For example, it is important to cover the most common respiratory, food, venom, and other allergen sources by a representative collection of allergen molecules. In this context it should be noted that there are different opinions regarding the inclusion of certain allergen molecules in screening assays. For example, some argue that the inclusion of venom allergens may create ethical and legal issues because in case of a positive or negative test result one cannot predict or exclude a sting reaction. However, this is in principle applicable for every IgE test result which needs to be considered in the context of clinical information and/or results from provocation testing. Therefore others think that the inclusion of venom allergens in screening tests is not a problem provided the patients are adequately informed about the relevance of the IgE test results.
The addition of allergen-derived peptides may be interesting for example in the diagnosis of food allergy where sequential IgE epitopes play a role and for the monitoring of AIT-induced IgG responses. Since only small amounts of allergen are immobilized on microarrays it is important to use highly pure allergens of high quality to detect also low specific IgE responses. Natural allergen preparation containing impurities in terms of unrelated allergens or cross-reactive carbohydrates may give rise to unclear, false-positive test results. Although carbohydrates are highly cross-reactive, it seems that patients mount quite specific and selective IgE to certain carbohydrates which cannot be completely inhibited by pre-incubation of sera with a single carbohydrate (92) so that unclear background reactivity may remain. This is of particular relevance because IgE-reactive carbohydrates have been shown to have little or no clinical relevance. Accordingly it is important to establish a library of high quality allergen molecules and peptides which can be reproduced according to defined protocols. The allergen molecules should be preferentially made as recombinant, non-glycosylated allergens to avoid unclear results due to carbohydrate-specific IgE.
This allergen library should be as complete as possible to pick up every relevant IgE sensitization in a given population. For example it has been shown, that the MeDALL allergen chip, a customized allergen array based on the ImmunoCAP ISAC platform containing more than 170 allergen molecules (83), was more sensitive in picking up IgE sensitizations than traditional allergy tests based on comprehensive panels of allergen extracts for skin testing or IgE serology (93). In order to refine the panel of allergen molecules on a chip it will be necessary to investigate molecular IgE sensitization profiles in different populations in different countries and continents to define the allergen repertoire of a microarray suitable for allergy diagnosis in the whole world. Such a complete representation of allergen molecules seems very important because due to the high mobility of the world population allergen arrays representing only a local allergen repertoire will not be sufficient for diagnosis. The production of microarrays containing subsets of allergen molecules does not seem to have any advantages because the microarray technology does not set limitations regarding the numbers of molecules which can be immobilized, and the costs of goods for production are low due to the low amounts of allergen needed. However, one must consider that costs for quality control, validation, and registration of complex assays may increase costs considerably.
New Materials May Increase the Quality and Versatility of Microarrays
There are basically two types of multiplex diagnostic platforms available. One contains allergen molecules adsorbed to microbeads, whereas the other platform is based on allergens which are immobilized on chips by micro-spotting. Microbead-based multiplex assay usually can accommodate only a limited number of less than 50 different allergen molecules in a single test and require quite expensive instruments such as Luminex readers or FACS-based technology for read out (94, 95). By contrast, microarrays allow measuring specific antibody reactivity to more than hundred different allergen molecules at the same time. There are currently two major types of allergen arrays available, the ImmunoCAP ISAC platform, using allergen molecules immobilized onto glass (Thermo Fisher ImmunoCAP ISAC Immuno-solid-phase Allergen Chip which contains 112 allergens from 51 allergen sources) (38) and an allergen macro-array prepared on the basis of a nitrocellulose membrane (e.g., MADx Allergen Explorer ALEX; containing 282 allergens: 156 extracts and 126 components) (39). Both systems allow reasonable detection of allergen-specific IgE but one may consider increasing the quality of the arrays by selecting different materials for allergen immobilization. Already in the original patent application made for the ImmunoCAP ISAC technology, silicon-based surfaces have been considered as alternative to glass (96).
In fact, the high sensitivity of protein assays on microarray silicon slides has then been demonstrated (97, 98). The latter studies demonstrated optimized layers of thermally grown silicon oxide with highly reproducible thickness, low roughness, and fluorescence background which yielded fluorescence intensification due to the constructive interference between the incident and reflected waves of the fluorescence radiation. Furthermore, the studies suggested that by combining an optimized reflective substrate with a high performance surface chemistry may strongly improve the quality of diagnostic protein array by obtaining a 5–10 fold enhancement of the fluorescence signals when compared to glass surfaces. The favorable features of silicon slides have been further demonstrated for peptide arrays (99) and for the field of food allergy diagnosis (100).
Below, we have evaluated allergen microarrays based on silicon oxide surfaces and compared them with glass slides currently used in the ImmunoCAP ISAC platform confirming the higher sensitivity of the silicon technology. We are then also discussing advantages of this technology for the versatile production of different formats of allergen microarrays.
Affordability of Microarray-Based Diagnosis
Currently available microarray-based allergy tests are relatively expensive although one has to consider that a single microarray test result provides more than hundred individual test results. For example costs for one array may range between 60 and 100 Euro depending on prices requested by different manufacturers in different countries, and additional costs for the processing of one sample may vary considerably 50–300 Euro depending on costs of laboratory facilities and personnel. Therefore, it will be necessary to decrease the costs of production for the micro-arrays and the costs for the test procedure which is currently performed by hand pipetting.
Different Formats of Microarrays for Different Needs
As mentioned above currently available microarrays are made for manual operation and thus individual testing requires wet laboratory facilities and relatively expensive scanning equipment for the analysis of results. The available formats thus can be used for manual analysis of relatively limited numbers of samples and require trained personnel. Unfortunately, no automatization for the processing of the available allergen arrays is available which would allow large scale and fully automated analysis of large numbers of patients. Therefore, there is still an unmet need for different formats of allergen arrays allowing different applications such as the occasional analysis of few or single serum samples yielding fast results with a minimum of equipment, the medium scale analysis of several serum samples and the fully automated analysis of large numbers of sera (Figure 2).
Furthermore, allergen arrays should allow the analysis not only of allergen-specific IgE but also of other immunoglobulin isotypes as well as the visualization of the competitive activity of allergen-specific IgG on IgE binding for the monitoring of AIT.
Interpretation, Reporting and Clinical Decision Making
Since allergen microarrays deliver test results for more than 100 different allergen molecules it is important that doctors who see allergic patients and wish to correctly interpret the sometimes complex test results keep themselves updated by continuous medical education. The transition of allergy diagnosis from the use of allergen extracts to allergen molecules requires knowledge regarding the characteristics of the individual allergen molecules. Thus molecular allergy diagnosis may be compared a bit with the switching from previous old telephones to mobile phones which offer many different additional applications that need to be explored by the user. The challenges of interpreting allergen microarray results may be met by machine learning approaches and other diagnostic algorithms in addition to continuous medical education (10, 101).
