Summary
De novo germline variation in POLR2A was recently reported to associate with a neurodevelopmental disorder. We report twelve individuals harboring putatively pathogenic de novo or inherited variants in POLR2A, detail their phenotypes, and map all known variants to the domain structure of POLR2A and crystal structure of RNA polymerase II. Affected individuals were ascertained from a local data lake, pediatric genetics clinic, and an online community of families of affected individuals. These include six affected by de novo missense variants (including one previously reported individual), four clinical laboratory samples affected by missense variation with unknown inheritance—with yeast functional assays further supporting altered function—one affected by a de novo in-frame deletion, and one affected by a C-terminal frameshift variant inherited from a largely asymptomatic mother. Recurrently observed phenotypes include ataxia, joint hypermobility, short stature, skin abnormalities, congenital cardiac abnormalities, immune system abnormalities, hip dysplasia, and short Achilles tendons. We report a significantly higher occurrence of epilepsy (8/12, 66.7%) than previously reported (3/15, 20%) (p value = 0.014196; chi-square test) and a lower occurrence of hypotonia (8/12, 66.7%) than previously reported (14/15, 93.3%) (p value = 0.076309). POLR2A-related developmental disorders likely represent a spectrum of related, multi-systemic developmental disorders, driven by distinct mechanisms, converging at a single locus.
Keywords: POLR2A, Pol II, RNA polymerase II, Mendelian disease, developmental disorder, phenotypic expansion, RNAPII, RPB1, genocentric, transcriptopathy
We report twelve pathogenic de novo or inherited variants in POLR2A, detail associated phenotypes, and map all known pathogenic variants to the domain and crystal structure of RNA polymerase II. POLR2A-related developmental disorders likely represent a spectrum of related, multi-systemic disorders, driven by distinct mechanisms, converging at a single locus.
Introduction
The human enzyme DNA-directed RNA polymerase II (EC 2.7.7.6) transcribes all nuclearly encoded messenger RNA (mRNA). It is a large enzyme composed of twelve subunits, the largest of which—the 220-kDa subunit A—is encoded by POLR2A (MIM: 180660). POLR2A contains essential domains of the RNA polymerase II enzyme, including the catalytic core and a C-terminal heptapeptide repeat, the differential phosphorylation of which is critical for regulating transcriptional dynamics.1,2 Numerous structural and mutational studies in various systems have been conducted, revealing a spectrum of genetic variants differentially impacting distinct dimensions of transcription (i.e., initiation, elongation, etc.).3,4 Indeed, the function of RNA polymerase II has been extensively studied for decades.
Despite its centrality within the central dogma of molecular biology and its extensive study over decades, POLR2A was not implicated in human disease until 2016, when Clark et al.5 reported multiple distinct, recurrent somatic mutations in the gene as causative for a clinically unique subset of meningiomas. Very recently the first report of pathogenic germline mutations in POLR2A was published, describing a phenotypically heterogeneous neurodevelopmental syndrome with hypotonia (MIM: 618603).6 Here, we report additional clinical and molecular evidence strengthening the case for POLR2A dysfunction as a multi-systemic, phenotypically heterogeneous Mendelian disorder.
Material and methods
DNA sequencing and genotyping
For individuals 1, 6, and 8–13, DNA capture and sequencing of exomes was carried out as previously described by Hansen et al.7 at either the Baylor Genetics (BG) laboratories or at the Baylor College of Medicine Human Genome Sequencing Center (HGSC). Sequencing and analysis for individuals 2 (genome sequencing) and 5 (exome sequencing [ES]) were provided by the Broad Institute of MIT and Harvard Center for Mendelian Genomics (Broad CMG). ES and analysis for individuals 3, 4, and 7 was performed at other commercial clinical laboratories. Chromosomal microarray analysis (CMA) for individual 8 was performed at BG. CMA for individuals 1–8, 10, and 12 were performed at other commercial clinical laboratories. It is unknown whether CMA was performed for individuals 9 and 11.
NGS analysis
Initially, a local data lake containing ES data for approximately 20,000 individuals with suspected Mendelian disorders (Hadoop ARchitecture LakE of Exomes [HARLEE]) was utilized to discover POLR2A as a candidate Mendelian disease-associating gene. Within this dataset, fastq files were aligned to hg19, and variants were called with Atlas2 (v1.4.3) and annotated with VEP.7 High-quality ultra-rare (MAF < 1/10,000) variants observed in individuals within HARLEE were prioritized. This analysis resulted in the discovery of ultra-rare, potentially pathogenic POLR2A variants in individuals 1, 6, and 8–13. Individuals 2–5 and 7 were ascertained for this study after the initial published discovery of a POLR2A-related developmental disorder, with analysis conducted by the Broad CMG or other commercial clinical laboratories.6
For all individuals, region-specific intolerance to missense variants is calculated with the Missense Tolerance Ratio (MTR) score,8 with scores < 1.0 indicating a lower-than-expected ratio of missense to synonymous variants in the ExAC dataset9 for the 31-bp window surrounding an amino acid residue. Estimates of residue-level conservation were obtained from GERP++10 via the UCSC Genome Browser.11
Phenotyping
Phenotyping is described in the Supplemental material and methods, with all phenotypes summarized in Table 1.
Table 1.
