Abstract
Background
Serological confirmation of coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is critical for understanding the dynamics of the pandemic and determining seroprevalence rates within afflicted communities. Common challenges with SARS-CoV-2 serological assays include poor analytical specificity and sensitivity and lack of a serological standard for quantitative assessment of antibody titers.
Methods
To overcome these obstacles, we developed a quantitative enzyme-linked immunosorbent assay based on an optimized 2-dimensional screening assay that utilizes SARS-CoV-2 receptor binding domain (RBD) of spike protein and SARS-CoV-2 spike S1 subunit.
Results
A total of 4 SARS-CoV-2-reactive monoclonal antibodies were evaluated for use as serum standards for calibrating assays performed on different days or by different laboratories. This approach provided quantitative analysis of hospitalized reverse transcription polymerase chain reaction–confirmed COVID-19 cases that in some cases reached >100 μg/mL. The assay demonstrated 72% sensitivity based on time points ranging from 2 to 52 days post–symptom onset, with 100% sensitivity at time points measured ≥13 days post–symptom onset and 100% specificity.
Conclusions
Using these optimized reagents and serological standards, we believe this approach will be useful for sensitive and specific determination of seroconversion rates and quantitatively measuring the durability of antiviral antibody responses following SARS-CoV-2 infection or vaccination.
Keywords: coronavirus disease 2019, coronavirus, COVID-19, diagnostics, ELISA, IgG, SARS-CoV-2, severe acute respiratory syndrome coronavirus 2
Coronavirus disease 2019 (COVID-19) has spread rapidly across the globe, and as of January 20, 2020, there have been over 24 million cases reported in the United States and over 95 million confirmed cases reported worldwide (https://coronavirus.jhu.edu/map.html). During this same period, more than 403 000 COVID-19-related deaths have been identified in the United States, with nearly 2.1 million COVID-19-related deaths reported worldwide.
It has been just over a year since SARS-CoV-2 began spreading in the human population, and COVID-19 seroepidemiology studies are mainly still in their infancy [1–7]. At the individual level, serology tests may allow retrospective diagnosis of prior infection, and this may be particularly useful in cases where virological testing was not performed during the window of early infection or was not available at the time of exposure. If an immune correlate is eventually identified, then quantitative serology tests could also prove valuable for determining immune status. However, the use of serology tests to determine immune status has been viewed with caution in low-incidence settings. For example, if the true seroprevalence of a population is 5%, then an assay with 95% specificity would have a high false-positive rate resulting in only 50% positive predictive value (PPV) and would be unsuitable for clinical decision-making or determining potential risk for infection [8]. Serologic assays may also be used to characterize COVID-19 convalescent donor plasma by providing a quantitative estimate of antibody titers before blood donation [8]. At the population level, seroepidemiology studies can be used to characterize transmission within COVID-19 clusters as well as during larger outbreaks, in addition to determining the extent of disease burden and/or immunity within a particular community at a given point in time. However, interpretation of many serology studies has been complicated by poor assay sensitivity and specificity, leading some authors to note that seroconversion rates may be driven by the serological test performance characteristics themselves [5] rather than correctly identifying true seropositive and seronegative individuals. Moreover, because of ongoing SARS-CoV-2 transmission, serosurveys performed in the first few months of the pandemic are no longer representative of the current population. For instance, although an early study [5] identified a 6.9% seroconversion rate in New York City using samples collected from March 23 to April 1, 2020, another study conducted a few weeks later found seroconversion rates as high as 22.7% [4]. For these reasons, development of quantitative serological assays with high sensitivity and specificity will be important for future COVID-19 research.
In the studies described here, we have optimized a SARS-CoV-2 serological assay that is based on antiviral antibodies binding to 2 antigen substrates (SARS-CoV-2 receptor binding domain [RBD] and spike protein subunit S1) and provides both high sensitivity and specificity. We have also characterized 4 SARS-CoV-2-reactive monoclonal antibodies for use as enzyme-linked immunosorbent assay (ELISA) standards. The use of a monoclonal immunoglobulin G (IgG) standard allows for the quantitative assessment of antiviral antibody levels over time and provides an approach for different laboratories to compare results across assay platforms by using the same readily available antibody reagents. Together, this work provides an important new tool for the assessment of humoral immunity following SARS-CoV-2 infection including longitudinal studies on immunological memory as well as serology studies to determine transmission dynamics, seroprevalence, and estimated burden of disease.
METHODS
Patient Consent Statement
Samples were obtained from reverse transcription polymerase chain reaction (RT-PCR)–confirmed hospitalized COVID-19 patients who provided informed written consent. If subjects were unable to provide written consent, then written consent was obtained from their legally authorized representative (LAR). In some cases, de-identified samples were obtained from a biorepository that included hospitalized COVID-19 patients at Oregon Health and Science University (OHSU) who tested positive for SARS-CoV-2 by nasopharyngeal swab and subsequent RT-PCR. The 23 COVID-19 patients were an average (range) of 64 (20–88) years old and 52.2% female, 30.4% required mechanical ventilation, and 8.7% required extracorporeal membrane oxygenation (ECMO). A total of 50 plasma samples from the COVID-19 patients were used in these experiments, and prepandemic plasma or serum samples obtained between November 1989 and August 2019 from another 300 adults were included in the study as negative controls. The study was approved by the institutional review board of Oregon Health & Science University.
