Peptide-binding assays suggest that SARS-CoV-2 receptors engage the endocytosis and autophagy machinery.
SARS-CoV-2: From entry to autophagy?
SARS-CoV-2, the virus that causes COVID-19, enters cells through endocytosis upon binding to the cell surface receptor ACE2 and potentially others, including integrins. Using bioinformatics, Mészáros et al. predicted the presence of short amino acid sequences, called short linear motifs (SLiMs), in the cytoplasmic tails of ACE2 and various integrins that may engage the endocytic and autophagic machinery. Using affinity binding assays, Kliche et al. not only confirmed that many of these predicted SLiMs interacted with target peptides in various components of the endocytosis and autophagy machinery, but also found that these interactions were regulated by the phosphorylation of SLiM-adjacent amino acids. Together, these findings have identified a potential link between autophagy and integrin signaling and could lead to new ways to prevent viral infection.
Abstract
The spike protein of SARS-CoV-2 binds the angiotensin-converting enzyme 2 (ACE2) on the host cell surface and subsequently enters host cells through receptor-mediated endocytosis. Additional cell receptors may be directly or indirectly involved, including integrins. The cytoplasmic tails of ACE2 and integrins contain several predicted short linear motifs (SLiMs) that may facilitate internalization of the virus as well as its subsequent propagation through processes such as autophagy. Here, we measured the binding affinity of predicted interactions between SLiMs in the cytoplasmic tails of ACE2 and integrin β3 with proteins that mediate endocytic trafficking and autophagy. We validated that a class I PDZ-binding motif mediated binding of ACE2 to the scaffolding proteins SNX27, NHERF3, and SHANK, and that a binding site for the clathrin adaptor AP2 μ2 in ACE2 overlaps with a phospho-dependent binding site for the SH2 domains of Src family tyrosine kinases. Furthermore, we validated that an LC3-interacting region (LIR) in integrin β3 bound to the ATG8 domains of the autophagy receptors MAP1LC3 and GABARAP in a manner enhanced by LIR-adjacent phosphorylation. Our results provide molecular links between cell receptors and mediators of endocytosis and autophagy that may facilitate viral entry and propagation.
INTRODUCTION
A previously unknown coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), was identified as the pathologic agent for an outbreak of pneumonia in Wuhan, China in December 2019 (1). The virus, which is causing the new lung disease termed coronavirus disease 19 (COVID-19), has infected 40 million people and claimed over 1.1 million fatalities as of October 2020. It is hence posing an immediate health and socioeconomic threat with worldwide impact (2). Understanding how the virus infects and replicates within the host cell is crucial for the development of targeted therapies. Consequently, several large-scale mass spectrometry efforts have been focused on uncovering intracellular interaction partners of viral proteins (3–5) and on understanding how signaling pathways are affected by viral infection (6). Genome-wide CRISPR screening has also been used to identify essential host factors and pathways (7). Despite these studies, much remain unknown in terms of the molecular interactions necessary for viral infection and propagation.
On the extracellular side, SARS-CoV-2 uses the angiotensin-converting enzyme 2 (ACE2) as a human host receptor for cell attachment and subsequent receptor-mediated endocytosis (8, 9). Structural information on the interaction has been provided by cocrystallization of the receptor-binding domain of the SARS-CoV-2 spike protein with the ACE2 ectodomain and a cryo–electron microscopy structure with full-length ACE2 (10, 11). However, analysis of ACE2 expression across different human tissues reveals unexpectedly poor expression in the lungs as compared to, for instance, high expression in the kidney and heart (12, 13). The extreme damage that the virus can inflict on infected lungs indicates the requirement to use alternative receptors for host cell attachment and entry. In line with this observation, neuropilin-1 (NRP1), which is abundantly expressed in the olfactory epithelium, enhances SARS-CoV-2 infection and has been confirmed as a host receptor (14). The role of C-type lectins as interactors of the SARS-CoV-2 spike protein on innate immune cells has additionally been reported (15). Furthermore, integrins have been suggested as SARS-CoV-2 receptors due to the presence of a known integrin-binding RGD motif in the spike protein (16). An ACE2-integrin interaction seems structurally feasible based on a bioinformatic analysis performed by Mészáros et al. (17). Integrin-mediated endocytosis is used by a variety of viruses for cell attachment and cell entry (18, 19). In addition, integrin tissue distribution matches SARS-CoV-2 tropism more closely, which includes functional importance of integrin β1 expression on type 2 alveolar epithelial cells (20). Furthermore, there are, as of now, two case reports of COVID-19 patients with concomitant multiple sclerosis background, who were treated with natalizumab, an antibody that effectively blocks integrin α4 binding (21, 22). Both reports support a positive correlation between natalizumab treatment and mild COVID-19 progression. Integrins thus appear as plausible co-receptors of ACE2.
Considerable research has been focused on the interaction between the SARS-CoV-2 spike protein and cell surface receptor proteins, particularly ACE2 (8, 9, 14–16). Further insight into the viral entry mechanism may be provided by investigating the downstream, intracellular, effects of the host cell receptor binding event. Taking a bioinformatical approach, Mészáros et al. (17) analyzed the cytoplasmic tails of ACE2 and integrins and predicted them to contain a number of short linear motifs (SLiMs) that may link host receptor binding to endocytosis, autophagy, and signaling. SLiMs are typically 3– to 10–amino acid–long stretches found in the intrinsically disordered regions of the proteome. They may serve as binding sites and engage in dynamic protein-protein interactions (23). SLiMs are divided into different classes based on their functions and target domains. Well-known SLiMs mediating protein-protein binding include the class I PSD-95/Dlg/ZO-1 (PDZ)–binding motif x[ST]xΦ-COO− (where x indicates any amino acid and Φ indicates a hydrophobic amino acid) that directs PDZ domain–containing proteins to the C-terminal tails of target proteins (24); the endocytic trafficking motif YxxΦ that binds to the clathrin adaptor AP2 μ2 (25); and the LC3-interacting region (LIR) motif, which mediates interaction with the autophagy-related protein 8 (ATG8) domain–containing proteins MAP1LC3s (microtubule-associated protein 1 light chain 3 isoforms, also called LC3s) and GABARAPs (γ-aminobutyric acid receptor–associated proteins) (Fig. 1A) (26).
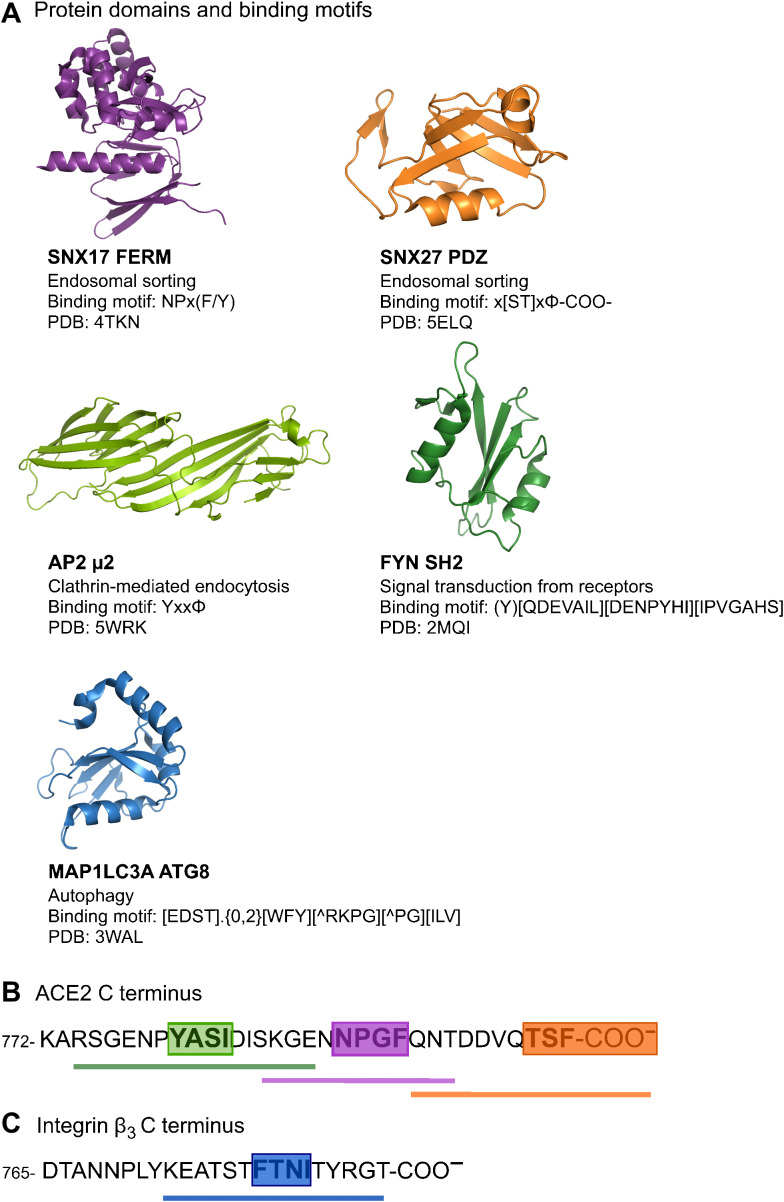
Fig. 1. Overview of domains and peptides tested for binding.
(A to C) Representative structures of peptide-binding domain assessed for their potential interactions with peptide sequences from the cytoplasmic tails of ACE2 (B) and integrin β3 (C). Green: Region containing predicted overlapping binding sites for NCK SH2 domain, the ATG8 domains of MAP1LC3As and GABARAPs, and AP2 μ2. Magenta: Predicted PTB binding site. Orange: Predicted class I PDZ-binding site. Blue: Predicted ATG8 binding site in integrin β3. PDB, Protein Data Bank.
Using the information on SLiMs stored in the Eukaryotic Linear Motif (ELM) database (27), Mészáros et al. performed a detailed bioinformatic analysis of potential SLiMs in ACE2 and integrins. On the basis of the presence of conserved consensus sequences that match previously established binding SLiMs, they predicted that ACE2 contains SLiMs that bind to class I PDZ domains, to phosphotyrosine-binding (PTB) domains, to AP2 μ2, to the Src homology 2 (SH2) domain, and to the ATG8 domains of MAP1LC3s and GABARAPs (Fig. 1, A and B). These SLiMs, if functional, would act as molecular platforms linking the receptor to the various biological processes associated with the interacting proteins. AP2 μ2, for example, is part of the clathrin adaptor complex formed during clathrin-dependent endocytosis (28). The PDZ and PTB domains, on the other hand, are well-established scaffolding domains implicated in the assembly of protein complexes and in receptor trafficking (29, 30). Hence, SLiMs on the cytosolic side of the host receptor proteins may allow the recruitment of proteins that enable and mediate receptor and concomitant virus particle internalization.
