Abstract
The coronavirus, Severe Acute Respiratory Syndrome (SARS)-CoV-2, responsible for the ongoing coronavirus disease 2019 (COVID-19) pandemic, has emphasized the need for a better understanding of the evolution of virus-host interactions. ORF3a in both SARS-CoV-1 and SARS-CoV-2 are ion channels (viroporins) implicated in virion assembly and membrane budding. Using sensitive profile-based homology detection methods, we unify the SARS-CoV ORF3a family with several families of viral proteins, including ORF5 from MERS-CoVs, proteins from beta-CoVs (ORF3c), alpha-CoVs (ORF3b), most importantly, the Matrix (M) proteins from CoVs, and more distant homologs from other nidoviruses. We present computational evidence that these viral families might utilize specific conserved polar residues to constitute an aqueous pore within the membrane-spanning region. We reconstruct an evolutionary history of these families and objectively establish the common origin of the M proteins of CoVs and Toroviruses. We also show that the divergent ORF3 clade (ORF3a/ORF3b/ORF3c/ORF5 families) represents a duplication stemming from the M protein in alpha- and beta-CoVs. By phyletic profiling of major structural components of primary nidoviruses, we present a hypothesis for their role in virion assembly of CoVs, ToroVs, and Arteriviruses. The unification of diverse M/ORF3 ion channel families in a wide range of nidoviruses, especially the typical M protein in CoVs, reveal a conserved, previously under-appreciated role of ion channels in virion assembly and membrane budding. We show that M and ORF3 are under different evolutionary pressures; in contrast to the slow evolution of M as core structural component, the ORF3 clade is under selection for diversification, which suggests it might act at the interface with host molecules and/or immune attack.
Keywords: SARS-CoV-2, COVID-19, nidoviruses, matrix protein, ORF3a, ion channels
1. Introduction
The recent outbreak of the human coronavirus disease 2019 (COVID-19) has generated a global health crisis (Mehta et al. 2020). It is the seventh human disease caused by coronaviruses, after Severe Acute Respiratory Syndrome (SARS) in 2003 (Marra et al. 2003), Middle Eastern Respiratory Syndrome (MERS) in 2012 (Zaki et al. 2012), and four less-severe infections caused by human coronaviruses 229E (hCoV-229E) (Macnaughton and Hilary 1978), hCoV-OC43 (Lau et al. 2011), hCoV-NL63 in 2004 (Van Der Hoek et al. 2004), and hCoV-HKU1 in 2004 (Woo et al. 2005). Of these, SARS-CoV-2, SARS-CoV/SARS-CoV-1, MERS-CoV, hCoV-OC43, and hCoV-HKU1 belong to the beta coronavirus clade, while hCoV-229E and hCoV-NL63 belong to the alpha coronavirus clade. Although the broad genomic structure and core gene-composition of these viruses is similar, the pathology and severity of these viruses, including SARS-CoV-2, are markedly distinct. According to the WHO report, as of February 22, 2021, there have been over 110 million of confirmed cases with over 2 million deaths from COVID-19 globally. Therefore, the need for a better understanding of the biology and evolution of SARS-CoV-2 is a major desideratum to combat and prevent the disease.
Coronaviruses possess a large positive-sense single-stranded RNA genome with two-thirds of the genome coding for the ORF1a/ORF1ab polyprotein. This is followed by several ORFs encoding so-called structural and accessory proteins, a subset of which might be variable between CoVs (Marra et al. 2003). The ORF1-derived proteins are involved in polyprotein cleavage (the peptidase domain) (Graham et al. 2008), viral replication, viral RNA-processing (e.g. xEndoU endoRNase domain) and countering of defenses centered on NAD+/ADP-ribose (Macro domains) (Ricagno et al. 2006). The structural and accessory proteins contribute to virion structure and assembly, virulence and immune manipulation and invasion (Marra et al. 2003; Lu et al. 2020). However, despite concerted experimental studies, the structural understanding of many of these viral proteins is still lacking; for example, in SARS-CoV-2, these include ORF3a, ORF3b, M, ORF6, ORF8, ORF9b, ORF9c, ORF10, and certain domains of ORF1a/b. Here, we utilize a domain-centric computational strategy to systematically study the function and structure of CoV proteins. In our recent work, we have demonstrated that the mysterious SARS-CoV-2 protein, ORF8, belongs to a novel family of the immunoglobulin fold and it is one of the fast-evolving genes in the SARS-CoV-2 genome (Tan et al. 2020). Based on its inferred structural fold and enhanced evolutionary rate, we further predicted that ORF8 is likely to be involved in disrupting the host immune responses (Tan et al. 2020). Our computational predictions of both ORF8 structure and function have been recently confirmed by wet-lab studies (Flower et al. 2020; Zhang et al. 2020). In this study, we present results on the function and evolution of novel ion channel proteins in CoVs and other nidoviruses.
Viral ion channels (viroporins) represent an expanding functional class of proteins that have been identified in different animal viruses, such as the human immunodeficiency virus, hepatitis C virus, and influenza A virus (Nieva, Madan, and Carrasco 2012). These proteins are shown to operate at several steps in the viral life cycle including regulation of replication compartment, viroplasm formation, and virion budding and viral infection (Nieva, Madan, and Carrasco 2012). CoVs also code for their own ion channels. The SARS-CoV ORF3a was found to function as a potassium-specific channel promoting virus release (Lu et al. 2006). Thereafter, several other CoV proteins were shown to display ion channel activities, including porcine epidemic diarrhea virus (PEDV) ORF3 (Wang et al. 2012), hCoV-229E ORF4a (Zhang et al. 2014), and SARS-CoV envelope (E) protein (Li et al. 2014; Surya, Yan, and Jaume 2018). Among them, SARS-CoV ORF3a, PEDV ORF3, and hCoV-229E ORF4a, appear to utilize their three transmembrane (3-TM) region to constitute an ion channel either as a dimer or a tetramer, whereas SARS-CoV E protein with a 1TM region forms an ion channel as a pentamer (Li et al. 2014; Surya, Yan, and Jaume 2018). Recently, a similar ion channel activity was also observed in SARS-CoV-2 ORF3a and its structure was determined (Kern et al. 2020). This prompted us to systematically identify other ion channel proteins in coronavirus and related viruses by using sensitive profile-based homology detection and structural modeling methods. As a result, we have identified several homologous protein families, including ORF5 from MERS-CoV, many proteins from beta-CoVs, ORF3b from alpha-CoVs, and importantly, the well-known Matrix (M) proteins from CoVs, as well as more distant homologs from other nidoviruses. We present evolutionary and structural evidence that the newly identified families have preserved family-specific residues to constitute the ion conducting pore in the membrane. Using both phylogenetic analysis and phyletic profiling, we show that the M proteins are the most conserved structural components for CoVs, ToroVs, and Arteriviruses. By contrast, the ORF3a/ORF5/OR3b families appear to have emerged via a duplication from the M proteins at the base of alpha- and beta-CoVs and have undergone constantly rapid diversification, indicating they might be involved in host-virus interactions. Thus, our results have (1) expanded the repertoire of ion channel proteins across a large subset of nidoviruses (including all CoVs); (2) suggested an evolutionarily conserved role for the M protein in these viruses that might operate via the formation of a potential aqueous channel in the membrane; and (3) pointed to potential new functions of the ORF3-like families in host-virus interactions which might again result from transmembrane-associate pore formation.
2. Materials and methods
2.1 Homologous sequence searches and remote relationship detection
We utilized two protein sequence profile-based methods for homology searches and remote relationship detection. The first one is the iterated PSSM (profile)-based method, PSI-BLAST (Position-Specific Iterated BLAST) (Altschul et al. 1997). For most searches, a cut-off e-value of 0.005 was used as the significance threshold. In each iteration, the newly detected sequences that had e-values lower than the above cutoff were examined for being false-positives and the search was continued with the same e-value threshold only if the profile was uncorrupted. The second method is the HHsearch program (Söding 2005), which is used for the sequence-profile and profile–profile comparisons. HHsearch detects remote relationships between domains by comparing a Hidden Markov Model (HMM) constructed from a PSI-BLAST search and a pre-computed library of profile HMMs compiled from the Pfam domains (El-Gebali et al. 2019) and our own domains. The significance was evaluated by probabilities.