Enzyme-Linked Immunosorbent Assay
SARS-CoV-2 recombinant antigens used in the ELISA included the receptor binding domain (Cat #230-30162, Ray Biotech), S1 subunit of the spike protein (spike S1, Cat#40591-V08H, Sino Biologicals), full-length spike protein (spike, Cat#40589-V08B1, Sino Biological), and nucleocapsid protein (NP, Cat#40588-V08B, Sino Biologicals). Recombinant spike proteins from seasonal human coronaviruses (HCoVs) included HCoV-NL63 (Cat#40604-V08B, Sino Biologicals), HCoV-OC43 (Cat #40607-V08B, Sino Biologicals), HCoV-229E (Cat#40605-V08B, Sino Biologicals), and HCoV-HKU1 (Cat #40606-V08B Sino Biologicals).
The 96-well ELISA plates (Cat#9018, Corning) were coated with 100 μL of each antigen at a concentration of 1 μg/mL prepared in phosphate-buffered saline (PBS), and the plates were incubated overnight at 4°C and then stored frozen at –20°C until use. Plates were thawed at room temperature (RT), the coating antigen was removed, and plates were blocked for 1 hour at RT with 5% Omniblok (Cat#AB10109-01000, American Bio) prepared in PBS-Tween containing 0.05% Tween (ie, dilution buffer). Plates were washed once with PBST containing 0.05% Tween (ie, wash buffer), and for diagnostic screening, 50 μL of dilution buffer was added to each well, along with 50 μL of a 1:50 dilution of heat-inactivated serum or plasma (1:100 dilution final). For quantitative analysis, samples were serially 3-fold diluted in dilution buffer. Plates were incubated at RT for 1 hour, followed by the addition of 50 μL of 10% hydrogen peroxide and further incubation for 30 minutes at RT. The plates were washed 3 times with wash buffer, and 100 μL of 1:2000 dilution of antihuman IgG-HRP (Cat#555788, BD Pharmingen) detection antibody was added and incubated at RT for 1 hour. After washing the plates 3 times with wash buffer, 100 μL of colorimetric detection reagent containing 0.4 mg/mL of o-phenylenediamine and 0.01% hydrogen peroxide in 0.05-M citrate buffer (pH 5) was added, and the reaction was stopped after 20 minutes by the addition of 100 μL of 1-M HCl. Optical density (OD) at 490 nm was measured using a VersaMax ELISA plate reader (Molecular Devices). Antibody titers were determined by logarithmic transformation of the linear portion of the curve, with 0.1 OD units used as the end point before converting to final values. A standard curve was generated using 1 of 4 SARS-CoV2-reactive monoclonal antibodies including ABMX-002 (Cat#10–2005, Abeomics), Sanyou (Cat# AHA001, Sanyou Biopharmaceuticals), CR3022 (Cat# NR-52392, BEI resources), or DA0002 (Cat# A19215, ABClonal).
RESULTS
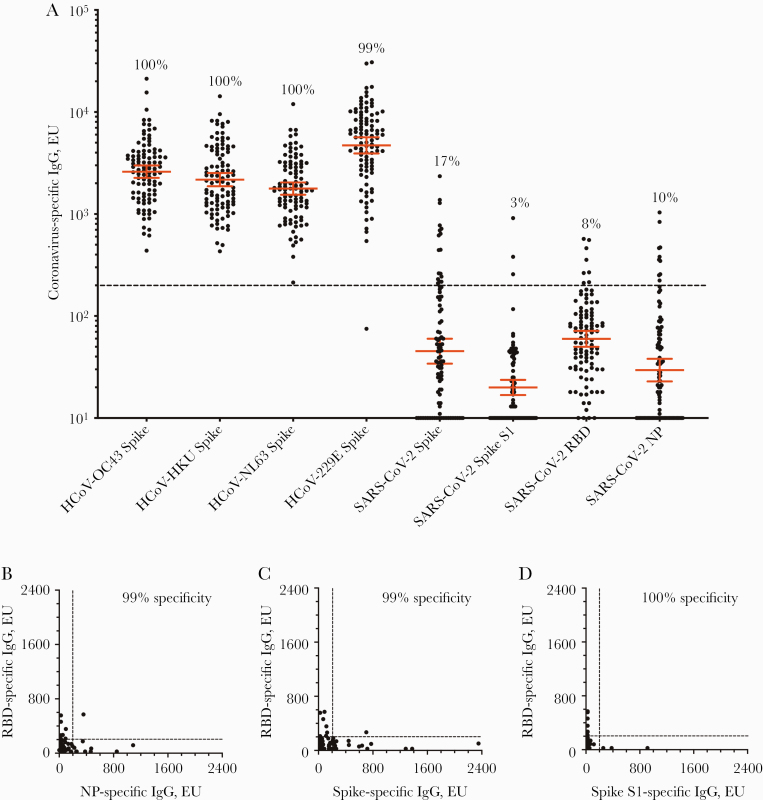