Adding to the complexity, phosphorylation of Ser-Thr-Tyr residues may enable, disable, or tune a given interaction (31, 32). In the case of the ACE2 tail, Tyr781 has, for example, emerged as a reproducible phosphosite (17). Phosphorylation of Tyr781, which is a core residue of the predicted endocytic sorting motif, is expected to disable the interaction with AP2 μ2 and simultaneously create a putative binding site for phospho-Tyr binding SH2 domains. Mészáros et al. (17) initially suggested the site to be bound by the SH2 domain of the adaptor protein noncatalytic region of tyrosine kinase (NCK) proteins. The authors further proposed the presence of a phospho-modulated LIR autophagy motif in both ACE2 and the integrin β3 tail (Fig. 1) (17). If functional, such a motif would provide a link between the receptors and the potential viral entry route to the endocytic-lysosomal pathway. Moreover, the β-integrin tails have a Ser-Thr–rich sequence that precedes the predicted LIR autophagy motif. This stretch has been proposed to be phosphorylated by the kinases Akt and PDK1 (3-phosphoinositide–dependent protein kinase 1) (33, 34), which would likely modulate the proposed ATG8 domain binding. As rewiring of phospho-signaling has been found as one of the primary host responses to SARS-CoV-2 infection (35), it is important to explore the phospho-regulation of protein-protein interactions in relation to viral entry and hijacking of the cellular machinery.
In this study, we set out to test predicted binding motifs in ACE2 and integrin β3 (17) and the potential phospho-regulation of the interactions by affinity measurements. Our results confirm the functionality of a subset of the bioinformatically predicted binding motifs and shed light on the intracellular molecular recognition events in which ACE2 and integrin β3 engage that, with further work, may contribute to the understanding of how SARS-CoV-2 enters the host cell. More broadly, identification of a functional phospho-dependent LIR motif in the tail of integrin β3 reveals a potentially critical link from integrin signaling to autophagy.
RESULTS
We aimed to test the predicted SLiMs in the cytoplasmic tails of ACE2 and integrin β3 for binding to PDZ, PTB, and SH2 domains, the μ2 subunit from the clathrin adaptor AP2, as well as the ATG8 domains of MAP1LC3s and GABARAPs. We therefore expressed and purified a representative collection of protein domains and used them to explore the affinities for synthetic peptides representing the proposed binding motifs (Fig. 1). Affinities were determined by fluorescence polarization (FP) using fluorescein isothiocyanate (FITC)–labeled peptides as direct binders [saturation curves; KD (dissociation constant) values] or reporters of binding [displacement curves; KI (inhibition constant) values].
NHERF3 PDZ1, SHANK1, and SNX27 PDZ domains bind the ACE2 C-terminus
The presence of a putative classical class I PDZ-binding motif in the cytoplasmic tail of ACE2 prompted us to assess direct binding of the C-terminal region of ACE2 to a selection of eight PDZ domains, namely, the PDZ domains from SH3 and multiple ankyrin repeat domains 1 (SHANK1), syntrophin 1 (SNTA1), sorting nexin 27 (SNX27), and Tax1-binding protein 3 (TAX1BP3); the first PDZ domains (PDZ1) from the membrane-associated guanylate kinase (MAGI) and the scaffold Scribble (SCRIB); and PDZ1 and PDZ2 from Na+/H+ exchanger regulatory factor 3 (NHERF3). These domains are known to interact with class I PDZ-binding motifs (36) and are found in proteins expressed in the lung based on UniProt annotations.
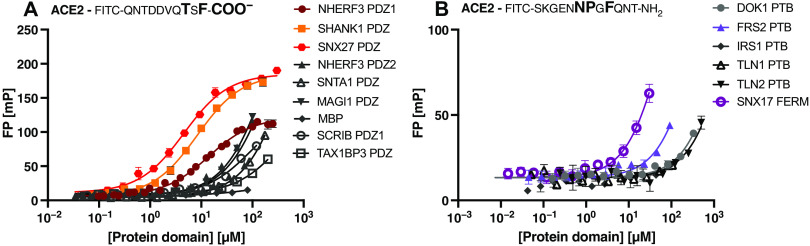
The FITC-labeled C-terminal peptide of ACE2796–805 (FITC-QNTDDVQTSF-COO−) was titrated with increasing concentrations of PDZ domain (Fig. 2A). The change in FP signal upon ligand binding was measured and plotted against the protein concentration. Among the eight tested domains, five PDZ domains were classified as nonbinders or low-affinity binders of ACE2, given that there was no saturation within the tested concentration intervals (0 to 100 or 200 μM, depending on the protein domain). In contrast, the first PDZ domain of NHERF3 and the PDZ domains of SHANK1 and SNX27 were found to bind ACE2 with relatively high affinities in respect of PDZ domain interactions (KD values of 8, 14, and 5 μM, respectively; Fig. 2A and Table 1).
Fig. 2. Assessment of PDZ and PTB domain binding to the ACE2 cytoplasmic tail.
(A and B) Saturation curves obtained by fluorescence polarization (FP) experiments for selected class I PDZ domains (A) and a selection of PTB domains and SNX17 FERM (B). Curves were obtained by titrating the protein domains against the respective FITC-labeled ACE2 peptide, containing either a class I PDZ or a predicted PTB binding motif. Data are means ± SEM; n = 3 experiments.
Table 1. Affinities of PDZ, PTB, FERM, SH2, and ATG8 domains as well as AP2 μ2 for the ACE2 C-terminal peptide as determined by FP.
Equilibrium dissociation constant (KD) values of the tested domains for FITC-labeled ACE2 C-terminal peptides. Indicated errors are the errors of the mean (SEM); n = 3 replicates. N.B., no or low affinity (KD >100 μM).
| Peptide sequence | Name | Modification | KD ± SEM (μM) |
| ACE2796-805* FITC-QNTDDVQTSF-COO− |
MAGI1 PDZ1 | Unphos. | N.B. |
| NHERF3 PDZ1 | 13.9 ± 0.3 | ||
| NHERF3 PDZ3 | N.B. | ||
| SCRIB PDZ1 | N.B. | ||
| SHANK1 PDZ | 8.3 ± 0.2 | ||
| SNTA1 PDZ | N.B. | ||
| SNX27 PDZ | 4.7 ± 0.3 | ||
| TAX1BP3 PDZ | N.B. | ||
| ACE2787–798 FITC-SKGENNPGFQNT-NH2 |
DOK1 PTB | Unphos. | N.B. |
| FRS2 PTB | N.B. | ||
| IRS1 PTB | N.B. | ||
| TLN1 PTB | N.B. | ||
| TLN2 PTB | N.B. | ||
| SNX17 FERM | (100)† | ||
| ACE2775–790 FITC-RSGENPYASIDISKGE-NH2 |
AP2 μ2 | Unphos. | (60)† |
| pTyr781 | N.B. | ||
| pSer783 | (30)† | ||
| FYN SH2 | Unphos. | N.B. | |
| pTyr781 | 7.0 ± 0.5 | ||
| pSer783 | N.B. | ||
| NCK1 SH2 | Unphos. | N.B. | |
| pTyr781 | N.B. | ||
| pSer783 | N.B. | ||
| LYN SH2 | Unphos. | N.B. | |
| pTyr781 | (60) | ||
| pSer783 | N.B. | ||
| MAP1LC3A ATG8 | Unphos. | N.B. | |
| pTyr781 | N.B. | ||
| pSer783 | N.B. | ||
| MAP1LC3B ATG8 | Unphos. | N.B. | |
| pTyr781 | N.B. | ||
| pSer783 | N.B. | ||
| GABARAP ATG8 | Unphos. | N.B. | |
| pTyr781 | N.B. | ||
| pSer783 | N.B. | ||
| GABARAPL2 ATG8 | Unphos. | N.B. | |
| pTyr781 | N.B. | ||
| pSer783 | N.B. |
*MBP was tested as a control for binding (N.B.) due to the PDZ domains being MBP-tagged. †Values in brackets indicate estimated affinities due to incomplete saturation curves.
The putative PTB binding motif of ACE2 binds to the SNX FERM domains with low affinity
We next tested the interactions of the predicted PTB binding motif (NPxF) in the C-terminal tail of ACE2. We attempted to obtain saturation curves for the FITC-labeled ACE2787–798 peptide containing the predicted motif (FITC-SKGENNPGFQNT-NH2) and the PTB domains from docking protein 1 (DOK1), insulin receptor substrate 1 (IRS1), and fibroblast growth factor receptor substrate 2 (FRS2); the PTB-like lobe of the FERM (4.1 protein/ezrin/radixin/moesin) domains of Talin-1 and Talin-2 (TLN1 and TLN2); and the full FERM domain from sorting nexin 17 (SNX17). The proteins have low tissue specificity based on Human Protein Atlas annotations (37) and were selected to sample different types of PTB-like domains. However, none of the tested domains was found to be a high-affinity binder of the ACE2787–798 peptide (Fig. 2B); thus, we classified the PTB domains of DOK1, FRS2, TLN1/2, and IRS1 as nonbinders. Given that about 60 human proteins contain PTB domains (30), a systematic study of PTB domain specificities would be needed to identify the relevant binder.
In contrast, we obtained a promising result for the interaction between the SNX17 FERM domain and the ACE2787–798 peptide (Fig. 2B), for which we estimate the KD to be in the range of 100 μM. The experiment was, however, hampered by the low solubility of the protein domain, and the attempt to use the glutathione S-transferase (GST)–tagged version of the domain did not yield higher protein concentrations. In line with an interaction between the NPxF motif of ACE2, we note that the SNX17 FERM domain binds to NPx(F/Y)-containing peptides. It has further been crystallized in complex with an NPxF-containing peptide, similar to the one tested here, of the microtubule-associated Krev interaction trapped protein 1 (KRIT1) (38). An interaction between the SNX17 FERM domain appears reasonable, given that the protein is involved in endosomal recycling of various proteins, including integrins (38, 39). The paralogous protein SNX27 has—in addition to the PDZ domain that binds to the ACE2 C-terminus—also a FERM domain that is homologous to the SNX17 FERM domain. Both the SNX27 PDZ domain and the FERM domain are involved in cargo recognition for the SNX27-retromer complex, which is a major regulator of recycling of transmembrane cargos from the endosome to the plasma membrane (38, 40). We thus hypothesized that the SNX27 FERM domain may also bind to the NPxF motif and attempted to validate the interaction. However, because of the challenge to concentrate the SNX27 FERM domain, we could not complement our dataset by this affinity measurement.
