Fighting zoonotic coronaviruses
In the past 20 years, three betacoronaviruses thought to have originated in bats have caused devastating disease in humans. The global pandemic caused by the latest such virus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), highlights the need to protect against other strains that could present a threat to humans. Cohen et al. constructed nanoparticles displaying the protein domain that binds the host cell receptor (receptor-binding domain or RBD), either a homotypic SARS-CoV-2 particle or mosaic particles displaying RBDs from four or eight different betacoronaviruses. In mice, antibodies to the SARS-CoV-2 RBD were elicited just as well by mosaic particles as by homotypic nanoparticles. The mosaic nanoparticles elicited antibodies that, beyond recognizing the strains displayed, also recognized mismatched strains.
Science, this issue p. 735
Immunizing with nanoparticles that display diverse coronavirus receptor binding domains elicits cross-reactive and neutralizing antibody responses.
Abstract
Protection against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and SARS-related emergent zoonotic coronaviruses is urgently needed. We made homotypic nanoparticles displaying the receptor binding domain (RBD) of SARS-CoV-2 or co-displaying SARS-CoV-2 RBD along with RBDs from animal betacoronaviruses that represent threats to humans (mosaic nanoparticles with four to eight distinct RBDs). Mice immunized with RBD nanoparticles, but not soluble antigen, elicited cross-reactive binding and neutralization responses. Mosaic RBD nanoparticles elicited antibodies with superior cross-reactive recognition of heterologous RBDs relative to sera from immunizations with homotypic SARS-CoV-2–RBD nanoparticles or COVID-19 convalescent human plasmas. Moreover, after priming, sera from mosaic RBD–immunized mice neutralized heterologous pseudotyped coronaviruses as well as or better than sera from homotypic SARS-CoV-2–RBD nanoparticle immunizations, demonstrating no loss of immunogenicity against particular RBDs resulting from co-display. A single immunization with mosaic RBD nanoparticles provides a potential strategy to simultaneously protect against SARS-CoV-2 and emerging zoonotic coronaviruses.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a newly emergent betacoronavirus, resulted in a global pandemic in 2020, infecting millions and causing the respiratory disease COVID-19 (1, 2). Two other zoonotic betacoronaviruses, SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV), have also resulted in outbreaks within the past 20 years (3). All three viruses presumably originated in bats (4), with SARS-CoV and MERS-CoV adapting to intermediary animal hosts before jumping to humans. SARS-like viruses circulate in bats, and serological surveillance of people living near caves where bats carry diverse coronaviruses demonstrates direct transmission of SARS-like viruses with pandemic potential (5). This finding suggests that a pan-coronavirus vaccine is needed to protect against future outbreaks and pandemics. In particular, the bat WIV1 and SHC014 strains are thought to represent an ongoing threat to humans (6, 7).
Most current SARS-CoV-2 vaccine candidates include the spike trimer (S), the viral protein that mediates target cell entry after one or more of its receptor binding domains (RBDs) adopts an “up” position to bind a host receptor (Fig. 1A). The RBDs of human coronaviruses SARS-CoV-2, SARS-CoV, and HCoV-NL63, as well as those of the related animal coronaviruses WIV1 and SHC014, use angiotensin-converting enzyme 2 (ACE2) as their host receptor (1, 8, 9), whereas other coronaviruses use receptors such as dipeptidyl peptidase 4 (10) or sialic acids (11, 12). Consistent with its function in viral entry, S is the primary target of neutralizing antibodies (13–22), with many targeting the RBD (14–18, 21–26).
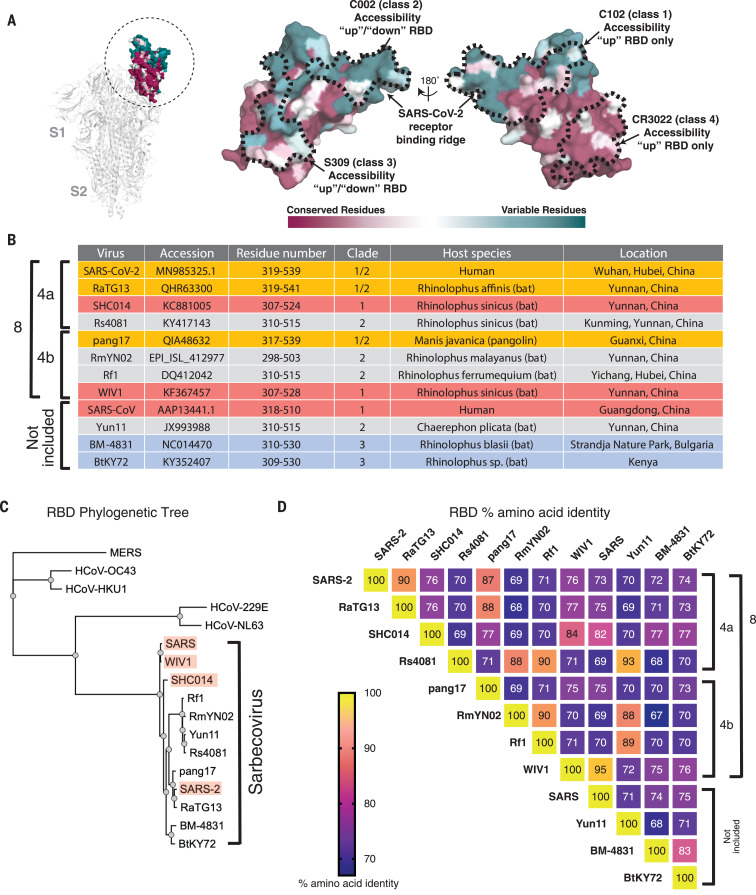
Fig. 1. Properties of RBDs chosen for this study.
(A) Left: Structure of SARS-CoV-2 S trimer (PDB 6VXX) with one RBD (dashed circle) in an “up” position. Center and right: Sequence conservation of 12 RBDs calculated by the ConSurf Database (49) plotted on a surface representation of the RBD structure (PDB 7BZ5). Epitopes for representatives from defined classes of RBD-binding antibodies (classes 1 to 4) (24) are indicated by dashed lines. (B) Summary of properties of the viral strains from which the 12 sarbecovirus RBDs were derived. (C) Phylogenetic tree of human and selected other coronaviruses based on RBD protein sequences. Red shading indicates strains known to use ACE2 as a receptor. (D) Heat map showing percent amino acid sequence identities among 12 sarbecovirus RBDs.
