Abstract
In this paper, we review recent human respiratory virus epidemics, their zoonotic nature, and our current inability to identify future prepandemic threats. We propose a cost-efficient, One Health surveillance strategy that will be more efficient and more sustainable than previous efforts.
Keywords: emerging respiratory viruses, adenoviruses, coronaviruses, influenza viruses, enteroviruses
The coronavirus disease 2019 (COVID-19) pandemic has preoccupied health care and public health professionals for more than a year. We are assured that the future holds new emergent respiratory virus threats that could be equally devastating. What virus types should we anticipate? Where do we conduct surveillance for them? How do we better mitigate such prepandemic threats before they cause worldwide morbidity?
Spillover and Emergence of New Threats
In addition to the widely recognized novel influenza A and coronavirus threats, other respiratory viruses have epidemic if not pandemic potential. These include human metapneumovirus [1, 2], human respirovirus 3 (formerly parainfluenza virus 3) [1], rhinovirus C [3], respiratory syncytial virus, and multiple human adenovirus species [4]. Many of these threats are now recognized as zoonotic (here we equate anthroponosis or reverse zoonosis with zooanthroponosis because zoonoses are often recognized as bidirectional).
The zoonotic nature of some of these viruses was not always appreciated. Human respirovirus 3 and human rhinovirus 3 had not previously been thought to be zoonotic. Cross-species spillovers of metapneumovirus [5] and adenoviruses [6] have been previously postulated, but until recently [1, 2] were largely discounted [7, 8]. There is growing evidence that adenoviruses [9] and enteroviruses [10] are, at least occasionally, jumping species. The zoonotic nature of such pathogens matters because the viruses responsible for the 2009 H1N1 pandemic and the current COVID-19 pandemic came from animals and have both abjectly surprised public health officials with their rapid spread and high morbidity. We argue that their emergence was not expected because sparse surveillance was being performed at the human–animal interface.
Novel Respiratory Viruses’ Impact on Humans
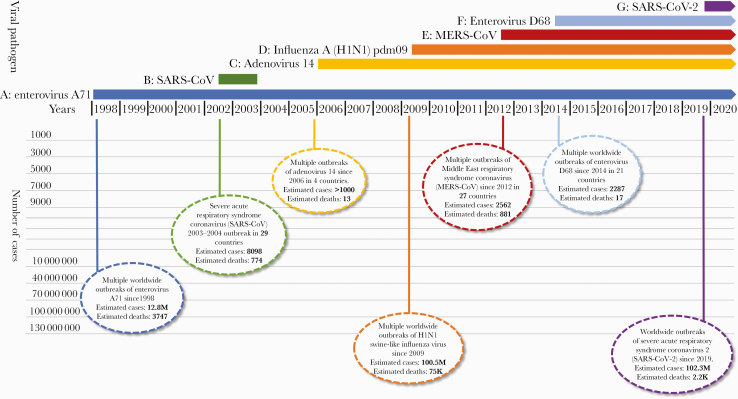
The impact of novel respiratory virus infections in humans is not trivial (Figure 1). Between 2002 and 2004, severe acute respiratory syndrome coronavirus (SARS-CoV) emerged in China and rapidly spread to 29 countries before it was controlled with aggressive public health and hospital infection control measures. In 2006, a novel human adenovirus 14 strain (Ad14) emerged in the United States and spread to other countries, causing at least 1000 illnesses and 13 deaths. While surveillance for this “killer cold virus” is sparse, available data suggest that it spread to at least Ireland, Canada, and China and still circulates today. In 2009, a triple reassortant H1N1 swine-like influenza virus emerged in Mexico and quickly spread throughout the world, causing an estimated 60.8 million illnesses and 12 469 deaths in the United States alone [11]. This deadly influenza strain continues to circulate today. In 1969, enterovirus A71 (EV-A71) was first isolated in California, with outbreaks from 1972 to 1990 identified in 6 countries [12]. In the Asian-Pacific region, EV-A71 became endemic in the 1990s, affecting thousands of children to manifest what is recognized as hand, foot, and mouth disease [12]. While surveillance is still sparse, the virus has caused at least 12.8M infections and 3747 deaths in Asia alone, with major outbreaks continuing to occur every 3–4 years [12]. Enterovirus D68 was first isolated in California in 1962, and since then, there have been infrequent cases in the United States and minor outbreaks in Europe, Africa, and Southeast Asia. In 2014, a novel clade of a second enterovirus, D68 (EV-D68), was implicated in a series of outbreaks of respiratory disease in 21 countries. There were at least 2287 EV-D68 illnesses and 17 deaths attributed to this virus [13]. The virus has also caused outbreaks of severe respiratory disease in Asia and was recently implicated as the cause of acute flaccid myelitis [14].