The ACE2 tail harbors an overlapping phospho-regulated AP2 μ2 and SH2 domain binding motif
The ACE2 tail contains a nine–amino acid stretch predicted to contain overlapping binding motifs for the adaptor proteins AP2 μ2, for the SH2 domain of NCK1, and for the ATG8 domains of MAP1LC3s and GABARAPs (Fig. 1) (17). In addition, the region contains two phosphosites (Tyr781 and Ser783) that are proposed to serve as bimolecular switches. We aimed at testing the interactions and the predicted phospho-switches in ACE2 binding by measuring the affinities of AP2 μ2, NCK1 SH2, and four ATG8 domains (from MAP1LC3A, MAP1LC3B, MAP1LC3C, and GABARAPL2) for unphosphorylated and phosphorylated (pTyr781 and pSer783) ACE2 peptides through peptide displacement experiments.
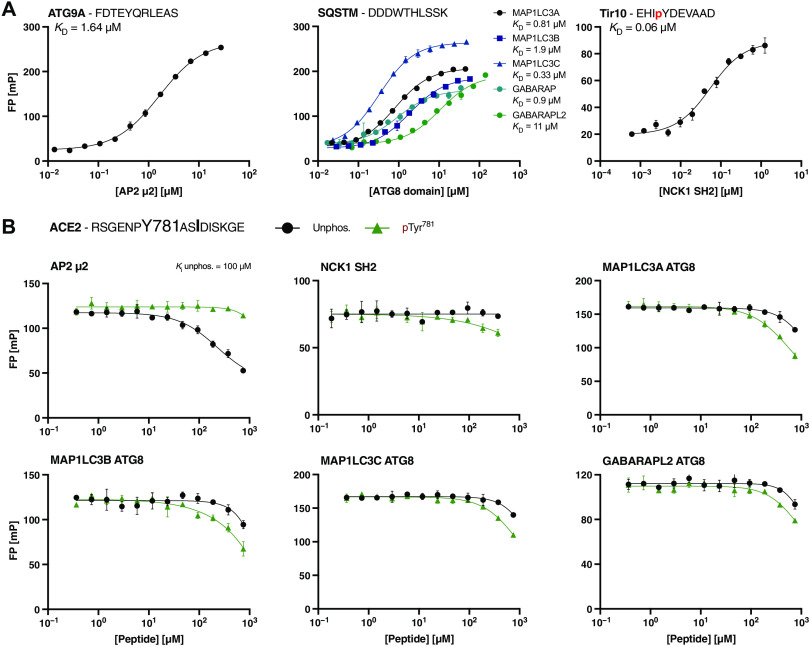
First, to determine appropriate protein domain concentrations for displacement experiments, saturation curves for the different protein domains with respective FITC-labeled probe peptides were obtained (Fig. 3A and Table 2). The labeled peptides for AP2 μ2 (ATG9A4–14) and the ATG8 (SQSTM1335–344) domains were chosen on the basis of in-house proteomic peptide phage display data, whereas the FITC-labeled peptide (Tir10471–480) for NCK1 SH2 is a reported high-affinity binder (41). Subsequently, the ACE2775–790 peptides were titrated against the varying complexes of FITC-labeled peptides and domains (Fig. 3B), from which IC50 (half-maximal inhibitory concentration) values were derived and KI values could be calculated.
Fig. 3. Testing the predicted overlapping SH2, ATG8, and AP2 μ2 domain binding motifs in the ACE2 tail (ACE2775–790) by displacement experiments.
(A) Saturation curves of AP2 μ2, the ATG8 domains, and NCK1 SH2 for the respective FITC-labeled peptides (ATG9A, SQSTM1, and Tir10). (B) Displacement curves of FP experiments using a peptide from the cytoplasmic ACE2 tail predicted to contain AP2 μ2, ATG8, and NCK SH2 domain binding motifs. Preferential binding of the different domains for unphosphorylated and pTyr781 ACE2 peptide was tested. Data are means ± SEM; n = 3 experiments.
Table 2. Affinities of AP2 μ2, ATG8 domains of GABARAPs and MAP1LC3s, and NCK1 SH2 domains for probe peptides as determined by FP.
Equilibrium dissociation constant (KD) values of AP2 μ2, the ATG8 domains, and NCK1 SH2 for the respective FITC-labeled peptides (ATG9A, SQSTM1, and Tir10). Indicated errors are the errors of the mean (SEM); n = 3 replicates.
| Peptide sequence | Protein domains | |
| Protein domain | KD ± SEM (μM) | |
| ATG9A4–14 FITC-FDTEYQRLEAS-NH2 |
AP2 μ2 | 1.64 ± 0.07 |
| SQSTM1335–344 FITC-DDDWTHLSSK-NH2 |
GABARAP ATG8 | 0.9 ± 0.1 |
| GABARAPL2 ATG8 | 11 ± 1 | |
| MAP1LC3A ATG8 | 0.81 ± 0.04 | |
| MAP1LC3B ATG8 | 1.9 ± 0.1 | |
| MAP1LC3C ATG8 | 0.33 ± 0.01 | |
| Tir10371–280 FITC-EHIpYDEVAAD-NH2 |
NCK1 SH2 | 0.055 ± 0.006 |
Through these peptide displacement experiments, we confirmed an interaction between AP2 μ2 and the ACE2 peptide, with preferential binding to unphosphorylated ACE2 peptide (KI value = 101 ± 5 μM; Fig. 3B). Phosphorylation at Tyr781 disabled the interaction with AP2 μ2, which is consistent with previous reports (42, 43). We thus confirmed the presence of an endocytic sorting signal in the ACE2 C-terminal tail that is negatively regulated by Tyr781 phosphorylation. We further attempted to determine the KI value for the pSer783 ACE2775–790 peptide, but we were unable to obtain reliable results due to the presence of contaminations that hindered accurate measurement, despite ordering the peptide from two different companies.
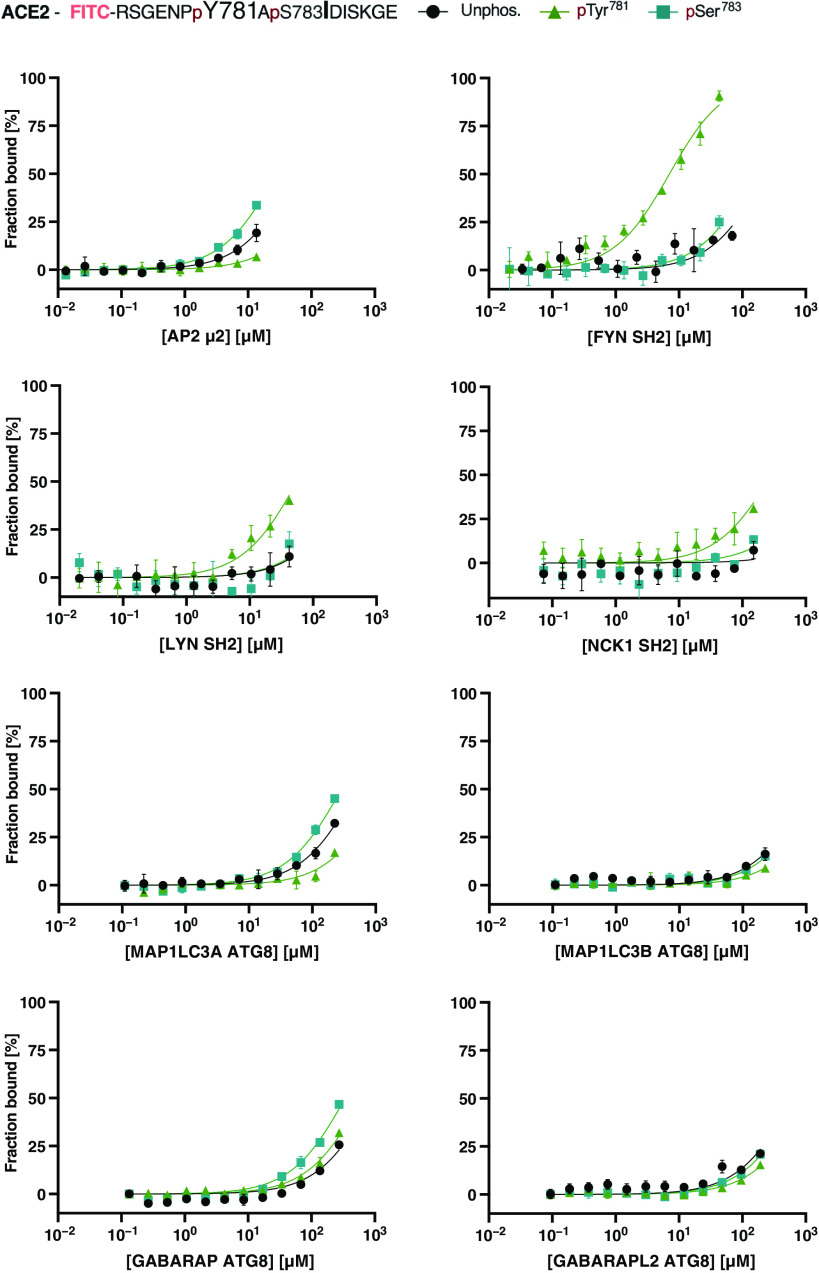
To circumvent the contamination issues of the unlabeled pSer783 ACE2775–790 peptide, we complemented our dataset by assessing direct binding of the tested domains to the corresponding FITC-labeled ACE2775–790 peptides (Fig. 4 and Table 1). For appropriate comparison of the three FITC-labeled peptides, we normalized the millipolarization (mP) signal by taking the different baselines and the amplitudes of the domain types into account. Plotting the fraction bound (%) against the varying protein domain concentrations allowed the retrieval of the corresponding KD values (Fig. 4 and Table 1). This analysis confirmed an interaction between AP2 μ2 and ACE2, which is negatively affected by Tyr781 phosphorylation. In contrast, we found that the pSer783 ACE2775–790 peptide enhanced the affinity of the interaction by twofold (estimated KD = 30 μM).