Populations that have low rates of SARS-CoV-2 infection can be problematic for seroepidemiology studies unless the diagnostic assay has sufficiently high specificity. Bearing this in mind, we performed routine optimization of assay conditions (time, temperature, antigen concentration, blocking buffer, and wash buffer formulations) and prepared an initial prepandemic serology panel from 100 individuals and tested this panel for antibodies to seasonal human coronaviruses in addition to SARS-CoV-2 antigens (Figure 1). Coating ELISA plates with purified full-length spike protein from each of the 4 seasonal human coronaviruses (HCoV-OC43, HCoV-HKU, HCoV-NL63, and HCoV-229E) revealed high seroconversion rates that ranged from 99% (HCoV-229E) to 100% (HCoV-OC43, HCoV-HKU, and HCoV-NL63) with high geometric mean antibody titers that ranged between 1774 and 4710 ELISA units (EU) (Figure 1A). In contrast, cross-reactive antibodies to the SARS-CoV-2 spike protein were low (geometric mean; 45 EU), with only 17% of the prepandemic control samples scoring above the detection threshold (200 EU). Narrowing the viral antigen from the 1213–amino acid full-length SARS-CoV-2 spike protein to either the SARS-CoV-2 spike S1 subunit (685 amino acids) or to the smaller SARS-CoV-2 receptor binding domain (222 amino acids) further reduced serological cross-reactivity from 17% cross-reactivity to 3% and 8%, respectively. Similarly, serological cross-reactivity to SARS-CoV-2 nucleocapsid protein (NP) was observed among 10% of the prepandemic samples. Together, these results indicate that preexisting cross-reactive antibody responses to SARS-CoV-2 antigens are low or undetectable among people who have not been exposed to the virus and are consistent with prior studies showing a lack of cross-reactive neutralizing antibodies to SARS-CoV-2 [9–12]. However, based on these results, none of the individual SARS-CoV-2 antigens would provide sufficient specificity for seroepidemiology studies in low-incidence settings unless the detection threshold was raised from 200 EU to >1000 EU in order to achieve 100% specificity. Although this type of approach would improve assay specificity, it would also decrease assay sensitivity, which is a critical parameter for detecting low antiviral antibody titers in COVID-19 serology studies [13]. Since SARS-CoV-2 RBD is an important target for neutralizing antibodies and spike/RBD ELISA titers correlate well with neutralizing titers [7, 14–17], we focused on RBD seroreactivity for further diagnostic assay development. To improve specificity without reducing sensitivity, we tested a 2-dimensional screening approach by comparing RBD vs NP (Figure 1B), RBD vs full-length spike (Figure 1C), and RBD vs spike S1 (Figure 1D). Using a detection threshold of 200 EU for each antigen, we found that this technique greatly improved the assay, with results that reached 99% specificity for RBD vs NP, 99% specificity for RBD vs full-length spike, and 100% specificity when using RBD vs spike S1 (Figure 1D). Overall, this indicates that a combination screening assay based on RBD and spike S1 provided the best diagnostic approach for these studies.
Figure 1.