Figure 1.
Recent outbreaks of emerging respiratory viruses in humans. A, Enterovirus A71 worldwide outbreak. Presented cases are from the Asian-Pacific region only, 1998–2018 [12]. B, Severe acute respiratory syndrome coronavirus (SARS-CoV) November 2002–July 2003 outbreak. This virus is zoonotic in origin [15]. C, Adenovirus 14 multiple outbreaks [16]. D, Influenza A (H1N1) pdm09 virus. Presented cases are for the years 2009–2018. This virus is zoonotic in origin [11]. E, Middle East respiratory syndrome coronavirus. Presented cases include data from September 2012 to November 2020. This virus is thought to be of zoonotic origin [17]. F, Enterovirus D68 multiple outbreaks. Cases presented here are for 2014–2015 [13]. G, Severe acute respiratory syndrome coronavirus 2 pandemic. Cases presented here are counted through January 2021. This virus is zoonotic in origin [18].
It is unfortunate that our public health response to recent emergent novel pathogen events has largely been transient. Former Centers for Disease Control and Prevention Director Julie Gerberding, MD, addressed this concern in her November 2019 testimony [19] before the US Senate Armed Services Hearing:
When health crises strike—measles, MERS, Zika, dengue, Ebola, pandemic flu—the American people grow alarmed and U.S. policymakers spring into action, rushing to allocate resources in response. Yet all too often, when the crisis fades and public attention subsides, urgency morphs into complacency. Investments dry up, attention shifts, and a false sense of security takes hold.
Novel Respiratory Viruses Also Threaten Domestic Animals and Wildlife
Novel respiratory pathogens are not only emerging among humans, but also among animals. The livestock–wildlife interface has played an important role in the spillover and the virulence of viral pathogens. For instance, transmission of avian influenza viruses from migratory aquatic birds to domestic poultry has been commonly reported. In addition, new threats of reverse spillover have recently been identified. Surveillance of avian respiratory viruses in wild birds indicates the spillover of mutant viral vaccine strains avian paramyxovirus serotype 1 (APMV1) and avian coronaviruses (ACoV) from commercial poultry to wild birds. These strains have an enhanced ability to transmit across species [20]. Interactions between humans, domestic animals, and wildlife create inevitable opportunities for the emergence of novel strains.
Economic Burden of Novel Respiratory Viruses
Beyond causing high morbidity and mortality, emerging respiratory viruses have also had major impacts upon international travel, trade, and economies. In a 2018 report [21], The World Bank estimated that the SARS epidemic cost the affected nations US$30–50 billion, and the H1N1 pandemic US$45–55 billion. An incursion of MERS-CoV in South Korea during June 2015–June 2016 was estimated to cost that nation US$2.6 billion [22]. At this juncture, it is clear that the economic impact of COVID-19 will exceed all recent respiratory disease outbreaks.
Where and How Should We Conduct Surveillance for Novel Respiratory Viruses
Most of these novel emergent respiratory viruses come from 4 different viral families (Coronaviridae, Adenoviridae, Orthomyxoviridae, and Picornaviridae) and are thought to have originated in animals before crossing over to infect humans. Epidemiologists, ecologists, and modelers have attempted to predict novel pathogen emergence. Studies have been conducted to identify risk factors associated with emerging influenza A viruses [23], emerging infectious diseases [24], and emerging zoonotic diseases [25]. The identified risk factors for specific viruses are complex and sometimes disparate. However, these factors often point to densely populated geographical areas in Asia as areas of high emerging infectious disease risk. They also point to encounters with live animals, especially around markets where animals are sold [26, 27], as a source of virus mixing and human infection risk.