Fig. 4. Testing predicted overlapping SH2, ATG8, and AP2 μ2 domain binding motifs in the ACE2 tail by direct binding.
Saturation curves of AP2 μ2, SH2, and ATG8 domains of the indicated proteins for the respective FITC-labeled ACE2 peptide (key above). Data are means ± SEM; n = 3 experiments.
We next tested pTyr781 ACE2775–790–NCK1 SH2 interaction predicted by Mészáros et al. (17) using the peptide displacement experiment but could only confirm a very low affinity interaction (in the millimolar KI range). Our negative result prompted Mészáros et al. (17) to revisit their prediction of potential SH2 domain binding to ACE2 and, consequently, to suggest that the pTyr781 ACE2775–790 peptide binds to SH2 domains of the Src family tyrosine kinases (such as FYN, LYN, and SRC) and other SH2 domains with very similar binding specificities. We therefore determined the affinities of the FYN and LYN SH2 domains for the FITC-labeled ACE2 peptides, this time through direct binding. We found that the FYN SH2 domain binds the pTyr781 (KD of 7 μM; Table 1 and Fig. 4) but not the unphosphorylated ACE2775–790 peptide. Our data further indicate weak binding of LYN SH2 to the pTyr781 ACE2775–790 peptide, although the affinity of 60 μM is only an estimation due to incomplete saturation of the measurement. The results thus suggest that the Src family tyrosine kinases may be targeted to ACE2 upon phosphorylation. That being said, the SH2 domain family is one of the largest families of modular domains (44), and it is likely that also other SH2 domains with similar or overlapping specificities may bind to the motif given that the proteins are expressed in the same tissue.
We further attempted to validate the predicted phospho-regulated interactions between the ACE2775–790 peptide and the ATG8 domains. However, the ATG8 domains bound neither the unphosphorylated nor the pTyr781 ACE2775–790 peptide (Fig. 3B; estimated KI values in millimolar range), arguing against a functional LIR in the ACE2 tail. In agreement with the negative results of the displacement experiments, neither unphosphorylated nor pTyr781 FITC–ACE2775–790 peptides were found to be bound by the tested ATG8 domains in direct binding experiment (Fig. 4). A tendency of binding could be detected for the pSer783 ACE2775–790 peptide for MAP1LC3A and GABARAP ATG8 domain, which indicates that the phosphosite interspersed in the predicted LIR motif may enhance affinity. However, we judged the interactions with the pSer783 ACE2775–790 peptide to be of too low affinity to be of any functional relevance.
We validated overlapping binding motifs for AP2 μ2 and the SH2 domains in the ACE2 tail, wherein phosphorylation of Tyr781 emerged as a molecular switch that changes the specificity of ACE2 for the different binding domains. We further disproved the predicted interaction between ACE2 and the ATG8 domains, although we note that pSer783 ACE2775–790 binds with low affinity, an interaction that potentially may gain affinity within the context of a longer peptide, as the LIR is a fairly long motif (Fig. 1).
The integrin β3 tail contains a phospho-regulated LIR motif
After the evaluation of the proposed SLiMs in the ACE2 tail, we assessed the predicted LIR motifs in integrin tails, using the integrin β3 tail as a model system. We determined KI values for unphosphorylated peptide and phospho-peptides spanning the region of the potential binding motif by displacement experiments. The hydrophobic motif is represented by Phe780 and Ile783 and interspaced by two variable amino acids (Fig. 1C). Preceding the motif, there is a Ser-Thr–rich sequence (residues 777 to 779) that is suggested to be subjected to phosphorylation by the PDK1 (34). Downstream of the LIR motif, there is also Tyr785 that emerges as a potentially relevant phosphosite (17) and has been reported to be phosphorylated by the tyrosine kinases Src and Syk (45). We therefore explored if the integrin β3 tail has a functional LIR motif and if the interactions with the ATG8 domains were tuned by phosphorylation of residues upstream and downstream of the core hydrophobic motif.
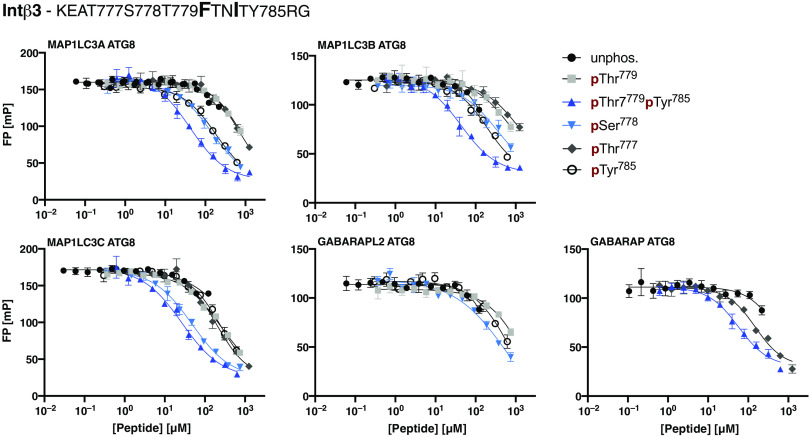
Displacement curves were obtained in the same fashion as described above, and KI values were concomitantly calculated (Fig. 5 and Table 3). We found that the unphosphorylated integrin β3 peptide was essentially not bound by the tested ATG8 domains. However, we found that phosphorylation of Ser778 located upstream of the hydrophobic motif strengthened ATG8 binding for all domains tested, while Thr779 phosphorylation enhanced the affinity for MAP1LC3C (KI = 82 μM) but had no effect on the binding of the other ATG8 domains. Phosphorylation of Tyr785 downstream of the LIR motif conferred improved affinity in all tested cases (e.g., MAP1LC3C, KI = 83 μM). Concomitant phosphorylation of Thr779 and Tyr785 (double phosphorylated integrin β3 peptide) promoted ATG8 domain binding and increased the binding affinity of MAP1LC3C even further (KI = 8 μM), indicating a synergistic effect of the two phosphosites (Fig. 5). Our results thus demonstrate a phospho-dependent interaction between the ATG8 proteins and integrin β3 at the biophysical level. The functional relevance of the LIR remains to be tested in a cellular setting.
Fig. 5. Investigating the phospho-regulation of the LIR motif in the integrin β3 tail.
Fluorescence polarization displacement curves obtained using unphosphorylated and phosphorylated peptides representing the cytoplasmic integrin β3 tail. Peptide sequence and sampled phosphosites are indicated in the figure. Data are means ± SEM; n = 3 experiments.
Table 3. Affinities of ATG8 domains for the integrin β3 peptide.
KI values for ATG8 domains calculated from displacement experiments using unphosphorylated integrin β3 peptide and phosphorylated peptides (pThr777, pSer778, pThr779, pTyr785, or pThr779 pTyr785). Indicated error is the error of the mean (SEM); n = 3 replicates. N.B., no or low affinity (KI > 100 μM); N.M., not measured.
| Peptide sequences | KI values ± SEM (μM) | ||||
|
MAP1LC3A ATG8 |
MAP1LC3B ATG8 |
MAP1LC3C ATG8 |
GABARAPL2 ATG8 |
GABARAP ATG8 |
|
| Unphosphorylated CH3CO- KEATSTFTNITYRG-NH2 |
N.B. | N.B. | N.B. | N.B. | N.B. |
| pThr777 CH3CO- KEApTSTFTNITYRG-NH2 |
N.B. | N.B. | 73 ± 4 | N.M. | 47 ± 3 |
| pSer778 CH3CO- KEATpSTFTNITYRG-NH2 |
29.4 ± 0.5 | N.B. | 16.1 ± 0.5 | 80 ± 5 | N.M. |
| pThr779 CH3CO- KEATSpTFTNITYRG-NH2 |
N.B. | N.B. | 82 ± 3 | N.B. | N.M. |
| pTyr785 CH3CO- KEATSTFTNITpYRG-NH2 |
38 ± 2 | 81 ± 2 | 83 ± 4 | 89 ± 12 | N.M. |
| pThr779/pTyr785 CH3CO- KEATSpTFTNITpYRG-NH2 |
8.5 ± 0.5 | 15 ± 1 | 8 ± 2 | N.M. | 17.8 ± 0.8 |
DISCUSSION
Coronaviruses, including SARS-CoV-2, are proposed to use either the endocytic or non-endosomal pathway for cell entry (46). Our study provides biophysical evidence for the functionality of SLiMs in the cytoplasmic tails of ACE2 and integrin β3, which provide possible molecular links between the established (ACE2) and putative (integrin beta3) SARS-CoV-2 receptors and mediators of endocytosis and autophagy. Our finding that an endocytic AP2 μ2 binding motif exists in the ACE2 C-terminus and that such binding is negatively regulated by Tyr781 phosphorylation supports the possibility of clathrin-dependent endocytosis of SARS-CoV-2 upon receptor binding. It will, of course, be necessary to assess the biological relevance of the ACE2-AP2 μ2 interaction, particularly in light of its low affinity. Concerning the latter, we note that while affinities of AP2 core complexes for endocytic sorting signals are reported in the nanomolar range, measured KD values of the AP2 μ2 subunit alone vary between 10 and 70 μM (47–49). Those reports contextualize the here reported affinity (KI value = 100 μM by displacement experiments, estimated KD = 60 μM by direct binding) to a reasonable range. In support of AP2-dependent endocytosis as a part of SARS-CoV-2 internalization, a report published during the revision of this manuscript found that mutation of the endocytic motif that interacts with AP2 μ2 reduced the sensitivity of ACE2-expressing HeLa cells to infection by SARS-CoV-2 pseudovirus (50). Thus, there is convincing evidence that AP2 μ2 is a crucial host factor for coronaviral entry.
While phosphorylation of Tyr781 abolished the interaction of ACE2 with AP2 μ2, it enabled its binding to the SH2 domain of the kinase FYN. FYN belongs to the SRC family of protein tyrosine kinases that are implicated in the regulation of signal transduction through cross-talk with receptor tyrosine kinases and G protein–coupled receptors as well as through cytoskeletal rearrangements (51). Linking back to SARS-CoV-2, remodeling of the actin cytoskeleton could possibly facilitate viral entry after formation of the endocytic vesicles, with phosphorylation of Tyr781 acting as binding switch.