Multivalent display of antigen enhances B cell responses and can provide longer-lasting immunity than monovalent antigens (27, 28); thus, protein-based vaccine candidates often involve a nanoparticle that enables antigen multimerization. Many nanoparticles and coupling strategies have been explored for vaccine design (29), with “plug and display” strategies being especially useful (30, 31). In one such approach, multiple copies of an engineered protein domain called SpyCatcher fused to subunits of a virus-like particle form spontaneous isopeptide bonds to purified antigens tagged with a 13-residue SpyTag (29–32). The SpyCatcher-SpyTag system was used to prepare multimerized SARS-CoV-2 RBD or S trimer that elicited high titers of neutralizing antibodies (33, 34). Although promising for protection against SARS-CoV-2, coronavirus reservoirs in bats suggest future cross-species transmission (6, 7, 35), necessitating a vaccine that protects against emerging coronaviruses as well as SARS-CoV-2. Here, we prepared SpyCatcher003-mi3 nanoparticles (31, 36) simultaneously displaying SpyTagged RBDs from human and animal coronaviruses to evaluate whether mosaic particles can elicit cross-reactive antibody responses, as previously demonstrated for influenza head domain mosaic particles (37). We show that mice immunized with homotypic or mosaic nanoparticles produced broad binding and neutralizing responses, in contrast to plasma antibodies elicited in humans by SARS-CoV-2 infection. Moreover, relative to homotypic SARS-CoV-2 nanoparticles, mosaic nanoparticles showed enhanced heterologous binding and neutralization properties against human and bat SARS-like betacoronaviruses (sarbecoviruses).
We used a study of sarbecovirus RBD receptor usage and cell tropism (38) to guide our choice of RBDs for co-display on mosaic particles. From 29 RBDs that were classified into distinct clades (clades 1, 2, 1/2, and 3) (38), we identified diverse RBDs from SARS-CoV, WIV1, and SHC014 (clade 1); SARS-CoV-2 (clade 1/2); Rs4081, Yunnan 2011 (Yun11), and Rf1 (clade 2); and BM-4831 (clade 3). Of these, SARS-CoV-2 and SARS-CoV are human coronaviruses and the rest are bat viruses originating in China or Bulgaria (BM-4831). We also included RBDs from the GX pangolin clade 1/2 coronavirus (referred to here as pang17) (39); RaTG13, the bat clade 1/2 virus most closely related to SARS-CoV-2 (40); RmYN02, a clade 2 bat virus from China (41); and BtKY72, a Kenyan bat clade 3 virus (42). Mapping of the sequence conservation across selected RBDs showed varying degrees of sequence identity (68 to 95%), with highest sequence variability in residues corresponding to the SARS-CoV-2 ACE2 receptor binding motif (Fig. 1, A to D, and fig. S1). We chose 8 of the 12 RBDs as sources for three types of mosaic nanoparticles—mosaic-4a (coupled to SARS-2, RaTG13, SHC014, and Rs4081 RBDs); mosaic-4b (coupled to pang17, RmYN02, Rf1, and WIV1 RBDs); and mosaic-8 (coupled to all eight RBDs)—and compared them with homotypic mi3 particles constructed from SARS-CoV-2 RBD alone (homotypic SARS-2). RBDs from SARS, Yun11, BM-4831, and BtKY72, which were not coupled to mosaic particles, were used to evaluate sera for cross-reactive responses.
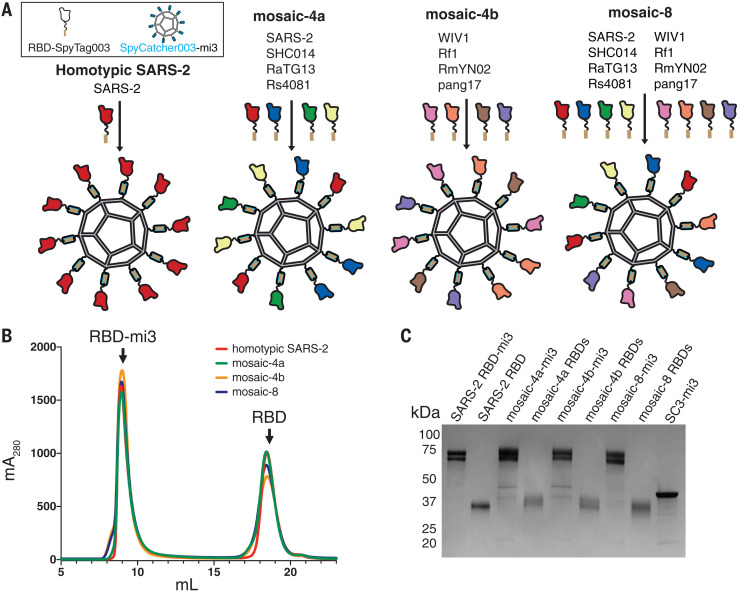
SpyTag003-RBDs were coupled to SpyCatcher003-mi3 (60 potential conjugation sites) (36, 43) to make homotypic and mosaic nanoparticles (Fig. 2A). Particles were purified by size exclusion chromatography (SEC) and analyzed by SDS–polyacrylamide gel electrophoresis (PAGE), revealing monodisperse SEC profiles and nearly 100% conjugation (Fig. 2, B and C). Representative RBDs were conjugated to SpyCatcher003-mi3 with similar or identical efficiencies (fig. S2), which suggests that mosaic particles contained approximately equimolar mixtures of different RBDs.
Fig. 2. Construction of RBD nanoparticles.
(A) Left: SpyTagged RBDs were attached to SpyCatcher003-mi3 to make a homotypic particle and three mosaic particles. There are 60 potential coupling sites on mi3; only 10 are shown for clarity. (B) SEC profile showing separation of RBD nanoparticles and free RBD proteins. (C) Coomassie-stained SDS-PAGE of RBD-coupled nanoparticles, free RBD proteins, and uncoupled SpyCatcher003-mi3 particles (SC3-mi3).