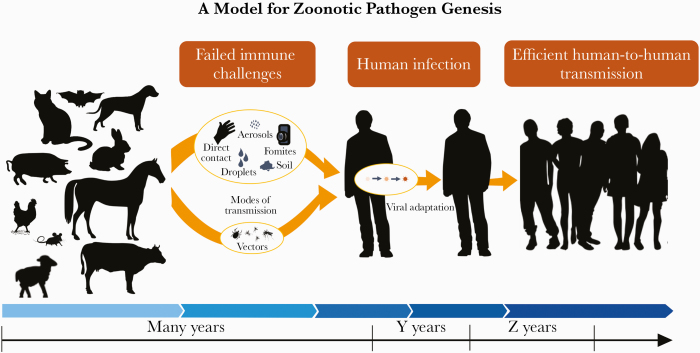
The time and process for viruses to transition or adapt from an animal virus to a human zoonotic pathogen are unknown, but it seems logical that it takes many years and varies with each zoonotic pathogen (Figure 2). For instance, viral evolutionary data suggest that the coronavirus lineage giving rise to SARS-CoV-2 departed from the nearest similar virus in bats after ~1969 [28]. While the virus likely circulated in bats or in an intermediate animal host during some of this time, it seems likely that the virus took quite some time in adapting to humans and longer still to become the pathogen we face today. These long transition periods provide an important opportunity to mitigate novel respiratory virus threats before they fully adapt to humans and become highly transmissible.
Figure 2.
Here we illustrate how a animal pathogen may become a zoonotic human pathogen. Evolutionary studies suggest that such viral adaption to man is a progression of events, each step of which that may take multiple years. Illustration by Emily Robie, images from publicdomainvectors.org. Adapted from Dr. V. Stalin Raj’s graphic: http://www.iisertvm.ac.in/faculty/stalin/research_areas?%2Ffaculties%2Fstalin%2Fresearch_areas=.
If we focus novel pathogen surveillance at the human–animal nexus (Figure 2), we are likely to be more efficient in detecting pathogens that threaten humans, as compared with other more ambitious surveillance strategies that focus upon the comprehensive study of all the viruses that reside in the world’s animals. In embracing the latter approach, programs such as USAID’s PREDICT I and II programs and the new Global Virome Project have been criticized as expensive and not likely to find pathogens that will be human threats. Their previous costs (PREDICT >$169M since 2009), future proposed costs (Global Virome Project $3.4B), and ambitious plans have been questioned [29–33]. Similarly, leaders at the US National Institute of Allergy and Infectious Diseases have proposed 20 years of large human cohort studies and intensive immunology and pathology research involving 120 pathogen groups [34–36], yet they freely admit that the associated cost is prohibitive [34].
There is a growing demand for innovative and cost-effective approaches to surveil for the next novel respiratory virus threat. We need to mitigate novel respiratory virus threats before they fully adapt to humans and become highly transmissible. The most strategic approach would be directing periodic, novel respiratory virus surveillance at the human–animal interface, studying both animal workers and their animals [37]. Alternatively, if surveillance cannot be conducted at the human–animal interface, surveillance for novel respiratory viruses among human pneumonia patients should be conducted in geographical areas known to be at high risk of novel respiratory virus emergence. We have recently used these methods and found evidence of novel zoonotic coronaviruses among humans hospitalized with pneumonia [38], a zoonotic adenovirus infection in a patient with respiratory illness [39], evidence of human enteroviruses in pigs [39], and first evidence of influenza D virus in poultry [40]. We have found success through conducting monthly periodic surveillance both in live animal environments (industrialized farms, live animal markets, etc.) and among pneumonia patients in large hospitals for about $25 000 per year per site.
To conduct such effective surveillance, developed countries need to rethink current global security strategies and form better global health partnerships, embracing the interdisciplinary One Health approach to novel respiratory virus surveillance. Surveillance at the human–animal nexus is likely to be better focused and less expensive than some of the other strategies currently being contemplated.
Acknowledgments
Financial support. This work was funded by the US Naval Medical Research Center-Asia and Vysnova Partners (SC-2016-SABER-003-002, SC-2017- SABER-010-001), and Professor Gregory Gray’s discretionary funds from Duke University.
Potential conflicts of interest. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Patient consent. This study does not include factors necessitating patient consent.