A Comparison of Different Surfaces for Microarrays: Glass Versus Silicon
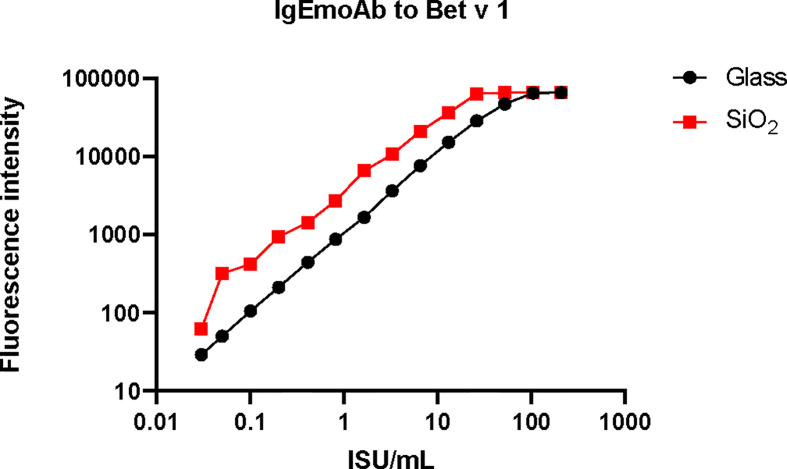
In order to compare the glass surface which currently is used for ImmunoCAP ISAC with silicon slides (97–100) a panel of important respiratory allergens was spotted on the two surfaces and allergic patients’ IgE and IgG reactivity was assessed. A set of 24 allergens containing mite allergens (Der p 1, Der p 2, Der p 4, Der p 5, Der p 7, Der p 10, Der p 15, Der p 18, Der p 20, Der p 21, Der p 23, Der p 37, Blo t 5, Blo t 12 and Blot 21), cat allergens (Fel d 1, Fel d 2, Fel d 3, Fel d 4 and Fel d 6), and PR10 allergens (Bet v 1, Gly m 4, Ara h 8 and Pru p 4) were spotted in triplicates on glass and silicon wafers in the order described (Figure 3) (Supplementary Materials and Methods). In the first experiment we determined the sensitivity of IgE reactivity to Bet v 1 immobilized to glass and silicon chips using a human monoclonal chimeric IgE antibody (IgEmoAb) (102) (Figure 4). A two-fold serial dilution of IgEmoAb corresponding to 208–0.025 ISU/ml was used to detect Bet v 1. Silicon microarrays showed a five-fold higher fluorescence intensity of IgE reactivity of IgEmoAb in the range of 52–0.025 ISU/ml to Bet v 1 than the glass surface (Figure 4). The silicon surface allowed measuring the Bet v 1-specific IgE down to 0.025 ISU/ml which is much lower than the cut-off used in currently available microarray tests (0.3 ISU/ml) and the detection limit 0.1 ISU/ml used for certain research purposes.
Figure 3.
Outlay of prototype allergen microarrays made by printing on glass slides and assembled silicon elements. Order of house dust mite, mite, cat, and PR10 allergens microarrayed in triplicates on glass slides (left) and precut and assembled silicon chips-derived elements.
Figure 4.
Sensitivity of the reactivity of a human monoclonal Bet v 1-specific IgE antibody to Bet v 1 printed on glass versus silicon. Shown are the fluorescence light intensities (y-axis) corresponding to different concentrations (x-axis) of the monoclonal human Bet v 1-specific IgE antibody (IgEmoAb).
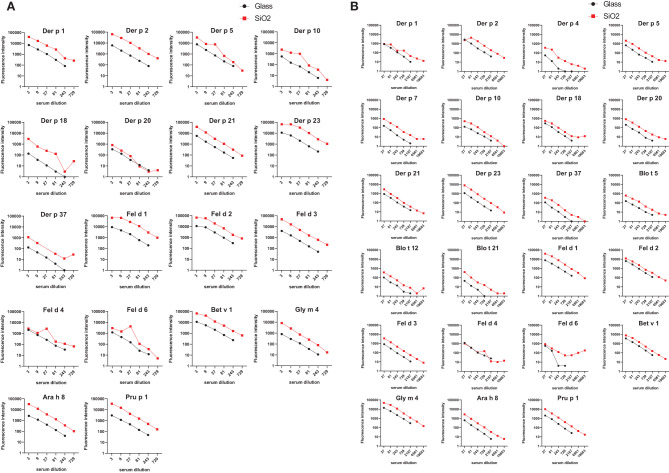
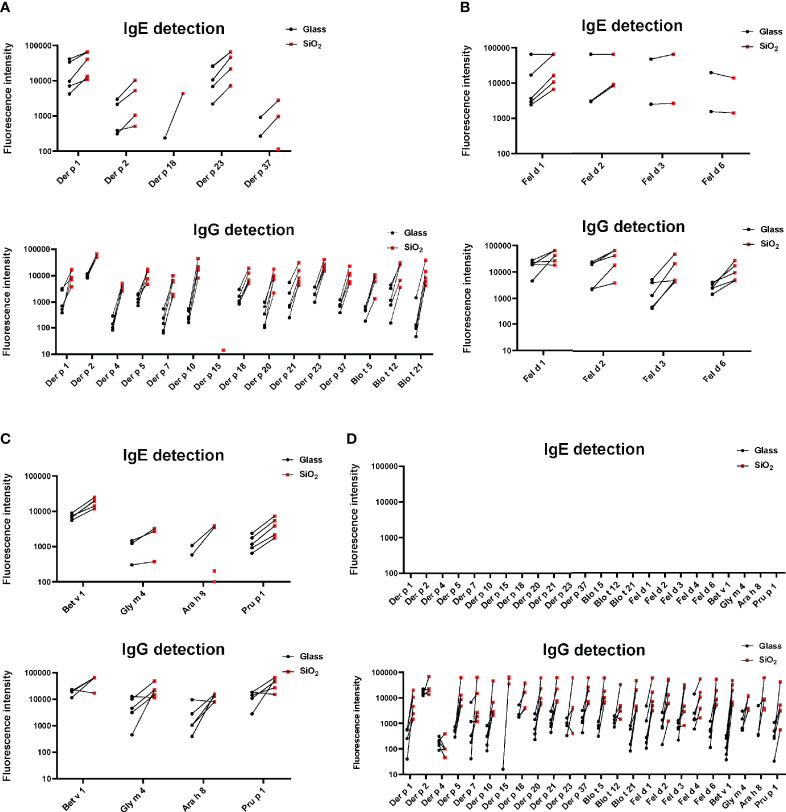
Figure 5 shows the comparison of IgE and IgG reactivity to allergens spotted on glass and silicon microarrayed chips determined with a serum pool from allergic patients with IgE reactivity against the tested allergen panel that was diluted 3- to 729-fold for IgE detection (Figure 5A) and 27- to 19,683-fold for IgG detection (Figure 5B). Figure 5A shows that the silicon surface yielded an approximately 10-fold higher IgE binding according to fluorescence intensity to all but one (i.e., Der p 20) allergen compared to the glass surface. Figure 5B demonstrates that silicon was superior to glass also regarding IgG detection showing approximately five-fold higher IgG signals, and specific binding was detectable even at a dilution of 1:19,683 of the serum pool on silicon microarrays.