Serological analysis of prepandemic antibodies to seasonal HCoVs vs SARS-CoV-2. ELISA titers from prepandemic samples (obtained before 2011) from 100 human subjects were tested by ELISA. A, Plates were coated with full-length spike proteins from each of the 4 seasonal human coronaviruses (HCoV-OC43, HCoV-HKU, HCoV-NL63, and HCoV-229E), full-length SARS-CoV-2 spike protein, the SARS-CoV-2 spike S1 subunit, the SARS-CoV-2 RBD of the spike protein, and the SARS-CoV-2 nucleocapsid protein. The numbers above each group of symbols indicate the percentage seropositivity rate, and the geometric mean antibody titer ±95% CI is shown for each group. To increase assay specificity, ELISA titers from different antigen combinations were graphed with (B) RBD vs NP, (C) RBD vs full-length spike, and (D) RBD vs spike S1. The dashed line (200 EU) indicates the cutoff for a seropositive antibody titer. Abbreviations: ELISA, enzyme-linked immunosorbent assay; EU, ELISA units; HCoV, human coronavirus; NP, nucleocapsid protein; RBD, receptor binding domain; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
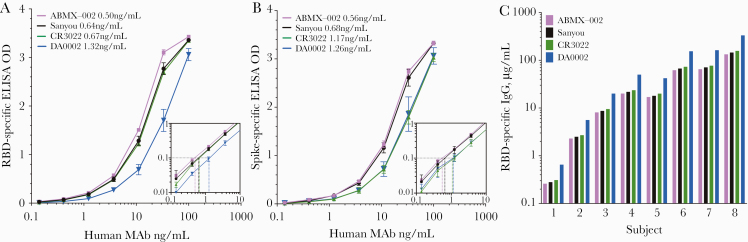
There is currently no international serum standard approved for SARS-CoV-2. This can be problematic for making direct comparisons of antibody titers published by individual research groups because each laboratory may obtain disparate antibody titers based on the use of different reagents and assay conditions. Use of an antibody assay standard not only makes it easier to normalize results between experiments performed on different days or performed by different operators within the same laboratory, but this also provides an important bridge for comparing results between independent research groups. As a step toward the development of a serological SARS-CoV-2 standard for quantitation of antiviral antibody titers, we compared 4 human SARS-CoV-2-reactive IgG monoclonal antibodies with respect to their binding profiles to RBD and spike S1 (Figure 2). The monoclonal antibodies ABMX-002, Sanyou, and DA0002 are derived from SARS-CoV-2 infection, whereas CR3022 is a SARS-CoV-1-specific clone that cross-reacts with SARS-CoV-2. Three of these monoclonals had similar RBD-binding curves (Figure 2A) and similar limits of detection (ABMX-002: 0.50 ng/mL; Sanyou: 0.64 ng/mL; and CR3022: 0.67 ng/mL), whereas the DA0002 antibody showed ~2-fold lower binding (limit of detection; 1.32 ng/mL) in comparison with the other monoclonal antibodies. Similar limits of detection were also observed with spike S1-binding analysis (ABMX-002: 0.56 ng/mL; Sanyou: 0.68 ng/mL; CR3022: 1.17 ng/mL; and DA0002: 1.26 ng/mL) (Figure 2B), although in this case both the CR3022 and DA0002 monoclonal antibodies showed ~2-fold lower binding than the ABMX-002 and Sanyou clones. It is interesting that these 3 monoclonal antibodies (ABMX-002, Sanyou, DA0002) bound both RBD and spike S1 antigens equally well, whereas the CR3022 clone demonstrated nearly 2-fold lower binding to spike S1 compared with RBD. This indicates that if the CR3022 monoclonal antibody is used as an ELISA standard, then the experimentally derived antibody titers for SARS-CoV-2 samples will differ markedly (~2-fold) depending on whether the RBD or the spike S1 antigen is used in the assay.
Figure 2.
Analysis of SARS-CoV-2-reactive human monoclonal antibodies as ELISA standards. ELISA antigen binding curves of 4 monoclonal antibodies to (A) SARS-CoV-2 RBD and (B) SARS-CoV-2 spike S1 subunit were determined by end point dilution series. Inset graphs provide detailed resolution of binding curves near the end point dilution of 0.1 OD after log-log transformation. The limit of detection ranged between 0.50 and 1.32 ng/mL for RBD and 0.56 and 1.26 ng/mL for spike S1 antigens. C, Antibody titers from 8 RT-PCR-confirmed COVID-19 cases were calculated based on 1 of each of the 4 monoclonal antibody standards shown in (A). Although monoclonal antibodies ABMX-002, Sanyou, and CR3022 provided comparable results, antibody titers calculated using the DA0002 monoclonal antibody as the standard were 2–2.5-fold higher than the other 3 virus-specific monoclonal IgG standards. Abbreviations: ELISA, enzyme-linked immunosorbent assay; COVID-19, coronavirus disease 2019; EU, ELISA units; IgG, immunoglobulin G; OD, optical density; RBD, receptor binding domain; RT-PCR, reverse transcription polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Based on the antibody binding characteristics described in Figure 2A and B, the choice of monoclonal antibody used as the ELISA standard was anticipated to impact the calculated magnitude of antiviral antibody titers. To address this question, RBD-specific antibody levels were calculated from 8 COVID-19 cases based on each monoclonal standard (Figure 2C). As expected, use of the monoclonal antibodies ABMX-002, Sanyou, or CR3022 provided similar results. This indicates that these monoclonal antibodies may be used interchangeably for quantitating SARS-CoV-2-specific antibody titers as long as the RBD antigen is used in the ELISA. In contrast, when the DA0002 monoclonal antibody was used as the standard, the calculated antibody titers were much higher. For example, when SARS-CoV-2-specific antibody titers from Subject #8 were measured, the antiviral IgG levels were determined to be ~145 μg/mL when using ABMX-002, Sanyou, or CR3022 but were estimated at 331 μg/mL when calibrated against the DA0002 monoclonal antibody. Together, this indicates that the DA0002 monoclonal standard resulted in inflated antibody scores that were ~2–2.5-fold higher than those obtained with the other 3 monoclonal standards, and in terms of providing conservative estimates of antibody titer, we recommend ABMX-002, Sanyou, or CR3022 as a serological standard when using SARS-CoV-2 RBD to quantitate antiviral antibody levels. If SARS-CoV-2 spike S1 is used in the ELISA, then either ABMX-002 or Sanyou is recommended to quantitate antiviral antibody levels.