2.2 Sequence clustering and multiple sequence alignment
The collected sequence homologs of each protein family were subjected to a similarity-based clustering that was conducted by BLASTCLUST, a BLAST score-based single-linkage clustering method (ftp://ftp.ncbi.nih.gov/blast/documents/blastclust.html). This was used to remove highly similar sequences in the dataset. Multiple sequence alignments (MSAs) were built using the KALIGN (Lassmann and Erikk 2005), MUSCLE (Edgar 2004), and PROMALS3D (Pei, Kim, and Grishin 2008) programs. Based on prior benchmarks with divergent proteins, these programs can produce accurate MSA for protein families (as assessed by structural comparisons) when their sequence identity is high (>25%). However, when they are used to make super-alignments that contain multiple highly divergent domain families, they typically tend to introduce obvious mis-alignments. We tackle this problem using multiple steps to make a good MSA. Specifically, we use both KALIGN and MUSCLE to generate the MSA for each domain family. We choose the alignments which have fewer gaps in the core structural elements that determine the fold, that is alpha-helices and beta-sheets. We then use PROMALS3D to generate the super-alignment, which will have both errors within each family and between families. For each family, we use the prior MSAs generated by either MUSCLE or KALIGN to correct mis-alignments introduced by PROMALS3D. We correct those misalignments that occur between families by using the predicted secondary structure information and profile–profile alignments generated by HHsearch (Söding 2005). Finally, the generated super-alignment can be sampled randomly for each family and checked by superimposing the sequence against known structures to ascertain contiguity of secondary structure elements.
2.3 Conservation analysis using PSSM and consensus methods
Conservation analysis is conducted by comparing the individual alignment of each family and the superalignment of all members. Each family has its own conservation patterns which we compute as a PSSM profile and more simply a sequence consensus. The PSSM captures the position-specific frequencies of all 20 amino acids across the alignment (Altschul et al. 1997; Schaffer et al. 1999) corrected for their background frequency in the alignment, which we present in a two-dimensional plot (Supplementary Figs S8 and S9). The PSSM is computed with pseudocounts using the following process:
1. Frequency determination with pseudocounts for an MSA column j with n sequences
where and are the observed frequency of the amino acids in column j and their respective background frequencies across the alignment for pseudocount correction.
2. These are converted to the initial PSSM score thus:
The score is then rescaled as , where is the maximum value of the score across all columns. The conservation metric at each column of the PSSM can be calculated taking the inverse of the mean of the PSSM scores and centering them. This metric for each column is then multiplied by , where is the frequency of gaps in that column to down-weight gapped columns.
For the consensus method, we generate consensus sequence based on different categories of amino acid properties, a classification developed by Taylor (Taylor, 1986). In this method, different amino acids are classified into 11 groups according to their shared physico-chemical properties, including:
hydrophobic amino acids (labeled in ‘h’, including Ala, Cys, Phe, Ile, Leu, Met, Val, Try, Tyr),
aliphatic group (labeled in ‘l’, including Ile, Leu, Val),
aromatic group (labeled in ‘a’, including Phe, His, Trp, Tyr),
big amino acids group (labeled in ‘b’, including Glu, Phe, Ile, Lys, Leu, Met, Gln, Arg, Trp, Tyr),
small amino acids (labeled in ‘s’, including Ala, Cys, Asp, Gly, Asn, Pro, Ser, Thr, Val),
amino acids containing alcohol (labeled in ‘o’, including Ser, Thr),
negative amino acids (labeled in ‘-‘, including Asp, Glu),
positive amino acids (labeled in ‘+’, including His, Lys, Arg),
charged amino acids (labeled in ‘c’, including Asp, Glu, His, Lys, Arg),
polar group (labeled in ‘p’, including Cys, Asp, Glu, His, Lys, Asn, Gln, Arg, Ser, Thr),
tiny amino acids (labeled in ‘u’, including Ala, Gly, Ser)
Consensus is calculated by examining each column of the MSA to determine whether an above-threshold fraction of the amino acids belongs to a group defined previously. For each MSA, we generated a series of consensus sequences based on threshold from 70 per cent, 80 per cent, 90 per cent to 100 per cent. We colored the MSA using the CHROMA program (32) based on the consensus sequence calculated from a threshold (either 90% or 80%) and further modified using Adobe Illustrator or Microsoft Word.
Comparison of either the profiles or the consensus between different families in a superfamily allows one to identify the specific versus general conservation patterns. The general conservation pattern captured as a sequence profile or the consensus for the superalignment primarily reveals a pattern of just hydrophobic residues that form the folding core of the multiple domains. However, the family-specific consensus or profiles reveal residues which are conserved only in a given family and constitute their unique conservation pattern.
2.4 Entropy analysis
Position-wise Shannon entropy (H) for a given column of the MSA was calculated using the equation:
P is the fraction of residues of amino acid type i, and M is the number of amino acid types.
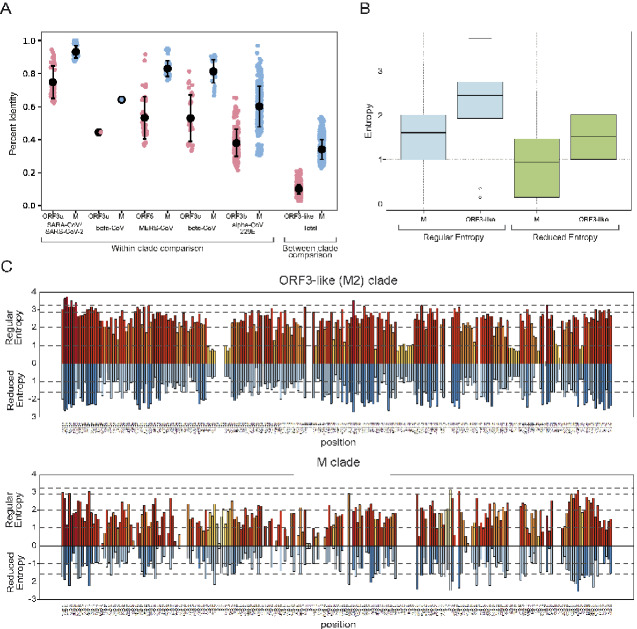
Two distinct alphabets used to calculate the column-wise Shannon entropy. The first is the regular 20 amino acid alphabet. The Shannon entropy for the ith position in the alignment ranges from 0 (only one residue at that position) to 4.32 (all 20 residues equally represented at that position) in a 20 letter alphabet, which is shown on the positive y-axis with the x-axis being the position of the column along the multiple alignment (Fig. 4C). The second is in a reduced alphabet of eight symbols that groups the amino acids into non-overlapping categories based on related sidechain properties. For example, in the reduced alphabet both D and E are represented by a single alphabet (acidic category). This is plotted downwards, i.e., along the negative y-axis with magnitude equal to the entropy in the reduced alphabet for the same column. Comparing the entropies in the regular 20 aa alphabet versus the reduced alphabet helps discern positions that show genuine diversifying pressures. For instance, hydrophobic positions can show high entropy in the regular entropy if they present multiple hydrophobic residues. However, high entropy in this scenario is not biochemically very meaningful especially for membrane protein because the different hydrophobic residues perform an equivalent role. This is effectively filtered by the reduced alphabet that brings the focus on genuinely variable positions with different sidechain characteristics. The way the graph is to be understood is by looking at the overall tendency for the heights of the bars in each alphabet—it can be seen that the ORF3-like families show higher amplitude of the bars on average than the genomically coupled M proteins. This is statistically quantified and found to be significantly higher in both alphabets for the former families (shown as boxplot the P-values are provided in the text).
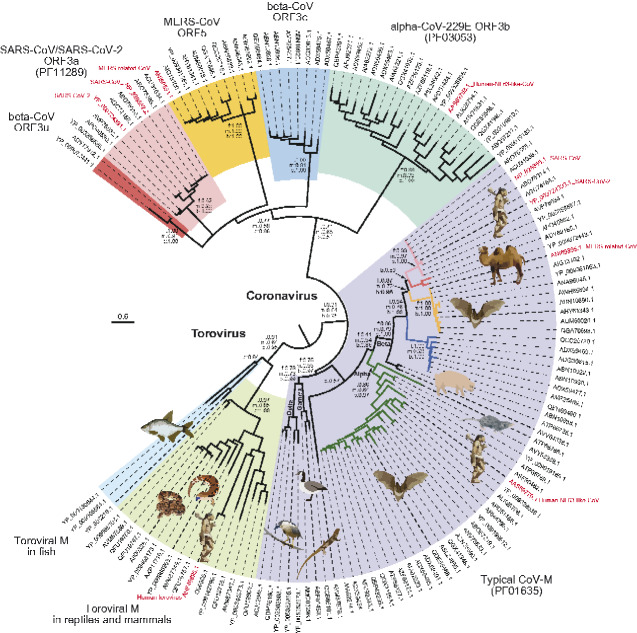
Figure 4.
Evolutionary relationship of M/ORF3 ion channel families of CoVs and ToroVs. The ML tree with the highest log likelihood (-16934.18) is shown with supporting values for the major branches from three phylogenetic methods: f for FastTree SH-like local support value, m for MEGA-ML bootstrap value and b for BI posterior value. Protein sequences are represented by the NCBI accession IDs and those from human viruses are further labeled with the virus name in red. The cartoon of several viral hosts is shown; its correspondence with the sequence is indicated by the bold dashed lines. Each family is highlighted in a different background. For the sequence within the M family, if they are coupled with one of the ORF3 families, their branches are colored with the same theme, accordingly.