Figure 5.
Comparison of IgE and IgG reactivity to HDM/mite, cat, and PR10 allergens which had been microarrayed on glass and silicon. A serum pool containing IgE and IgG antibodies against each of the tested allergens was tested for (A) IgE and (B) IgG reactivity to the individual allergens in different dilutions (x-axes). Fluorescence intensities corresponding to bound antibodies are shown on the y-axes.
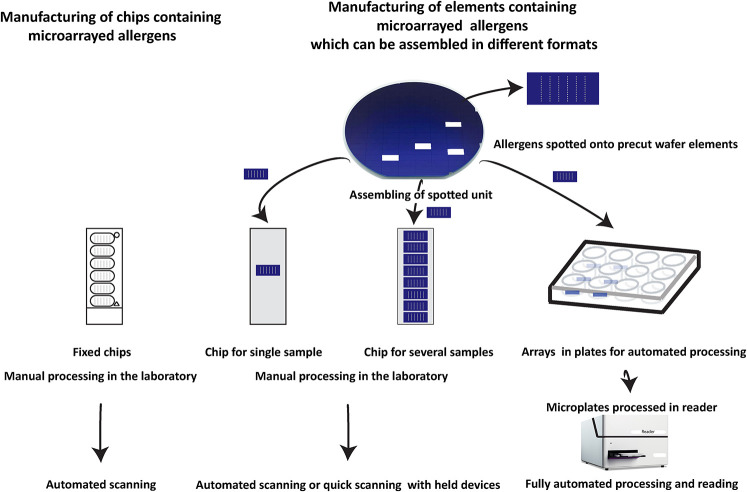
Next, we tested sera from HDM and mite- (Figure 6A), cat- (Figure 6B), birch pollen allergic patients (Figure 6C) and non-allergic subjects (Figure 6D) for IgE and IgG reactivity to allergens microarrayed on silicon and glass. This experiment confirmed for almost all tested sera and allergens that allergens immobilized on silicon have higher IgE and IgG detection signals. Importantly, IgE detection was highly specific for glass and silicon because none of the non-allergic subjects showed detectable allergen-specific IgE reactivity (Figure 6D). Our results thus indicate that allergen microarrays based on silicon are superior to glass for IgE and IgG detection of allergens. It should be also noted that another advantage of silicon arrays is that spotting can be performed on small, precut silicon elements which can be assembled in different formats (Figure 7, right upper part).
Figure 6.
IgE- and IgG-reactivity (y-axes: fluorescence intensities corresponding to bound antibodies) of allergic patients to (A) microarrayed HDM/mite allergens, (B) cat allergens and (C) PR10 allergens (x-axes) and of non-allergic subjects to all tested allergens (D). Glass: black dots; Silicon: red dots.
Figure 7.
Different assembly of silicon-based microarray elements generates different microarray formats for different diagnostic needs. Currently available allergen chips represent a single format which cannot adapt to different needs as shown for ImmunoCAP ISAC. By contrast, silicon-based microarray elements can be assembled in different formats. For example, chips containing only one array for individualized rapid testing can be produced which due to the high sensitivity can be read in simple scanners or by using mobile phone-based cameras. Alternatively, chips containing up to eight microarrays can be assembled which allow manual testing of medium scale numbers of sera and subsequent analysis by inexpensive automated scanners. For large scale automated testing of large numbers of sera, silicon elements can be mounted in ELISA-type plates and subjected to automated testing in a closed ELISA-based instrument containing an incubation, washing and detection unit followed by a scanning unit.
Manufacturing of MicroArray Elements and Subsequent Assembly Meets Different Needs in Allergy Diagnosis
In Figure 7 we try to provide an overview of how microarrays based on silicon elements may contribute to innovation in allergy testing as compared to currently available chips and arrays used for IgE serology. Currently available allergen arrays are manufactured in one predetermined format which is a chip, containing one or several identically prepared allergen arrays (Figure 7, left part) which then need to be processed in the laboratory by hand pipetting.
Disadvantages of chips containing more than one array are that the spotting of the microarrays is performed directly on chips which are relatively large in comparison to a single silicon element. However, the single silicon elements can be arranged in much shorter distance close to each other for the spotting than preformed chips. Accordingly the spotting machine (microarray printer) makes much shorter movements when spotting closely assembled silicon elements as compared to premade chips which should speed up the production by shortening the production time. Another disadvantage of chips containing several microarrays is that one array of bad quality will lead to the discarding of a complete chip although the other arrays may meet the quality criteria. By contrast, when single silicon elements are used only the few poor quality elements will be discarded keeping the loss of material low. However, the most important advantage of microarrays based on single silicon elements is that after spotting, the single elements can be assembled in different formats for different uses. This allows producing chips containing only one microarray for fast testing of single serum samples. Furthermore, chips containing several silicon elements for testing of several sera can be assembled. Importantly, silicon elements can be also assembled in plates (e.g., ELISA plate format) for automated processing of samples which can be incubated with samples, washed, developed, and read in machines without requiring hand pipetting. Thus microarrays printed on silicon elements allow assembling of different devices for testing based on one standardized element. Furthermore, silicon surfaces give 5–10-fold higher sensitivity as compared to glass which should allow detecting also low allergen-specific IgE levels with high precision, and the measurement can be done with very simple and inexpensive detection devices.
Conclusion
Since the first description of allergen microarrays for allergy diagnosis almost 20 years ago, these multi-allergen tests have been successfully used to answer many research questions and have proved highly valuable for allergy diagnosis in multiple applications. However, several needs for improvement have remained unmet until today limiting the broad application of microarray-aided allergy diagnosis. We introduce here a novel concept for improving allergen microarray technology by showing that microarrays prepared on silicon offer higher sensitivity for the detection of specific IgE than the currently used glass surfaces and other surfaces with similar sensitivity as glass. Instead of spotting allergen arrays on preformed, inflexible devices we propose manufacturing of flexible silicon elements containing microarrays which then can be assembled in different formats. This allows addressing the different needs of allergy diagnosis ranging from manual testing of single or few sera to fully automated processing of large numbers of sera. Microarrays based on silicon elements are versatile arrays that can be easily produced. Using this technology it should be possible to decrease the costs of microarray testing to make the technology affordable to the health care systems. The heart of the allergen array is a library of high-quality allergen molecules. Microarrays utilizing low amounts of immobilized allergens allowing visualizing the interplay of allergen-specific IgE and IgG mimicking the in vivo situation and thus should deliver serological test results resembling the clinical sensitivity. Our vision for microarray-aided allergy diagnosis is to make available to the doctor the complete IgE reactivity profile of the patient already at the initial visit or during teleconsultation to achieve a complete diagnosis and personalized treatment without need for multiple time-consuming visits for the benefit of the patient and to reduce the costs for health care in allergy.