Author contributions. G.C.G. conceived the idea and drafted the manuscript. A.A. assisted in drafting the manuscript. Both authors approved this article for publication.
References
- 1. Negrey JD, Reddy RB, Scully EJ, et al. . Simultaneous outbreaks of respiratory disease in wild chimpanzees caused by distinct viruses of human origin. Emerg Microbes Infect 2019; 8:139–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Slater OM, Terio KA, Zhang Y, et al. . Human metapneumovirus infection in chimpanzees, United States. Emerg Infect Dis 2014; 20:2115–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Scully EJ, Basnet S, Wrangham RW, et al. . Lethal respiratory disease associated with human rhinovirus C in wild chimpanzees, Uganda, 2013. Emerg Infect Dis 2018; 24:267–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Medkour H, Amona I, Akiana J, et al. . Adenovirus infections in African humans and wild non-human primates: great diversity and cross-species transmission. Viruses 2020; . 12:657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Velayudhan BT, Nagaraja KV, Thachil AJ, et al. . Human metapneumovirus in turkey poults. Emerg Infect Dis 2006; 12:1853–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xiang Z, Li Y, Cun A, et al. . Chimpanzee adenovirus antibodies in humans, Sub-Saharan Africa. Emerg Infect Dis 2006; 12:1596–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. van den Hoogen BG, de Jong JC, Groen J, et al. . A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med 2001; 7:719–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Benkő M, Harrach B, Kremer EJ. Do nonhuman primate or bat adenoviruses pose a risk for human health? Future Microbiol 2014; 9:269–72. [DOI] [PubMed] [Google Scholar]
- 9. Borkenhagen LK, Fieldhouse JK, Seto D, Gray GC. Are adenoviruses zoonotic? A systematic review of the evidence. Emerg Microbes Infect 2019; 8:1679–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fieldhouse JK, Wang X, Mallinson KA, et al. . A systematic review of evidence that enteroviruses may be zoonotic. Emerg Microbes Infect 2018; 7:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shrestha SS, Swerdlow DL, Borse RH, et al. . Estimating the burden of 2009 pandemic influenza A (H1N1) in the United States (April 2009-April 2010). Clin Infect Dis 2011; 52(Suppl 1):S75–82. [DOI] [PubMed] [Google Scholar]
- 12. Puenpa J, Wanlapakorn N, Vongpunsawad S, Poovorawan Y. The history of enterovirus A71 outbreaks and molecular epidemiology in the Asia-Pacific region. J Biomed Sci 2019; 26:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Holm-Hansen CC, Midgley SE, Fischer TK. Global emergence of enterovirus D68: a systematic review. Lancet Infect Dis 2016; 16:e64–75. [DOI] [PubMed] [Google Scholar]
- 14. Kujawski SA, Midgley CM, Rha B, et al. . Enterovirus D68–associated acute respiratory illness—new vaccine surveillance network, United States, July–October, 2017 and 2018. MMWR Morb Mortal Wkly Rep 2019; 68:277–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Centers for Disease Control and Prevention. Revised US surveillance case definition for severe acute respiratory syndrome (SARS) and update on SARS cases—United States and worldwide, December 2003. MMWR Morb Mortal Wkly Rep 2003; 52:1202–6. [PubMed] [Google Scholar]
- 16. Gautret P, Gray GC, Charrel RN, et al. . Emerging viral respiratory tract infections—environmental risk factors and transmission. Lancet Infect Dis 2014; 14:1113–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. World Health Organization. Middle East respiratory syndrome coronavirus (MERS-CoV). 2019. Available at: http://www.who.int/emergencies/mers-cov/en/files/1628/en.html. Accessed 1 November 2020.
- 18.Johns Hopkins Coronavirus Resource Center. COVID-19 map. 2020. Available at: https://coronavirus.jhu.edu/map.html. Accessed 29 January 2021.
- 19.Center for Strategic and International Studies, Commission on Strengthening America’s Health Security. Biological threats to U.S. national security: testimony before the senate armed services subcommittee on emerging threats and capabilities. 2019. Available at: https://healthsecurity.csis.org/events/biological-threats-to-u-s-national-security-testimony-before-the-senate-armed-services-subcommittee-on-emerging-threats-and-capabilities/. Accessed 1 November 2020.