The Kullback–Leibler entropy, also called the Kullback–Leibler divergence (or relative entropy), was computed for each column as described (Manning and Hinrich 1999) by the equation:
where is the observed frequency of amino acid (AA) x in the column and is its background frequency in the entire alignment. The value D is then centered by the mean taken across all columns and normalized by the range defined by its maximum and minimum values across the alignment to identify functionally unique positions of each family.
Analysis of the entropy values was performed using the R language.
2.5 Protein structure prediction and analysis
Secondary structure was predicted using the JPRED program (Drozdetskiy et al. 2015). The transmembrane regions were predicted using the TMHMM Server v. 2.0 (Krogh et al. 2001).
The MODELLER (version 9v1) program (Webb and Andrej 2016) was utilized for homology modeling of the tertiary structures of SARS-CoV-2 M protein, MERS-CoV ORF5, human NL63-like-CoV ORF3b and Bat-CoV HKU9-2 ORF3c by using the SARS-CoV ORF3a (6xdc) as a template. The dimeric status was modeled according to the template. The sequence identity between the template and the targets is very low, from 15 per cent between the template and the SARS-CoV-2 M protein, 19 per cent with the MERS-CoV ORF5, 15 per cent with the NL63-like-CoV ORF3b, to 22 per cent with the Bat-CoV HKU9-2 ORF3c. Since in these low sequence-identity cases, sequence alignment is the most important factor affecting the quality of the model (Cozzetto and Anna, 2004), alignments used in this analysis have been carefully built and cross-validated based on the information from HHsearch and edited manually using the secondary structure information. For each protein, we generated five models and selected the one that had the highest model accuracy P value (ranging from 0.06 to 0.013) and global model quality score (ranging from 0.34 to 0.38) as assessed by ModFOLD6 online server (Maghrabi and McGuffin 2017). Structural analysis and comparison were conducted using the molecular visualization program PyMOL (DeLano 2002).
2.6 Molecular phylogenetic analysis
Based on the super-alignment of the β-sandwich domains of nine M/ORF3 families (Supplementary Dataset S1), we conducted phylogenetic analysis using three robust methods, including the Maximum Likelihood (ML) analysis implemented in the MEGA7 program (Kumar, Stecher, and Tamura 2016), an approximately-maximum-likelihood method implemented in the FastTree 2.1 program (Price, Dehal, and Arkin 2009), and Bayesian Inference implemented in the BEAST 1.8.3 program (Suchard et al. 2018). For ML analysis, initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. A discrete Gamma distribution was used to model evolutionary rate differences among sites (four categories). The rate variation model allowed for some sites to be evolutionarily invariable. A bootstrap analysis with 100 repetitions was performed to assess the significance of phylogenetic grouping. For FastTree analysis, default parameters were applied, which include the WAG evolutionary model and the discrete gamma model with 20 rate categories. For Bayesian inference, a JTT amino acid substitution model with a discrete Gamma distribution (four rate categories) was used to model evolutionary rate differences among sites. Markov chain Monte Carlo (MCMC) duplicate runs of 10 million states each, sampling every 10,000 steps was computed. Logs of MCMC runs were examined using Tracer 1.7.1. Burn-ins were set to be 4 per cent of iterations.
The tree with the highest log likelihood from the ML analysis was visualized using the FigTree 1.4.4 program of the BEAST package (Suchard et al. 2018). The ML-bootstrapping value, FastTree SH-like local support value and Bayesian Posterior value are shown next to the branches.
2.7 Genome organization analysis
Open reading frames of viral genomes used in this study were extracted from NCBI GenBank files (Benson et al. 2018). Protein sequences were subjected to similarity-based clustering by BLASTCLUST with -S at 0.4 and -L at 0.4. Protein clusters were further annotated with conserved domains which are identified by the hmmscan searching against Pfam (Eddy 1998; El-Gebali et al. 2019) and our own curated domain profiles. For previously unknown domains, we used sequence searches, MSA analysis, and further sequence-profile searches (Söding 2005) to study their sequence and structural features. All sequence alignments can be found in the supplementary data.
3. Results
3.1 Unification of M/ORF3 ion channel families in CoVs and other nidoviruses
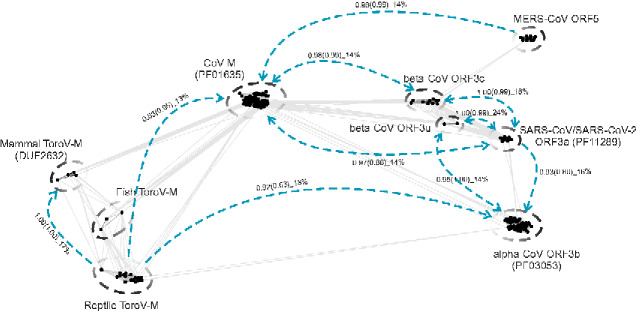
Examination of the structure of the SARS-CoV-2 ORF3a (Genbank: YP_009724391.1) reveals two distinct domains, namely a N-terminal 3-transmembrane (3-TM) region and a C-terminal β-sandwich domain. We first identified other viral homologs of ORF3a by conducting iterative sequence searches using PSIBLAST against the NCBI NR database (Altschul et al. 1997). We then prepared an MSA to identify the evolutionarily conserved residues, and the majority of them line the ion channel pore of the 3-TM structure (Supplementary Fig. S1). Interestingly, when we used HMM profile-based homology detection against Pfam profiles via HHsearch (Söding 2005), we found two other coronavirus protein families as the significant hits: the coronavirus M protein (Pfam ID: PF01635) and the alpha-coronavirus ORF3b (Pfam ID: PF03053) (Fig. 1) which were not known to be related to ORF3a. As many coronavirus proteins are not covered by Pfam profiles, we performed a BLAST bit-score based single-linkage clustering analysis of a comprehensive collection of coronavirus proteins (excluding the ORF1a/b) and selected the representatives, for a series of HHsearch searches. This procedure uncovered three other viral protein families that are related to ORF3a and M families, prototyped by ORF5 of MERS-CoVs (NCBI accession number: YP_009047208.1), ORF4 of 229E-related bat CoV (NCBI accession number: ALK28794.1) and ORF3 of Eidolon bat coronavirus/Kenya/KY24/2006 (NCBI accession number: ADX59467.1) (Fig. 1). Further, we extended our clustering analysis together with profile searches to other nidoviruses beyond coronaviruses, leading to the identification of numerous M proteins from fish, reptile, and mammal ToroVs as homologs (Fig. 1). Figure 1 summarizes the above homology detection searches and the viral protein families that were uncovered by them. In this figure, representative viral proteins were clustered using all-against-all BLASTP comparisons and are presented nodes of a two-dimensional graph arranged using the Fruchterman and Reingold algorithm (Fruchterman and Reingold 1991) implemented in the CLANS program (Frickey and Lupas 2004), which are linked by edges of denoting the detected sequence similarity. The viral proteins belonging to the same family segregate as a dense sub-graph, while the families themselves are linked together by edges mostly derived from profile-profile analysis (blue dashed lines in Fig. 1).
Figure 1.
Graphic two-dimensional representation of sequence similarity between the divergent M/ORF3 proteins of CoVs and ToroVs. The similarities were detected by both CLANS and HHsearch programs. Each node corresponds to a protein sequence. Straight lines indicate significant high-scoring segment pairs (HSPs) detected by all-against-all BLASTP searches with scoring matrix BLOSUM45 and e-value cutoff of 0.02. The graph was generated by CLANS, which uses the Fruchterman and Reingold graph drawing algorithm (Frickey and Lupas 2004). The gapped circles indicate individual protein families. Blue dashed lines indicate the remote relationship between families detected by the HHsearch program where the arrow indicates the search direction. The probabilities of the HHsearch comparisons with full-length protein sequences and C-terminal cytoplasmic β-sandwiches (in brackets) are indicated, followed by the percentage sequence identity.
To examine the relationship of the nine viral protein families identified above more closely, we generated MSAs of each family, and predicted their potential secondary structures (Supplementary Figs S2–S7). Importantly, secondary structure (Drozdetskiy et al. 2015) and transmembrane region prediction (Krogh et al. 2001) revealed that all nine viral protein families share a congruent domain architecture, with a N-terminal 3-TM region and a C-terminal β-sandwich domain. As TM regions tend to have a biased composition with an enrichment in hydrophobic residues, we next investigated if the above observed profile–profile similarity between the families could be recapitulated by their globular β-sandwich domains. Accordingly, we extracted these domains from above families and conducted a comparable HHsearch as above. These searches recovered significant similarities between families as with the searches with the full-length proteins (Fig. 1). This was further supported by a super-alignment and secondary structure prediction of the β-sandwich domains from the different families (Fig. 2): despite their low sequence identity (10-20%), they all share a comparable eight β-stranded predicted secondary structure, with several conserved hydrophobic amino acids forming the predicted folding cores of the β-strands (Fig. 2). Thus, the sensitive sequence comparisons and alignments using either full-length proteins or β-sandwich domains revealed previously undetected relationships between the SARS-CoV ORF3a ion-channel proteins and several distinct viral protein families in CoVs and other nidoviruses. Collectively, we term these unified families the viral M/ORF3 superfamily.