Variant information and phenotypes of individuals with ultra-rare POLR2A variants
| Individual ID | 1 | 2 | 3 | 4 | 5 | 6 | 7 (Haijes et al.6individual 15) | 8 | 9 | 10 | 11 | 12 | Summary |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Consented to sharing of individual-level phenotypes? | yes | yes | yes | yes | yes | yes | yes | yes | no | no | no | no | NA |
| Sex | F | M | M | M | F | F | F | F | M | F | M | M | NA |
| Age (in years, at requisition or most recent phenotyping) | 0.32 | 4 | 3.5 | 10 | 21 | 6.75 | 11 | 14 | 1.13 | 16.12 | 1.49 | 4.36 | NA |
| Molecular | |||||||||||||
| Previous/additional Molecular Diagnosis | unsolved | unsolved | unsolved | unsolved | unsolved | unsolved | mitochondrial complex IV deficiency indicated via skeletal muscle enzyme analysis; mtDNA sequencing was negative | unsolved | unsolved | unsolved | unsolved | unsolved | NA |
| Chromosome | 17 | 17 | 17 | 17 | 17 | 17 | 17 | 17 | 17 | 17 | 17 | 17 | NA |
| Position | 7401503 | 7411610 | 7411736 | 7412890 | 7412890 | 7415280 | 7416391 | 7417020 | 7388166 | 7399625 | 7399813 | 7411604 | NA |
| Genomic variant | GACCTTC>G | C>T | C>T | A>G | A>G | G>A | G>A | CCA>C | C>G | G>A | C>T | C>T | NA |
| cDNA variant | c.1314_1319del | c.3281C>T | c.3407C>T | c.3752A>G | c.3752A>G | c.4252G>A | c.4808G>A | c.5440_5441del | c.83C>G | c.323G>A | c.418C>T | c.3275C>T | NA |
| Amino acid change | p.Leu438_His439del | p.Ser1094Phe | p.Thr1136Ile | p.Asn1251Ser | p.Asn1251Ser | p.Gly1418Arg | p.Arg1603His | p.Gln1814Valfs99ter | p.Pro28Arg | p.Arg108His | p.Arg140Trp | p.Ala1092Val | NA |
| GERP++ score (conservation) | mean = 4.64 (0.89–5.6; SD = 1.9; 6 bases) | 5.36 | 5.23 | 5.11 | 5.11 | 5.06 | 3.93 | mean = 1.68 (−8.25–4.13; SD = 3.06; 476 bases) | 5.82 | 5.57 | 5.57 | 5.36 | NA |
| MTR score (31 bp) (missense constraint) | mean = 0.675 (0.637–0.678; 2 amino acids) | 0.477 | 0.172 | 0.516 | 0.516 | 0.301 | 1.031 | mean = 0.7 (0.448–0.968; SD = 0.131; 158 amino acids) | 0.598 | 0.649 | 0.673 | 0.528 | NA |
| Zygosity | het | het | het | het | het | het | het | het | het | het | het | het | NA |
| De novo in proband? | yes | yes | yes | yes | yes | yes | yes | no; inherited from mother with mild difficulty learning | ? | ? | ? | ? | NA |
| Yeast variant phenotype | transcription defect (weak) (yeast Ile424Δ corresponds to human p.Leu438del) | ? | ? | ? | ? | transcription defect (slight) | ? | ? | protein defect (strong) | protein defect (weak) | protein defect (strong) | transcription defect (weak) | NA |
| Phenotypes | |||||||||||||
| Short stature (HP:0004322) | + | − | − | − | − | − | 5th percentile | − | 3/4 | 5/12 | |||
| Head and neck | |||||||||||||
| Dysmorphic features (HP:0001999) | + | high anterior hairline; mild downslanting palpebral fissures; strabismus | high anterior hairline; epicanthus; downslanting palpebral fissures; long lateral palpebral fissures; tented upper lip vermilion; prominent ears | prominent supraorbital ridges; high posterior hairline; upslanted palpebral fissures; fair complexion; deeply set eyes; large ear lobes; high palate | broad forehead; bilateral epicanthus; prominent supraorbital ridges; fair complexion; deeply set eyes; strabismus (when tired) | − | prominent ears; thin upper lip vermilion; smooth philtrum | NP | 3/4 | 9/12 | |||
| Abnormal brain MRI (HP:0012443) | polymicrogyria (HP:0002126), ventriculomegaly (HP:0002119), hydrocephalus (HP:0000238), hypomyelination (HP:0006808) | Rathke cleft cyst | − | thin corpus collosum (HP:0002079), mild pontine and interior vermian hypoplasia (HP:0012110, HP:0007068), enlargement of the fourth ventricle (HP:0002198), delayed myelination (HP:0012448) | prominent third and lateral ventricles at 1 year old (HP:0007082, HP:0006956) | − | thin corpus collosum (HP:0002079), ventriculomegaly (HP:0002119), 7 mm cyst in temporal parietal junction, bilateral loss of white matter (HP:0012429) | polymicrogyria involving large portions of left cerebral hemisphere including perisylvian region and portions of the left frontal, parietal, and temporal lobes (HP:0002126); left cerebral atrophy (HP:0002059); small enhancing developmental venous anomaly at right parieto-occipital junction (HP:0012481) | 1/4 | 7/12 | |||
| Microcephaly/ macrocephaly | NP | − | − | significant microcephaly (<2 percentile) | − | − | microcephaly | − | macrocephaly 1/4 | 3/12 | |||
| Visual impairment (HP:0000505) | NP | − | astigmatism, both eyes | bilateral elongation of ocular globes noted on MRI | − | cortical | irregular astigmatism, both eyes | wears glasses since age 2 years, no astigmatism reported | NP | 5/12 | |||
| Hearing impairment (HP:0000365) | NP | − | − | − | − | sensorineural | − | right-side hearing deficit attributed to hemiplegia | NP | 2/12 | |||
| Headache (HP:0002315) | NP | − | − | reported by parents; cannot confirm as individual is non-verbal | − | − | reported by parents; cannot confirm as individual is non-verbal | daily severe headaches | NP | 1/10 | |||
| Central nervous system | |||||||||||||
| Developmental delay (HP:0012758) | + | + | + | + | speech and motor (HP:0000750, HP:0001270) | + | + | speech and motor (HP:0000750, HP:0001270) | 4/4 | 12/12 | |||
| Developmental regression (HP:0002376) | NP | three separate 6-month episodes of regression and plateau | − | possible recent regression | regression at age 16 years attributed to worsening anxiety, with new aggressive behaviors | − | − | no regression, but developed personality changes (HP:0000751) and memory problems (HP:0002354) at age 8 years | NP | 4/12 | |||
| Seizures (HP:0001250) | NP | EEG revealed right centroparietal epileptiform discharges | − | generalized tonic-clonic and absence | during childhood, with no discernable seizures since puberty, no longer on anticonvulsants | absence seizures | + | started at 13 years old | 2/4 | 8/12 | |||
| Intellectual disability (HP:0001249) | NP | + | + | + | + | + | + | mild | 1/4 | 8/12 | |||
| Autism spectrum disorder (HP:0000729) | NP | + | + | + | − | + | + | − | 1/4 | 6/12 | |||
| Hypotonia (HP:0001252) | + | + | + | + | + | + | + | − | 1/4 | 8/12 | |||
| Dystonia (HP:0001332) | NP | − | + | + | − | − | − | hypertonia/spasticity of the right extremities due to hemiplegia (HP:0001276, HP:0004374) | NP | 3/12 | |||
| Neck and back arching in infancy | NP | + | + | − | + | NP | − | − | NP | 3/12 | |||
| Abnormal movements (HP:0100022) | NP | hand flapping | hand flapping, kicking, rubs side of head | head banging | occasional hand flapping | hand flapping | head hitting, stimming arms and legs | − | 1/4 | 7/12 | |||
| Pica (HP:0011856) | NP | eats hair, paper, feces | NP | frequently puts objects in mouth | NP | NP | chews on hands, toes, clothes, paper, furniture, toys | NP | NP | 3/12 | |||
| Ataxia (HP:0001251) | NP | + | + | + | ataxic high-stepping gait | + | + | ataxic gait | NP | 7/12 | |||