Through our small-scale sampling, guided by tissue distribution and PDZ-binding motif preferences, we further demonstrate that the PDZ domains of SNX27 and SHANK1 and the PDZ1 domain of NHERF3 bind to the C-terminus of ACE2 with similar affinities. Between these, the SHANK1 interaction may be less physiologically relevant, given their role in the structural organization of the postsynaptic density (52), whereas NHERF3 serves as a scaffold protein that regulates the surface expression of plasma membrane proteins in the apical domain of epithelial cells (53, 54). This aligns with the fact that ACE2 is enriched on the apical surface of conducting airway epithelia (55). Moreover, NHERF3 interacts with the glutamate transporter excitatory amino acid carrier (56), which harbors—apart from the PDZ-binding motif, and similar to ACE2—an endocytic sorting signal that mediates AP2 μ2–dependent endocytosis. Hence, it is possible that an equivalent mechanism orchestrates ACE2 stabilization (PDZ-mediated) and internalization (AP2-mediated). In turn, SNX27 is involved in retrograde transport from endosome to plasma membrane and, hence, in the recycling of internalized transmembrane proteins (57). It is also a host factor required for efficient entry of an engineered SARS-CoV-2 variant, the spike protein of which contains a deletion at the S1/S2 subunit cleavage site (58). Thus, it is plausible that SNX27 is involved in recycling ACE2 to the plasma membrane, which would, by extension, promote viral entry. The high-affinity SNX27 PDZ–mediated interaction may be reinforced by an interaction between the upstream NPxF motif and the SNX27 FERM domain, but this awaits further experimental validation. We further note that SNX27 is involved in preventing lysosomal degradation of β integrins by binding to an NPxY motif in their cytoplasmic tails (39).
Although limited by the set of PDZ domains tested, our analysis suggests a specificity of ACE2-PDZ interactions, among which NHERF3 and SNX27 emerge as biologically most relevant to SARS-CoV-2 biology. However, we note that there are more than 260 human PDZ domains with partially overlapping specificities (36) and that there might be other class I PDZ domains with similar binding preferences. Alternative methods, such as the hold-up assay against the full PDZ domain collection (59), may shed further light on the PDZ specificity profile of the ACE2 C-terminus in the future.
Our finding that ATG8 domains (namely, of MAP1LC3A/B/C, GABARAP, and GABARAPL2) bind the cytosolic tail of integrin β3 in a phospho-dependent and synergistic fashion supports and extends previous reports of phospho-regulation of the LIR motif (60). The functionality of the LIR binding motif in integrin β3 provides a direct link between the receptor and the autophagic machinery, establishing a possible mechanism of how SARS-CoV-2 could hijack this pathway for its propagation. This hypothesis appears particularly feasible in light of the fact that ATG8 domains act as adaptors for recruiting LIR-containing proteins to phagosomal membranes (60).
How the autophagic machinery is deployed during coronavirus infection is still under debate, with most of the research conducted on viruses such as SARS-CoV and mouse hepatitis virus (46). Recent interest is driven by, for example, the finding that a number of autophagy inhibitors reduce the cytopathic effect of SARS-CoV-2 infection in primate kidney epithelial Vero-E6 cells (61). Furthermore, a study repurposing approved compounds identified apilimod, a PIKfyve kinase inhibitor (62), as a potent inhibitor of viral replication in an ex vivo lung culture system (63). PIKfyve kinase activity has been linked to critical membrane trafficking events due to the generation of the second-messenger lipid phosphatidylinositol 3,5-bisphosphate [PI(3, 5)P2] (64) and its inhibition by apilimod to induce secretory autophagy (65). Our data presented here may hence shed light on the interconnection between infection and the autophagic pathway. Redistribution of membranes that are important for viral replication, as implicated by apilimod treatment, may sensitively affect viral entry and replication. However, follow-up studies in cells are required to test whether the virus deploys the potential protein-protein interactions of integrin β3.
From a more general perspective, an inverse relationship between autophagy and integrin-dependent anchorage signaling has been proposed, implying that the autophagic machinery counteracts anoikis (detachment-induced apoptosis) (66). For instance, down-regulation of integrin β3 results in up-regulation of autophagy markers (67). Likewise, blockage of αVβ3 signaling results in inhibition of adhesion and concomitant activation of the autophagic machinery, manifested in increased MAP1LC3B expression (68). Conclusive evidence linking integrin signaling and autophagy has thus been reported, whereas the molecular basis underlying vesicular rearrangements and the trafficking of integrins has not yet been established.
The phospho-dependence of the LIR motif, which we demonstrate here, might explain the conditional regulation of the interaction. Shc has been identified as a key mediator of anoikis, and deletion of its PTB domain is reported to result in loss of anoikis activity (69). Furthermore, binding of its PTB domain to the integrin β3 tail is dependent on the phosphorylation of Tyr785 (70, 71), whereas double phosphorylation of Thr779 and Tyr785 abolishes the interaction (34). Thr779 was further reported to be phosphorylated in vitro by PDK1 and Akt/PKB (protein kinase B) (34). As we could show that the double phosphorylation synergistically enables binding of MAP1LC3s and GABARAP, we hypothesize that the integrin β3 tail serves as a molecular signaling platform for induction either of anoikis or of protective autophagy.
Our study opens the way to researching the mechanistic details of the interplay between autophagy and integrin-mediated signaling. It would be interesting to assess whether the remainder of the β-integrin tails contain phospho-modulated LIR motifs as well. At this point, the coupled involvement of both integrins and the autophagic process in SARS-CoV-2 biology still awaits experimental validation, but one can hypothesize from our data that the virus might deploy autophagosomes for viral propagation. Contextualizing the biological relevance of these confirmed SLiM-based interactions is the next logical step.
MATERIALS AND METHODS
Peptides
Peptides were obtained from GL Biochem (Shanghai, China) or Genecust (Boynes, France). FITC-labeled peptides were dissolved in dimethyl sulfoxide, whereas unlabeled peptides were dissolved in 50 mM sodium phosphate buffer (pH 7.4). Either 50 mM sodium phosphate buffer (pH 7.4) or phosphate-buffered saline (PBS) (pH 7.4) with 3 mM dithiothreitol (DTT) was used and supplemented with 0.05% Tween for diluting labeled peptides during FP experiments.
Protein expression and purification
Protein domains (Table 4) were expressed in Escherichia coli BL21-Gold (DE3), grown in 2YT (yeast extract, 10 g/l; tryptone, 16 g/l; and NaCl, 5 g/l), and induced at OD600 (optical density at 600 nm) = 0.7 with 1 mM isopropyl-β-d-thiogalactopyranoside followed by 16-hour incubation at 18°C. All domains were GST-tagged, except for the PDZ domains, which were expressed with a maltose binding protein (MBP) tag. The GST-tagged constructs were purified with glutathione beads (Cytiva), whereas ion metal affinity chromatography (IMAC) using Ni2+ beads (Cytiva) was used for PDZ domains. For affinity measurements, the GST tag was removed overnight at 4°C by protease cleavage with either human rhinovirus (HRV) 3C (SH2 domains), tobacco etch virus (TEV) protease (SNX17 FERM), or thrombin (remainder of domains). Thrombin cleavage was on beads in PBS (pH 7.4), 1 mM β-mercaptoethanol, whereas proteins were eluted [50 mM tris (pH 8.0), 10 mM glutathione] and dialyzed (3.5-kDa cutoff, SnakeSkin, Pierce) for HRV 3C [50 mM tris (pH 8.0), 150 mM NaCl, 1 mM EDTA, and 1 mM β-mercaptoethanol] and TEV [50 mM tris (pH 8.0), 0.5 mM EDTA, and 1 mM β-mercaptoethanol] cleavage. Reverse IMAC [50 mM tris (pH 8.0)] and reverse chromatography with glutathione sepharose beads (PBS, pH 7.4) were performed to remove HRV 3C, and TEV proteases as well as GST. For removal of thrombin, a benzamidine column (Cytiva) was applied [20 mM tris (pH 7.4), 500 mM NaCl]. Fluorescence polarization experiments of the PDZ domains were performed on the MBP-tagged protein domains. Protein purity was assessed by SDS–polyacrylamide gel electrophoresis using Mini-PROTEAN TGX Stain-Free Protein Gels (Bio-Rad). After purification, protein domains were dialyzed (3.5-kDa cutoff, SnakeSkin, Pierce) in 50 mM sodium phosphate buffer (pH 7.4), 1 mM DTT or PBS (pH 7.4), 3 mM DTT (PDZ domains). For the complex in displacement experiments, proteins were supplemented with 0.05% Tween.
Table 4. Constructs used for expression of protein domains.
Protein domains and the encoding plasmids. Species is human unless otherwise stated.
| Name | UniProt | Plasmid |
| AP2 μ2 (160–435) | Q96CW1 | pGEX-4 T1 |
| DOK1 PTB (151–256) | Q99704 | pH1003 |
| FRS2 PTB (8–136) | Q8WU20 | pH1003 |
| FYN SH2 (142–251) | P06241 | pGEX |
| GABARAP ATG8 (2-115) | O95166 | pGEX-4 T1 |
| GABARAPL2 ATG8 (2–115) | P60520 | pGEX-4 T1 |
| IRS1 PTB (157–267) | P35568 | pH1003 |
| LYN SH2 (106–213) | P07948 | pGEX |
| MAGI1 PDZ1 (1–106) | Q96QZ7 | pETM41 |
| MAP1LC3A ATG8 (2–119) | Q9H492 | pGEX-4 T1 |
| MAP1LC3B ATG8 (2–119) | Q9GZQ8 | pGEX-4 T1 |
| MAP1LC3C ATG8 (2–125) | Q9BXW4 | pGEX-4 T1 |
| Mouse SNX17 FERM (109–388) | Q15036 | pMCSG10 |
| NCK1 SH2 (275–377) | P16333 | pGEX-2 T |
| NHERF3 PDZ1 (2–107) | Q5T2W1 | pETM41 |
| NHERF3 PDZ2 (132–224) | Q5T2W1 | pETM41 |
| SCRIB PDZ1 (718–814) | Q14160 | pETM41 |
| SHANK1 PDZ (655–761) | Q9Y566 | pETM41 |
| SNX27 PDZ (40–141) | Q96L92 | pETM41 |
| SNTA1 PDZ (84–171) | Q13424 | pETM41 |
| TAX1B3 PDZ (7–124) | O14907 | pETM41 |
| TLN1 PTB (442–770) | Q9Y490 | pH1003 |
| TLN2 PTB (309–401) | Q9Y4G6 | pH1003 |
FP experiments
FITC-labeled peptides were used as probes (excitation, 485 nm; emission, 535 nm), and the emitted light was measured in the SpectraMax iD5 Multi-Mode Microplate Reader (Molecular Devices) to calculate the mP signal. Experiments were performed at room temperature using nonbinding black half-area 96-well microplates (Corning) (50 μl per well). Saturation experiments were conducted using 5 nM FITC-labeled peptide (25 μl) and a dilution series of respective protein domain (25 μl) in a total volume of 50 μl. Measurements were performed directly without preincubation, and the reduced data were fitted to the quadratic equation as described previously (72) using Prism, which allowed to retrieve corresponding KD values. Displacement curves were obtained by using a premixed complex of protein domain ([protein domain] = one to twofold the KD of FITC-labeled probe, 25 μl) and FITC-labeled peptide (10 nM, 25 μl) and a dilution series of displacing peptide in a total volume of 50 μl. All measurements were performed in technical triplicates, except for MAP1LC3B and GABARAPL2 ATG8 domain binding to ACE2 pTyr785 peptide for which a duplicate of the displacement was recorded. KI values were calculated as described elsewhere (73).