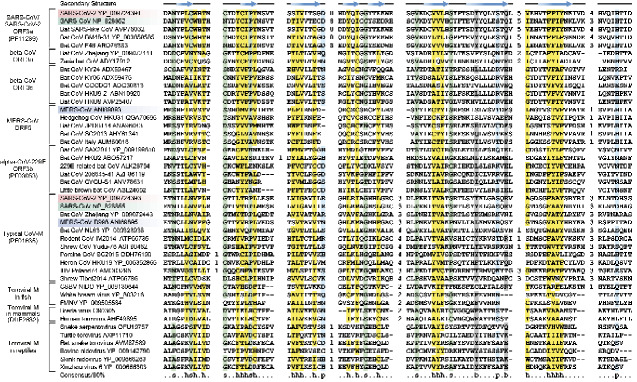
Figure 2.
MSA of β-sandwich domains of nine M/ORF3 protein families. The secondary structure is shown above the alignment and the consensus is shown below the alignment, where h stands for hydrophobic residues, s for small residues, b for big residues, and p for polar residues. The numbers are indicative of the excluded residues from sequences. Sequences from three human-infected CoVs are highlighted (SARS-CoV-2 in red, SARS-CoV in green and MERS-CoV in blue).
3.2 Novel M/ORF3 families might potentially form membrane pores or function as ion channels
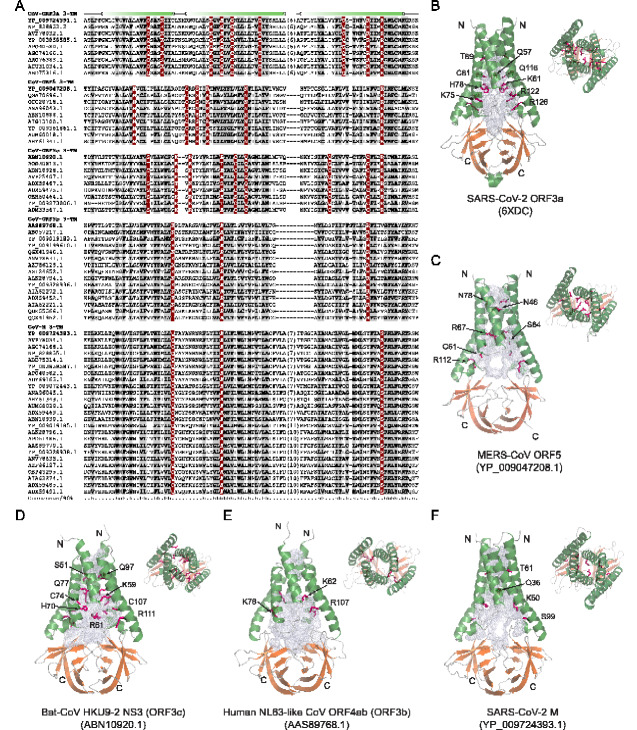
Both SARS-CoV and SARS-CoV-2 ORF3a proteins are ion channels that form a polar ion-conducting cavity through dimerization or tetramerization (Lu et al. 2006; Kern et al. 2020). To explore the functions of the other viral families which we unified into the M/ORF3 superfamily, we generated an MSA of the 3-TM regions of several CoV-M/ORF3 families (Fig. 3A). We used this to examine their sequence conservation by generating both position-specific-score matrices (PSSMs) (Supplementary Fig. S8) (Schaffer et al. 1999) and conservation consensus (Fig. 3A, Supplementary Figs S1–S5) (32). Both these approaches revealed several conserved positions across the superfamily (Fig. 3A, Supplementary Figs S1–S5 and S8). The majority of them are hydrophobic residues and are located on the TM helices (the columns with grey background in Fig. 3A), indicating the TM domain of this superfamily is likely to adopt a similar structure. Further, the profile of conserved hydrophobic residues also indicated that other members of the M/ORF3 superfamily might form similar membrane-embedded multimers as ORF3a.
Figure 3.
(A) MSA of 3-TM regions from five CoV ORF3-like (M2) families. The secondary structure is shown above the alignment and the consensus is shown below the alignment, where h stands for hydrophobic residues, s for small residues, b for big residues, and p for polar residues. The numbers are indicative of the excluded residues from sequences. (B–F) Dimeric structures of SARS-CoV-2 ORF3a and homology models of SARS-CoV-2 M, human-NL63-like-CoV ORF3b, bat-CoV HKU9-2 ORF3c, and MERS-CoV ORF5. Both the side-view and top-view are presented. Alpha-helices are colored in green, β-sheets of the β-sandwich domains in orange, and loops in grey. The channel cavities are colored in grey. The polar residues, which are conserved in each family and located on the cavity surface, are shown in a purple stick model. The PDB and NCBI ids are shown in brackets.
We next investigated if they might also form transmembrane pores: we reasoned that if the capacity to form aqueous transmembrane pores is present more generally across proteins of this superfamily, then the selective pressure would favor retention of specific polar residues in their inferred ion-conducting cavity. Further, the presence of such residues could also indicate the capacity to transport of ions through the pore thereby resulting in channel activity. However, the above analysis did not identify any universally conserved polar residues across the M/ORF3 superfamily. This is not entirely unexpected for divergence of distinct families with a superfamily, given that they might have acquired distinct family-specific functions (Zhang, Iyer, et al. 2014). Therefore, using the same consensus and PSSM methods as above, we examined each of the families of M/ORF3 superfamily separately. Notably, this analysis identified several family-specific, conserved polar positions, mostly basic residues (Fig. 3A). We inferred their positions using the ORF3a structure (PDB: 6XDC) (Kern et al. 2020), and found them to be located at the inner surface of the cavity forming the TM pore as well as just outside and inside the membrane. As a proof of concept, we examined the location of the conserved residues we had identified in the ORF3a family (Fig. 3A) and found them to constitute the inner surface of the ion-transporting cavity in SARS-CoV-2 ORF3a (PDB: 6XDC; Fig. 3B) (Kern et al. 2020). Those polar residues that are located just outside or on the inner side of the membrane might be involved in maintaining TM polarity or in mediating ion movement in the vicinity of the channel (Supplementary Fig. S1).
As no structure is available for other M/ORF3 families, we utilized the MODELLER program (Webb and Sali 2016) to generate homology models of the dimeric form for representatives of other families (Fig. 3C–F) by using the SARS-CoV-2 ORF3a structure as the template. These models strongly supported our proposal that several of the above-identified evolutionarily conserved polar residues specific to each family line a TM cavity equivalent to the ion-conducting channel of the ORF3a viroporin (Fig. 3 and Supplementary Figs S2–S5). This suggests that even though M and the remaining newly identified viral M/ORF3 families might not possess the same pattern of conserved polar residues as ORF3a, they do possess intra-TM polar residues that line a comparable pore cavity. This implies that, at the very least, they might form an aqueous pore in the membrane and could also potentially function as ion channels. In contrast to the TM domain, no universally conserved or family-specific polar residues are seen in the C-terminal β-sandwich domains indicating that they are neither enzymatic domains nor are likely to play a role in ion transport (Fig. 2). However, it is possible that they function as adaptor domains that undergo conformational changes induced by ion conduction and might help recruit partner proteins in a structural context.