Author Contributions
RV and HJH wrote the manuscript. RV, HJH and TS designed the figures and tables. HJH, RC, OA, MC, KR, AnK, OE, EF, AL, KN, ESa, and TS performed experiments and/or contributed materials. RC, OA, MC, ESa, AnK, KR, AlK, OE, EF, AL, ESm, MK, KN, SV, TS, HJH and RV critically read and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This review was supported by the Danube Allergy Research Cluster funded by the Country of Lower Austria, by the MCCA PhD program and grant No P29398 of the Austrian Science Fund (FWF), by the Russian Academic Excellence Project 5-100, by a Megagrant of the Government of the Russian Federation, grant No 14.W03.31.0024, and by grant from HVD Life Science, Vienna, Austria.
Conflict of Interest
RV has received research grants from HVD Life Science, Vienna, Austria, and Viravaxx, Vienna, Austria, and serves as a consultant for Viravaxx.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
The authors acknowledge the support of the Medical University of Vienna, Austria, for the research infrastructure.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2020.594978/full#supplementary-material
Abbreviations
IgE, Immunoglobulin E; IgG, Immunoglobulin G; Ab, antibody; AIT, allergen-specific immunotherapy; Ig, immunoglobulin; RAST, radioallergosorbent test; GP, general practitioner; OAS, oral allergy syndrome; RV, rhinovirus; HDM, house dust mite; Der p, Dermatophagoides pteronyssinus; Blo t, Blomia tropicalis; Fel d, Felis domesticus; PR10, pathogen-related protein 10; Bet v, Betula verrucosa; Gly m, Glycine max; Ara h, Arachis hypogaea; Pru p, Prunus persica.
References
- 1. Valenta R, Karaulov A, Niederberger V, Gattinger P, van Hage M, Flicker S, et al. Molecular aspects of allergens and allergy. Adv Immunol (2018) 138:195–256. 10.1016/bs.ai.2018.03.002 [DOI] [PubMed] [Google Scholar]
- 2. Hofmaier S, Hatzler L, Rohrbach A, Panetta V, Hakimeh D, Bauer CP, et al. “Default” versus “pre-atopic” IgG responses to foodborne and airborne pathogenesis-related group 10 protein molecules in birch-sensitized and nonatopic children. J Allergy Clin Immunol (2015) 135(5):1367–74.e1-8. 10.1016/j.jaci.2014.09.048 [DOI] [PubMed] [Google Scholar]
- 3. van Neerven RJ, Knol EF, Ejrnaes A, Wurtzen PA. IgE-mediated allergen presentation and blocking antibodies: regulation of T-cell activation in allergy. Int Arch Allergy Immunol (2006) 141(2):119–29. 10.1159/000094714 [DOI] [PubMed] [Google Scholar]
- 4. Eckl-Dorna J, Villazala-Merino S, Linhart B, Karaulov AV, Zhernov Y, Khaitov M, et al. Allergen-specific antibodies regulate secondary allergen-specific immune responses. Front Immunol (2018) 9:3131. 10.3389/fimmu.2018.03131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Anto JM, Bousquet J, Akdis M, Auffray C, Keil T, Momas I, et al. Mechanisms of the Development of Allergy (MeDALL): Introducing novel concepts in allergy phenotypes. J Allergy Clin Immunol (2017) 139(2):388–99. 10.1016/j.jaci.2016.12.940 [DOI] [PubMed] [Google Scholar]
- 6. Ishizaka K, Ishizaka T, Hornbrook MM. Physicochemical properties of reaginic antibody. V. Correlation of reaginic activity wth gamma-E-globulin antibody. J Immunol (1966) 97(6):840–53. [PubMed] [Google Scholar]
- 7. Wide L, Bennich H, Johansson SG. Diagnosis of allergy by an in-vitro test for allergen antibodies. Lancet (7526) 1967) 2:1105–7. 10.1016/s0140-6736(67)90615-0 [DOI] [PubMed] [Google Scholar]
- 8. Blackley CH. Experimental researches on the causes and nature of catarrhus aestivus (hay-fever or hay-asthma). London: Dawson’s of Pall Mall; (1959) p. 1959–2161. [Google Scholar]
- 9. Coombs RRA, Gell PGH. Classification of allergic reactionsresponsible for clinical hypersensitity and disease. In Clin Aspect Immunol Oxford Blackwell Sci Publications (1975) 575–96p.