| Difficulty sleeping (HP:0002360) | NP | obstructive sleep apnea prior to removal of tonsils and adenoids; sleeps >12 h and exhibits daytime sleepiness | − | + | woke up 10× per night in childhood, current symptoms are occasional and mild | NP | no sleep study, but reportedly very light sleeper | insomnia, takes clonidine for it | NP | 5/12 | |||
| Musculoskeletal | |||||||||||||
| Skeletal abnormality (HP:0000924) | NP | − | − | mild pectus excavatum (HP:0000767), hip dysplasia (HP:0001385), sixth lumbar vertebrae (HP:0008416) | short Achilles tendons have caused foot distortion and difficulty walking (HP:0001771) | NP | hip dysplasia (HP:0001385), pes planus (HP:0001763), leg length discrepancy (HP:0100559) | contractures on right side (HP:0001371), two Achilles tendon release surgeries (HP:0001771) | 1/4 | 5/12 | |||
| Scoliosis (HP:0002650) | NP | − | − | − | − | neurogenic | − | − | NP | 1/12 | |||
| Joint hypermobility (HP:0001382) | NP | + | + | + | − | hyperextensible elbows and knees | + | − | 1/4 | 6/12 | |||
| GI system | |||||||||||||
| Failure to thrive (HP:0001508)/ feeding difficulty (HP:0011968) | + | + | − | poor weight gain but normal linear growth | + | − | + | − | 2/4 | 7/12 | |||
| Uses gastrostomy tube | NP | planned but not yet inserted | − | inserted at 9 years old | − | inserted at 5 years old | − | NP | 3/12 | ||||
| GI reflux (HP:0002020) | NP | + | − | + | + | − | − | + | 1/4 | 5/12 | |||
| Constipation (HP:0002019)/ diarrhea (HP:0002014) | NP | constipation | constipation | constipation | − | − | constipation and diarrhea | − | diarrhea 1/4 | 5/12 | |||
| Other | |||||||||||||
| Urogenital system (HP:0000119) | NP | incontinence | incontinence | neurogenic bladder; incontinence; swollen left kidney after birth; small scrotum, retractile testes | toilet trained at age 6 years, still some incontinence | NP | incontinence | history of several urinary tract infections (HP:0000010) | kidney abnormality 1/4 | 7/12 | |||
| Skin abnormality (HP:0000951) | NP | easy scarring, keratosis pilaris | − | easy scarring | easy scarring | − | keratosis pilaris, facial flushing | − | unspecified abnormality 1/4 | 5/12 | |||
| Immune system (HP:0002715) | NP | neutropenia (HP:0001875); granulocytopenia (HP:0001913); high lymphocyte percentage (HP:0100827) | − | intermittent leukocytosis (HP:0001974) | − (no antibiotics needed for >10 years) | NP | − | − | unspecified abnormality 1/4 | 3/12 | |||
| Fever (HP:0001954) | NP | chronic (HP:0001955) | − | fever of unknown etiology (HP:0001955) | fever of unknown etiology in childhood (HP:0001955) | − | − | − | chronic fever 1/4 | 4/12 | |||
| Cardiac abnormality (HP:0001627) | dilated cardiomyopathy (HP:0001644) | − | Bicuspid aortic valve (HP:0001647) | Atrial septal defect (HP:0001631), congestive heart failure in infancy (HP:0001635) | − | NP | − | − | NP | 3/12 | |||
| Lung abnormality (HP:0002086) | immature lungs (HP:0006703) | recurrent upper respiratory infections (HP:0002788); frequent nebulizer use (budesonide and albuterol) | recurrent upper respiratory infections (HP:0002788); uses inhaler and nebulizer | poor airway clearance requiring vest therapy, suctioning, and cough assist; recurrent upper respiratory infections (HP:0002788) | − | − | recurrent upper respiratory infections (HP:0002788); hospitalized four times for pneumonia, once for bronchiolitis | − | NP | 5/12 | |||
| Congenital diaphragmatic hernia (HP:0000776) | NP | − | − | − | − | − | − | − | 1/4 | 1/12 | |||
| Other | NP | hypothyroidism (HP:0000821); tooth misalignment (HP:0000692); consistently elevated BUN/creatine ratio (HP:0040081), alkaline phosphatase (HP:0004379), and prolactin (HP:0040086) | − | history of angioedema with levetiracetam use (HP:0100665); hemangioma (HP:0001028); 2-vessel umbilical cord (HP:0001195); adverse reaction to therapeutic botulinum toxin; enlarged liver that resolved after birth (HP:0006564); hyponatremia (HP:0002902) | psychosis induced by fluoxetine; agitation with sedatives and general anesthesia | NP | intolerant of wheat and dairy (not allergic); reduced fetal movement (HP:0001558) | − | NP | 4/12 | |||
All variants are considered likely pathogenic. Phenotypes are indicated as either positive, negative, or not phenotyped or reported (NP) for each individual. For research-consented individuals, individual-level phenotypic information is presented. For the four other clinical individuals (individuals 9–12), phenotypic information is presented in aggregate. The summary column indicates how many individuals were positive for a given phenotype. All cDNA variants are based on the NM_000937.4 transcript definition. All genomic coordinates are based on the hg19 genome build. Human Phenotype Ontology (HPO) IDs are indicated for phenotypes where appropriate. +, positive; −, negative; BUN, blood urea nitrogen; het, heterozygous; GI, gastrointestinal; NA, not applicable.
Structural domains and alignment
POLR2A structural domains were derived from the yeast RNA polymerase II (Pol II) crystal structure, which shares a remarkably high level of conservation with POLR2A.3 Human POLR2A domain coordinates were derived by alignment as performed with MAFFT FFT-NS-2 (v7.305b).12
Functional evaluation of variants in yeast
Thirteen ultra-rare variants in POLR2A were identified in clinical samples at the time the yeast experiments were initiated. Yeast studies were conducted in the yeast ortholog of POLR2A, RPO21 (generally referred to as RPB1), and are summarized in Table 2. Detailed experimental methods are described in the Supplemental material and methods.
Table 2.
Phenotypic analyses of potentially pathogenic POLR2A variants in yeast
| RPB1 alleles | POLR2A (Human) | Individual with variant | YPD 30°C | YPD 37°C | SC-Leu MPA (20μg/ml) | YPD + Formamide (3%) | SC-Lys (Spt-) | YPRaf/Gal | SC-His |
|---|---|---|---|---|---|---|---|---|---|
| WT | WT | NA | + | + | MPAR | FormamideR | - | Gals | - |
| Pro24Arg | Pro28Arg | 9 | -/+ | - | MPAR | FormamideS | - | Gals | - |
| Glu26Val | Glu30Val | NA | + | + | MPAR | FormamideR | - | Gals | - |
| Glu104His | Arg108His | 10 | + | +/- | MPAR | FormamideR | - | Gals | - |
| Glu104Leu | Arg108Leu | NA | +/- | --/+ | MPAR | Weak FormamideS | - | Gals | - |
| Arg134Trp | Arg140Trp | 11 | -/+ | - | MPAR | FormamideS | - | Gals | - |
| Arg175Trp | Arg192Trp | NA | + | + | MPAR | FormamideR | - | Gals | - |
| Gln313Cys | Arg327Cys | NA | + | + | MPAR | FormamideR | - | Gals | - |
| Leu388Val | Leu402Val | NA | + | + | MPAR | FormamideR | - | Gals | - |
| Asp423ΔIle424Δ | Asp437_Leu438del | NA | + | + | MPAR | Weak FormamideS | - | GalR | - |
| Asp423Δ | Asp437del | NA | + | + | MPAR | Weak FormamideS | - | GalR | - |
| Ile424Δ | Leu438del | 1 | + | + | MPAR | FormamideR | - | GalR | - |
| Ala1069Val | Ala1092Val | 12 | + | + | MPAR | FormamideR | - | GalR | - |