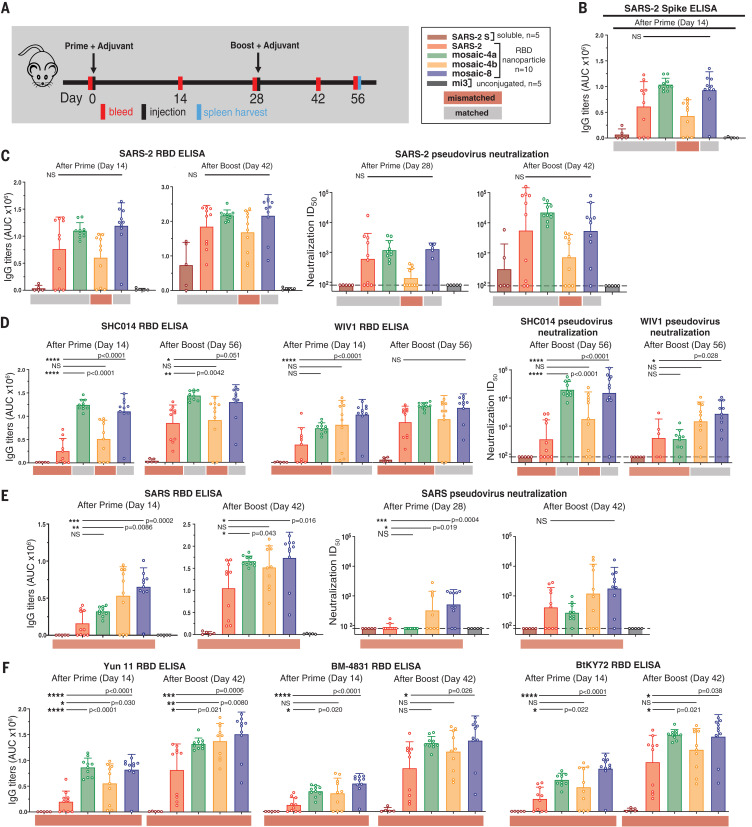
We immunized mice with soluble SARS-CoV-2 spike trimer (SARS-2 S), nanoparticles displaying only SARS-2 RBD (homotypic SARS-2), nanoparticles co-displaying RBDs (mosaic-4a, mosaic-4b, or mosaic-8), or unconjugated nanoparticles (mi3). Immunoglobulin G (IgG) responses were evaluated after prime or boost immunizations (Fig. 3A) by enzyme-linked immunosorbent assay (ELISA) against SARS-2 S (Fig. 3B) or a panel of RBDs (Fig. 3, C to F, and fig. S3). Sera from unconjugated nanoparticle-immunized animals (Fig. 3 and fig. S3, black) showed no responses above background. Anti–SARS-2 S trimer and anti–SARS-2 RBD serum responses were similar (Fig. 3, B and C), demonstrating that antibodies elicited against RBDs can access their epitopes on SARS-2 S trimer. We also conducted in vitro neutralization assays using a pseudotyped virus assay that quantitatively correlates with authentic virus neutralization (44) for strains known to infect 293TACE2 target cells (SARS-CoV-2, SARS, WIV1, and SHC104). Neutralization and ELISA titers were significantly correlated (fig. S4), which implies that ELISAs are predictive of neutralization results when viral entry receptor usage prevents accurate pseudotyped neutralization assays.
Fig. 3. RBD nanoparticles induce cross-reactive IgG responses in immunized mice.
Red and gray rectangles below ELISA and neutralization data represent mismatched strains (red; RBD from that strain was not present on the immunized particle) or matched strains (gray; RBD was present on the immunized particle). (A) Left: Immunization schedule; adjuvant is AddaVax (Invivogen). Right: Key for immunizations; number of mice in each cohort is indicated. (B to F) Neutralization and/or binding data for serum IgGs for recognition of (B) SARS-2 spike trimer, (C) SARS-2 RBD and SARS-2 pseudovirus, (D) SHC014 and WIV1 RBDs and corresponding pseudoviruses, (E) SARS RBD and SARS pseudovirus, and (F) Yun11, BM-4831, and BtKY72 RBDs. Mice were immunized with soluble SARS-CoV-2 S trimer (SARS-2 S; brown bars) or the following nanoparticles: homotypic SARS-2 (red), mosaic-4a (green), mosaic-4b (yellow), mosaic-8 (blue), or unconjugated SpyCatcher003-mi3 (mi3; black). ELISA data from serum IgG responses to SARS-2 spike trimer (B) or RBDs [(C) to (F)] are shown as area under the curve (AUC). For (C) to (E), neutralization potencies are presented as half-maximal inhibitory dilutions (ID50 values) of sera against the pseudoviruses from the indicated coronavirus strains. Dashed horizontal lines correspond to the lowest dilution representing the limit of detection. Each dot represents serum from one animal, with means and SDs for vaccinated cohorts denoted by rectangles and horizontal lines, respectively. Significant differences between groups linked by horizontal lines are indicated by asterisks and P values. NS, not significant.
Mice immunized with soluble SARS-2 S trimer showed no binding or neutralization except for autologous responses against SARS-2 after boosting (Fig. 3, C to F, brown bars). By contrast, sera from RBD nanoparticle–immunized animals exhibited binding to all RBDs (Fig. 3, C to F, red, green, yellow, and blue bars; fig. S3A) and neutralization against all four strains after boosting (Fig. 3, C to E), consistent with increased immunogenicities of multimerized antigen on nanoparticles versus soluble antigen (27, 28). Homotypic SARS-2 nanoparticles, but not soluble SARS-2 trimer, induced heterologous responses to zoonotic RBDs and neutralization of heterologous coronaviruses (Fig. 3, D to F). To address whether co-display of SARS-2 RBD along with other RBDs on mosaic-4a and mosaic-8 versus homotypic display of SARS-2 RBD (homotypic SARS-2) diminished anti–SARS-2 responses, we compared SARS-2–specific ELISA and neutralization titers for mosaic versus homotypic immunizations (Fig. 3C); there were no significant differences in IgG anti–SARS-2 titers for animals immunized with homotypic (Fig. 3C, red) versus mosaic nanoparticles (Fig. 3C, green and blue). Thus, in terms of the magnitude of immune response against SARS-2, there was no advantage of immunization with a homotypic RBD nanoparticle versus a mosaic nanoparticle that included SARS-2 RBD.
We next compared serum responses against matched RBDs (RBDs present on an injected nanoparticle) versus mismatched RBDs (RBDs not present on an injected nanoparticle) (Fig. 3 and fig. S3, gray and red horizontal shading, respectively). Although SARS-2 RBD was not presented on mosaic-4b, antibody titers elicited by mosaic-4b immunization (yellow) were not significantly different from titers elicited by matched nanoparticle immunizations [homotypic SARS-2 (red), mosaic-4a (green), and mosaic-8 (blue)], and sera from boosted mosaic-4b–immunized mice neutralized SARS-2 pseudovirus (Fig. 3C). In other matched versus mismatched comparisons, sera showed binding and neutralization of SHC014 and WIV1 regardless of whether these RBDs were included on the injected nanoparticle (Fig. 3D); this result implies sharing of common epitopes among RBDs (Fig. 1A).
In an experiment that demonstrated the advantages of mosaic versus homotypic SARS-2 nanoparticles, sera from mosaic-8–immunized mice bound SHC014 and WIV1 RBDs significantly better after priming than sera from homotypic SARS-2–immunized mice and retained better binding to SHC014 RBD after boosting (Fig. 3D). Thus, the potential increased avidity of the homotypic SARS-2 nanoparticle displaying only one type of RBD over the mosaic-8 nanoparticles did not confer increased breadth. Moreover, mosaic-8–immunized and boosted sera were more potent than sera from homotypic SARS-2–immunized animals in neutralizing SHC014 and WIV1 (Fig. 3D). Neutralization of the SHC014 and WIV1 pseudoviruses by mosaic-8 sera suggests that combining RBDs on a mosaic nanoparticle does not diminish the immune response against a particular RBD, as also suggested by ELISA binding of sera to Rs4081 and RaTG13 (fig. S3, A and B).