- 10. Matricardi PM, Kleine-Tebbe J, Hoffmann HJ, Valenta R, Hilger C, Hofmaier S, et al. EAACI molecular allergology user’s guide. Pediatr Allergy Immunol (2016) 27(Suppl 23):1–250. 10.1111/pai.12563 [DOI] [PubMed] [Google Scholar]
- 11. Linhart B, Freidl R, Elisyutina O, Khaitov M, Karaulov A, Valenta R. Molecular approaches for diagnosis, therapy and prevention of cow s milk allergy. Nutrients (2019) 11(7):1492. 10.3390/nu11071492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brunetto B, Tinghino R, Braschi MC, Antonicelli L, Pini C, Iacovacci P. Characterization and comparison of commercially available mite extracts for in vivo diagnosis. Allergy (2010) 65(2):184–90. 10.1111/j.1398-9995.2009.02150.x [DOI] [PubMed] [Google Scholar]
- 13. Klimek L, Hoffmann HJ, Renz H, Demoly P, Werfel T, Matricardi PM, et al. Diagnostic test allergens used for in vivo diagnosis of allergic diseases are at risk: a European Perspective. Allergy (2015) 70(10):1329–31. 10.1111/all.12676 [DOI] [PubMed] [Google Scholar]
- 14. Valenta R, Karaulov A, Niederberger V, Zhernov Y, Elisyutina O, Campana R, et al. Allergen extracts for In vivo diagnosis and treatment of allergy: Is there a future? J Allergy Clin Immunol Pract (2018) 6(6):1845–55.e2. 10.1016/j.jaip.2018.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Klimek L, Hoffmann HJ, Kalpaklioglu AF, Demoly P, Agache I, Popov TA, et al. In-vivo diagnostic test allergens in Europe: A call to action and proposal for recovery plan-An EAACI position paper. Allergy (2020) Sep 75(9):2161–9. 10.1111/all.14329 [DOI] [PubMed] [Google Scholar]
- 16. Fang KS, Vitale M, Fehlner P, King TP. cDNA cloning and primary structure of a white-face hornet venom allergen, antigen 5. Proc Natl Acad Sci USA (1988) 85(3):895–9. 10.1073/pnas.85.3.895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Breiteneder H, Pettenburger K, Bito A, Valenta R, Kraft D, Rumpold H, et al. The gene coding for the major birch pollen allergen Betv1, is highly homologous to a pea disease resistance response gene. EMBO J (1989) 8(7):1935–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chua KY, Stewart GA, Thomas WR, Simpson RJ, Dilworth RJ, Plozza TM, et al. Sequence analysis of cDNA coding for a major house dust mite allergen, Der p 1. Homology with cysteine proteases. J Exp Med (1988) 167(1):175–82. 10.1084/jem.167.1.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Valenta R, Duchene M, Vrtala S, Birkner T, Ebner C, Hirschwehr R, et al. Recombinant allergens for immunoblot diagnosis of tree-pollen allergy. J Allergy Clin Immunol (1991) 88(6):889–94. 10.1016/0091-6749(91)90245-j [DOI] [PubMed] [Google Scholar]
- 20. Valenta R, Vrtala S, Ebner C, Kraft D, Scheiner O. Diagnosis of grass pollen allergy with recombinant timothy grass (Phleum pratense) pollen allergens. Int Arch Allergy Immunol (1992) 97(4):287–94. 10.1159/000236135 [DOI] [PubMed] [Google Scholar]
- 21. Larsen JN, Stroman P, Ipsen H. PCR based cloning and sequencing of isogenes encoding the tree pollen major allergen Car b I from Carpinus betulus, hornbeam. Mol Immunol (1992) 29(6):703–11. 10.1016/0161-5890(92)90180-6 [DOI] [PubMed] [Google Scholar]
- 22. Breiteneder H, Ferreira F, Hoffmann-Sommergruber K, Ebner C, Breitenbach M, Rumpold H, et al. Four recombinant isoforms of Cor a I, the major allergen of hazel pollen, show different IgE-binding properties. Eur J Biochem (1993) 212(2):355–62. 10.1111/j.1432-1033.1993.tb17669.x [DOI] [PubMed] [Google Scholar]
- 23. Valenta R, Lidholm J, Niederberger V, Hayek B, Kraft D, Gronlund H. The recombinant allergen-based concept of component-resolved diagnostics and immunotherapy (CRD and CRIT). Clin Exp Allergy (1999) 29(7):896–904. 10.1046/j.1365-2222.1999.00653.x [DOI] [PubMed] [Google Scholar]
- 24. Hiller R, Laffer S, Harwanegg C, Huber M, Schmidt WM, Twardosz A, et al. Microarrayed allergen molecules: diagnostic gatekeepers for allergy treatment. FASEB J (2002) 16(3):414–6. 10.1096/fj.01-0711fje [DOI] [PubMed] [Google Scholar]
- 25. Harwanegg C, Laffer S, Hiller R, Mueller MW, Kraft D, Spitzauer S, et al. Microarrayed recombinant allergens for diagnosis of allergy. Clin Exp Allergy (2003) 33(1):7–13. 10.1046/j.1365-2222.2003.01550.x [DOI] [PubMed] [Google Scholar]
- 26. Asher MI, Keil U, Anderson HR, Beasley R, Crane J, Martinez F, et al. International Study of Asthma and Allergies in Childhood (ISAAC): rationale and methods. Eur Respir J (1995) 8(3):483–91. 10.1183/09031936.95.08030483 [DOI] [PubMed] [Google Scholar]
- 27. Matricardi PM, Dramburg S, Alvarez-Perea A, Antolín-Amérigo D, Apfelbacher C, Atanaskovic-Markovic M, et al. The role of mobile health technologies in allergy care: An EAACI position paper. Allergy (2020) 75(2):259–72. 10.1111/all.13953 [DOI] [PubMed] [Google Scholar]
- 28. Peveri S, Pattini S, Costantino MT, Incorvaia C, Montagni M, Roncallo C, et al. Molecular diagnostics improves diagnosis and treatment of respiratory allergy and food allergy with economic optimization and cost saving. Allergol Immunopathol (Madr) (2019) 47(1):64–72. 10.1016/j.aller.2018.05.008 [DOI] [PubMed] [Google Scholar]
- 29. Flores Kim J, McCleary N, Nwaru BI, Stoddart A, Sheikh A. Diagnostic accuracy, risk assessment, and cost-effectiveness of component-resolved diagnostics for food allergy: A systematic review. Allergy (2018) 73(8):1609–21. 10.1111/all.13399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zuberbier T, Lötvall J, Simoens S, Subramanian SV, Church MK. Economic burden of inadequate management of allergic diseases in the European Union: a GA(2) LEN review. Allergy (2014) 69(10):1275–9. 10.1111/all.12470 [DOI] [PubMed] [Google Scholar]
- 31. Kulthanan K, Chusakul S, Recto MT, Gabriel MT, Aw DCW, Prepageran N, et al. Economic Burden of the Inadequate Management of Allergic Rhinitis and Urticaria in Asian Countries Based on the GA²LEN Model. Allergy Asthma Immunol Res (2018) 10(4):370–8. 10.4168/aair.2018.10.4.370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hochwallner H, Alm J, Lupinek C, Johansson C, Mie A, Scheynius A, et al. Transmission of allergen-specific IgG and IgE from maternal blood into breast milk visualized with microarray technology. J Allergy Clin Immunol (2014) 134(5):1213–5. 10.1016/j.jaci.2014.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Berings M, Arasi S, De Ruyck N, Perna S, Resch Y, Lupinek C, et al. Reliable mite-specific IgE testing in nasal secretions by means of allergen microarray. J Allergy Clin Immunol (2017) 140(1):301–3.e8. 10.1016/j.jaci.2016.11.047 [DOI] [PubMed] [Google Scholar]