| Thr1272Ala | Thr1297Ala | NA | + | + | MPAR | FormamideR | - | Gals | -/+ |
| Gly1388Arg | Gly1418Arg | 6 | + | + | MPAR | FormamideR | - | Slight GalR | - |
| Gly1388Val | Gly1418Val | NA | + | + | MPAR | FormamideR | - | Slight GalR | - |
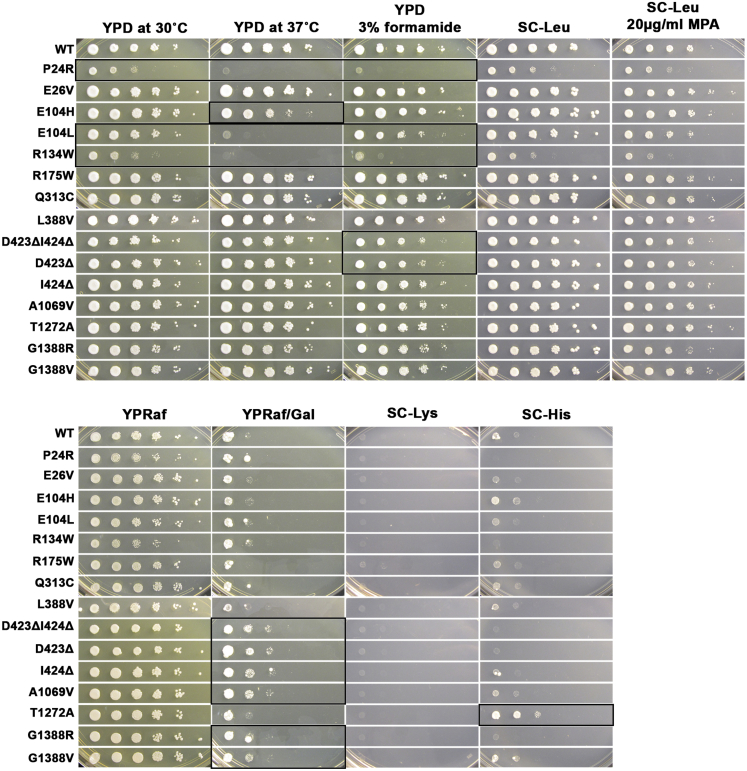
Table summarizing phenotypic analysis of potentially pathogenic POLR2A variants in yeast by spot assay (Figure 3). General (growth at standard propagation temperature on standard medium YPD 30°C) or conditional growth defects (YPD 37°C), YPD in presence of 3% formamide (a protein-folding stressor), or minimal media that enable detection of transcription-related phenotypes are shown: addition of drug mycophenolic acid (MPA) detecting defective IMD2 expression, medium lacking lysine (SC-Lys) for detection of altered transcription due to Suppressor of Ty phenotype (Spt-) at the lys2-128∂ reporter allele, rich medium YP with 2% raffinose and 1% galactose for detection of altered transcription apparent as suppression of the galactose toxicity (GalS) imposed by the gal10Δ56 reporter allele (YP Raf/Gal), medium lacking histidine for detection of altered transcription allowing constitutive expression of the imd2Δ::HIS3 transcriptional reporter (SC-His). Shaded cells indicate a phenotype different from WT, with darker shading indicating a stronger phenotype than lighter shading. Strength of growth is indicated as robust growth (+), reduced growth (+/−), weak growth (−/+), severe growth defect (−−/+), no growth (−).
Ethics statement
Data for individuals 1–8 were collected after written informed consent in conjunction with the Baylor Hopkins Center for Mendelian Genomics (CMG) (H-29697) study with approval by the institutional review board at Baylor College of Medicine. Other clinical samples (individuals 9–12) were from the Baylor College of Medicine clinical testing laboratories, now incorporated as BG; these data were studied in aggregate for the purpose of improving the diagnostic assay, under protocol H-41191. The procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national), and proper informed consent was obtained.
Results
Utilizing the HARLEE data lake,7 a total of seven clinical exomes were originally identified with ultra-rare (MAF ≤ 1/10,000) suspected pathogenic variants in POLR2A. Attempts were made to contact all individuals and recruit to a research protocol (see Material and methods). After the initial report of de novo variation in POLR2A causing a Mendelian disorder,6 five additional individuals were subsequently identified through the genetics clinic at Texas Children’s Hospital and a social media-based support group for families of individuals diagnosed with pathogenic variants in POLR2A (Table 1), including one previously published individual for whom amended and additional phenotypic information is provided (individual 7; Haijes et al.,6 individual 15). The family of individual 7 reports that she is positive for a few phenotypes previously reported as negative: feeding difficulty/failure to thrive, decreased endurance, and decreased fetal movement. Phenotypic data from clinical samples detected in HARLEE for individuals without research consent are reported in aggregate, in accordance with institutional review board (IRB)-approved protocols (see Ethics statement).
Clinical ES or genome sequencing revealed no other genetic diagnosis for any of these individuals. CMA reportedly did not reveal any findings for individuals 1–8 and 12. It is unknown whether CMA was performed for individuals 9 and 11. CMA for individual 10 reportedly identified a variant of uncertain significance (VUS) gain in Xp22.13. Thus, in total, twelve individuals with ultra-rare, putatively pathogenic variants in POLR2A are reported. Pathogenicity for all of these variants is supported by the extreme degree of constraint for missense (missense intolerance Z score = 8.59) and loss-of-function (pLI = 1.0) variants observed in healthy individuals from the ExAC dataset.9 All variants occur at highly conserved residues (as indicated by a GERP++ score ≥ 2.0), in regions further constrained for missense variation in the ExAC dataset (Table 1).8,10 Of these twelve individuals, there are ten affected by missense variants (including one previously published individual and two individuals affected by the same variant). We also report one individual affected by an in-frame deletion and one family affected by a C-terminal frameshift variant. Variants are distributed throughout the length of the protein product, with no obvious association between severity of phenotype and affected protein domain (Figure 1). We report one variant located within the Clamp core domain, two variants within the Clamp head, one variant within the Dock, three variants within the Cleft, one variant within the Trigger loop, one variant within the Jaw (observed in two individuals), and one frameshift variant in the C-terminal domain (CTD), which is likely to escape nonsense-mediated decay as it occurs within the last exon of the gene.13
Figure 1.
POLR2A allelic series
(A) The x axis represents the entire length of the POLR2A protein, spanning 1,970 amino acids. Potentially pathogenic variants observed in individuals enrolled in this study are shown along the top in blue, with each circle representing an individual harboring the indicated variant. Previously reported pathogenic variants are shown along the bottom, including both those associated with Mendelian (green) and somatic (red) disease. Each variant is mapped to its corresponding structural domain, as indicated by the legend. Previously reported Mendelian variants are limited to those occurring in the Active site, Pore 1, Funnel, Cleft, Trigger Loop, Jaw, and C-terminal domain (CTD). Variants observed in this study additionally include those affecting the Clamp core, Clamp head, and Dock. Variant p.Asn1251Ser is the only variant recurrently reported to date and is observed in three unrelated individuals. Variant p.His439_Leu440del is the only variant reported in both somatic (meningioma) and Mendelian (POLR2A-related congenital transcriptopathy) disorders.
(B) Structural view of yeast Pol II (amalgam of RNA polymerase and TFIIS from PDB 1Y1V and nucleic acid DNA and RNA components from PDF 2E2H). Non-Rpb1 (POLR2A ortholog) subunits are shown in transparent surface view. Rpb1 is shown as cartoon within transparent surface. Domains of Rpb1 are color coded to match diagram in (A), and implicated residues are labeled and shown as colored spheres. On left is an oblique “front” view of the Pol II complex; on right, the structure is rotated to a “top” view. Generally speaking, pathogenic variants are widely distributed throughout the structure, without any obvious patterns of clustering, potentially consistent with multiple, distinct mechanisms of pathogenicity.