- 20. Rohaim MA, El Naggar RF, Helal AM, et al. . Reverse spillover of avian viral vaccine strains from domesticated poultry to wild birds. Vaccine 2017; 35:3523–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. World Bank Group. One Health Operational Framework for Strengthening Human, Animal, and Environmental Public Health Systems at Their Interface. Washington, DC: The World Bank; 2018. [Google Scholar]
- 22. Joo H, Maskery BA, Berro AD, et al. . Economic impact of the 2015 MERS outbreak on the Republic of Korea’s tourism-related industries. Health Secur 2019; 17:100–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fuller TL, Gilbert M, Martin V, et al. . Predicting hotspots for influenza virus reassortment. Emerg Infect Dis 2013; 19:581–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jones KE, Patel NG, Levy MA, et al. . Global trends in emerging infectious diseases. Nature 2008; 451:990–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Allen T, Murray KA, Zambrana-Torrelio C, et al. . Global hotspots and correlates of emerging zoonotic diseases. Nat Commun 2017; 8:1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Su S, Bi Y, Wong G, et al. . Epidemiology, evolution, and recent outbreaks of avian influenza virus in China. J Virol 2015; 89:8671–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhou P, Ma J, Lai A, et al. . Avian influenza A(H7N9) virus and mixed live poultry-animal markets in Guangdong province: a perfect storm in the making? Emerg Microbes Infect 2015; 4:e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Boni MF, Lemey P, Jiang X, et al. . Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat Microbiol 2020; 5:1408–17. [DOI] [PubMed] [Google Scholar]
- 29. Holmes EC, Rambaut A, Andersen KG. Pandemics: spend on surveillance, not prediction. Nature 2018; 558:180–2. [DOI] [PubMed] [Google Scholar]
- 30. Geoghegan JL, Holmes EC. Predicting virus emergence amid evolutionary noise. Open Biol 2017; 7:170189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yong E. Is it possible to predict the next pandemic? The Atlantic. 25 October 2017. Available at: https://www.theatlantic.com/science/archive/2017/10/pandemic-prediction-challenge/543954/. Accessed 1 November 2020.
- 32. Morrison J. Can virus hunters stop the next pandemic before it happens? Smithsonian Magazine. 25 January 2018. Available at: https://www.smithsonianmag.com/science-nature/how-to-stop-next-animal-borne-pandemic-180967908/. Accessed 1 November 2020. [Google Scholar]
- 33. Robbins J. Before the next pandemic, an ambitious push to catalog viruses in wildlife. Yale Environment 360. 22 April 2020. Available at: https://e360.yale.edu/features/before-the-next-pandemic-an-ambitious-push-to-catalog-viruses-in-wildlife#:. Accessed 1 November 2020. [Google Scholar]
- 34. Graham B. Issue 15: COVID-19: a wake-up call for more fundamental research in immunology. Human Vaccines Project. 5 August 2020. Available at: https://www.humanvaccinesproject.org/covid-post/issue-15-covid-19-a-wake-up-call-for-more-fundamental-research-in-immunology/. Accessed 1 November 2020. [Google Scholar]
- 35. Graham BS, Corbett KS. Prototype pathogen approach for pandemic preparedness: world on fire. J Clin Invest 2020; 130:3348–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Graham BS, Sullivan NJ. Emerging viral diseases from a vaccinology perspective: preparing for the next pandemic. Nat Immunol 2018; 19:20–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Anderson BD, Ma MJ, Wang GL, et al. . Prospective surveillance for influenza. virus in Chinese swine farms. Emerg Microbes Infect 2018; 7:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xiu L, Binder RA, Alarja NA, et al. . A RT-PCR assay for the detection of coronaviruses from four genera. J Clin Virol 2020; 128:104391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fieldhouse JK, Bailey ES, Toh TH, et al. . Panspecies molecular assays detect viral pathogens missed by real-time PCR/reverse-transcriptase PCR among pneumonia patients, Sarawak, Malaysia. Trop Dis Travel Med Vaccines 2020; 6:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bailey ES, Fieldhouse JK, Alarja NA, et al. . First sequence of influenza D virus identified in poultry farm bioaerosols in Sarawak, Malaysia. Trop Dis Travel Med Vaccines 2020; 6:5. [DOI] [PMC free article] [PubMed] [Google Scholar]