3.3 Evolutionary relationships of divergent M/ORF3 families
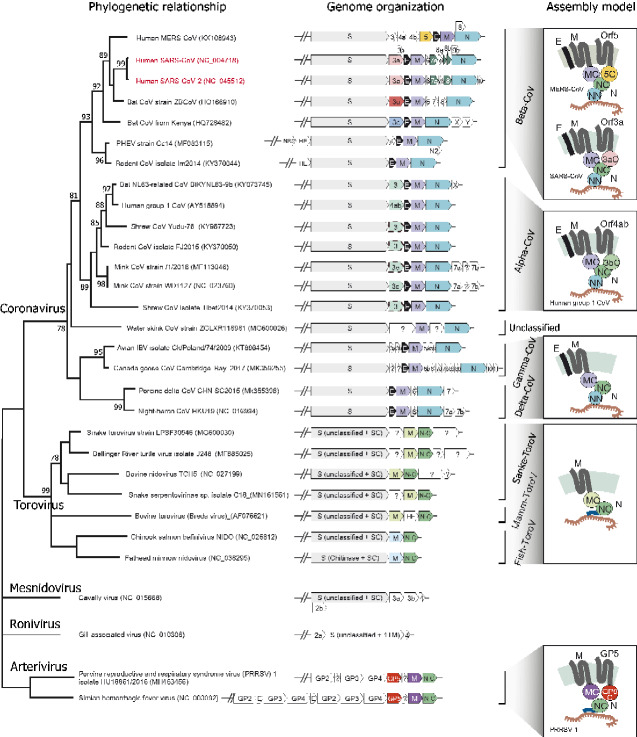
We next examined the evolutionary relationship of the families within the M/ORF3 superfamily. Using a super-alignment of the β-sandwich domains (Supplementary Dataset S1), we conducted phylogenetic analyses using rapid approximately-maximum-likelihood inference (FastTree), robust Maximum Likelihood (ML), and Bayesian inference (BI) methods (Suchard et al. 2018; Price, Dehal, and Arkin 2009; Kumar, Stecher, and Tamura 2016). All three methods produced similar tree topologies, especially in terms of the higher-order relationships between the ToroV M families, CoV M family and CoV ORF3-like families. The only differences in topology between the methods are seen within the CoV ORF3-like families and within reptilian and mammal Toroviral M families. This situation is typical for divergent families where certain branches might differ in positions due to their distinct evolutionary rates. The relationship of M/ORF3 families mirror the relationship of viruses that contain them (Fig. 4). The M protein families from ToroVs cluster together as a strongly supported clade. They form a sister group to all coronavirus families. Among these CoV families, the CoV-M protein family forms a distinct clade, which can be further divided into several subfamilies in Delta-, Gamma-, Alpha-, Beta-CoVs and a new unclassified group typified by a CoV from the Guangdong Chinese water skink (NCBI accession number: AVM87576; Fig. 4). All other CoV families form a second major clade, indicating that they diverged from a common ancestor that split from the CoV-M clade. This relationship is in accordance with the presence of an N-terminal ecto-region in these CoV ORF3-like families (Supplementary Figs S1–S4). In terms of their distribution, ORF3b is only present in alpha-CoVs whereas ORF3c, MERS-CoV ORF5, ORF3u and SARS-CoV/SARS-CoV-2 ORF3a are present in different beta-CoV subgroups. Collectively, due to their relationship with CoV M proteins, we propose to name these ORF3a-like proteins as CoV Matrix 2 (M2) proteins while the typical CoV M proteins as M1.
3.4 The coupled ORF3-like (M2) and M proteins display different evolutionary rates
Examination of the genome organization of the viruses that code for both M1 and M2 clade proteins (see next section for details) showed the genes for the two families to be strictly coupled, in an M2-E-M1 order, where ORF3 represents one of the M2 clade families, E is the envelope protein, and M1 is the typical matrix protein of CoVs (M). The inter-relationships of the alpha- and beta-CoV M2 families are congruent to the relationships of the corresponding coupled M1 clade proteins from the same genome (Fig. 4). This indicates that both the M2 (ORF3-like) and M proteins have been vertically inherited from the common ancestor of the alpha- and beta-CoVs. However, we found that the sequence identity between the ORF3-like proteins is always considerably lower than that of the coupled M proteins (Fig. 5A): the average percent identity of the M1 clade proteins is 34 per cent, whereas the average percent identity of the M2 clade proteins is only 10 per cent (Fig. 5A). This was also supported by the significantly higher number of above-average conservation positions in the PSSM of the M1 clade as opposed to the M2 clade (t-test, P = 1.23 × 10−5, Supplementary Fig. S9). This suggested that the M2 clade proteins are diverging much faster than the cognate M proteins in the same genome.
Figure 5.
(A) Percent identity comparison between the coupled M and ORF3-like (M2) proteins within different viral clades and between different clades. (B) A boxplot comparing the distribution column-wise Shannon entropy between the M and ORF3-like (M2) proteins from the same set of viral genomes based on regular alphabet of 20 amino acids and reduced alphabet of 8 symbols. The box in the plot indicates the first quartile, the second quartile (median), and the third quartile. The whiskers indicate the min and max entropy values excluding outliers; and if a value is lower or greater than the distance of 1.5 times the interquartile range, they are plotted as outliers shown as dots in the plot. (C) The column-wise entropy values for each column in the alignment of the M and ORF3-like (M2) proteins from the same set of viral genomes. Shannon entropy computed based on the regular amino acid alphabet (20 amino acids) is shown above the zero line in shades of orange, while Shannon entropy computed based on a reduced alphabet of 8 symbols are shown below the zero line in shades of blue. Those positions showing high entropy in both alphabets are under potential diversifying selection for amino acids of different chemical character.
To better understand this divergence, we performed a column-wise Shannon entropy analysis (Fig. 5B and C) that objectively quantifies the conservation in each column (Vinga 2014; Krishnan et al. 2018). When using the regular 20-amino-acid alphabet, we found that across the length of the whole alignment, ORF3-like (M2) proteins have significantly higher mean column-wise entropy than the M1 proteins from the same set of viral genomes (2.28 as opposed to 1.49; P = 1.6 × 10−16 for the H0 of congruent means by t-test). When using a reduced eight-letter alphabet (where amino acids are grouped based on similar side chain chemistries), we found a similar result (1.52 for the M2 clade proteins as opposed to 0.91 for the coupled M1 clade proteins; P = 2.2 × 10−11). The significantly higher mean column-wise entropy of the ORF3-like (M2) clade in the reduced alphabet indicates that there is a much greater tendency in these proteins to contain positions that differ drastically in side chain chemistry (e.g. substitution of a charged or polar residue for a hydrophobic residue). To explore this further, we computed the normalized Kulback–Leibler entropy (see Section 2) which measures the divergence of the observed residue distribution in a column from the background residue distribution across the entire alignment of both the M1 and ORF3-like (M2) clades. Thus, this measure accurately picks out those columns in the MSA of a clade that define its unique sequence conservation profile (Supplementary Figs S10 and S11). We then examined the top ranked quartile of these columns from the M1 and M2 clades to see if they contained amino acids with the same side chain chemistry or not. We found a dramatic difference between the two clades—whereas only 16 out 63 top Kulback–Leibler entropy columns in the M1 clade had residues with different side-chain properties, 37 out of 63 showed this trend in the M2 clade (proportions test p-value = 3 × 10−4).
In conclusion, these results suggest that the M1 is under stronger selection for retention of conserved positions (purifying selection); conversely, the M2 clade proteins appear to be under diversifying selection.
3.5 Evolution of genomic coupling of the virion structure and host-interaction genes in CoVs, ToroVs, and other related nidoviruses
Other than the M1 and M2 clade proteins, there are several key structural proteins of the mature coronavirus particle. These include (1) the spike protein S, which is involved in cellular receptor binding and internalization (Walls et al. 2020); (2) E, which is a 1-TM ion channel protein critical for envelope formation and membrane budding (Ruch and Machamer 2012); (3) N, which is essential for viral genome packaging and linking the viral ribonucleoprotein to the membrane (Kuo, Koetzner, and Masters 2016; Masters 2019). Hence, we investigated their phyletic patterns relative to that of the M/ORF3 proteins. We conducted a genomic organization analysis by using sequence similarity-based clustering and domain analysis for CoVs, ToroVs, and other nidoviruses such as roniviruses, mesnidoviruses and arteriviruses (Fig. 6).
Figure 6.
Genomic organization of representative CoVs, ToroVs and other related nidoviruses. The detailed species tree of the CoV-ToroV clade was adapted from the Fig. 4. The genomic regions encoding for structural and accessory proteins are illustrated right to the terminal nodes of the phylogenetic tree. The gene names are shown according to their NCBI genome annotation, but the color of the gene box corresponds to different protein families: all CoV Spike proteins in grey, SARS-CoV/SARS-CoV-2 ORF3a in pink, beta-CoV ORF3u in red, MERS-CoV ORF5 in yellow, beta-CoV ORF3c in sky-blue, alpha-CoV-229E ORF3b in pale-green, typical CoV-M proteins in light-purple, reptile and mammal ToroV M protein in light-green, fish ToroV M proteins in light-blue, envelope (E) protein in black, SARS-CoV ORF7a-Ig and ORF8-Ig in dark-green, nucleocapsid (N) protein in blue-green, nucleocapsid (N) protein C-terminal dimerization domain in green, Arterivirus ORF3-like GP5 protein in vermilion and Arterivirus M protein in purple. Several previously unannotated alpha-CoV ORF3b genes are recovered and highlighted in dashed boxes. The predicted assembly models (without Spike) are shown on the right side of genomic organization. Protein abbreviations used in the model are as follows: MC: M protein C-terminal β-sandwich domain; NN: N protein N-terminal RBD domain; NC: N protein C-terminal dimerization domain; SC: Spike protein C-terminal domain; 5C: MERS-ORF5 C-terminal β-sandwich domain; 3aC: SARS-CoV/SARS-CoV-2 ORF3a C-terminal β-sandwich domain; 3bC: alpha-CoV-229E ORF3b C-terminal β-sandwich domain; GP5C: Arterivirus GP5 protein C-terminal β-sandwich domain.