Missense variants
Of the ten individuals harboring missense variants, four non-research-consented individuals have phenotypes reported in aggregate (individuals 9–12) rather than at an individual level. Six of these ten missense variants were confirmed to be inherited de novo. The remaining four were observed in clinical samples for which parental ES was not performed, but their pathogenicity is further supported by functional evidence and phenotypic similarity. Commonly reported phenotypes among these individuals include developmental delay (10/10), intellectual disability (7/7), seizures (≥7/10), hypotonia (≥7/10), abnormal movements (≥7/10), ataxia (≥6/10), autism spectrum disorder (ASD) (≥6/7), failure to thrive/feeding difficulty (≥6/10), joint hypermobility (≥6/10), abnormal brain MRI (≥5/10), incontinence (≥5/10), skin abnormalities including keratosis pilaris and easy scarring (≥5/10), visual impairment (≥4/10), short stature (≥4/10), difficulty sleeping (≥4/10), skeletal abnormalities (≥4/10), recurrent upper respiratory infections (≥4/10), and cool distal extremities (≥4/10). (Diagnoses of intellectual disability and autism spectrum disorder cannot be ruled out for three clinical samples, as phenotyping for these individuals is limited to what was included on the ES requisition, ordered for these individuals at an age too young to typically diagnose intellectual disability or ASD.)
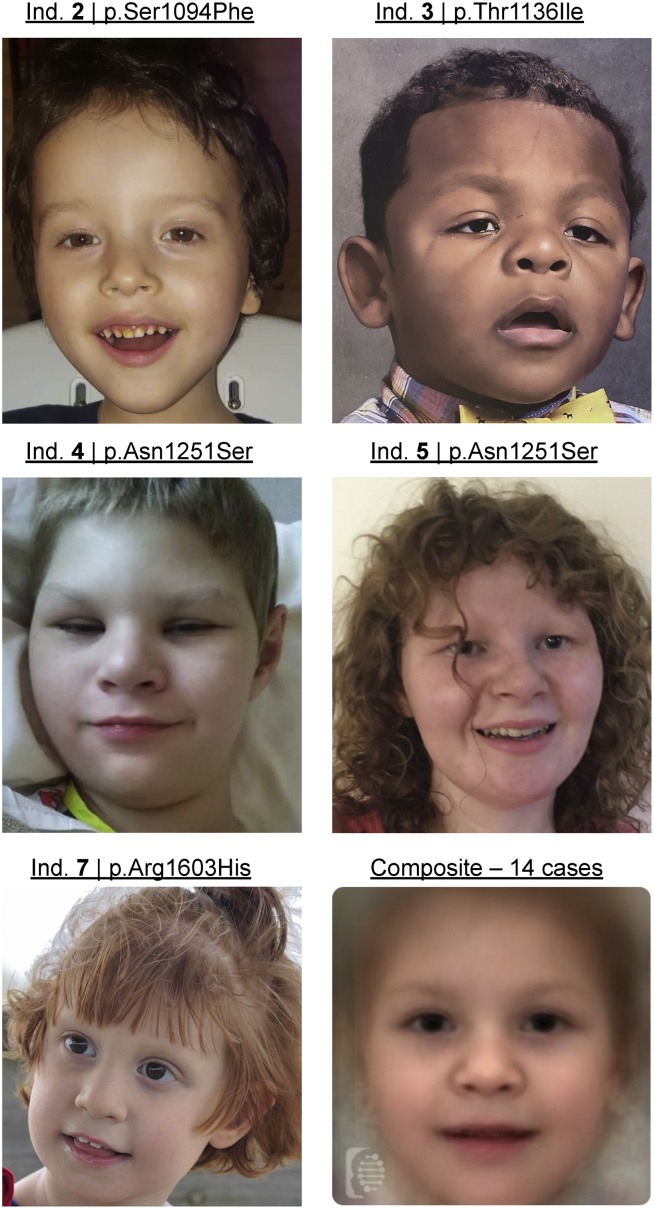
Individuals 4 and 5 are of note, sharing an identical variant with a previously reported individual: c.3752A>G (p.Asn1251Ser; Haijes et al.6) Individual 14 was reported as a 6-year-old girl with hypotonia, strabismus, frog position in infancy, decreased endurance, feeding difficulties, recurrent respiratory tract infections, disturbed sleeping, gastro-esophageal reflux, failure to thrive, microcephaly, brachyplagiocephaly, decreased fetal movements, aggressive behavior, pectus excavatum, walking at 5.5 years of age, and mega cisterna magna.6 A detailed clinical description of individuals 4 and 5 is included in the supplement of this manuscript (Supplemental note). There is considerable variability in age of walking across these three individuals, reported as 3.5 (individual 5), 4.5 (individual 4), and 5.5 (Haijes et al.,6 individual 14) years of age. Other notable phenotypic differences include the presence of a cardiac abnormality (atrial septal defect) in individual 4 and the presence of recurrent respiratory infections in individual 4 and Haijes et al.6 individual 14, but not in individual 5. Facial dysmorphology is relatively similar for individuals 4 and 5 (Figure 2).
Figure 2.
Photographs of individuals with putatively pathogenic POLR2A variants
All research subjects consenting to publication of photographs are included. High forehead, mild downslanting palpebral fissures, and strabismus are observed in individual 2. High forehead, epicanthus, downslanting palpebral fissures, tented mouth, and simplified and prominent ears are observed in individual 3. Prominent supraorbital ridges, high posterior hairline, upslanted palpebral fissures, large, fleshy ears, deep-set eyes, and high palate are observed in individual 4; individual 5, affected by the same variant as individual 4, exhibits remarkable similarity to individual 4, with broad forehead, bilateral epicanthus, prominent supraorbital ridges, and deep-set eyes. Prominent ears, thin upper lip, tubular nose, and smooth philtrum are observed in individual 7. In the lower-right panel, a composite image was created using the Face2Gene tool, compiled using the five individuals reported here together with photos of individuals previously reported by Haijes et al.6 All photos published by Haijes et al. were included except for two photos with individuals wearing glasses (Haijes et al. individuals 7 and 13) and an additional photo (Haijes et al. individual 2) unable to be processed by Face2Gene, likely due to poor lighting. The composite image reveals no consistent, remarkably distinctive facial dysmorphology observed across all individuals affected by potentially pathogenic variants in POLR2A.
