Abstract
As the coronavirus disease 2019 (COVID-19) pandemic continues to rise and second waves are reported in some countries, serological test kits and strips are being considered to scale up an adequate laboratory response. This study provides an update on the kinetics of humoral immune response to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection and performance characteristics of serological protocols (lateral flow assay [LFA], chemiluminescence immunoassay [CLIA] and ELISA) used for evaluations of recent and past SARS-CoV-2 infection. A thorough and comprehensive review of suitable and eligible full-text articles was performed on PubMed, Scopus, Web of Science, Wordometer and medRxiv from 10 January to 16 July 2020. These articles were searched using the Medical Subject Headings terms ‘COVID-19’, ‘Serological assay’, ‘Laboratory Diagnosis’, ‘Performance characteristics’, ‘POCT’, ‘LFA’, ‘CLIA’, ‘ELISA’ and ‘SARS-CoV-2’. Data from original research articles on SARS-CoV-2 antibody detection ≥second day postinfection were included in this study. In total, there were 7938 published articles on humoral immune response and laboratory diagnosis of COVID-19. Of these, 74 were included in this study. The detection, peak and decline period of blood anti-SARS-CoV-2 IgM, IgG and total antibodies for point-of-care testing (POCT), ELISA and CLIA vary widely. The most promising of these assays for POCT detected anti-SARS-CoV-2 at day 3 postinfection and peaked on the 15th day; ELISA products detected anti-SARS-CoV-2 IgM and IgG at days 2 and 6 then peaked on the eighth day; and the most promising CLIA product detected anti-SARS-CoV-2 at day 1 and peaked on the 30th day. The most promising LFA, ELISA and CLIA that had the best performance characteristics were those targeting total SARS-CoV-2 antibodies followed by those targeting anti-SARS-CoV-2 IgG then IgM. Essentially, the CLIA-based SARS-CoV-2 tests had the best performance characteristics, followed by ELISA then POCT. Given the varied performance characteristics of all the serological assays, there is a need to continuously improve their detection thresholds, as well as to monitor and re-evaluate their performances to assure their significance and applicability for COVID-19 clinical and epidemiological purposes.
Keywords: COVID-19 serology, diagnostics, laboratory tests, SARS-CoV-2
Introduction
The coronavirus disease 2019 (COVID-19) pandemic has caused an unprecedented global health emergency and economic uncertainty. As the incidence of COVID-19 continues to rise, many countries have sought to develop or procure serological test kits and strips with the plan of scaling up laboratory investigations into the COVID-19 pandemic.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the etiological agent of COVID-19. It is one of the three highly pathogenic members of the family of coronaviridae.1 Infection with SARS-CoV-2 has been associated with a range of hallmarks that progress from mild to severe clinical presentations before terminating in death in less than 10% of cases.2
The WHO has recommended RT-PCR as the gold standard protocol for screening individuals with typical symptoms who are suspected of having COVID-19. Although appropriate use of RT-PCR provides very accurate results, test reagents and consumables are mostly in short supply. Besides, this protocol is laborious, expensive to operate, requiring technical expertise and it has a long test turnaround time. Also, one of the major technical drawbacks in using RT-PCR is the significant number of cases of false-negative results, despite patients having clinical features and radiologic findings that are highly suspicious of SARS-CoV-2 infection. The false-negative results could be due to wrong sampling, where SARS-CoV-2 might have been present in the lower respiratory tract rather than upper respiratory tract samples often collected for laboratory diagnosis. This poses a challenge in the proper evaluation of some SARS-CoV-2-infected people.3
It has been observed that the transmission dynamics of COVID-19 have made it an arduous task and challenge in the control of the pandemic, despite WHO-proposed measures having already been introduced.4 Consequently, the COVID-19 pandemic has seriously challenged the operation of the entire healthcare system, including hospitals, laboratory diagnosis, the management of patients and every other aspect of human endeavor.5,6
In the quest to augment several lapses in the use of RT-PCR testing for COVID-19, serological assays that detect and /or measure antibodies (immunoglobulins) against SARS-CoV-2 have been developed and evaluated for performance by many institutions and private biotechnology firms. Global efforts to scale up the testing and diagnosis of COVID-19 has led to the commercial production of serological kits and devices. Some of these products have gained executive approval in some countries. For instance, the US Food and Drug Administration (FDA) gave expedient approval for some COVID-19 serological kits based on their accuracy and reliability.7
Instances have arisen where massive production and the use of finger-prick assays and in vitro testing have been encouraged in the UK and the USA to scale up COVID-19 surveillance through rapid testing and measurement of either antigens or antibodies to SARS-CoV-2. These rapid testing protocols adopted in these countries are point-of-care testing (POCT), which are designed as lateral flow devices (colloid gold-based immunochromatographic cassettes or test strips) with a diverse range of performance characteristics. These devices require a small sample volume (in microliters), are conducted within a short period (a few seconds to minutes), and are easier to perform as their use requires less technical expertise and equipment compared with protocols that detect nucleic acid.8
The transmission dynamics of the COVID-19 pandemic make it very challenging to control despite measures put in place in various countries of Africa and elsewhere outside the continent. Adequate laboratory diagnosis of COVID-19 plays a highly significant role in the control and prevention of the pandemic. However, some of the emerging challenges of testing for SARS-CoV-2 generally include sourcing personal protective equipment, low human capacity, scaling up testing, overwhelming contact tracing and inadequate hospital capacity to accommodate COVID-19 patients, resulting in increased morbidity and mortality. Hence, improved testing capacity, adequate provision of human and material resources, combined with innovative ways of scaling up contact tracing and improved testing capacity, are essential in the control of the COVID-19 pandemic.