- 34. Castelli S, Arasi S, Pawankar R, Matricardi PM. Collection of nasal secretions and tears and their use in allergology. Curr Opin Allergy Cl (2018) 18(1):1–9. 10.1097/Aci.0000000000000412 [DOI] [PubMed] [Google Scholar]
- 35. Castelli S, Arasi S, Tripodi S, Villalta D, Martelli P, Conte M, et al. IgE antibody repertoire in nasal secretions of children and adults with seasonal allergic rhinitis: A molecular analysis. Pediatr Allergy Immunol (2020) 31(3):273–80. 10.1111/pai.13148 [DOI] [PubMed] [Google Scholar]
- 36. Garib V, Rigler E, Gastager F, Campana R, Dorofeeva Y, Gattinger P, et al. Determination of IgE and IgG reactivity to more than 170 allergen molecules in paper-dried blood spots. J Allergy Clin Immun (2019) 143(1):437–40. 10.1016/j.jaci.2018.08.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shamji MH, Kappen JH, Akdis M, Jensen-Jarolim E, Knol EF, Kleine-Tebbe J, et al. Biomarkers for monitoring clinical efficacy of allergen immunotherapy for allergic rhinoconjunctivitis and allergic asthma: an EAACI Position Paper. Allergy (2017) 72(8):1156–73. 10.1111/all.13138 [DOI] [PubMed] [Google Scholar]
- 38. Jakob T, Forstenlechner P, Matricardi P, Kleine-Tebbe J. Molecular allergy diagnostics using multiplex assays: methodological and practical considerations for use in research and clinical routine. Part 21 of the Series Molecular Allergology. Allergo J (2015) 24(8):42–+. 10.1007/s15007-015-0982-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Heffler E, Puggioni F, Peveri S, Montagni M, Canonica GW, Melioli G. Extended IgE profile based on an allergen macroarray: a novel tool for precision medicine in allergy diagnosis. World Allergy Organ J (2018) 11(1):7. 10.1186/s40413-018-0186-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lupinek C, Hochwallner H, Johansson C, Mie A, Rigler E, Scheynius A, et al. Maternal allergen-specific IgG might protect the child against allergic sensitization. J Allergy Clin Immun (2019) 144(2):536–48. 10.1016/j.jaci.2018.11.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Curin M, Swoboda I, Wollmann E, Lupinek C, Spitzauer S, van Hage M, et al. Microarrayed dog, cat, and horse allergens show weak correlation between allergen-specific IgE and IgG responses. J Allergy Clin Immun (2014) 133(3):918–21. 10.1016/j.jaci.2013.10.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Huang X, Tsilochristou O, Perna S, Hofmaier S, Cappella A, Bauer CP, et al. Evolution of the IgE and IgG repertoire to a comprehensive array of allergen molecules in the first decade of life. Allergy (2018) 73(2):421–30. 10.1111/all.13269 [DOI] [PubMed] [Google Scholar]
- 43. Siroux V, Lupinek C, Resch Y, Curin M, Just J, Keil T, et al. Specific IgE and IgG measured by the MeDALL allergen-chip depend on allergen and route of exposure: The EGEA study. J Allergy Clin Immun (2017) 139(2):643–+. 10.1016/j.jaci.2016.05.023 [DOI] [PubMed] [Google Scholar]
- 44. Lupinek C, Marth K, Niederberger V, Valenta R. Analysis of serum IgE reactivity profiles with microarrayed allergens indicates absence of de novo IgE sensitizations in adults. J Allergy Clin Immunol (2012) 130(6):1418–20.e4. 10.1016/j.jaci.2012.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cabauatan CR, Lupinek C, Scheiblhofer S, Weiss R, Focke-Tejkl M, Bhalla PL, et al. Allergen microarray detects high prevalence of asymptomatic IgE sensitizations to tropical pollen-derived carbohydrates. J Allergy Clin Immunol (2014) 133(3):910–4.e5. 10.1016/j.jaci.2013.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Garib V, Wollmann E, Djambekova G, Lemell P, Kmenta M, Berger U, et al. Possible effect of landscape design on IgE recognition profiles of two generations revealed with microarrayed allergens. Allergy (2017) 72(10):1579–82. 10.1111/all.13169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Custovic A, Sonntag HJ, Buchan IE, Belgrave D, Simpson A, Prosperi MCF. Evolution pathways of IgE responses to grass and mite allergens throughout childhood. J Allergy Clin Immunol (2015) 136(6):1645–52.e8. 10.1016/j.jaci.2015.03.041 [DOI] [PubMed] [Google Scholar]
- 48. Howard R, Belgrave D, Papastamoulis P, Simpson A, Rattray M, Custovic A. Evolution of IgE responses to multiple allergen components throughout childhood. J Allergy Clin Immunol (2018) 142(4):1322–30. 10.1016/j.jaci.2017.11.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Westman M, Lupinek C, Bousquet J, Andersson N, Pahr S, Baar A, et al. Early childhood IgE reactivity to pathogenesis-related class 10 proteins predicts allergic rhinitis in adolescence. J Allergy Clin Immunol (2015) 135(5):1199–206.e1-11. 10.1016/j.jaci.2014.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Posa D, Perna S, Resch Y, Lupinek C, Panetta V, Hofmaier S, et al. Evolution and predictive value of IgE responses toward a comprehensive panel of house dust mite allergens during the first 2 decades of life. J Allergy Clin Immunol (2017) 139(2):541–9.e8. 10.1016/j.jaci.2016.08.014 [DOI] [PubMed] [Google Scholar]
- 51. Wickman M, Lupinek C, Andersson N, Belgrave D, Asarnoj A, Benet M, et al. Detection of IgE reactivity to a handful of allergen molecules in early childhood predicts respiratory allergy in adolescence. EBioMedicine (2017) 26:91–9. 10.1016/j.ebiom.2017.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Westman M, Aberg K, Apostolovic D, Lupinek C, Gattinger P, Mittermann I, et al. Sensitization to grass pollen allergen molecules in a birth cohort-natural Phl p 4 as an early indicator of grass pollen allergy. J Allergy Clin Immun (2020) 145(4):1174–+. 10.1016/j.jaci.2020.01.006 [DOI] [PubMed] [Google Scholar]
- 53. Szepfalusi Z, Bannert C, Ronceray L, Mayer E, Hassler M, Wissmann E, et al. Preventive sublingual immunotherapy in preschool children: first evidence for safety and pro-tolerogenic effects. Pediatr Allergy Immunol (2014) 25(8):788–95. 10.1111/pai.12310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tulaeva I, Kratzer B, Campana R, Curin M, van Hage M, Karsonova A, et al. Preventive allergen-specific vaccination against allergy: mission possible? Front Immunol (2020) 11:1368. 10.3389/fimmu.2020.01368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dramburg S, Matricardi PM. Molecular diagnosis of allergy: The pediatric perspective. Front Pediatr (2019) 7:369. 10.3389/fped.2019.00369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hood L, Flores M. A personal view on systems medicine and the emergence of proactive P4 medicine: predictive, preventive, personalized and participatory. N Biotechnol (2012) 29(6):613–24. 10.1016/j.nbt.2012.03.004 [DOI] [PubMed] [Google Scholar]