To further address whether RBD nanoparticles elicited antibodies that recognized totally mismatched strains and SARS-CoV-2 RBD mutants, we evaluated sera for binding to SARS, Yun11, BM-4831, and BtKY72 RBDs (Fig. 3, E and F); SARS-2 RBD mutants (fig. S3C); and MERS-CoV RBD (fig. S3D), as well as for neutralization in SARS pseudovirus assays (Fig. 3E). We found no reductions in SARS-2 RBD binding as a result of mutations [Y453F, the “Danish mink variant” (45), or a Q493K/Q498Y/P499T triple mutant (46)] (fig. S3C), no binding of any elicited sera to MERS-CoV RBD (fig. S3D), and higher and more cross-reactive antibody responses for mosaic immunizations than for homotypic SARS-2 immunizations (e.g., mosaic-8–primed and boosted animals showed significantly higher titers against SARS RBD than did sera from homotypic SARS-2–immunized mice) (Fig. 3E). After priming, sera from the homotypic SARS-2–immunized animals did not neutralize SARS, whereas the mosaic-4b and mosaic-8 sera were neutralizing (Fig. 3E), perhaps because these nanoparticles included WIV1 RBD, which is related by 95% amino acid identity to SARS RBD (Fig. 1D). After boosting, SARS-2 and mosaic-4a sera were also neutralizing, although titers were lower than for mosaic-8–immunized animals by a factor of ~4 (Fig. 3E). ELISA titers against other mismatched RBDs (Yun11, BM-4831, and BtKY72) were significantly higher for sera collected after mosaic-8 priming than for sera from homotypic SARS-2 priming, and heightened binding was retained after boosting (Fig. 3F). Thus, relative to homotypic SARS-2 nanoparticles, mosaic nanoparticles (particularly mosaic-8) induce higher antibody titers against mismatched RBDs, This is another finding that favors the co-display approach for inducing broader anti-coronavirus responses, especially after a single prime.
Using flow cytometry, we investigated the potential for cross-reactive recognition—specifically, whether B cell receptors on IgG+ splenic B cells from RBD nanoparticle–boosted animals could simultaneously recognize RBDs from SARS-2 and Rs4081 (related by 70% sequence identity) (Fig. 1D and fig. S5). Whereas control animals were negative, all other groups showed B cells that recognized SARS-2 and Rs4081 RBDs simultaneously, suggesting the existence of antibodies that cross-react with both RBDs (fig. S5E).
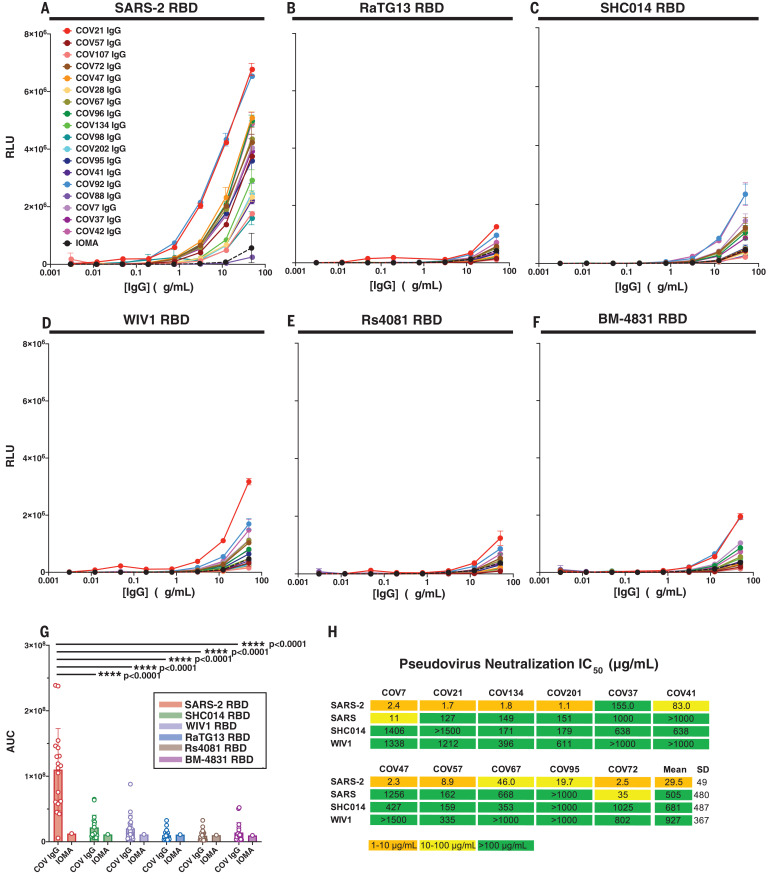
To compare antibodies elicited by RBD nanoparticle immunization to antibodies elicited by SARS-CoV-2 infection, we repeated ELISAs against the RBD panel using IgGs from COVID-19 plasma donors (47) (Fig. 4). Most of the convalescent plasmas showed detectable binding to SARS-2 RBD (Fig. 4A). However, binding to other sarbecovirus RBDs (RaTG13, SHC014, WIV1, Rs4081, and BM-4831) was significantly weaker than binding to SARS 2 RBD, with many human plasma IgGs showing no binding above background (Fig. 4, B to G). In addition, although convalescent plasma IgGs neutralized SARS-CoV-2 pseudoviruses, they showed weak or no neutralization of SARS, SHC014, or WIV1 pseudoviruses (Fig. 4H). These results are consistent with little to no cross-reactive recognition of RBDs from zoonotic coronavirus strains resulting from SARS-CoV-2 infection in humans.
Fig. 4. IgGs from convalescent COVID-19 plasma show little to no cross-reactive responses.
(A to F) Plasma IgG (18, 24) responses were evaluated by ELISA [data shown as binding curves with plasma names (18) listed] against RBDs from (A) SARS-2, (B) RaTG13, (C) SHC014, (D) WIV1, (E) Rs4081, and (F) BM-4831. Data points are plotted as means ± SD of duplicate measurements. IOMA, an anti–HIV-1 IgG (50), was used as a control. (G) ELISA results from (A) to (F), presented as AUC; each dot represents one plasma sample, with means ± SD shown as colored bars. Significant differences between groups linked by horizontal lines are indicated by asterisks and P values. (H) IC50 values for pseudotyped neutralization assays using IgGs from COV7, COV21, and COV72 plasmas (18) (evaluated at top concentrations of 1500 μg/ml) against the indicated strains. Mean = arithmetic mean IC50.