Acknowledgments
We thank B. Mészáros, L. Chemes, T. Gibson, and coauthors for useful discussions and constructive comments on the manuscript. For the gifts of reagents, we thank A. Ernst for the MAP1LC3 constructs (74), S. S. Sidhu (University of Toronto) for the PTB domain constructs, R. Vincentelli and C. Fontes for the PDZ domain constructs (75), B. Mayer for the NCK SH2 domain (Addgene plasmid # 46457), and B. M. Collins for the SNX17 FERM domain construct (38). Funding: This work was supported by grants from the Swedish Research Council (2016-04965 to Y.I.) and the Swedish Foundation for Strategic Research (SB16-0039 to Y.I.). Author contributions: Y.I. and J.K. conceived the experiments. H.K., M.A., and J.K. performed the experiments and analyzed the results. J.K. and Y.I. wrote the manuscript. All authors approved the final version. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions herein are present in the paper.
REFERENCES AND NOTES
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G. F., Tan W.; China Novel Coronavirus Investigating; Research Team , A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 382, 727–733 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization, Weekly Epidemiological Update (World Health Organization, 2020). [Google Scholar]
- 3.Gordon D. E., Hiatt J., Bouhaddou M., Rezelj V. V., Ulferts S., Braberg H., Jureka A. S., Obernier K., Guo J. Z., Batra J., Kaake R. M., Weckstein A. R., Owens T. W., Gupta M., Pourmal S., Titus E. W., Cakir M., Soucheray M., Gregor M. M., Cakir Z., Jang G., O’Meara M. J., Tummino T. A., Zhang Z., Foussard H., Rojc A., Zhou Y., Kuchenov D., Hüttenhain R., Xu J., Eckhardt M., Swaney D. L., Fabius J. M., Ummadi M., Tutuncuoglu B., Rathore U., Modak M., Haas P., Haas K. M., Naing Z. Z. C., Pulido E. H., Shi Y., Barrio-Hernandez I., Memon D., Petsalaki E., Dunham A., Marrero M. C., Burke D., Koh C., Vallet T., Silvas J. A., Azumaya C. M., Billesbølle C., Brilot A. F., Campbell M. G., Diallo A., Dickinson M. S., Diwanji D., Herrera N., Hoppe N., Kratochvil H. T., Liu Y., Merz G. E., Moritz M., Nguyen H. C., Nowotny C., Puchades C., Rizo A. N., Schulze-Gahmen U., Smith A. M., Sun M., Young I. D., Zhao J., Asarnow D., Biel J., Bowen A., Braxton J. R., Chen J., Chio C. M., Chio U. S., Deshpande I., Doan L., Faust B., Flores S., Jin M., Kim K., Lam V. L., Li F., Li J., Li Y.-L., Li Y., Liu X., Lo M., Lopez K. E., Melo A. A., Moss F. R. III, Nguyen P., Paulino J., Pawar K. I., Peters J. K., Pospiech T. H. Jr., Safari M., Sangwan S., Schaefer K., Thomas P. V., Thwin A. C., Trenker R., Tse E., Tsui T. K. M., Wang F., Whitis N., Yu Z., Zhang K., Zhang Y., Zhou F., Saltzberg D.; QCRG Structural Biology Consortium, Hodder A. J., Shun-Shion A. S., Williams D. M., White K. M., Rosales R., Kehrer T., Miorin L., Moreno E., Patel A. H., Rihn S., Khalid M. M., Vallejo-Gracia A., Fozouni P., Simoneau C. R., Roth T. L., Wu D., Karim M. A., Ghoussaini M., Dunham I., Berardi F., Weigang S., Chazal M., Park J., Logue J., Grath M. M., Weston S., Haupt R., Hastie C. J., Elliott M., Brown F., Burness K. A., Reid E., Dorward M., Johnson C., Wilkinson S. G., Geyer A., Giesel D. M., Baillie C., Raggett S., Leech H., Toth R., Goodman N., Keough K. C., Lind A. L.; Zoonomia Consortium, Klesh R. J., Hemphill K. R., Carlson-Stevermer J., Oki J., Holden K., Maures T., Pollard K. S., Sali A., Agard D. A., Cheng Y., Fraser J. S., Frost A., Jura N., Kortemme T., Manglik A., Southworth D. R., Stroud R. M., Alessi D. R., Davies P., Frieman M. B., Ideker T., Abate C., Jouvenet N., Kochs G., Shoichet B., Ott M., Palmarini M., Shokat K. M., García-Sastre A., Rassen J. A., Grosse R., Rosenberg O. S., Verba K. A., Basler C. F., Vignuzzi M., Peden A. A., Beltrao P., Krogan N. J., Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science 370, eabe9403 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gordon D. E., Jang G. M., Bouhaddou M., Xu J., Obernier K., White K. M., O’Meara M. J., Rezelj V. V., Guo J. Z., Swaney D. L., Tummino T. A., Hüttenhain R., Kaake R. M., Richards A. L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B. J., Braberg H., Fabius J. M., Eckhardt M., Soucheray M., Bennett M. J., Cakir M., McGregor M. J., Li Q., Meyer B., Roesch F., Vallet T., Kain A. M., Miorin L., Moreno E., Naing Z. Z. C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I. T., Melnyk J. E., Chorba J. S., Lou K., Dai S. A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C. J. P., Perica T., Pilla K. B., Ganesan S. J., Saltzberg D. J., Rakesh R., Liu X., Rosenthal S. B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X. P., Liu Y. F., Wankowicz S. A., Bohn M., Safari M., Ugur F. S., Koh C., Savar N. S., Tran Q. D., Shengjuler D., Fletcher S. J., O’Neal M. C., Cai Y., Chang J. C. J., Broadhurst D. J., Klippsten S., Sharp P. P., Wenzell N. A., Kuzuoglu-Ozturk D., Wang H. Y., Trenker R., Young J. M., Cavero D. A., Hiatt J., Roth T. L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R. M., Frankel A. D., Rosenberg O. S., Verba K. A., Agard D. A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., d’Enfert C., Mukherjee S., Jacobson M., Malik H. S., Fujimori D. G., Ideker T., Craik C. S., Floor S. N., Fraser J. S., Gross J. D., Sali A., Roth B. L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., García-Sastre A., Shokat K. M., Shoichet B. K., Krogan N. J., A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583, 459–468 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.A. Stukalov, V. Girault, V. Grass, V. Bergant, O. Karayel, C. Urban, D. A. Haas, Y. Huang, L. Oubraham, A. Wang, S. M. Hamad, A. Piras, M. Tanzer, F. M. Hansen, T. Enghleitner, M. Reinecke, T. M. Lavacca, R. Ehmann, R. Wölfel, J. Jores, B. Kuster, U. Protzer, R. Rad, J. Ziebuhr, V. Thiel, P. Scaturro, M. Mann, A. Pichlmair, Multi-level proteomics reveals host-perturbation strategies of SARS-CoV-2 and SARS-CoV. bioRxiv 2020.06.17.156455 [Preprint]. 17 June 2020. 10.1101/2020.06.17.156455. [DOI]
- 6.Bouhaddou M., Memon D., Meyer B., White K. M., Rezelj V. V., Correa Marrero M., Polacco B. J., Melnyk J. E., Ulferts S., Kaake R. M., Batra J., Richards A. L., Stevenson E., Gordon D. E., Rojc A., Obernier K., Fabius J. M., Soucheray M., Miorin L., Moreno E., Koh C., Tran Q. D., Hardy A., Robinot R., Vallet T., Nilsson-Payant B. E., Hernandez-Armenta C., Dunham A., Weigang S., Knerr J., Modak M., Quintero D., Zhou Y., Dugourd A., Valdeolivas A., Patil T., Li Q., Hüttenhain R., Cakir M., Muralidharan M., Kim M., Jang G., Tutuncuoglu B., Hiatt J., Guo J. Z., Xu J., Bouhaddou S., Mathy C. J. P., Gaulton A., Manners E. J., Félix E., Shi Y., Goff M., Lim J. K., McBride T., O’Neal M. C., Cai Y., Chang J. C. J., Broadhurst D. J., Klippsten S., de wit E., Leach A. R., Kortemme T., Shoichet B., Ott M., Saez-Rodriguez J., tenOever B. R., Mullins R. D., Fischer E. R., Kochs G., Grosse R., García-Sastre A., Vignuzzi M., Johnson J. R., Shokat K. M., Swaney D. L., Beltrao P., Krogan N. J., The global phosphorylation landscape of SARS-CoV-2 infection. Cell 182, 685–712.e19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daniloski Z., Jordan T. X., Wessels H. H., Hoagland D. A., Kasela S., Legut M., Maniatis S., Mimitou E. P., Lu L., Geller E., Danziger O., Rosenberg B. R., Phatnani H., Smibert P., Lappalainen T., tenOever B. R., Sanjana N. E., Identification of required host factors for SARS-CoV-2 infection in human cells. Cell , (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L., A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T. S., Herrler G., Wu N. H., Nitsche A., Müller M. A., Drosten C., Pöhlmann S., SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271–280.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F., Structural basis of receptor recognition by SARS-CoV-2. Nature 581, 221–224 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q., Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 367, 1444–1448 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li M.-Y., Li L., Zhang Y., Wang X.-S., Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infect. Dis. Poverty 9, 45 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hikmet F., Méar L., Edvinsson Å., Micke P., Uhlén M., Lindskog C., The protein expression profile of ACE2 in human tissues. Mol. Syst. Biol. 16, e9610 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.L. Cantuti-Castelvetri, R. Ojha, L. D. Pedro, M. Djannatian, J. Franz, S. Kuivanen, K. Kallio, T. Kaya, M. Anastasina, T. Smura, L. Levanov, L. Szirovicza, A. Tobi, H. Kallio-Kokko, P. Österlund, M. Joensuu, F. A. Meunier, S. Butcher, M. S. Winkler, B. Mollenhauer, A. Helenius, O. Gokce, T. Teesalu, J. Hepojoki, O. Vapalahti, C. Stadelmann, G. Balistreri, M. Simons, Neuropilin-1 facilitates SARS-CoV-2 cell entry and provides a possible pathway into the central nervous system. bioRxiv 2020.06.07.137802 [Preprint]. 7 June 2020. 10.1101/2020.06.07.137802. [DOI]