Consequently, we found that the S proteins of three major lineages of nidoviruses such as CoVs, ToroVs, and mesnidoviruses show a conserved C-terminal region (SC; Pfam domain: PF01601) while their N-terminal regions, which are cleaved upon engaging host receptors by peptidase domains, are greatly variable (Fig. 6). The M proteins from both CoVs and ToroVs share a similar architecture with N-terminal 3TM and C-terminal β-sandwich domains. Interestingly, we found that Arterivirus contains two proteins with distinct 3TM domains, so-called M and GP5 (Supplementary Figures S12 and S13). Both of their core 3TM domains share similarity with the M/ORF3-3TM domain and display conserved polar residues. However, their C-terminal cytoplasmic domain, while β-strand rich, is shorter with just six β-strands (Supplementary Figs S12 and S13). This indicates that the 3TM domains of the CoVs and ToroVs, on the one hand, and Arterviruses, on the other, share a common ancestry, but we cannot currently detect statistically significant relationships between their respective coupled β-strand-rich C-terminal domains. The N proteins of all CoVs contain an N-terminal RNA-binding domain (RBD) and a C-terminal dimerization domain (NC in Fig. 6). Beyond the CoVs, the NC domain of the N protein is present more widely in ToroVs and Arteriviruses but these proteins do not possess the RBD of CoV-N; instead, they carry a long N-terminal region that contains a stretch of basic residues (Supplementary Fig. S14). Therefore, we predict that this basic region functions equivalently in binding the negatively-charged viral genomic RNA.
Tracing the genomic organization of these genes across nidoviruses indicates that the juxtaposition of the M and NC genes is present in arteriviruses, ToroVs and CoV (Fig. 6). This suggests that the M-NC coupling was an ancestral feature present in the ancient nidoviral particle with the dyad playing a role in anchoring the genomic RNA to the membranous envelop. However, these are absent in the currently available roniviruses and mesonidoviruses suggesting that this mechanism was secondarily lost in these viruses (Fig. 6). Nevertheless, the latter two viral clades share the spike protein S with ToroVs and CoVs. Indeed, in the common ancestor the ToroVs and CoVs, we can infer an S-M-NC gene triad indicating that ancient M-NC dyad was joined by the S protein with rapidly evolving N-terminal regions that became central to the invasion process. At the base of the CoV clade, the above gene-triad was joined by the E gene inserted between S and M resulting the ancestral S-E-M-N order. This represents the first incorporation of a new viral ion channel into the system in CoVs. Finally, the common ancestor of the Alpha- and Beta-CoVs saw the split of the M1 and M2 clade with members of the M2 clade acquiring channel functions in the host-virus interface. A similar process might have independently occurred in the Arteriviruses, where the paralogous 3TM proteins GP5 and M might respectively function as a viroporin and a pore-forming structural component of the envelope.
Beyond these, we also observed several cases of accretion of lineage-specific genes in the S-M-N genomic region across diverse nidoviruses (Fig. 6). For example, the SARS-related clade acquired several new genes including ORF6, ORF7 and ORF8 that were inserted between M and N genes (Tan et al. 2020), while the MERS-CoV clade, has several genes that were inserted between Spike and ORF5 genes. Similar uncharacterized genes or genes coding for proteins with domains playing a predicted role in pathogenesis (e.g. the hemagglutinin-esterase domain in Bovine Toroviruses) are seen inserted between the known genes in the corresponding genomic regions of Ateriviruses and Toroviruses. Together, these observations suggest that components of the membrane-structural complex (e.g. M, N) are frequently coupled to (e.g. S or E and the lineage-specific ORFs) or gave rise to (e.g. M2 clade) proteins that are directly part of the host-virus interface.
4. Discussion
Coronaviruses have emerged as a major threat to human health in the past two decades as the causative agents of several severe infectious diseases, namely SARS, MERS, and the currently ongoing COVID-19 pandemic. The rapid spread, severity of these diseases, as well as the potential re-emergence of other CoV-associated diseases emphasize the need for a thorough understanding of the biology and evolution of these viruses. Viral ion channel proteins exemplified by ORF3a are structurally distinct from all previously characterized ion channels (Flower et al. 2020) and represent a new functional theme in coronavirus biology. In the current study, we use sensitive profile-based sequence analysis and homology modeling methods to study these channels and their homologs. We have unified several divergent families of viral ion channel proteins, including SARS-CoV/SARS-CoV-2 ORF3a, MERS-CoV ORF5, proteins from other beta-CoVs (ORF3c) and alpha-CoVs (ORF3b), the CoV M proteins, and many distant homologs from other nidoviruses such as ToroVs and Arterivirus into a single superfamily of viral membrane proteins. We present a natural classification of proteins in this superfamily and provide computational evidence that these viral protein families have preserved several family-specific polar residues that can form a TM aqueous pore with potential ion-conducting capacity. Thus, our study has greatly expanded the repertoire of viral ion channel-related proteins in CoVs and other nidoviruses and provides several insights into the function and evolution of these viruses.
4.1 Roles for potential transmembrane ion transport in assembly of nidoviral particles
CoVs, ToroVs and arterivirus are nidoviruses, which feature an envelope derived from the host cell membranes with embedded viral proteins (Neuman and Buchmeier 2016). M proteins are a key structural component of the viral envelope. Although their roles in promoting virus assembly and membrane-budding are well documented (Neuman et al. 2011), their mechanism of action remains unknown. The results of our study raise the possibility that the M proteins might possess the capacity to form a TM-pore and potentially possess ion-channel activity comparable to their viroporin homologs. If this were the case, then it is conceivable that the establishment of ionic gradients by M plays a role in regulating the interactions of proteins in the process of virion assembly and membrane budding.
Indeed, several previous studies have demonstrated that M promotes viral assembly and membrane fusion by multimerization and interactions with E, S, and N proteins (de Haan et al. 1999; de Haan, Vennema, and Rottier 2000; Narayanan et al. 2000; Siu et al. 2008). Importantly, these interactions are related to the two conformations of the M protein, as revealed by cryo-electron microscopy (Neuman et al. 2011). The elongated conformation of the M protein is associated with a rigid structural state that associates with clusters of S proteins and imparts a spherical membrane curvature of about 5–6 degrees per M dimer. On the other hand, the compact conformation is associated with a flexible state, low S protein density, and does not appear to impart membrane curvature. The same study also showed that a conversion between the elongated conformation and the compact conformation can be induced by a transient acidification, weakening the M interactions with other viral proteins. Therefore, it was proposed that the formation of elongated conformation of M drives the membrane budding process (Neuman et al. 2011). Our results complement this model—the proposal that the M protein itself might mediate ion transport could be key to the observed response to pH changes that might occur in the intracellular membranous compartments during virion assembly.
In addition to virus assembly, previous research on several animal viruses have shown that viral ion channels can also promote membrane fusion and regulate viral replication and/or packaging genomic RNA into viral particles (Ciampor et al. 1995; Nieto-Torres et al. 2015). It is possible that the M and ORF3-like proteins may also contribute to these processes through their ion transport activity (Yount et al. 2005; Castano-Rodriguez et al. 2018).
4.2 M2-clade (ORF3-like) families might be at the interface of host–virus interactions
We unified five highly divergent ORF3-like families from alpha- and beta-CoVs into the M2 clade of the M/ORF3 superfamily. All five families of the M2 clade share a common ancestor that split from the M protein at the base of alpha- and beta-CoVs (Fig. 4). The incorporation of ORF3-like proteins has been observed in the virions of SARS-CoV (containing the ORF3a family) (Shen et al. 2005; Ito et al. 2005; Huang et al. 2006) and human CoV NL63 (containing the ORF3b family) (Muller et al. 2010). This is consistent with ORF3-like proteins partly retaining the ancestral structural association with the envelope as seen with M.
Deletion of ORF3a in SARS-CoV is associated with a reduction in virus growth (Yount et al. 2005) indicating that it is not redundant with M and has evolved a distinct function. This is also supported by the different metrics that suggest selection for diversification in the M2-like clade as opposed to purifying selection in the M1 clade. This rapid diversification is comparable to that observed in the proteins that are involved in host-virus interactions, such as S, that binds the host receptor, and ORF8, a viral immunoglobulin protein that interferes with the host immune system (Tan et al. 2020; Zhang et al. 2020). This suggests that the M2 clade families, like S and ORF8, might be under diversifying selection due to either the need to interact with correspondingly diversifying host molecules or host immune attack. Two lines of evidence support our hypothesis: (1) Half of patients recovered from SARS have developed antibodies against the OR3a N-terminal peptide (Zhong et al. 2006), suggesting that ORF3a might be a target of the host immune system. (2) During the outbreak of both SARS-CoV and SARS-CoV-2, positive selection was observed in ORF3a along with S and ORF8, suggesting that the it might be evolving in response to the pressure from the host immune attack (Velazquez-Salinas et al. 2020; Yeh et al. 2004). Therefore, we propose that beyond a structural role in the envelope, the M2 clade proteins might be directly at the interface of CoV-host conflicts. In this context it remains to be seen if their ion channel activity might play a role in directly modulating or hijacking host membrane permeability, glycoprotein trafficking or vesicular transport.