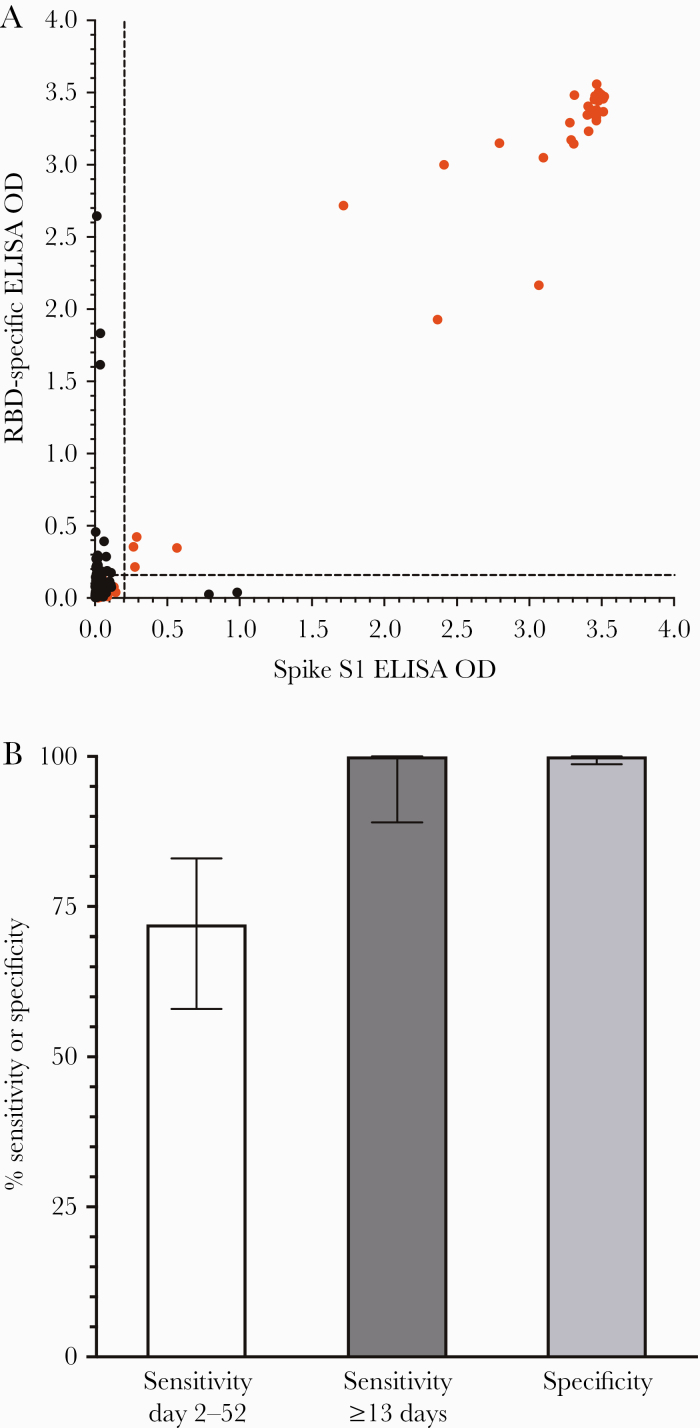
Having optimized ELISA protocol parameters to detect as little as 0.5 ng/mL of purified SARS-CoV-2-specific monoclonal IgG (Figure 2), we next determined the sensitivity and specificity of this diagnostic approach based on 50 samples from hospitalized RT-PCR-confirmed COVID-19 cases as well as 300 prepandemic negative control samples obtained before SARS-CoV-2 circulation in the United States (Figure 3). Samples were screened against SARS-CoV-2 RBD and spike S1 in a 2-dimensional plot (Figure 3A). A diagnostic detection threshold set at 120 ng/mL allows detection of low-level antiviral IgG while still maintaining 100% specificity (95% CI, 98.7%–100%) (Figure 3B). Although a small number of prepandemic samples scored above the detection threshold for either RBD (Y-axis) or spike S1 (X-axis) alone, none of the 300 negative controls scored positive on both antigens. Screening samples obtained between 2 and 52 days after symptom onset resulted in 36/50 samples scoring positive against both RBD and spike S1 antigens, for an overall sensitivity of 72% (95% CI, 58%–83%) (Figure 3B). Antiviral antibodies are most likely to reach detectable levels by ~2 weeks after symptom onset [14], and for samples obtained at ≥13 days, we observed 100% sensitivity (95% CI, 89%–100%) (Figure 3B).
Figure 3.