Other variants
We report one frameshift variant that is potentially pathogenic—g.7417023_7417024del (GenBank: NC_000017.10) (c.5440_5441del [NM_000937.4]; p.Gln1814Valfs99ter)—observed in individual 8, with a remarkably more mild presentation than the individuals affected by missense variants. This variant occurs in the last exon of the gene (29/29) and leads to a premature termination near the C-terminal end of the protein, after amino acid residue 1912/1970, and an alteration of 99 amino acids. This alteration would result in truncation of the CTD by 20 heptapeptide repeats and 1 partial out of 52. Truncation of CTD repeats can confer phenotypes in model organisms.14, 15, 16, 17, 18, 19, 20 Furthermore, given its position in the gene, it is predicted to escape nonsense-mediated decay, with a fully expressed, yet truncated, protein product. Individual 8 presented at 8 years of age with hypertonia/spasticity of the right extremities, recent worsening headache, memory problems and personality changes, hemiplegia, and delayed speech and motor milestones. MRI of the brain revealed left hippocampal atrophy, and an electroencephalogram (EEG) revealed a focus of spike activity in the left central region. Clinical ES initially failed to detect any pathogenic variants consistent with phenotypic presentation. At most recent follow-up, individual 8 was reported to have developed seizures at age 13 years, recurrent urinary tract infections, and a history of two Achilles tendon release surgeries. Daily severe headaches were reported, as well as difficulty sleeping. Aggregate genocentric reanalysis through HARLEE7 revealed individual 8 as positive for the ultra-rare C-terminal frameshift variant in POLR2A. Sanger sequencing of proband and maternal saliva samples revealed the variant to be maternally inherited. The mother of individual 8 reports no significant related medical history other than delayed speech and mild learning difficulty. Maternal family history is also positive for a son who was born at 35 weeks and had hypoplastic left heart, brain anomaly, and failure to thrive. He had three open heart surgeries and died at 7 months of age. He was reported to have a chromosome 6p duplication with an unknown POLR2A genotype.
Harboring an in-frame deletion of two amino acids, individual 1 presented with the most severe phenotype of all evaluated individuals. Clinical ES was ordered at 3 months of age, with reported phenotypes of immature lungs, dilated cardiomyopathy, failure to thrive, hypotonia, developmental delay, dysmorphic features, and abnormal brain MRI findings including polymicrogyria, ventriculomegaly, hydrocephalus, and hypomyelination. ES failed to detect any known pathogenic variants consistent with phenotypic presentation. Individual 1 reportedly died during infancy. As above, genocentric reanalysis identified an ultra-rare variant in POLR2A—chr17:g.7401503GACCTTC>G (NM_000937.4) (c.1314_1319del; p.His439_Leu440del). The average GERP score across the six deleted nucleotides is 4.64, indicative of high evolutionary conservation. Notably, this same variant, when present as a developmental somatic mutation, has been established as causal for a subset of meningiomas.5 Meningioma was not reported in individual 1. Sanger sequencing of parental saliva samples failed to detect the variant. Sanger sequencing in individual 1 confirmed the presence of the heterozygous variant.
Functional studies
Functional assays were conducted in yeast to further evaluate the pathogenicity of observed POLR2A variants in clinical and research samples (Figure 3). As noted above, the large subunit of Pol II (encoded by POLR2A in human, RPB1 in yeast) is highly conserved in sequence and structure. We previously established a number of plate phenotypes highly predictive of transcription defects due to specific alterations to Pol II catalytic activity in yeast.21,22 Residues analogous to some identified in individuals had been identified previously as being mutated in genetic screens for yeast transcription mutants; rpb1 Pro24Ser was identified as rpb1-915 (analogous residue to POLR2A p.Pro28), while rpb1 Gly1388Val was identified as sua8-423 (analogous to POLR2A p.Gly1418). Here we employed these tests to interrogate conserved residues impacted by missense variants observed in humans for growth defects in yeast. The yeast strains utilized were the same as in Haijes et al.,6 as their strains were derived from the Kaplan lab. Mutant plasmids encoding variants in conserved residues identified in a subset of individuals were introduced into yeast as the sole copy of RPB1 and phenotyped on a number of growth media. We observed conditional growth defects as well as phenotypes related to altered transcription for a subset of mutants (Table 2). Conditional defects such as temperature sensitivity or formamide sensitivity are consistent with protein folding or assembly defects exacerbated by heat or solvent, and these were observed for yeast rpb1 Pro24Arg, Glu104Leu, and Arg134Trp (analogous to p.Pro28Arg, p.Arg108Leu, and p.Arg140Trp, respectively). Other subsets of alleles show suppression of the gal10Δ56 transcriptional reporter (Asp423delIle424Δ, Asp423Δ, Ile424Δ, Asp1069Val, Gly1388Arg, and Gly1388Val) or constitutive expression of the imd2promoter::HIS3 transcriptional reporter due to altered transcription start selection (Thr1272Ala) (see Table 2 for corresponding human variants).21,22,24 Each of these phenotypes has been linked to altered Pol II transcription, usually due to decrease in Pol II catalytic function.21,22,24,25 These phenotypic effects are relatively minor and would be consistent with subtle alterations to Pol II function.
Figure 3.
Phenotypes of yeast rpb1 mutants analogous to human POLR2A alleles
10-fold serial dilution of saturated cultures of rpb1 mutants or RPB1 wild-type (WT) control were spotted onto different media and phenotypes determined. Growth on YPD, a rich medium containing dextrose as carbon source, serves as control, against which growth phenotypes on YPD at 37°C and 3% formamide are compared. Growth sensitivity to 20 μg/mL of the drug mycophenolic acid (MPA) added to a synthetic defined medium lacking leucine (SC-Leu) is compared to growth on SC-Leu. SC-Leu serves as a selection medium for LEU2 expressing plasmids. The presence of gal10Δ56 in cells confers galactose toxicity that can be suppressed by mutants that alter transcription and is apparent as resistance to galactose, as evidenced by comparing YPRaf/Gal (containing galactose and raffinose as carbon source) to YPRaf (containing only raffinose). Growth of cells possessing the lys2-128∂ on SC-Lys results in Lys+/Spt− phenotype, wherein WT cells are Lys−; this medium can detect a subset of Pol II alleles with increased catalytic activity. In cells containing imd2Δ::HIS3, aberrant constitutive expression from the IMD2 promoter can be detected by growth on medium lacking histidine, as this allele replaces the IMD2 open reading frame with HIS3, meaning HIS3 is now under control of the IMD2 promoter; mutants defective for Pol II catalytic activity can confer this phenotype. Strains possessing imd2Δ::HIS3 have been derived from CKY865, whereas all the other mutants have been tested in CKY283 (Table S1). For all assays and mutants (Table S2), serial dilutions demonstrating abnormal phenotypes as compared to WT are indicated with a black box.
Discussion
Herein, we confirm the recent discovery of association between pathogenic germline variation in POLR2A and a phenotypically heterogeneous neurodevelopmental disorder.6 We report the transmission of a potentially pathogenic POLR2A variant within a family—individual 8, inheriting a p.Gln1814Valfs99ter variant from a mother with a remarkably mild presentation of delayed speech and mild learning difficulties. We observe several previously unreported phenotypes (as compared to Haijes et al.6) in individuals with POLR2A-related disorders, including ataxia (observed in 7/12, or 58.3% of individuals), joint hypermobility (6/12, 50%), short stature (5/12, 41.7%), skin abnormalities including easy scarring and keratosis pilaris (5/12, 41.7%), recurrent febrile illness of unknown etiology (4/12, 33.3%), congenital cardiac abnormalities (3/12, 25%), immune system abnormalities (3/12, 25%), hip dysplasia (2/12, 16.7%), and short Achilles tendons (2/12, 16.7%). We also report a significantly higher proportion of individuals with epilepsy (8/12, 66.7%) than previously reported (3/15, 20%) (p value = 0.014196; chi-square test) and a somewhat lower proportion of individuals with hypotonia (8/12, 66.7%) than previously reported (14/15, 93.3%) (p value = 0.076309).6 We describe the facial dysmorphology of a subset of affected individuals, which is generally mild and nonspecific across individuals with different variants but remarkably similar for the two reported individuals sharing the same variant (individuals 4-5) (Figure 2).