- 15.C. Gao, J. Zeng, N. Jia, K. Stavenhagen, Y. Matsumoto, H. Zhang, J. Li, A. J. Hume, E. Mühlberger, I. van Die, J. Kwan, K. Tantisira, A. Emili, R. D. Cummings, SARS-CoV-2 spike protein interacts with multiple innate immune receptors. bioRxiv 2020.07.29.227462 [Preprint]. 30 July 2020. 10.1101/2020.07.29.227462. [DOI]
- 16.Sigrist C. J. A., Bridge A., Mercier P. L., A potential role for integrins in host cell entry by SARS-CoV-2. Antivir. Res. 177, 104759 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mészáros B., Sámano-Sánchez H., Alvarado-Valverde J., Čalyševa J., Martínez-Pérez E., Alves R., Shields D. C., Kumar M., Rippmann F., Chemes L. B., Gibson T. J., Short linear motif candidates in the cell entry system used by SARS-CoV-2 and their potential therapeutic implications. Sci. Signal. 14, eabd0334 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torres-Gomez A., Cabañas C., Lafuente E. M., Phagocytic integrins: Activation and signaling. Front. Immunol. 11, 738 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hussein H. A. M., Walker L. R., Abdel-Raouf U. M., Desouky S. A., Montasser A. K. M., Akula S. M., Beyond RGD: Virus interactions with integrins. Arch. Virol. 160, 2669–2681 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plosa E. J., Benjamin J. T., Sucre J. M., Gulleman P. M., Gleaves L. A., Han W., Kook S., Polosukhin V. V., Haake S. M., Guttentag S. H., Young L. R., Pozzi A., Blackwell T. S., Zent R., β1 Integrin regulates adult lung alveolar epithelial cell inflammation. JCI Insight 5, e129259 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borriello G., Ianniello A., COVID-19 occurring during Natalizumab treatment: A case report in a patient with extended interval dosing approach. Mult. Scler. Relat. Disord. 41, 102165 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aguirre C., Meca-Lallana V., Barrios-Blandino A., Del Río B., Vivancos J., Covid-19 in a patient with multiple sclerosis treated with natalizumab: May the blockade of integrins have a protective role? Mult. Scler. Relat. Disord. 44, 102250 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davey N. E., Van Roey K., Weatheritt R. J., Toedt G., Uyar B., Altenberg B., Budd A., Diella F., Dinkel H., Gibson T. J., Attributes of short linear motifs. Mol. BioSyst. 8, 268–281 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Ivarsson Y., Plasticity of PDZ domains in ligand recognition and signaling. FEBS Lett. 586, 2638–2647 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boll W., Ohno H., Songyang Z., Rapoport I., Cantley L. C., Bonifacino J. S., Kirchhausen T., Sequence requirements for the recognition of tyrosine-based endocytic signals by clathrin AP-2 complexes. EMBO J. 15, 5789–5795 (1996). [PMC free article] [PubMed] [Google Scholar]
- 26.Sora V., Kumar M., Maiani E., Lambrughi M., Tiberti M., Papaleo E., Structure and dynamics in the ATG8 family from experimental to computational techniques. Front. Cell Dev. Biol. 8, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar M., Gouw M., Michael S., Sámano-Sánchez H., Pancsa R., Glavina J., Diakogianni A., Valverde J. A., Bukirova D., Čalyševa J., Palopoli N., Davey N. E., Chemes L. B., Gibson T. J., ELM-the eukaryotic linear motif resource in 2020. Nucleic Acids Res. 48, D296–D306 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaksonen M., Roux A., Mechanisms of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 19, 313–326 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Lee H. J., Zheng J. J., PDZ domains and their binding partners: Structure, specificity, and modification. Cell Commun. Signal. 8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uhlik M. T., Temple B., Bencharit S., Kimple A. J., Siderovski D. P., Johnson G. L., Structural and evolutionary division of phosphotyrosine binding (PTB) domains. J. Mol. Biol. 345, 1–20 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Tompa P., Davey N. E., Gibson T. J., Babu M. M., A million peptide motifs for the molecular biologist. Mol. Cell 55, 161–169 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Nishi H., Hashimoto K., Panchenko A. R., Phosphorylation in protein-protein binding: Effect on stability and function. Structure 19, 1807–1815 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Legate K. R., Fässler R., Mechanisms that regulate adaptor binding to β-integrin cytoplasmic tails. J. Cell Sci. 122, 187–198 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Kirk R. I., Sanderson M. R., Lerea K. M., Threonine phosphorylation of the β3 integrin cytoplasmic tail, at a site recognized by PDK1 and Akt/PKB in vitro, regulates Shc binding. J. Biol. Chem. 275, 30901–30906 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Bouhaddou M., Memon D., Meyer B., White K. M., Rezelj V. V., Correa Marrero M., Polacco B. J., Melnyk J. E., Ulferts S., Kaake R. M., Batra J., Richards A. L., Stevenson E., Gordon D. E., Rojc A., Obernier K., Fabius J. M., Soucheray M., Miorin L., Moreno E., Koh C., Tran Q. D., Hardy A., Robinot R., Vallet T., Nilsson-Payant B. E., Hernandez-Armenta C., Dunham A., Weigang S., Knerr J., Modak M., Quintero D., Zhou Y., Dugourd A., Valdeolivas A., Patil T., Li Q., Hüttenhain R., Cakir M., Muralidharan M., Kim M., Jang G., Tutuncuoglu B., Hiatt J., Guo J. Z., Xu J., Bouhaddou S., Mathy C. J. P., Gaulton A., Manners E. J., Félix E., Shi Y., Goff M., Lim J. K., McBride T., O’Neal M. C., Cai Y., Chang J. C. J., Broadhurst D. J., Klippsten S., de wit E., Leach A. R., Kortemme T., Shoichet B., Ott M., Saez-Rodriguez J., tenOever B. R., Mullins R. D., Fischer E. R., Kochs G., Grosse R., García-Sastre A., Vignuzzi M., Johnson J. R., Shokat K. M., Swaney D. L., Beltrao P., Krogan N. J., The global phosphorylation landscape of SARS-CoV-2 infection. Cell 182, 685–712.e19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tonikian R., Zhang Y., Sazinsky S. L., Currell B., Yeh J.-H., Reva B., Held H. A., Appleton B. A., Evangelista M., Wu Y., Xin X., Chan A. C., Seshagiri S., Lasky L. A., Sander C., Boone C., Bader G. D., Sidhu S. S., A specificity map for the PDZ domain family. PLOS Biol. 6, e239 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uhlén M., Fagerberg L., Hallström B. M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., Olsson I. M., Edlund K., Lundberg E., Navani S., Szigyarto C. A.-K., Odeberg J., Djureinovic D., Takanen J. O., Hober S., Alm T., Edqvist P.-H., Berling H., Tegel H., Mulder J., Rockberg J., Nilsson P., Schwenk J. M., Hamsten M., von Feilitzen K., Forsberg M., Persson L., Johansson F., Zwahlen M., von Heijne G., Nielsen J., Pontén F., Tissue-based map of the human proteome. Science 347, 1260419 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Ghai R., Bugarcic A., Liu H., Norwood S. J., Skeldal S., Coulson E. J., Li S. S.-C., Teasdale R. D., Collins B. M., Structural basis for endosomal trafficking of diverse transmembrane cargos by PX-FERM proteins. Proc. Natl. Acad. Sci. U.S.A. 110, E643–E652 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steinberg F., Heesom K. J., Bass M. D., Cullen P. J., SNX17 protects integrins from degradation by sorting between lysosomal and recycling pathways. J. Cell Biol. 197, 219–230 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gallon M., Clairfeuille T., Steinberg F., Mas C., Ghai R., Sessions R. B., Teasdale R. D., Collins B. M., Cullen P. J., A unique PDZ domain and arrestin-like fold interaction reveals mechanistic details of endocytic recycling by SNX27-retromer. Proc. Natl. Acad. Sci. U.S.A. 111, E3604–E3613 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frese S., Schubert W. D., Findeis A. C., Marquardt T., Roske Y. S., Stradal T. E. B., Heinz D. W., The phosphotyrosine peptide binding specificity of Nck1 and Nck2 Src homology 2 domains. J. Biol. Chem. 281, 18236–18245 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Shiratori T., Miyatake S., Ohno H., Nakaseko C., Isono K., Bonifacino J. S., Saito T., Tyrosine phosphorylation controls internalization of CTLA-4 by regulating its interaction with clathrin-associated adaptor complex AP-2. Immunity 6, 583–589 (1997). [DOI] [PubMed] [Google Scholar]
- 43.De Franceschi N., Arjonen A., Elkhatib N., Denessiouk K., Wrobel A. G., Wilson T. A., Pouwels J., Montagnac G., Owen D. J., Ivaska J., Selective integrin endocytosis is driven by interactions between the integrin α-chain and AP2. Nat. Struct. Mol. Biol. 23, 172–179 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tinti M., Kiemer L., Costa S., Miller M. L., Sacco F., Olsen J. V., Carducci M., Paoluzi S., Langone F., Workman C. T., Blom N., Machida K., Thompson C. M., Schutkowski M., Brunak S., Mann M., Mayer B. J., Castagnoli L., Cesareni G., The SH2 domain interaction landscape. Cell Rep. 3, 1293–1305 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Law D. A., Nannizzi-Alaimo L., Phillips D. R., Outside-in integrin signal transduction. J. Biol. Chem. 271, 10811–10815 (1996). [DOI] [PubMed] [Google Scholar]