SARS-CoV-2 serological assay performance. A, ELISA titers from hospitalized RT-PCR + COVID-19 cases (red symbols, n = 50) and prepandemic naïve controls (black symbols, n = 300) were assayed on SARS-CoV-2 RBD (y-axis) vs SARS-CoV-2 spike S1 (x-axis), with samples that scored >200 EU/120 ng/mL on each axis considered seropositive and samples that scored <200 EU/120 ng/mL on 1 or both antigens considered seronegative. B, Diagnostic sensitivity was 72% for all samples obtained from days 2–52 after symptom onset and reached 100% sensitivity for detecting virus-specific antibodies at time points ≥13 days after symptom onset, with 100% specificity based on 300 prepandemic negative controls. Error bars represent 95% CIs. Abbreviations: ELISA, enzyme-linked immunosorbent assay; COVID-19, coronavirus disease 2019; EU, ELISA units; OD, optical density; RBD, receptor binding domain; RT-PCR, reverse transcription polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
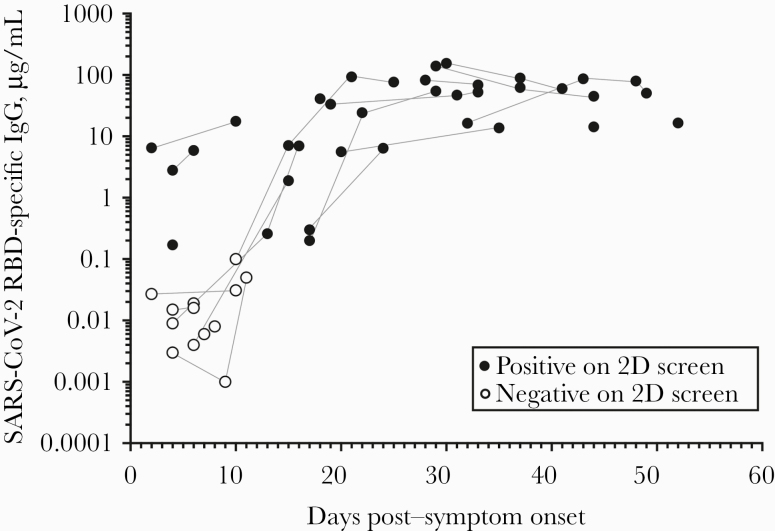
SARS-CoV-2 specific antibodies were measured as a function of time post-symptom onset (Figure 4). These studies indicated that antibody titers increased rapidly during the first 2–3 weeks and reached titers that, in 2 of the severe hospitalized cases (requiring mechanical ventilation or ECMO for respiratory support), exceeded 100 μg/mL within 4 weeks after symptom onset. Antiviral antibody titers remained stable out to 52 days after symptom onset, indicating that antiviral immunity may be long-lived, and more long-term longitudinal studies are currently in progress. Together, these results indicate that we have developed a sensitive and specific diagnostic approach for determining SARS-CoV-2 serostatus with the ability to quantitatively measure antiviral IgG titers over time.
Figure 4.
Kinetics and magnitude of antiviral antibody responses to SARS-CoV-2. Quantitative analysis of SARS-CoV-2-specific IgG shows that among hospitalized cases with COVID-19, these antibodies increase with time and plateau by ~4 weeks post–symptom onset. Open symbols represent samples that scored negative based on the 2-dimensional ELISA, and closed symbols represent samples that scored positive based on the 2-dimensional ELISA in Figure 3A. Samples obtained from the same study subject are connected by gray lines. Abbreviations: ELISA, enzyme-linked immunosorbent assay; COVID-19, coronavirus disease 2019; IgG, immunoglobulin G; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
DISCUSSION
In these studies, we developed a robust 2-dimensional diagnostic SARS-CoV-2 ELISA that at 13 days or more post–symptom onset provided up to 100% sensitivity and 100% specificity for serological detection of SARS-CoV-2 infection. High sensitivity was attained by optimizing the reagents and assay conditions to detect virus-specific monoclonal IgG at concentrations as low as 0.5 ng/mL. High specificity was attained by requiring that samples score above the detection threshold against not 1, but 2 closely related SARS-CoV-2 antigens (RBD and spike S1). Inclusion of a human monoclonal antibody standard in the assay is essential for providing a quantitative assessment of antiviral antibody titers, and this not only allows future SARS-CoV-2 serological studies by independent groups to be directly compared using the same calibrated unit values, but may eventually provide a framework for quantitation of an immunological correlate of immunity if one is eventually established for COVID-19.