4.3 Potential therapeutic significance of CoV M/ORF3-like superfamily proteins
Ion channels are major drug targets that account for ∼13 per cent of Food and Drug Administration-approved drugs (McManus 2014). Inhibition of host ion channels with multiple ion channel modulators has been repeatedly shown to affect virion entry and endosomal fusion (Hover et al. 2017). The recent identification of viral ion channels has allowed screening for novel virus-specific channel modulators, given the lack of homology between the viral and human ion channels. Successful products include the approved drugs amantadine and rimantadine which target the M2 ion channel in influenza A virus (Jefferson et al. 2004; Kozakov et al. 2010). As ORF3a and E are potential ion channels for SARS-CoV/SARS-CoV-2, they have been proposed as candidates of drug targets (McClenaghan et al. 2020; Alothaid et al. 2020; Dey, Borkotoky, and Banerjee 2020). Our results suggest that the M protein might be a potential candidate that might have even better prospects than the M2 clade proteins like ORF3a and E: (1) M2 clade proteins are fast-evolving; hence the virus is predicted to more easily develop resistance against the modulating compounds. In contrast, M appears to be under stronger selective constraint for conservation. (2) The presence of M in all CoVs makes it a better candidate for a wide spectrum drug. This is notable because human pathogenic CoVs have originated from different clades of CoVs. (3) In term of structural organization, M with 3-TM regions and multiple inter-TM loops offers more potential for drug-protein interactions than the other envelope-associated channel E that has only 1-TM forming a loosely packed pentamer (Mandala et al. 2020). Hence, our findings prioritize M as potential therapeutic target against CoVs.
Data availability
All genome and protein sequences analyzed in this study were downloaded from NCBI GenBank database. The accession numbers of the sequences can be found in Supplementary data.
Supplementary data
Supplementary data are available at Virus Evolution online.
Funding
Y.T., T. S., and D. Z. are supported by the Saint Louis University start-up fund and the Research Growth Fund—COVID-19 Rapid Response Award. L.A. is supported by the Intramural Research Program of the NIH, National Library of Medicine. P.K.S and M.B.C are supported by NIGMS-5R01GM127783.
Conflict of interest: None declared.
Supplementary Material
Contributor Information
Yongjun Tan, Department of Biology, College of Arts and Sciences, Saint Louis University, St. Louis, MO 63103, USA.
Theresa Schneider, Department of Biology, College of Arts and Sciences, Saint Louis University, St. Louis, MO 63103, USA.
Prakash K Shukla, Department of Radiation Oncology, Huntsman Cancer Institute, University of Utah School of Medicine, Salt Lake City, UT 84112, USA.
Mahesh B Chandrasekharan, Department of Radiation Oncology, Huntsman Cancer Institute, University of Utah School of Medicine, Salt Lake City, UT 84112, USA.
L Aravind, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, MD 20894, USA.
Dapeng Zhang, Department of Biology, College of Arts and Sciences, Saint Louis University, St. Louis, MO 63103, USA; Program of Bioinformatics and Computational Biology, College of Arts and Sciences, Saint Louis University, St. Louis, MO 63103, USA.
References
- Alothaid H. et al. (2020) ‘ Similarities between the Effect of SARS-CoV-2 and HCV on the Cellular Level, and the Possible Role of Ion Channels in COVID19 Progression: A Review of Potential Targets for Diagnosis and Treatment’, Channels, 14: 403–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F. et al. (1997) ‘ Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs’, Nucleic Acids Research, 25: 3389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson D. A. et al. (2018) ‘ GenBank’, Nucleic Acids Research, 46: D41–D47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castano-Rodriguez C. et al. (2018) ‘ Role of Severe Acute Respiratory Syndrome Coronavirus Viroporins E, 3a, and 8a in Replication and Pathogenesis’, mBio, 9: e02325–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciampor F. et al. (1995) ‘ 'Influenza Virus M2 Protein and Haemagglutinin Conformation Changes during Intracellular Transport’, Acta Virologica, 39: 171–81. [PubMed] [Google Scholar]
- Cozzetto D., Anna T. (2004) ‘ Relationship between Multiple Sequence Alignments and Quality of Protein Comparative Models’, Proteins: Structure, Function, and Bioinformatics, 58: 151–7. [DOI] [PubMed] [Google Scholar]
- de Haan C. A. et al. (1999) ‘ Mapping of the Coronavirus Membrane Protein Domains Involved in Interaction with the Spike Protein’, Journal of Virology, 73: 7441–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C. A., Vennema H., Rottier P. J. (2000) ‘ Assembly of the Coronavirus Envelope: Homotypic Interactions between the M Proteins’, Journal of Virology, 74: 4967–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano W. L. (2002) ‘ Pymol: An Open-Source Molecular Graphics Tool’, CCP4 Newsletter on Protein Crystallography, 40: 82–92. [Google Scholar]
- Dey D., Borkotoky S., Banerjee M. (2020) ‘ In Silico Identification of Tretinoin as a SARS-CoV-2 Envelope (E) protein Ion Channel Inhibitor’, Computers in Biology and Medicine, 127: 104063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozdetskiy A. et al. (2015) ‘ 'JPred4: A Protein Secondary Structure Prediction Server’, Nucleic Acids Research, 43: W389–W94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S. R. (1998) ‘ Profile Hidden Markov Models’, Bioinformatics, 14: 755–63. [DOI] [PubMed] [Google Scholar]
- Edgar R. C. (2004) ‘ MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput’, Nucleic Acids Research, 32: 1792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Gebali S. et al. (2019) ‘ The Pfam Protein Families Database in 2019’, Nucleic Acids Research, 47: D427–D32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flower T. G. et al. (2020) 'Structure of SARS-CoV-2 ORF8, a rapidly evolving immune evasion protein', Proceedings of the National Academy of Sciences of the United States of America, 118: e2021785118. [DOI] [PMC free article] [PubMed]
- Frickey T., Lupas A. (2004) ‘ CLANS: A Java Application for Visualizing Protein Families Based on Pairwise Similarity’, Bioinformatics, 20: 3702–4. [DOI] [PubMed] [Google Scholar]
- Fruchterman T. M., Reingold E. M. (1991) ‘ Graph Drawing by Force‐Directed Placement’, Software: Practice and Experience, 21: 1129–64. [Google Scholar]
- Graham R. L. et al. (2008) ‘ SARS Coronavirus Replicase Proteins in Pathogenesis’, Virus Research, 133: 88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hover S. et al. (2017) ‘ Viral Dependence on Cellular Ion Channels - an Emerging anti-Viral Target?’, Journal of General Virology, 98: 345–51. [DOI] [PubMed] [Google Scholar]
- Huang C. et al. (2006) ‘ Severe Acute Respiratory Syndrome Coronavirus 3a Protein is Released in Membranous Structures from 3a Protein-Expressing Cells and Infected Cells’, Journal of Virology, 80: 210–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito N. et al. (2005) ‘ Severe Acute Respiratory Syndrome Coronavirus 3a Protein is a Viral Structural Protein’, Journal of Virology, 79: 3182–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson T. et al. (2004) ‘ Amantadine and Rimantadine for Preventing and Treating Influenza a in Adults’, The Cochrane Database of Systematic Review, 2: CD001169. [DOI] [PubMed] [Google Scholar]
- Kern D. M. et al. (2020) ‘Cryo-EM structure of the SARS-CoV-2 3a ion channel in lipid nanodiscs’, BioRxiv, doi: 10.1101/2020.06.17.156554. [DOI]
- Kozakov D. et al. (2010) ‘ Where Does Amantadine Bind to the Influenza Virus M2 Proton Channel?’, Trends in Biochemical Sciences, 35: 471–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan A. et al. (2018) ‘ Diversification of AID/APOBEC-like Deaminases in Metazoa: Multiplicity of Clades and Widespread Roles in Immunity’, Proceedings of the National Academy of Sciences of the United States of America, 115: E3201–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A. et al. (2001) ‘ Predicting Transmembrane Protein Topology with a Hidden Markov Model: Application to Complete Genomes’, Journal of Molecular Biology, 305: 567–80. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016) ‘ MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets’, Molecular Biology and Evolution, 33: 1870–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo L., Koetzner C. A., Masters P. S. (2016) ‘ A Key Role for the Carboxy-Terminal Tail of the Murine Coronavirus Nucleocapsid Protein in Coordination of Genome Packaging’, Virology, 494: 100–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassmann T., Erik LL S. (2005) ‘ Kalign–an Accurate and Fast Multiple Sequence Alignment Algorithm’, BMC Bioinformatics, 6: 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S. K. P et al. (2011) ‘ Molecular Epidemiology of Human Coronavirus OC43 Reveals Evolution of Different Genotypes over Time and Recent Emergence of a Novel Genotype Due to Natural Recombination’, Journal of Virology, 85: 11325–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. et al. (2014) ‘ Structure of a Conserved Golgi Complex-Targeting Signal in Coronavirus Envelope Proteins’, The Journal of Biological Chemistry, 289: 12535–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R. et al. (2020) ‘ Genomic Characterisation and Epidemiology of 2019 Novel Coronavirus: Implications for Virus Origins and Receptor Binding’, The Lancet, 395: 565–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W.,B.-J. et al. (2006) ‘ Severe Acute Respiratory Syndrome-Associated Coronavirus 3a Protein Forms an Ion Channel and Modulates Virus Release’, Proceedings of the National Academy of Sciences of the United States of America, 103: 12540–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnaughton M. R., Hilary M. (1978) ‘ The Genome of Human Coronavirus Strain 229E’, Journal of General Virology, 39: 497–504. [DOI] [PubMed] [Google Scholar]