In this cohort, previously unreported neuroradiological anomalies include polymicrogyria (2/12, 16.7%) and various benign, congenital anomalies, which cannot yet be ruled out as unrelated to POLR2A dysfunction, each occurring in a single individual: Rathke cleft cyst, hemangioma, and a small, enhancing developmental venous anomaly (DVA). We also report one individual (individual 1) with a germline variant identical to a previously reported meningioma-causing somatic mutation (p.His439_Leu440del).5 As individual 1 died during infancy, the extent of correlation between germline inheritance of p.His439_Leu440del (or other pathogenic germline variants) and risk of developing meningioma remains unclear.
Due to the centrality of POLR2A in transcriptional networks and the wide range of ways in which its function is known to be regulated, it can be reasonably inferred that a spectrum of possible pathogenic genetic variants will present with differential phenotypic presentation and severity. Future efforts should focus on elucidating the molecular mechanisms of pathogenicity—common or distinct—across the spectrum of known pathogenic POLR2A variants. Phenotypes of tested mutants in yeast in most instances were relatively weak, though a subset are strongly predicted to have protein structural or stability defects. Known catalytic mutants identified in yeast cause widespread transcriptional defects when introduced into human cells and likely would not be viable in an organism.26, 27, 28 For example, a known slow-elongating variant has been introduced into mouse embryonic stem cells (Polr2a Arg749His, analogous to Arg726His in yeast), could not be transmitted through the germline, and caused early embryonic lethality.29 Mouse embryonic stem cells containing Arg749His showed defects upon neuronal differentiation, likely deriving from observed altered elongation rate, gene expression, and alternative splicing changes. Of interest, long genes, which are enriched among neuronally expressed genes, might be predicted to be especially sensitive to altered Pol II elongation or cotranscriptional splicing defects.30 Haijes et al.6 delineate two potential molecular mechanisms of disease: haploinsufficiency (with a relatively mild presentation) and a dominant-negative effect caused by aberrant Pol II elongation. We emphasize that despite the relatively mild presentation of all individuals harboring truncating and frameshift variants reported to date (p.Gln700∗, p.Gln735∗, p.Pro1767fs, and p.Gln1814fs),6 the potentially pathogenic CTD frameshift variants (p.Pro1767fs and p.Gln1814fs) most likely escape nonsense-mediated decay and thus exhibit a dominant-negative mechanism of pathogenicity. Therefore, while haploinsufficiency associated with p.Gln700∗ and p.Gln735∗ almost certainly constitutes a distinct mechanism of pathogenicity, expressed mutant POLR2A products can cause the full range of phenotypic severity observed in POLR2A-related disorders. Taken together, these molecular and phenotypic data suggest that these pathogenic variants constitute a spectrum of transcriptional dysfunction, with phenotypes likely explained by a combination of specific POLR2A variation in conjunction with the genetic burden across a potential variant- or domain-specific network of interacting partners. Such a model could explain both a degree of phenotypic convergence (i.e., similar facial dysmorphism in individuals 4 and 5) and variable expressivity (i.e., microcephaly, cardiac and immune abnormalities present in individual 4 but not in individual 5) or incomplete penetrance (i.e., the mother of individual 8 exhibiting only sub-clinical phenotypes) for a given disease-associating variant. To assess this model, global transcriptional profiling (via RNA-sequencing [RNA-seq], GRO-seq, etc.) could be evaluated across biologically relevant cell types or tissues using patient-derived induced pluripotent stem cell (IPSC) lines. The function of individual pathogenic variants—in the appropriate genetic background—could be assessed by transcriptional profile comparison of patient-derived cells against a split of the same cell line with a mutationally induced wild-type POLR2A. The function of different pathogenic variants could then be compared by normalizing their impact against their respective isogenic controls.
Acknowledgments
This work was supported in part by grants UM1 HG008898 from the National Human Genome Research Institute (NHGRI) to the Baylor College of Medicine Center for Common Disease Genetics and UM1 HG006542 from the National Heart, Lung, and Blood Institute (NHLBI) and NHGRI to the Baylor Hopkins Center for Mendelian Genomics. C.D.K. was supported by grants R01 GM097260 and R01 GM120450 National Institute of General Medical Sciences (NIGMS). A.W.H. was supported in part by NIH T32 GM08307-26, The Cullen Foundation, and the Baylor College of Medicine President’s Circle. D.P. was supported by Clinical Research Training Scholarship in Neuromuscular Disease partnered by the American Academy of Neurology (AAN), American Brain Foundation (ABF) and Muscle Study Group (MSG), and the International Rett Syndrome Foundation (IRSF grant #3701-1). S.M.W. is supported by the Victorian Government’s Operational Infrastructure Support Program. S.D. was supported by the National Human Genome Research Institute, the National Eye Institute, and the National Heart, Lung, and Blood Institute grant UM1 HG008900 and in part by National Human Genome Research Institute grant R01 HG009141. We thank Ali Jalali and Amy L. McGuire for the valuable discussions. Last, we thank the individuals participating in research and their families for their assistance and significant contributions to this research.
Declaration of interests
J.R.L. has stock ownership in 23andMe, is a paid consultant for Regeneron Pharmaceuticals, and is a co-inventor on multiple US and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, and bacterial genomic fingerprinting. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from clinical genetic testing offered in the Baylor Genetics Laboratory.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.xhgg.2020.100014.
Contributor Information
Adam W. Hansen, Email: awhphd@gmail.com.
Craig D. Kaplan, Email: craig.kaplan@pitt.edu.
Data and Code Availability
POLR2A variants are available on ClinVar: SUB7404119, accession pending review.