High assay sensitivity is important for detecting early SARS-CoV-2-specific antibody responses shortly after infection as well as at later time points when antibody levels have declined from their initial peaks. Furthermore, accurate antibody measurements will be critical following the introduction of vaccines in order to determine response rates and confirm antibody production. One limitation is that we measured antibody responses only from hospitalized COVID-19 cases, and further studies will be needed to determine assay sensitivity at different time points after mild or asymptomatic SARS-CoV-2 infection. Another limitation of our study is that we focused primarily on IgG measurements because we had IgG monoclonal antibodies available for direct reference/standardization/quantitation, but we did not measure IgM or IgA levels as we did not have monoclonal antibodies that matched these specific immunoglobulin isotypes. However, recent studies have shown nearly simultaneous seroconversion for IgG, IgM, and IgA isotypes, with the median seroconversion date for each isotype occurring around 11–12 days after symptom onset [14]. This suggests that the timing of antiviral IgG production after symptom onset does not lag appreciably behind IgM or IgA and so the impact on overall assay sensitivity in minimized. In those studies [14], predictive accuracy for identifying SARS-CoV-2 cases by serology using a high IgG detection threshold of 570 ng/mL indicated that SARS-CoV-2 RBD-specific IgG could be detected among 7% of cases at ≤7 days, 51% of cases between 8 and 14 days, and 95% of cases at >15 days post–symptom onset [14]. Our IgG detection threshold is 120 ng/mL, and using this approach we identified 31% seroconversion at ≤7 days, 44% seroconversion between 8 and 14 days, and 100% seroconversion at ≥13 days post–symptom onset. The differences between these 2 studies may be due in part to sample size or differences in COVID-19 disease severity, but we believe that the improved assay sensitivity observed at both the early and late time periods is likely to be attributable to having nearly a 5-fold lower limit of detection obtained through the use of the 2-dimensional ELISA screening (Figures 1C and 3A).
Over 50 serological assays have received Emergency Use Authorization (EUA) by the Food and Drug Administration [18], but problems with poor specificity have complicated many early COVID-19 seroepidemiology studies, especially in settings of low seroprevalence where the number of COVID-19 cases is lower than the false-positive rate of the diagnostic test itself. The Abbott Architect SARS-CoV-2 IgG Assay is believed to be one of the top-performing serological diagnostic tests for COVID-19. It utilizes a single antigen (NP), and in an analysis of 1020 prepandemic samples, the assay showed 99.9% specificity [1]. However, a more recent study that analyzed 2204 serum samples found what appears to be a much higher false-positive rate and substantial discordance with antibodies to RBD and spike S1 from the same samples, and this has raised concerns for its continued use in the United Kingdom [19]. Specificity may vary under different field conditions, and a recent study that used the Abbott test to determine seropositivity among 120 crewmembers on a Seattle-based fishing vessel provides further insight into this question [20]. Before departure, crewmembers were screened for COVID-19 by RT-PCR and serology. All individuals tested negative by RT-PCR, whereas 6 individuals tested seropositive by the Abbott Architect assay. However, in subsequent experiments, it was revealed that only 3/6 of these individuals had detectable antibodies to SARS-CoV-2 spike or RBD antigens, RBD/ACE2-blocking antibodies, or neutralizing antibodies to SARS-CoV-2 pseudotyped lentiviral particles. This indicates that the other 3/6 individuals who only had NP-binding antibodies were false-positives, resulting in just 50% PPV in this case. This work provides a cautionary note for relying upon some of the current serology assays for determining individual immune status or for obtaining an “immunity passport” [21] in settings of low COVID-19 seroprevalence.
Currently there is a wide range of SARS-CoV-2-specific serological assays and techniques that are based on different antigens such as full-length spike, spike S1, RBD, and/or NP that may each utilize different dilution-series quantitation calculations (midpoint titer vs end point titer), simple optical density measurements at a single sample dilution, or area under the curve measurements. Each of these approaches will result in a different titer for the magnitude of the antiviral antibody response, and it can be difficult to compare the results obtained by independent groups because of the differences in assay conditions and the various approaches used for their quantitative assessment. Here, we have provided a framework and comparative data supporting the use of specific SARS-CoV-2-reactive monoclonal antibodies as serological standards. By utilizing a defined serological standard in every assay, this approach allows one to normalize antibody titers from assays performed on different days as well as provide for more informative comparisons between individual operators within a given research group or between different research groups. This is important not only for quantitation of antibody titers after natural SARS-CoV-2 infection, but also for comparing the magnitude and durability of antiviral antibody responses induced by vaccination—a key question that will require considerable investigation in future studies as we build a better understanding of this ongoing pandemic. Together, these preliminary analyses and comparisons support the use of quantitative methods, including appropriate monoclonal IgG standards, in upcoming serological studies on COVID-19.
Acknowledgments
The following reagent was obtained through BEI Resources, NIAID, NIH: Monoclonal Anti-SARS Coronavirus Recombinant Human IgG1, Clone CR3022 (produced in Nicotiana benthamiana), NR-52392.
Financial support. This work was supported in part by the Oregon National Primate Research Center grant 8P51 OD011092 (M.K.S., D.N.S.), pilot project grants from the OHSU Foundation (M.K.S., D.E.H.), and a supplement to National Institutes of Health National Institute of Allergy and Infectious Diseases R01 AI145835 (W.B.M.). The funders of this research were not involved in the study design, data analysis, manuscript preparation, or decision to publish this work.