- 57. Valenta R. Early prevention instead of mending late damage in allergy? EBioMedicine (2019) 45:17–8. 10.1016/j.ebiom.2019.06.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Melioli G, Passalacqua G, Canonica GW, Baena-Cagnani CE, Matricardi P. Component-resolved diagnosis in pediatric allergic rhinoconjunctivitis and asthma. Curr Opin Allergy Clin Immunol (2013) 13(4):446–51. 10.1097/ACI.0b013e32836274d8 [DOI] [PubMed] [Google Scholar]
- 59. Fedenko E, Elisyutina O, Shtyrbul O, Pampura A, Valenta R, Lupinek C, et al. Microarray-based IgE serology improves management of severe atopic dermatitis in two children. Pediatr Allergy Immunol (2016) 27(6):645–9. 10.1111/pai.12572 [DOI] [PubMed] [Google Scholar]
- 60. Resch Y, Michel S, Kabesch M, Lupinek C, Valenta R, Vrtala S. Different IgE recognition of mite allergen components in asthmatic and nonasthmatic children. J Allergy Clin Immunol (2015) 136(4):1083–91. 10.1016/j.jaci.2015.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Elisyutina O, Fedenko E, Campana R, Litovkina A, Ilina N, Kudlay D, et al. Bet v 1-specific IgE levels and PR-10 reactivity discriminate silent sensitization from phenotypes of birch allergy. Allergy (2019) 74(12):2525–8. 10.1111/all.13931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Siroux V, Ballardini N, Soler M, Lupinek C, Boudier A, Pin I, et al. The asthma-rhinitis multimorbidity is associated with IgE polysensitization in adolescents and adults. Allergy (2018) 73(7):1447–58. 10.1111/all.13410 [DOI] [PubMed] [Google Scholar]
- 63. Douladiris N, Garib V, Piskou K, Focke-Tejkl M, Valenta R, Papadopoulos NG, et al. Molecular allergy diagnosis: A potential tool for the assessment of severity of grass pollen-induced rhinitis in children. Pediatr Allergy Immunol (2019) 30(8):852–5. 10.1111/pai.13107 [DOI] [PubMed] [Google Scholar]
- 64. Valenta R, Hochwallner H, Linhart B, Pahr S. Food allergies: the basics. Gastroenterology (2015) 148(6):1120–31.e4. 10.1053/j.gastro.2015.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wollmann E, Hamsten C, Sibanda E, Ochome M, Focke-Tejkl M, Asarnoj A, et al. Natural clinical tolerance to peanut in African patients is caused by poor allergenic activity of peanut IgE. Allergy (2015) 70(6):638–52. 10.1111/all.12592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kowalski ML, Ansotegui I, Aberer W, Al-Ahmad M, Akdis M, Ballmer-Weber BK, et al. Risk and safety requirements for diagnostic and therapeutic procedures in allergology: World Allergy Organization Statement. World Allergy Organ J (2016) 9(1):33. 10.1186/s40413-016-0122-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Steering Committee Authors; Review Panel Members . A WAO - ARIA - GA2LEN consensus document on molecular-based allergy diagnosis (PAMD@): Update 2020. World Allergy Organ J (2020) 13(2):100091. 10.1016/j.waojou.2019.100091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hochwallner H, Schulmeister U, Swoboda I, Balic N, Geller B, Nystrand M, et al. Microarray and allergenic activity assessment of milk allergens. Clin Exp Allergy (2010) 40(12):1809–18. 10.1111/j.1365-2222.2010.03602.x [DOI] [PubMed] [Google Scholar]
- 69. Santos AF, Barbosa-Morais NL, Hurlburt BK, Ramaswamy S, Hemmings O, Kwok M, et al. IgE to epitopes of Ara h 2 enhance the diagnostic accuracy of Ara h 2-specific IgE. Allergy (2020) 75(9):2309–18. 10.1111/all.14301 [DOI] [PubMed] [Google Scholar]
- 70. Asarnoj A, Hamsten C, Lupinek C, Melen E, Andersson N, Anto JM, et al. Prediction of peanut allergy in adolescence by early childhood storage protein-specific IgE signatures: The BAMSE population-based birth cohort. J Allergy Clin Immunol (2017) 140(2):587–90.e7. 10.1016/j.jaci.2016.12.973 [DOI] [PubMed] [Google Scholar]
- 71. Sastre-Ibanez M, Sastre J. Molecular allergy diagnosis for the clinical characterization of asthma. Expert Rev Mol Diagn (2015) 15(6):789–99. 10.1586/14737159.2015.1036745 [DOI] [PubMed] [Google Scholar]
- 72. Karaulov AV, Garib V, Garib F, Valenta R. Protein biomarkers in asthma. Int Arch Allergy Immunol (2018) 175(4):189–208. 10.1159/000486856 [DOI] [PubMed] [Google Scholar]
- 73. Fontanella S, Frainay C, Murray CS, Simpson A, Custovic A. Machine learning to identify pairwise interactions between specific IgE antibodies and their association with asthma: A cross-sectional analysis within a population-based birth cohort. PloS Med (2018) 15(11):e1002691. 10.1371/journal.pmed.1002691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Asarnoj A, Hamsten C, Waden K, Lupinek C, Andersson N, Kull I, et al. Sensitization to cat and dog allergen molecules in childhood and prediction of symptoms of cat and dog allergy in adolescence: A BAMSE/MeDALL study. J Allergy Clin Immunol (2016) 137(3):813–21.e7. 10.1016/j.jaci.2015.09.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Niespodziana K, Stenberg-Hammar K, Megremis S, Cabauatan CR, Napora-Wijata K, Vacal PC, et al. PreDicta chip-based high resolution diagnosis of rhinovirus-induced wheeze. Nat Commun (2018) 9(1):2382. 10.1038/s41467-018-04591-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Megremis S, Niespodziana K, Cabauatan C, Xepapadaki P, Kowalski ML, Jartti T, et al. Rhinovirus species-specific antibodies differentially reflect clinical outcomes in health and asthma. Am J Respir Crit Care Med (2018) 198(12):1490–9. 10.1164/rccm.201803-0575OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Niespodziana K, Borochova K, Pazderova P, Schlederer T, Astafyeva N, Baranovskaya T, et al. Toward personalization of asthma treatment according to trigger factors. J Allergy Clin Immunol (2020) 145(6):1529–34. 10.1016/j.jaci.2020.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kazemi-Shirazi L, Niederberger V, Linhart B, Lidholm J, Kraft D, Valenta R. Recombinant marker allergens: diagnostic gatekeepers for the treatment of allergy. Int Arch Allergy Immunol (2002) 127(4):259–68. 10.1159/000057742 [DOI] [PubMed] [Google Scholar]
- 79. Valenta R, Twaroch T, Swoboda I. Component-resolved diagnosis to optimize allergen-specific immunotherapy in the Mediterranean area. J Invest Allergol Clin Immunol (2007) 17(Suppl 1):36–40. [PubMed] [Google Scholar]