- Maghrabi A. H. A., McGuffin L. J. (2017) ‘ ModFOLD6: An Accurate Web Server for the Global and Local Quality Estimation of 3D Protein Models’, Nucleic Acids Research, 45: W416–W21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandala V. S. et al. (2020) ‘ Structure and Drug Binding of the SARS-CoV-2 Envelope Protein Transmembrane Domain in Lipid Bilayers’, Nature Structural & Molecular Biology, 27: 1202–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning C., Hinrich S. (1999) Foundations of Statistical Natural Language Processing. Cambridge, MA: MIT Press. [Google Scholar]
- Marra M. A., Jones S. J. M., et al. (2003) ‘ The Genome Sequence of the SARS-Associated Coronavirus’, Science, 300: 1399–404. [DOI] [PubMed] [Google Scholar]
- Masters P. S. (2019) ‘ Coronavirus Genomic RNA Packaging’, Virology, 537: 198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClenaghan C. et al. (2020) ‘ Coronavirus Proteins as Ion Channels: Current and Potential Research', Frontiers in Immunology, 11: 573339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus O. B. (2014) ‘ HTS Assays for Developing the Molecular Pharmacology of Ion Channels’, Current Opinion in Pharmacology, 15: 91–6. [DOI] [PubMed] [Google Scholar]
- Mehta P. et al. (2020) ‘ COVID-19: Consider Cytokine Storm Syndromes and Immunosuppression’, The Lancet, 395: 1033–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M. A. et al. (2010) ‘ Human Coronavirus NL63 Open Reading Frame 3 Encodes a Virion-Incorporated N-Glycosylated Membrane Protein’, Virology Journal, 7: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K. et al. (2000) ‘ Characterization of the Coronavirus M Protein and Nucleocapsid Interaction in Infected Cells’, Journal of Virology, 74: 8127–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B. W., Buchmeier M. J. (2016) ‘ Supramolecular Architecture of the Coronavirus Particle’, Advances in Virus Research, 96: 1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B. W. et al. (2011) ‘ A Structural Analysis of M Protein in Coronavirus Assembly and Morphology’, Journal of Structural Biology, 174: 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Torres J. L. et al. (2015) ‘ Relevance of Viroporin Ion Channel Activity on Viral Replication and Pathogenesis’, Viruses, 7: 3552–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieva J. L., Madan V., Carrasco L. (2012) ‘ Viroporins: Structure and Biological Functions’, Nature Reviews Microbiology, 10: 563–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J., Kim B.-H., Grishin N. V. (2008) ‘ 'PROMALS3D: A Tool for Multiple Protein Sequence and Structure Alignments’, Nucleic Acids Research, 36: 2295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M. N., Dehal P. S., Arkin A. P. (2009) ‘ FastTree: Computing Large Minimum Evolution Trees with Profiles instead of a Distance Matrix’, Molecular Biology and Evolution, 26: 1641–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricagno S. et al. (2006) ‘ Crystal Structure and Mechanistic Determinants of SARS Coronavirus Nonstructural Protein 15 Define an Endoribonuclease Family’, Proceedings of the National Academy of Sciences of the United States of America, 103: 11892–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruch T. R., Machamer C. E. (2012) ‘ The Coronavirus E Protein: Assembly and Beyond’, Viruses, 4: 363–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer A. A. et al. (1999) ‘ IMPALA: Matching a Protein Sequence against a Collection of PSI-BLAST-Constructed Position-Specific Score Matrices’, Bioinformatics, 15: 1000–11. [DOI] [PubMed] [Google Scholar]
- Shen S. et al. (2005) ‘ The Severe Acute Respiratory Syndrome Coronavirus 3a is a Novel Structural Protein’, Biochemical and Biophysical Research Communications, 330: 286–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu Y. L. et al. (2008) ‘ The M, E, and N Structural Proteins of the Severe Acute Respiratory Syndrome Coronavirus Are Required for Efficient Assembly, Trafficking, and Release of Virus-like Particles’, Journal of Virology, 82: 11318–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söding J. (2005) ‘ Protein Homology Detection by HMM–HMM Comparison’, Bioinformatics (Oxford, England), 21: 951–60. [DOI] [PubMed] [Google Scholar]
- Suchard M. A. et al. (2018) ‘ Bayesian Phylogenetic and Phylodynamic Data Integration Using BEAST 1.10’, Virus Evolution, 4: vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surya W., Yan L., Jaume T. (2018) ‘ Structural Model of the SARS Coronavirus E Channel in LMPG Micelles’, Biochimica et Biophysica Acta (Bba) - Biomembranes, 1860: 1309–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y. et al. (2020) ‘ Novel Immunoglobulin Domain Proteins Provide Insights into Evolution and Pathogenesis of SARS-CoV-2-Related Viruses’, mBio, 11: e00760–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor W. R. (1986) ‘ 'The Classification of Amino Acid Conservation’, Journal of Theoretical Biology, 119: 205–18. [DOI] [PubMed] [Google Scholar]
- Van Der Hoek L.,K. et al. (2004) ‘ Identification of a New Human Coronavirus’, Nature Medicine, 10: 368–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez-Salinas L. et al. (2020) ‘ Positive Selection of ORF1ab, ORF3a, and ORF8 Genes Drives the Early Evolutionary Trends of SARS-CoV-2 during the 2020 COVID-19 Pandemic’, Frontiers in Microbiology, 11:550674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinga S. (2014) ‘ Information Theory Applications for Biological Sequence Analysis’, Briefings in Bioinformatics, 15: 376–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A. C. et al. (2020) ‘ Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein’, Cell, 183: 1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K. et al. (2012) ‘ PEDV ORF3 Encodes an Ion Channel Protein and Regulates Virus Production’, FEBS Letters, 586: 384–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb B., Andrej S. (2016) ‘ Comparative Protein Structure Modeling Using MODELLER’, Current Protocols in Bioinformatics, 54: 5.6. 1–.6. 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo Patrick, C. Y. et al. (2005) ‘ Characterization and Complete Genome Sequence of a Novel Coronavirus, Coronavirus HKU1, from Patients with Pneumonia’, Journal of Virology, 79: 884–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh S. H., the National Taiwan University SARS Research Team. et al. (2004) ‘ Characterization of Severe Acute Respiratory Syndrome Coronavirus Genomes in Taiwan: Molecular Epidemiology and Genome Evolution’, Proceedings of the National Academy of Sciences of the United States of America, 101: 2542–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B. et al. (2005) ‘ Severe Acute Respiratory Syndrome Coronavirus Group-Specific Open Reading Frames Encode Nonessential Functions for Replication in Cell Cultures and Mice’, Journal of Virology, 79: 14909–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A. M. et al. (2012) ‘ Isolation of a Novel Coronavirus from a Man with Pneumonia in Saudi Arabia’, New England Journal of Medicine, 367: 1814–20. [DOI] [PubMed] [Google Scholar]
- Zhang D. et al. (2014a) ‘ Resilience of Biochemical Activity in Protein Domains in the Face of Structural Divergence’, Current Opinion in Structural Biology, 26: 92–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R. et al. (2014b) ‘ The ORF4a Protein of Human Coronavirus 229E Functions as a Viroporin That Regulates Viral Production’, Biochimica et Biophysica Acta (Bba) - Biomembranes, 1838: 1088–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. et al. (2020) ‘The ORF8 Protein of SARS-CoV-2 Mediates Immune Evasion through Potently Downregulating MHC-I’, BioRxiv, doi: 10.1101/2020.05.24.111823.
- Zhong X. et al. (2006) ‘ Amino Terminus of the SARS Coronavirus Protein 3a Elicits Strong, Potentially Protective Humoral Responses in Infected Patients’, Journal of General Virology, 87: 369–73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All genome and protein sequences analyzed in this study were downloaded from NCBI GenBank database. The accession numbers of the sequences can be found in Supplementary data.