Our results confirm that multimerization of RBDs on nanoparticles enhances immunogenicity relative to soluble antigen (33, 48). We found that homotypic SARS-2 nanoparticle immunization produces IgG responses that bind zoonotic RBDs and neutralize heterologous coronaviruses after boosting. By contrast, soluble SARS-2 S immunization and natural infection with SARS-CoV-2 resulted in weak or no heterologous responses in plasmas. Co-display of SARS-2 RBD along with diverse RBDs on mosaic nanoparticles showed no disadvantages for eliciting neutralizing antibodies against SARS-CoV-2 relative to homotypic SARS-2 nanoparticles; therefore, mosaic nanoparticles may represent a candidate vaccine to protect against COVID-19. Furthermore, relative to homotypic SARS-2 RBD particles, the mosaic co-display strategy demonstrated advantages for eliciting neutralizing antibodies against zoonotic sarbecoviruses, thus potentially also providing protection against emerging coronaviruses with human spillover potential. Neutralization of matched and mismatched strains was observed after mosaic priming; hence, a single injection of a mosaic RBD nanoparticle might be sufficient in a vaccine. Because COVID-19 convalescent plasmas showed little to no recognition of coronavirus RBDs other than SARS-CoV-2, COVD-19–induced immunity in humans may not protect against another emergent coronavirus. However, the mosaic nanoparticles described here could be used as described or easily adapted so that they present RBDs from newly discovered zoonotic coronaviruses.
Acknowledgments
We thank K. Brune (Genie Biotech) for advice about mi3 production; J. Bloom (Fred Hutchinson) and P. Bieniasz (Rockefeller University) for neutralization assay reagents; J. Vielmetter and Caltech’s Beckman Institute Protein Expression Center for protein production; A. Flyak for help with flow cytometry; M. Murphy for figures; COVID-19 plasma donors, B. Coller, S. Schlesinger, and the Rockefeller University Hospital Clinical Research Support Office and nursing staff; and A. Flyak and A. DeLaitsch for critical reading of the manuscript. Funding: Supported by NIH grant P01-AI138938-S1 (P.J.B. and M.C.N.), the Caltech Merkin Institute for Translational Research (P.J.B.), a George Mason University Fast Grant (P.J.B.), and the Medical Research Council (MR/P001351/1) (M.H.) (this UK-funded award is part of the EDCTP2 program supported by the European Union). M.C.N. is a Howard Hughes Medical Institute Investigator. Author contributions: A.A.C., C.O.B., and P.J.B. conceived and designed experiments; A.A.C., P.N.P.G., Y.E.L., P.R.H., S.O., and L.M.K. performed experiments; H.-J.W. generated and validated SpyCatcher003-mi3; M.H. supervised the generation and validation of SpyCatcher003-mi3; and A.A.C., J.R.K., A.P.W., C.O.B., M.C.N., and P.J.B. analyzed data and wrote the paper with contributions from other authors. Competing interests: M.H. is an inventor on a patent on SpyTag/SpyCatcher (EP2534484) and a patent application on SpyTag003:SpyCatcher003 (UK Intellectual Property Office 1706430.4), as well as a SpyBiotech cofounder, shareholder, and consultant. P.J.B. and A.A.C. are inventors on a provisional application from the California Institute of Technology that covers the mosaic nanoparticles described in this work. Data and materials availability: All data are available in the main text or the supplementary materials. Materials are available upon request to the corresponding author with a signed material transfer agreement. This work is licensed under a Creative Commons Attribution 4.0 International (CC BY 4.0) license, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. To view a copy of this license, visit https://creativecommons.org/licenses/by/4.0/. This license does not apply to figures/photos/artwork or other content included in the article that is credited to a third party; obtain authorization from the rights holder before using such material.
Supplementary Materials
science.sciencemag.org/content/371/6530/735/suppl/DC1
Materials and Methods
Figs. S1 to S5
MDAR Reproducibility Checklist
References and Notes
- 1.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L., A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020). 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E. C., Zhang Y.-Z., A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269 (2020). 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Wit E., van Doremalen N., Falzarano D., Munster V. J., SARS and MERS: Recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 14, 523–534 (2016). 10.1038/nrmicro.2016.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J. H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B. T., Zhang S., Wang L. F., Bats are natural reservoirs of SARS-like coronaviruses. Science 310, 676–679 (2005). 10.1126/science.1118391 [DOI] [PubMed] [Google Scholar]
- 5.Wang N., Li S.-Y., Yang X.-L., Huang H.-M., Zhang Y.-J., Guo H., Luo C.-M., Miller M., Zhu G., Chmura A. A., Hagan E., Zhou J.-H., Zhang Y.-Z., Wang L.-F., Daszak P., Shi Z.-L., Serological Evidence of Bat SARS-Related Coronavirus Infection in Humans, China. Virol. Sin. 33, 104–107 (2018). 10.1007/s12250-018-0012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Menachery V. D., Yount B. L. Jr.., Debbink K., Agnihothram S., Gralinski L. E., Plante J. A., Graham R. L., Scobey T., Ge X.-Y., Donaldson E. F., Randell S. H., Lanzavecchia A., Marasco W. A., Shi Z.-L., Baric R. S., A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat. Med. 21, 1508–1513 (2015). 10.1038/nm.3985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Menachery V. D., Yount B. L. Jr.., Sims A. C., Debbink K., Agnihothram S. S., Gralinski L. E., Graham R. L., Scobey T., Plante J. A., Royal S. R., Swanstrom J., Sheahan T. P., Pickles R. J., Corti D., Randell S. H., Lanzavecchia A., Marasco W. A., Baric R. S., SARS-like WIV1-CoV poised for human emergence. Proc. Natl. Acad. Sci. U.S.A. 113, 3048–3053 (2016). 10.1073/pnas.1517719113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li W., Moore M. J., Vasilieva N., Sui J., Wong S. K., Berne M. A., Somasundaran M., Sullivan J. L., Luzuriaga K., Greenough T. C., Choe H., Farzan M., Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426, 450–454 (2003). 10.1038/nature02145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T. S., Herrler G., Wu N.