Web resources
Baylor Genetics Laboratory, https://baylorgenetics.com
Face2Gene tool, https://www.face2gene.com
OMIM, https://www.omim.org
The Human Phenotype Ontology, https://hpo.jax.org/app/
Supplemental information
References
- 1.Jeronimo C., Collin P., Robert F. The RNA Polymerase II CTD: The Increasing Complexity of a Low-Complexity Protein Domain. J. Mol. Biol. 2016;428:2607–2622. doi: 10.1016/j.jmb.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 2.Harlen K.M., Churchman L.S. The code and beyond: transcription regulation by the RNA polymerase II carboxy-terminal domain. Nat. Rev. Mol. Cell Biol. 2017;18:263–273. doi: 10.1038/nrm.2017.10. [DOI] [PubMed] [Google Scholar]
- 3.Cramer P., Bushnell D.A., Kornberg R.D. Structural basis of transcription: RNA polymerase II at 2.8 ångstrom resolution. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 4.Cramer P. Organization and regulation of gene transcription. Nature. 2019;573:45–54. doi: 10.1038/s41586-019-1517-4. [DOI] [PubMed] [Google Scholar]
- 5.Clark V.E., Harmancı A.S., Bai H., Youngblood M.W., Lee T.I., Baranoski J.F., Ercan-Sencicek A.G., Abraham B.J., Weintraub A.S., Hnisz D., et al. Recurrent somatic mutations in POLR2A define a distinct subset of meningiomas. Nat. Genet. 2016;48:1253–1259. doi: 10.1038/ng.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haijes H.A., Koster M.J.E., Rehmann H., Li D., Hakonarson H., Cappuccio G., Hancarova M., Lehalle D., Reardon W., Schaefer G.B., et al. De Novo Heterozygous POLR2A Variants Cause a Neurodevelopmental Syndrome with Profound Infantile-Onset Hypotonia. Am. J. Hum. Genet. 2019;105:283–301. doi: 10.1016/j.ajhg.2019.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hansen A.W., Murugan M., Li H., Khayat M.M., Wang L., Rosenfeld J., Andrews B.K., Jhangiani S.N., Coban Akdemir Z.H., Sedlazeck F.J., et al. Task Force for Neonatal Genomics A Genocentric Approach to Discovery of Mendelian Disorders. Am. J. Hum. Genet. 2019;105:974–986. doi: 10.1016/j.ajhg.2019.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Traynelis J., Silk M., Wang Q., Berkovic S.F., Liu L., Ascher D.B., Balding D.J., Petrovski S. Optimizing genomic medicine in epilepsy through a gene-customized approach to missense variant interpretation. Genome Res. 2017;27:1715–1729. doi: 10.1101/gr.226589.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., et al. Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davydov E.V., Goode D.L., Sirota M., Cooper G.M., Sidow A., Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLoS Comput. Biol. 2010;6:e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katoh K., Misawa K., Kuma K., Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coban-Akdemir Z., White J.J., Song X., Jhangiani S.N., Fatih J.M., Gambin T., Bayram Y., Chinn I.K., Karaca E., Punetha J., et al. Baylor-Hopkins Center for Mendelian Genomics Identifying Genes Whose Mutant Transcripts Cause Dominant Disease Traits by Potential Gain-of-Function Alleles. Am. J. Hum. Genet. 2018;103:171–187. doi: 10.1016/j.ajhg.2018.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nonet M., Sweetser D., Young R.A. Functional redundancy and structural polymorphism in the large subunit of RNA polymerase II. Cell. 1987;50:909–915. doi: 10.1016/0092-8674(87)90517-4. [DOI] [PubMed] [Google Scholar]
- 15.Bartolomei M.S., Halden N.F., Cullen C.R., Corden J.L. Genetic analysis of the repetitive carboxyl-terminal domain of the largest subunit of mouse RNA polymerase II. Mol. Cell. Biol. 1988;8:330–339. doi: 10.1128/mcb.8.1.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scafe C., Martin C., Nonet M., Podos S., Okamura S., Young R.A. Conditional mutations occur predominantly in highly conserved residues of RNA polymerase II subunits. Mol. Cell. Biol. 1990;10:1270–1275. doi: 10.1128/mcb.10.3.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meisels E., Gileadi O., Corden J.L. Partial truncation of the yeast RNA polymerase II carboxyl-terminal domain preferentially reduces expression of glycolytic genes. J. Biol. Chem. 1995;270:31255–31261. doi: 10.1074/jbc.270.52.31255. [DOI] [PubMed] [Google Scholar]
- 18.Litingtung Y., Lawler A.M., Sebald S.M., Lee E., Gearhart J.D., Westphal H., Corden J.L. Growth retardation and neonatal lethality in mice with a homozygous deletion in the C-terminal domain of RNA polymerase II. Mol. Gen. Genet. 1999;261:100–105. doi: 10.1007/s004380050946. [DOI] [PubMed] [Google Scholar]
- 19.Chapman R.D., Conrad M., Eick D. Role of the mammalian RNA polymerase II C-terminal domain (CTD) nonconsensus repeats in CTD stability and cell proliferation. Mol. Cell. Biol. 2005;25:7665–7674. doi: 10.1128/MCB.25.17.7665-7674.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gibbs E.B., Lu F., Portz B., Fisher M.J., Medellin B.P., Laremore T.N., Zhang Y.J., Gilmour D.S., Showalter S.A. Phosphorylation induces sequence-specific conformational switches in the RNA polymerase II C-terminal domain. Nat. Commun. 2017;8:15233. doi: 10.1038/ncomms15233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaplan C.D., Jin H., Zhang I.L., Belyanin A. Dissection of Pol II trigger loop function and Pol II activity-dependent control of start site selection in vivo. PLoS Genet. 2012;8:e1002627. doi: 10.1371/journal.pgen.1002627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malik I., Qiu C., Snavely T., Kaplan C.D. Wide-ranging and unexpected consequences of altered Pol II catalytic activity in vivo. Nucleic Acids Res. 2017;45:4431–4451. doi: 10.1093/nar/gkx037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berroteran R.W., Ware D.E., Hampsey M. The sua8 suppressors of Saccharomyces cerevisiae encode replacements of conserved residues within the largest subunit of RNA polymerase II and affect transcription start site selection similarly to sua7 (TFIIB) mutations. Mol. Cell. Biol. 1994;14:226–237. doi: 10.1128/mcb.14.1.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaplan C.D. Basic mechanisms of RNA polymerase II activity and alteration of gene expression in Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2013;1829:39–54. doi: 10.1016/j.bbagrm.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiu C., Jin H., Vvedenskaya I., Llenas J.A., Zhao T., Malik I., Visbisky A.M., Schwartz S.L., Cui P., Čabart P., et al. Universal promoter scanning by Pol II during transcription initiation in Saccharomyces cerevisiae. Genome Biol. 2020;21:132. doi: 10.1186/s13059-020-02040-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fong N., Saldi T., Sheridan R.M., Cortazar M.A., Bentley D.L. RNA Pol II Dynamics Modulate Co-transcriptional Chromatin Modification, CTD Phosphorylation, and Transcriptional Direction. Mol. Cell. 2017;66:546–557.e3. doi: 10.1016/j.molcel.2017.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fong N., Kim H., Zhou Y., Ji X., Qiu J., Saldi T., Diener K., Jones K., Fu X.D., Bentley D.L. Pre-mRNA splicing is facilitated by an optimal RNA polymerase II elongation rate. Genes Dev. 2014;28:2663–2676. doi: 10.1101/gad.252106.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saldi T., Fong N., Bentley D.L. Transcription elongation rate affects nascent histone pre-mRNA folding and 3′ end processing. Genes Dev. 2018;32:297–308. doi: 10.1101/gad.310896.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maslon M.M., Braunschweig U., Aitken S., Mann A.R., Kilanowski F., Hunter C.J., Blencowe B.J., Kornblihtt A.R., Adams I.R., Cáceres J.F. A slow transcription rate causes embryonic lethality and perturbs kinetic coupling of neuronal genes. EMBO J. 2019;38 doi: 10.15252/embj.2018101244. e101244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zylka M.J., Simon J.M., Philpot B.D. Gene length matters in neurons. Neuron. 2015;86:353–355. doi: 10.1016/j.neuron.2015.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
POLR2A variants are available on ClinVar: SUB7404119, accession pending review.