Disclaimer. The opinions, results, and conclusions reported in this article are those of the authors and independent from the funding sources.
Potential conflicts of interest. The authors do not claim any financial conflicts of interest. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Author contributions. All authors contributed to the concept and study design. A.T. organized and analyzed the raw data, in addition to performing the ELISAs, preparing the figures, and performing the statistical analysis. W.B.M. enrolled COVID-19 cases and obtained medical histories and informed consent. D.E.H. organized the OHSU Biorepository for SARS-CoV-2. Z.L.L., Z.L., D.N.S., and S.C.K. were involved with sample procurement and data analysis. A.T. and M.K.S. wrote the manuscript, and W.B.M., D.E.H., D.N.S., Z.L.L., Z.L., and S.C.K. critically reviewed the manuscript before submission. Each of the authors had full access to the data included in the study. The corresponding author declares that all authors meet authorship criteria and that no others meeting the criteria have been omitted. M.K.S. had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
References
- 1. Bryan A, Pepper G, Wener MH, et al. Performance characteristics of the Abbott Architect SARS-CoV-2 IgG assay and seroprevalence in Boise, Idaho. J Clin Microbiol 2020; 58:e00941–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sood N, Simon P, Ebner P, et al. Seroprevalence of SARS-CoV-2-specific antibodies among adults in Los Angeles County, California, on April 10–11, 2020. JAMA 2020; 323:2425–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Biggs HM, Harris JB, Breakwell L, et al. ; CDC Field Surveyor Team . Estimated community seroprevalence of SARS-CoV-2 antibodies—two Georgia counties, April 28-May 3, 2020. MMWR Morb Mortal Wkly Rep 2020; 69:965–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rosenberg ES, Tesoriero JM, Rosenthal EM, et al. Cumulative incidence and diagnosis of SARS-CoV-2 infection in New York. Ann Epidemiol 2020; 48:23–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Havers FP, Reed C, Lim T, et al. Seroprevalence of antibodies to SARS-CoV-2 in 10 sites in the United States, March 23-May 12, 2020. JAMA Intern Med. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pickering S, Betancor G, Galão RP, et al. Comparative assessment of multiple COVID-19 serological technologies supports continued evaluation of point-of-care lateral flow assays in hospital and community healthcare settings. PLoS Pathog 2020; 16:e1008817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ripperger TJ, Uhrlaub JL, Watanabe M, et al. Orthogonal SARS-CoV-2 serological assays enable surveillance of low prevalence communities and reveal durable humoral immunity. Immunity 2020; 53:925–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cheng MP, Yansouni CP, Basta NE, et al. Serodiagnostics for severe acute respiratory syndrome-related coronavirus-2: a narrative review. Ann Intern Med 2020; 173:450–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Premkumar L, Segovia-Chumbez B, Jadi R, et al. The RBD of the spike protein of SARS-group coronaviruses is a highly specific target Of SARS-CoV-2 antibodies but not other pathogenic human and animal coronavirus antibodies. medRxiv 2020.05.06.20093377 [Preprint]. 10 May 2020. Available at: 10.1101/2020.05.06.20093377. Accessed 10 February 2021. [DOI] [Google Scholar]
- 10. Premkumar L, Segovia-Chumbez B, Jadi R, et al. The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci Immunol. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yuan M, Wu NC, Zhu X, et al. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science 2020; 368:630–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wec AZ, Wrapp D, Herbert AS, et al. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science 2020; 369:731–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yamaoka Y, Jeremiah SS, Miyakawa K, et al. Whole nucleocapsid protein of SARS-CoV-2 may cause false positive results in serological assays. Clin Infect Dis May 23, 2020; ciaa637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Iyer AS, Jones FK, Nodoushani A, et al. Persistence and decay of human antibody responses to the receptor binding domain of SARS-CoV-2 spike protein in COVID-19 patients. Sci Immunol. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Okba NMA, Müller MA, Li W, et al. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg Infect Dis 2020; 26:1478–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Amanat F, Stadlbauer D, Strohmeier S, et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med 2020; 26:1033–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Suthar MS, Zimmerman MG, Kauffman RC, et al. Rapid generation of neutralizing antibody responses in COVID-19 patients. Cell Rep Med 2020; 1:100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Food and Drug Administration. EUA authorized serology test performance. Available at: https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/eua-authorized-serology-test-performance. Accessed 19 October 2020.
- 19. Rosadas C, Randell P, Khan M, et al. Testing for responses to the wrong SARS-CoV-2 antigen? Lancet 2020; 396:e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Addetia A, Crawford KHD, Dingens A, et al. Neutralizing antibodies correlate with protection from SARS-CoV-2 in humans during a fishery vessel outbreak with high attack rate. J Clin Microbiol 2020; 58:e02107–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kofler N, Baylis F. Ten reasons why immunity passports are a bad idea. Nature 2020; 581:379–81. [DOI] [PubMed] [Google Scholar]