-H., Nitsche A., Müller M. A., Drosten C., Pöhlmann S., SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 181, 271–280.e8 (2020). 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raj V. S., Mou H., Smits S. L., Dekkers D. H. W., Müller M. A., Dijkman R., Muth D., Demmers J. A. A., Zaki A., Fouchier R. A. M., Thiel V., Drosten C., Rottier P. J. M., Osterhaus A. D. M. E., Bosch B. J., Haagmans B. L., Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 495, 251–254 (2013). 10.1038/nature12005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vlasak R., Luytjes W., Spaan W., Palese P., Human and bovine coronaviruses recognize sialic acid-containing receptors similar to those of influenza C viruses. Proc. Natl. Acad. Sci. U.S.A. 85, 4526–4529 (1988). 10.1073/pnas.85.12.4526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang X., Dong W., Milewska A., Golda A., Qi Y., Zhu Q. K., Marasco W. A., Baric R. S., Sims A. C., Pyrc K., Li W., Sui J., Human Coronavirus HKU1 Spike Protein Uses O-Acetylated Sialic Acid as an Attachment Receptor Determinant and Employs Hemagglutinin-Esterase Protein as a Receptor-Destroying Enzyme. J. Virol. 89, 7202–7213 (2015). 10.1128/JVI.00854-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fung T. S., Liu D. X., Human Coronavirus: Host-Pathogen Interaction. Annu. Rev. Microbiol. 73, 529–557 (2019). 10.1146/annurev-micro-020518-115759 [DOI] [PubMed] [Google Scholar]
- 14.Brouwer P. J. M., Caniels T. G., van der Straten K., Snitselaar J. L., Aldon Y., Bangaru S., Torres J. L., Okba N. M. A., Claireaux M., Kerster G., Bentlage A. E. H., van Haaren M. M., Guerra D., Burger J. A., Schermer E. E., Verheul K. D., van der Velde N., van der Kooi A., van Schooten J., van Breemen M. J., Bijl T. P. L., Sliepen K., Aartse A., Derking R., Bontjer I., Kootstra N. A., Wiersinga W. J., Vidarsson G., Haagmans B. L., Ward A. B., de Bree G. J., Sanders R. W., van Gils M. J., Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science 369, 643–650 (2020). 10.1126/science.abc5902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., Chai X., He R., Li X., Lv Q., Zhu H., Deng W., Xu Y., Wang Y., Qiao L., Tan Y., Song L., Wang G., Du X., Gao N., Liu J., Xiao J., Su X. D., Du Z., Feng Y., Qin C., Qin C., Jin R., Xie X. S., Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell 182, 73–84.e16 (2020). 10.1016/j.cell.2020.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kreer C., Zehner M., Weber T., Ercanoglu M. S., Gieselmann L., Rohde C., Halwe S., Korenkov M., Schommers P., Vanshylla K., Di Cristanziano V., Janicki H., Brinker R., Ashurov A., Krähling V., Kupke A., Cohen-Dvashi H., Koch M., Eckert J. M., Lederer S., Pfeifer N., Wolf T., Vehreschild M. J. G. T., Wendtner C., Diskin R., Gruell H., Becker S., Klein F., Longitudinal Isolation of Potent Near-Germline SARS-CoV-2-Neutralizing Antibodies from COVID-19 Patients. Cell 182, 843–854.e12 (2020). 10.1016/j.cell.2020.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu L., Wang P., Nair M. S., Yu J., Rapp M., Wang Q., Luo Y., Chan J. F.-W., Sahi V., Figueroa A., Guo X. V., Cerutti G., Bimela J., Gorman J., Zhou T., Chen Z., Yuen K.-Y., Kwong P. D., Sodroski J. G., Yin M. T., Sheng Z., Huang Y., Shapiro L., Ho D. D., Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 584, 450–456 (2020). 10.1038/s41586-020-2571-7 [DOI] [PubMed] [Google Scholar]
- 18.Robbiani D. F., Gaebler C., Muecksch F., Lorenzi J. C. C., Wang Z., Cho A., Agudelo M., Barnes C. O., Gazumyan A., Finkin S., Hägglöf T., Oliveira T. Y., Viant C., Hurley A., Hoffmann H.-H., Millard K. G., Kost R. G., Cipolla M., Gordon K., Bianchini F., Chen S. T., Ramos V., Patel R., Dizon J., Shimeliovich I., Mendoza P., Hartweger H., Nogueira L., Pack M., Horowitz J., Schmidt F., Weisblum Y., Michailidis E., Ashbrook A. W., Waltari E., Pak J. E., Huey-Tubman K. E., Koranda N., Hoffman P. R., West A. P. Jr.., Rice C. M., Hatziioannou T., Bjorkman P. J., Bieniasz P. D., Caskey M., Nussenzweig M. C., Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 584, 437–442 (2020). 10.1038/s41586-020-2456-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., Gao G., Hu X., Zhang Y., Tong Z., Huang W., Liu W. J., Wu G., Zhang B., Wang L., Qi J., Feng H., Wang F.-S., Wang Q., Gao G. F., Yuan Z., Yan J., A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature 584, 120–124 (2020). 10.1038/s41586-020-2381-y [DOI] [PubMed] [Google Scholar]
- 20.Zost S. J., Gilchuk P., Chen R. E., Case J. B., Reidy J. X., Trivette A., Nargi R. S., Sutton R. E., Suryadevara N., Chen E. C., Binshtein E., Shrihari S., Ostrowski M., Chu H. Y., Didier J. E., MacRenaris K. W., Jones T., Day S., Myers L., Eun-Hyung Lee F., Nguyen D. C., Sanz I., Martinez D. R., Rothlauf P. W., Bloyet L. M., Whelan S. P. J., Baric R. S., Thackray L. B., Diamond M. S., Carnahan R. H., Crowe J. E. Jr., Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. Nat. Med. 26, 1422–1427 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rogers T. F., Zhao F., Huang D., Beutler N., Burns A., He W. T., Limbo O., Smith C., Song G., Woehl J., Yang L., Abbott R. K., Callaghan S., Garcia E., Hurtado J., Parren M., Peng L., Ricketts J., Ricciardi M. J., Rawlings S. A., Smith D. M., Nemazee D., Teijaro J. R., Voss J. E., Andrabi R., Briney B., Landais E., Sok D., Jardine J. G., Burton D. R., Rapid isolation of potent SARS-CoV-2 neutralizing antibodies and protection in a small animal model. Science 369, 956–963 (2020). 10.1126/science.abc7520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seydoux E., Homad L. J., MacCamy A. J., Parks K. R., Hurlburt N. K., Jennewein M. F., Akins N. R., Stuart A. B., Wan Y.-H., Feng J., Whaley R. E., Singh S., Boeckh M., Cohen K. W., McElrath M. J., Englund J. A., Chu H. Y., Pancera M., McGuire A. T., Stamatatos L., Analysis of a SARS-CoV-2-Infected Individual Reveals Development of Potent Neutralizing Antibodies with Limited Somatic Mutation. Immunity 53, 98–105.e5 (2020). 10.1016/j.immuni.2020.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zost S. J., Gilchuk P., Case J. B., Binshtein E., Chen R. E., Nkolola J. P., Schäfer A., Reidy J. X., Trivette A., Nargi R. S., Sutton R. E., Suryadevara N., Martinez D. R., Williamson L. E., Chen E. C., Jones T., Day S., Myers L., Hassan A. O., Kafai N. M., Winkler E. S., Fox J. M., Shrihari S., Mueller B. K., Meiler J., Chandrashekar A., Mercado N. B., Steinhardt J. J., Ren K., Loo Y.