- 46.Yang N., Shen H. M., Targeting the endocytic pathway and autophagy process as a novel therapeutic strategy in COVID-19. Int. J. Biol. Sci. 16, 1724–1731 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grass I., Thiel S., Höning S., Haucke V., Recognition of a basic AP-2 binding motif within the C2B domain of synaptotagmin is dependent on multimerization. J. Biol. Chem. 279, 54872–54880 (2004). [DOI] [PubMed] [Google Scholar]
- 48.Höning S., Sosa M., Hille-Rehfeld A., Von Figura K., The 46-kDa mannose 6-phosphate receptor contains multiple binding sites for clathrin adaptors. J. Biol. Chem. 272, 19884–19890 (1997). [DOI] [PubMed] [Google Scholar]
- 49.Smith S. M., Baker M., Halebian M., Smith C. J., Weak molecular interactions in clathrin-mediated endocytosis. Front. Mol. Biosci. 4, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang P. G., Tang D. J., Hua Z., Wang Z., An J., Sunitinib reduces the infection of SARS-CoV, MERS-CoV and SARS-CoV-2 partially by inhibiting AP2M1 phosphorylation. Cell Discov. 6, 71 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parsons S. J., Parsons J. T., Src family kinases, key regulators of signal transduction. Oncogene 23, 7906–7909 (2004). [DOI] [PubMed] [Google Scholar]
- 52.Monteiro P., Feng G., SHANK proteins: Roles at the synapse and in autism spectrum disorder. Nat. Rev. Neurosci. 18, 147–157 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Seidler U., Singh A. K., Cinar A., Chen M., Hillesheim J., Hogema B., Riederer B., The role of the NHERF family of PDZ scaffolding proteins in the regulation of salt and water Transport. Ann. N. Y. Acad. Sci. 1165, 249–260 (2009). [DOI] [PubMed] [Google Scholar]
- 54.Broadbent D., Ahmadzai M. M., Kammala A. K., Yang C., Occhiuto C., Das R., Subramanian H., Roles of NHERF family of PDZ-binding proteins in regulating GPCR functions. Adv. Immunol. 136, 353–385 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Jia H. P., Look D. C., Shi L., Hickey M., Pewe L., Netland J., Farzan M., Wohlford-Lenane C., Perlman S., McCray P. B. Jr., ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J. Virol. 79, 14614–14621 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.D’Amico A., Soragna A., Cairano E. D., Panzeri N., Anzai N., Sacchi F. V., Perego C., The surface density of the glutamate transporter EAAC1 is controlled by interactions with PDZK1 and AP2 adaptor complexes. Traffic 11, 1455–1470 (2010). [DOI] [PubMed] [Google Scholar]
- 57.Lauffer B. E. L., Melero C., Temkin P., Lei C., Hong W., Kortemme T., von Zastrow M., SNX27 mediates PDZ-directed sorting from endosomes to the plasma membrane. J. Cell Biol. 190, 565–574 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Y. Zhu, F. Feng, G. Hu, Y. Wang, Y. Yu, Y. Zhu, W. Xu, X. Cai, Z. Sun, W. Han, R. Ye, H. Chen, Q. Ding, Q. Cai, D. Qu, Y. Xie, Z. Yuan, R. Zhang, The S1/S2 boundary of SARS-CoV-2 spike protein modulates cell entry pathways and transmission. bioRxiv 2020.08.25.266775 [Preprint]. 25 August 2020. 10.1101/2020.08.25.266775. [DOI]
- 59.Vincentelli R., Luck K., Poirson J., Polanowska J., Abdat J., Blémont M., Turchetto J., Iv F., Ricquier K., Straub M.-L., Forster A., Cassonnet P., Borg J.-P., Jacob Y., Masson M., Nominé Y., Reboul J., Wolff N., Charbonnier S., Travé G., Quantifying domain-ligand affinities and specificities by high-throughput holdup assay. Nat. Methods 12, 787–793 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Johansen T., Lamark T., Selective autophagy: ATG8 family proteins, LIR motifs and cargo receptors. J. Mol. Biol. 432, 80–103 (2020). [DOI] [PubMed] [Google Scholar]
- 61.K. Gorshkov, C. Z. Chen, R. Bostwick, L. Rasumussen, M. Xu, M. Pradhan, B. N. Tran, W. Zhu, K. Shamim, W. Huang, X. Hu, M. Shen, C. Klumpp-Thomas, Z. Itkin, P. Shinn, A. Simeonov, S. Michael, M. D. Hall, D. C. Lo, W. Zheng, The SARS-CoV-2 cytopathic effect is blocked with autophagy modulators. bioRxiv 2020.05.16.091520 [Preprint]. 28 May 2020. 10.1101/2020.05.16.091520. [DOI]
- 62.Cai X., Xu Y., Cheung A. K., Tomlinson R. C., Alcázar-Román A., Murphy L., Billich A., Zhang B., Feng Y., Klumpp M., Rondeau J. M., Fazal A. N., Wilson C. J., Myer V., Joberty G., Bouwmeester T., Labow M. A., Finan P. M., Porter J. A., Ploegh H. L., Baird D., de Camilli P., Tallarico J. A., Huang Q., PIKfyve, a class III PI kinase, is the target of the small molecular IL-12/IL-23 inhibitor apilimod and a player in toll-like receptor signaling. Chem. Biol. 20, 912–921 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Riva L., Yuan S., Yin X., Martin-Sancho L., Matsunaga N., Pache L., Burgstaller-Muehlbacher S., de Jesus P. D., Teriete P., Hull M. V., Chang M. W., Chan J. F. W., Cao J., Poon V. K. M., Herbert K. M., Cheng K., Nguyen T. T. H., Rubanov A., Pu Y., Nguyen C., Choi A., Rathnasinghe R., Schotsaert M., Miorin L., Dejosez M., Zwaka T. P., Sit K. Y., Martinez-Sobrido L., Liu W. C., White K. M., Chapman M. E., Lendy E. K., Glynne R. J., Albrecht R., Ruppin E., Mesecar A. D., Johnson J. R., Benner C., Sun R., Schultz P. G., Su A. I., García-Sastre A., Chatterjee A. K., Yuen K. Y., Chanda S. K., Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature 586, 113–119 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hasegawa J., Strunk B. S., Weisman L. S., PI5P and PI(3,5)P2: Minor, but essential phosphoinositides. Cell Struct. Funct. 42, 49–60 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hessvik N. P., Øverbye A., Brech A., Torgersen M. L., Jakobsen I. S., Sandvig K., Llorente A., PIKfyve inhibition increases exosome release and induces secretory autophagy. Cell. Mol. Life Sci. 73, 4717–4737 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vlahakis A., Debnath J., The Interconnections between autophagy and Integrin-mediated cell adhesion. J. Mol. Biol. 429, 515–530 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhu Y., Li L., Gong S., Yu Y., Dai H., Cai G., Yan J., ß3-Integrin inhibits lipopolysaccharide-induced autophagy in cardiomyocytes via the Akt signaling pathway. Cardiology 130, 249–259 (2015). [DOI] [PubMed] [Google Scholar]
- 68.Lino R. L. B., Santos P. K. D., Pisani G. F. D., Altei W. F., Cominetti M. R., Selistre-de-Araújo H. S., Alphavbeta3 integrin blocking inhibits apoptosis and induces autophagy in murine breast tumor cells. Biochim. Biophys. Acta Mol. Cell Res. 1866, 118536 (2019). [DOI] [PubMed] [Google Scholar]
- 69.Ma Z., Myers D. P., Wu R. F., Nwariaku F. E., Terada L. S., p66Shc mediates anoikis through RhoA. J. Cell Biol. 179, 23–31 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Deshmukh L., Gorbatyuk V., Vinogradova O., Integrin β3 phosphorylation dictates its complex with the Shc phosphotyrosine-binding (PTB) domain. J. Biol. Chem. 285, 34875–34884 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cowan K. J., Law D. A., Phillips D. R., Identification of Shc as the primary protein binding to the tyrosine-phosphorylated β3Subunit of αIIbβ3during outside-in integrin platelet signaling. J. Biol. Chem. 275, 36423–36429 (2000). [DOI] [PubMed] [Google Scholar]
- 72.Gianni S., Engström Å., Larsson M., Calosci N., Malatesta F., Eklund L., Ngang C. C., Travaglini-Allocatelli C., Jemth P., The kinetics of PDZ domain-ligand interactions and implications for the binding mechanism. J. Biol. Chem. 280, 34805–34812 (2005). [DOI] [PubMed] [Google Scholar]
- 73.Nikolovska-Coleska Z., Wang R., Fang X., Pan H., Tomita Y., Li P., Roller P. P., Krajewski K., Saito N. G., Stuckey J. A., Wang S., Development and optimization of a binding assay for the XIAP BIR3 domain using fluorescence polarization. Anal. Biochem. 332, 261–273 (2004). [DOI] [PubMed] [Google Scholar]
- 74.Stolz A., Putyrski M., Kutle I., Huber J., Wang C., Major V., Sidhu S. S., Youle R. J., Rogov V. V., Dötsch V., Ernst A., Dikic I., Fluorescence-based ATG 8 sensors monitor localization and function of LC 3/ GABARAP proteins. EMBO J. 36, 549–564 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Duhoo Y., Girault V., Turchetto J., Ramond L., Durbesson F., Fourquet P., Nominé Y., Cardoso V., Sequeira A. F., Brás J. L. A., Fontes C. M. G. A., Travé G., Wolff N., Vincentelli R., High-throughput production of a new library of human single and tandem PDZ domains allows quantitative PDZ-peptide interaction screening through high-throughput holdup assay. Methods Mol. Biol. 2025, 439–476 (2019). [DOI] [PubMed] [Google Scholar]