- 80. Pittner G, Vrtala S, Thomas WR, Weghofer M, Kundi M, Horak F, et al. Component-resolved diagnosis of house-dust mite allergy with purified natural and recombinant mite allergens. Clin Exp Allergy (2004) 34(4):597–603. 10.1111/j.1365-2222.2004.1930.x [DOI] [PubMed] [Google Scholar]
- 81. Douladiris N, Savvatianos S, Roumpedaki I, Skevaki C, Mitsias D, Papadopoulos NG. A molecular diagnostic algorithm to guide pollen immunotherapy in southern Europe: towards component-resolved management of allergic diseases. Int Arch Allergy Immunol (2013) 162(2):163–72. 10.1159/000353113 [DOI] [PubMed] [Google Scholar]
- 82. Eckl-Dorna J, Weber M, Stanek V, Linhart B, Ristl R, Waltl EE, et al. Two years of treatment with the recombinant grass pollen allergy vaccine BM32 induces a continuously increasing allergen-specific IgG4 response. EBioMedicine (2019) 50:421–32. 10.1016/j.ebiom.2019.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lupinek C, Wollmann E, Baar A, Banerjee S, Breiteneder H, Broecker BM, et al. Advances in allergen-microarray technology for diagnosis and monitoring of allergy: the MeDALL allergen-chip. Methods (2014) 66(1):106–19. 10.1016/j.ymeth.2013.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lupinek C, Wollmann E, Valenta R. Monitoring allergen immunotherapy effects by microarray. Curr Treat Options Allergy (2016) 3:189–203. 10.1007/s40521-016-0084-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. van Hage M, Hamsten C, Valenta R. ImmunoCAP assays: Pros and cons in allergology. J Allergy Clin Immunol (2017) 140(4):974–7. 10.1016/j.jaci.2017.05.008 [DOI] [PubMed] [Google Scholar]
- 86. Wollmann E, Lupinek C, Kundi M, Selb R, Niederberger V, Valenta R. Reduction in allergen-specific IgE binding as measured by microarray: A possible surrogate marker for effects of specific immunotherapy. J Allergy Clin Immun (2015) 136(3):806–9. 10.1016/j.jaci.2015.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Schmid JM, Wurtzen PA, Dahl R, Hoffmann HJ. Pretreatment IgE sensitization patterns determine the molecular profile of the IgG4 response during updosing of subcutaneous immunotherapy with timothy grass pollen extract. J Allergy Clin Immunol (2016) 137(2):562–70. 10.1016/j.jaci.2015.05.023 [DOI] [PubMed] [Google Scholar]
- 88. Sastre J, Landivar ME, Ruiz-Garcia M, Andregnette-Rosigno MV, Mahillo I. How molecular diagnosis can change allergen-specific immunotherapy prescription in a complex pollen area. Allergy (2012) 67(5):709–11. 10.1111/j.1398-9995.2012.02808.x [DOI] [PubMed] [Google Scholar]
- 89. Saltabayeva U, Garib V, Morenko M, Rosenson R, Ispayeva Z, Gatauova M, et al. Greater real-life diagnostic efficacy of allergen molecule-based diagnosis for prescription of immunotherapy in an area with multiple pollen exposure. Int Arch Allergy Immunol (2017) 173(2):93–8. 10.1159/000477442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Chen KW, Zieglmayer P, Zieglmayer R, Lemell P, Horak F, Bunu CP, et al. Selection of house dust mite-allergic patients by molecular diagnosis may enhance success of specific immunotherapy. J Allergy Clin Immun (2019) 143(3):1248–52. 10.1016/j.jaci.2018.10.048 [DOI] [PubMed] [Google Scholar]
- 91. Rodríguez-Domínguez A BM, Rohrbach A, Huang HJ, Curin M, Gevaert P, Matricardi PM, et al. Molecular profiling of allergen-specific antibody responses may enhance success of specific immunotherapy. J Allergy Clin Immunol (2020) 20):30479–86. 10.1016/j.jaci.2020.03.029 [DOI] [PubMed] [Google Scholar]
- 92. Gattinger P, Mittermann I, Lupinek C, Hofer G, Keller W, Bidovec Stojkovic U, et al. Recombinant glycoproteins resembling carbohydrate-specific IgE epitopes from plants, venoms and mites. EBioMedicine (2019) 39:33–43. 10.1016/j.ebiom.2018.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Skrindo I, Lupinek C, Valenta R, Hovland V, Pahr S, Baar A, et al. The use of the MeDALL-chip to assess IgE sensitization: a new diagnostic tool for allergic disease? Pediat Allerg Imm Uk (2015) 26(3):239–46. 10.1111/pai.12366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. King EM, Filep S, Smith B, Platts-Mills T, Hamilton RG, Schmechel D, et al. A multi-center ring trial of allergen analysis using fluorescent multiplex array technology. J Immunol Methods (2013) 387(1–2):89–95. 10.1016/j.jim.2012.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Pomponi D, Bernardi ML, Liso M, Palazzo P, Tuppo L, Rafaiani C, et al. Allergen micro-bead array for IgE detection: a feasibility study using allergenic molecules tested on a flexible multiplex flow cytometric immunoassay. PloS One (2012) 7(4):e35697. 10.1371/journal.pone.0035697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Hiller R, Harwanegg C, Müller MW. VBC Genomics Bioscience Research GmbH, assignee. Allergen Microarray Assay Patent WO2002029415 (2002).
- 97. Cretich M, di Carlo G, Longhi R, Gotti C, Spinella N, Coffa S, et al. High sensitivity protein assays on microarray silicon slides. Anal Chem (2009) 81(13):5197–203. 10.1021/ac900658c [DOI] [PubMed] [Google Scholar]
- 98. Cretich M, Breda D, Damin F, Borghi M, Sola L, Unlu SM, et al. Allergen microarrays on high-sensitivity silicon slides. Anal Bioanal Chem (2010) 398(4):1723–33. 10.1007/s00216-010-4077-x [DOI] [PubMed] [Google Scholar]
- 99. Cretich M, Damin F, Longhi R, Gotti C, Galati C, Renna L, et al. Peptide microarrays on coated silicon slides for highly sensitive antibody detection. Methods Mol Biol (2010) 669:147–60. 10.1007/978-1-60761-845-4_12 [DOI] [PubMed] [Google Scholar]
- 100. Sievers S, Cretich M, Gagni P, Ahrens B, Grishina G, Sampson HA, et al. Performance of a polymer coated silicon microarray for simultaneous detection of food allergen-specific IgE and IgG4. Clin Exp Allergy (2017) 47(8):1057–68. 10.1111/cea.12929 [DOI] [PubMed] [Google Scholar]
- 101. Dramburg S, Marchante Fernández M, Potapova E, Matricardi PM. The Potential of Clinical Decision Support Systems for Prevention, Diagnosis, and Monitoring of Allergic Diseases. Front Immunol (2020) 11:2116. 10.3389/fimmu.2020.02116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Karsonova A, Riabova K, Villazala-Merino S, Campana R, Niederberger V, Eckl-Dorna J, et al. Highly sensitive ELISA-based assay for quantification of allergen-specific IgE antibody levels. Allergy (2020) 75(10):2668–70. 10.1111/all.14325 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.