-M., Kallewaard N. L., McCune B. T., Keeler S. P., Holtzman M. J., Barouch D. H., Gralinski L. E., Baric R. S., Thackray L. B., Diamond M. S., Carnahan R. H., Crowe J. E. Jr., Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature 584, 443–449 (2020). 10.1038/s41586-020-2548-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barnes C. O., Jette C. A., Abernathy M. E., Dam K. A., Esswein S. R., Gristick H. B., Malyutin A. G., Sharaf N. G., Huey-Tubman K. E., Lee Y. E., Robbiani D. F., Nussenzweig M. C., West A. P. Jr.., Bjorkman P. J., SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 588, 682–687 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pinto D., Park Y.-J., Beltramello M., Walls A. C., Tortorici M. A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., Peter A., Guarino B., Spreafico R., Cameroni E., Case J. B., Chen R. E., Havenar-Daughton C., Snell G., Telenti A., Virgin H. W., Lanzavecchia A., Diamond M. S., Fink K., Veesler D., Corti D., Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 583, 290–295 (2020). 10.1038/s41586-020-2349-y [DOI] [PubMed] [Google Scholar]
- 26.Piccoli L., Park Y. J., Tortorici M. A., Czudnochowski N., Walls A. C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L. E., Bowen J. E., Acton O. J., Jaconi S., Guarino B., Minola A., Zatta F., Sprugasci N., Bassi J., Peter A., De Marco A., Nix J. C., Mele F., Jovic S., Rodriguez B. F., Gupta S. V., Jin F., Piumatti G., Lo Presti G., Pellanda A. F., Biggiogero M., Tarkowski M., Pizzuto M. S., Cameroni E., Havenar-Daughton C., Smithey M., Hong D., Lepori V., Albanese E., Ceschi A., Bernasconi E., Elzi L., Ferrari P., Garzoni C., Riva A., Snell G., Sallusto F., Fink K., Virgin H. W., Lanzavecchia A., Corti D., Veesler D., Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell 183, 1024–1042.e21 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.López-Sagaseta J., Malito E., Rappuoli R., Bottomley M. J., Self-assembling protein nanoparticles in the design of vaccines. Comput. Struct. Biotechnol. J. 14, 58–68 (2015). 10.1016/j.csbj.2015.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slifka M. K., Amanna I. J., Role of Multivalency and Antigenic Threshold in Generating Protective Antibody Responses. Front. Immunol. 10, 956 (2019). 10.3389/fimmu.2019.00956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brune K. D., Howarth M., New Routes and Opportunities for Modular Construction of Particulate Vaccines: Stick, Click, and Glue. Front. Immunol. 9, 1432 (2018). 10.3389/fimmu.2018.01432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brune K. D., Leneghan D. B., Brian I. J., Ishizuka A. S., Bachmann M. F., Draper S. J., Biswas S., Howarth M., Plug-and-Display: Decoration of Virus-Like Particles via isopeptide bonds for modular immunization. Sci. Rep. 6, 19234 (2016). 10.1038/srep19234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bruun T. U. J., Andersson A. C., Draper S. J., Howarth M., Engineering a Rugged Nanoscaffold To Enhance Plug-and-Display Vaccination. ACS Nano 12, 8855–8866 (2018). 10.1021/acsnano.8b02805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zakeri B., Fierer J. O., Celik E., Chittock E. C., Schwarz-Linek U., Moy V. T., Howarth M., Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl. Acad. Sci. U.S.A. 109, E690–E697 (2012). 10.1073/pnas.1115485109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.T. K. Tan et al., A COVID-19 vaccine candidate 1 using SpyCatcher multimerization of the SARS-CoV-2 spike protein receptor-binding domain induces potent neutralising antibody responses. bioRxiv 275701 [preprint]. 31 August 2020. [DOI] [PMC free article] [PubMed]
- 34.Zhang B., Chao C. W., Tsybovsky Y., Abiona O. M., Hutchinson G. B., Moliva J. I., Olia A. S., Pegu A., Phung E., Stewart-Jones G. B. E., Verardi R., Wang L., Wang S., Werner A., Yang E. S., Yap C., Zhou T., Mascola J. R., Sullivan N. J., Graham B. S., Corbett K. S., Kwong P. D., A platform incorporating trimeric antigens into self-assembling nanoparticles reveals SARS-CoV-2-spike nanoparticles to elicit substantially higher neutralizing responses than spike alone. Sci. Rep. 10, 18149 (2020). 10.1038/s41598-020-74949-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu B., Zeng L. P., Yang X. L., Ge X. Y., Zhang W., Li B., Xie J. Z., Shen X. R., Zhang Y. Z., Wang N., Luo D. S., Zheng X. S., Wang M. N., Daszak P., Wang L. F., Cui J., Shi Z. L., Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLOS Pathog. 13, e1006698 (2017). 10.1371/journal.ppat.1006698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rahikainen R., Rijal P., Tan T. K., Wu H. J., Andersson A. C., Barrett J. R., Bowden T. A., Draper S. J., Townsend A. R., Howarth M., Overcoming Symmetry Mismatch in Vaccine Nanoassembly through Spontaneous Amidation. Angew. Chem. Int. Ed. 60, 321–330 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanekiyo M., Joyce M. G., Gillespie R. A., Gallagher J. R., Andrews S. F., Yassine H. M., Wheatley A. K., Fisher B. E., Ambrozak D. R., Creanga A., Leung K., Yang E. S., Boyoglu-Barnum S., Georgiev I. S., Tsybovsky Y., Prabhakaran M. S., Andersen H., Kong W.-P., Baxa U., Zephir K. L., Ledgerwood J. E., Koup R. A., Kwong P. D., Harris A. K., McDermott A. B., Mascola J. R., Graham B. S., Mosaic nanoparticle display of diverse influenza virus hemagglutinins elicits broad B cell responses. Nat. Immunol. 20, 362–372 (2019). 10.1038/s41590-018-0305-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Letko M., Marzi A., Munster V., Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 5, 562–569 (2020). 10.1038/s41564-020-0688-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lam T. T.-Y., Jia N., Zhang Y.-W., Shum M. H.-H., Jiang J.-F., Zhu H.-C., Tong Y.-G., Shi Y.-X., Ni X.-B., Liao Y.-S., Li W.-J., Jiang B.-G., Wei W., Yuan T.-T., Zheng K., Cui X.-M., Li J., Pei G.-Q., Qiang X., Cheung W. Y.-M., Li L.-F., Sun F.-F., Qin S., Huang J.-C., Leung G. M., Holmes E. C., Hu Y.-L., Guan Y., Cao W.-C., Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature 583, 282–285 (2020). 10.1038/s41586-020-2169-0 [DOI] [PubMed] [Google Scholar]
- 40.Andersen K. G., Rambaut A., Lipkin W. I., Holmes E. C., Garry R. F., The proximal origin of SARS-CoV-2. Nat. Med. 26, 450–452 (2020). 10.1038/s41591-020-0820-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou H., Chen X., Hu T., Li J., Song H., Liu Y., Wang P., Liu D., Yang J., Holmes E. C., Hughes A. C., Bi Y., Shi W., A Novel Bat Coronavirus Closely Related to SARS-CoV-2 Contains Natural Insertions at the S1/S2 Cleavage Site of the Spike Protein. Curr. Biol. 30, 2196–2203.e3 (2020). 10.1016/j.cub.2020.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tao Y., Tong S., Complete Genome Sequence of a Severe Acute Respiratory Syndrome-Related Coronavirus from Kenyan Bats. Microbiol. Resour. Announce. 8, e00548-19 (2019). 10.1128/MRA.00548-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keeble A. H., Turkki P., Stokes S., Khairil Anuar I. N. A., Rahikainen R., Hytönen V. P., Howarth M., Approaching infinite affinity through engineering of peptide-protein interaction. Proc. Natl. Acad. Sci. U.S.A. 116, 26523–26533 (2019). 10.1073/pnas.1909653116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmidt F., Weisblum Y., Muecksch F., Hoffmann H.-H., Michailidis E., Lorenzi J. C. C., Mendoza P., Rutkowska M., Bednarski E., Gaebler C., Agudelo M., Cho A., Wang Z., Gazumyan A., Cipolla M., Caskey M., Robbiani D. F., Nussenzweig M. C., Rice C. M., Hatziioannou T., Bieniasz P. D., Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J. Exp. Med. 217, e20201181 (2020). 10.1084/jem.20201181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.L. van Dorp et al., Recurrent mutations in SARS-CoV-2 genomes isolated from mink point to rapid host-adaptation. bioRxiv 384743 [preprint]. 16 November 2020.384743
- 46.Leist S. R., Dinnon K. H. 3rd, Schäfer A., Tse L. V., Okuda K., Hou Y. J., West A., Edwards C. E., Sanders W., Fritch E. J., Gully K. L., Scobey T., Brown A. J., Sheahan T. P., Moorman N. J., Boucher R. C., Gralinski L. E., Montgomery S. A., Baric R. S., A Mouse-Adapted SARS-CoV-2 Induces Acute Lung Injury and Mortality in Standard Laboratory Mice. Cell 183, 1070–1085.e12 (2020). 10.1016/j.cell.2020.09.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barnes C. O., West A. P. Jr.., Huey-Tubman K. E., Hoffmann M. A. G., Sharaf N. G., Hoffman P. R., Koranda N., Gristick H. B., Gaebler C., Muecksch F., Lorenzi J. C. C., Finkin S., Hägglöf T., Hurley A., Millard K. G., Weisblum Y., Schmidt F., Hatziioannou T., Bieniasz P. D., Caskey M., Robbiani D. F., Nussenzweig M. C., Bjorkman P. J., Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies. Cell 182, 828–842.e16 (2020). 10.1016/j.cell.2020.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walls A. C., Fiala B., Schäfer A., Wrenn S., Pham M. N., Murphy M., Tse L. V., Shehata L., O’Connor M. A., Chen C., Navarro M. J., Miranda M. C., Pettie D., Ravichandran R., Kraft J. C., Ogohara C., Palser A., Chalk S., Lee E.-C., Guerriero K., Kepl E., Chow C. M., Sydeman C., Hodge E. A., Brown B., Fuller J. T., Dinnon K. H. 3rd, Gralinski L. E., Leist S. R., Gully K. L., Lewis T. B., Guttman M., Chu H. Y., Lee K. K., Fuller D. H., Baric R. S., Kellam P., Carter L., Pepper M., Sheahan T. P., Veesler D., King N. P., Elicitation of Potent Neutralizing Antibody Responses by Designed Protein Nanoparticle Vaccines for SARS-CoV-2. Cell 183, 1367–1382.e17 (2020). 10.1016/j.cell.2020.10.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Landau M., Mayrose I., Rosenberg Y., Glaser F., Martz E., Pupko T., Ben-Tal N., ConSurf 2005: The projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 33, W299–W302 (2005). 10.1093/nar/gki370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gristick H. B., von Boehmer L., West A. P. Jr.., Schamber M., Gazumyan A., Golijanin J., Seaman M. S., Fätkenheuer G., Klein F., Nussenzweig M. C., Bjorkman P. J., Natively glycosylated HIV-1 Env structure reveals new mode for antibody recognition of the CD4-binding site. Nat. Struct. Mol. Biol. 23, 906–915 (2016). 10.1038/nsmb.3291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., Higgins D. G., Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011). 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guindon S., Dufayard J.-F., Lefort V., Anisimova M., Hordijk W., Gascuel O., New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010). 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- 53.Hsieh C.-L., Goldsmith J. A., Schaub J. M., DiVenere A. M., Kuo H.-C., Javanmardi K., Le K. C., Wrapp D., Lee A. G., Liu Y., Chou C.-W., Byrne P. O., Hjorth C. K., Johnson N. V., Ludes-Meyers J., Nguyen A. W., Park J., Wang N., Amengor D., Lavinder J. J., Ippolito G. C., Maynard J. A., Finkelstein I. J., McLellan J. S., Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science 369, 1501–1505 (2020). 10.1126/science.abd0826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.A. A. Cohen et al., Construction, characterization, and immunization of nanoparticles that display a diverse array of influenza HA trimers. bioRxiv 911388 [preprint]. 19 January 2020. [DOI] [PMC free article] [PubMed]
- 55.Angeletti D., Gibbs J. S., Angel M., Kosik I., Hickman H. D., Frank G. M., Das S. R., Wheatley A. K., Prabhakaran M., Leggat D. J., McDermott A. B., Yewdell J. W., Defining B cell immunodominance to viruses. Nat. Immunol. 18, 456–463 (2017). 10.1038/ni.3680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Crawford K. H. D., Eguia R., Dingens A. S., Loes A. N., Malone K. D., Wolf C. R., Chu H. Y., Tortorici M. A., Veesler D., Murphy M., Pettie D., King N. P., Balazs A. B., Bloom J. D., Protocol and Reagents for Pseudotyping Lentiviral Particles with SARS-CoV-2 Spike Protein for Neutralization Assays. Viruses 12, 513 (2020). 10.3390/v12050513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.West A. P. Jr.., Scharf L., Horwitz J., Klein F., Nussenzweig M. C., Bjorkman P. J., Computational analysis of anti-HIV-1 antibody neutralization panel data to identify potential functional epitope residues. Proc. Natl. Acad. Sci. U.S.A. 110, 10598–10603 (2013). 10.1073/pnas.1309215110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Starr T. N., Greaney A. J., Hilton S. K., Ellis D., Crawford K. H. D., Dingens A. S., Navarro M. J., Bowen J. E., Tortorici M. A., Walls A. C., King N. P., Veesler D., Bloom J. D., Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell 182, 1295–1310.e20 (2020). 10.1016/j.cell.2020.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
science.sciencemag.org/content/371/6530/735/suppl/DC1
Materials and Methods
Figs. S1 to S5
MDAR Reproducibility Checklist