This study sought to provide an update on the kinetics of humoral immune response to SARS-CoV-2 infection and performance characteristics of serological protocols (lateral flow assay [LFA], chemiluminescence immunoassay [CLIA] and ELISA) used for evaluations of recent and past SARS-CoV-2 infection. Data from original research articles on SARS-CoV-2 antibody detection ≥second day postinfection were included in this study. Furthermore, this study examined whether these tests could be possible solutions that can ameliorate the constraints of underdiagnosis in resource-limited settings.
This review is conducted under the following sections:
Virology and structural organization of SARS CoV-2 useful in molecular and serological diagnosis.
Humoral immune response to SARS CoV-2.
COVID-19 serological assays.
Challenges of SARS-CoV-2 serological testing.
Accuracy and applicability of COVID-19 serological assays.
Performance characteristics of COVID-19 serological assays.
Article selection criteria
Search strategy
A thorough and comprehensive review of suitable and eligible full-text articles was performed on PubMed, Scopus, Web of Science, Wordometer and medRxiv from 10 January to 16 July 2020. These articles were searched using the MeSH terms ‘COVID-19’, ‘Serological assay’, ‘Laboratory Diagnosis’, ‘Performance characteristics’, ‘POCT’, ‘CLIA’, ‘ELISA’ and ‘SARS-CoV-2’.
Article evaluation and data extraction
Eight authors independently evaluated and scrutinized titles and abstracts to prospective studies to check for potentially eligible articles and to acquire full texts from credible databases. Articles that were unavailable, incomplete or contained duplicate data were excluded. Furthermore, data from review articles were not considered for computing the antibody kinetics and performance characteristics of the serological assays.
Data were extracted from all eligible studies using the following criteria: (1) author, title, published date, the countries where studies were conducted, study design, sampling technique, participant inclusion criteria, number of participants enrolled and number of participants with known and available results; (2) main data, consisting of the results of serologic tests and RT-PCR for COVID-19 (sensitivity, specificity, positive predictive value [PPV] and negative predictive value [NPV]), number of days after the onset of symptoms, days of detection, peak and decline of antibodies; and (3) the test protocol used for serology and SARS-CoV-2 RNA detection.
Search outcome
In total, there were 7938 published articles on humoral immune response and laboratory diagnosis of COVID-19. Of these, 74 were included in this study based on selection criteria.
Main findings
Structural organization of SARS-COV-2 useful in serological diagnosis
SARS-CoV-2 is a single-stranded RNA virus with positive polarity.9,10
The SARS-CoV-2 genome consists of 14 open reading frames (ORFs) that code for 27 viral proteins, where the longest ORF coding for the 15 non-structural proteins plays an important role in viral propagation and immune evasion; the ORF codings for structural and accessory proteins are located on the 5´ end and 3´ end, respectively.11 The first ORF code encompasses two-thirds of the viral genome and translates the polyproteins pp1a and pp1ab, which are implicated in the encoding of the 16 non-structural proteins. However, the remaining ORFs code for the viral structural and accessory proteins. The structural protein nucleocapsid (N) proteins, spike (S) glycoprotein, matrix (M) protein and small envelope (E) complete the remaining one third of the viral genome.12 These proteins and RNA-dependent RNA polymerase have been substantially harnessed for primers and antigens in the molecular and serological assays used for COVID-19, respectively.13
Humoral immune response to SARS-COV-2 infection
There is ongoing research into better understanding the viral genome assembly, replication and mutation of SARS-CoV-2. These viral attributes drastically influence the diagnostic performance of both molecular and serological assays as well as the transmissibility of SARS-CoV-2 and its immune responses.14
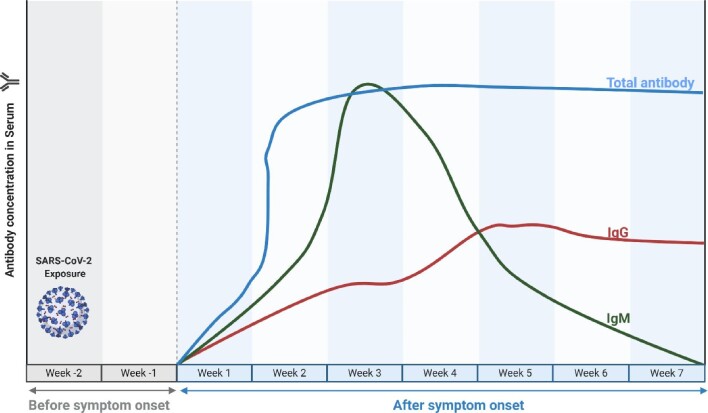
Prior to SARS-CoV-2 infection, an unexposed individual was expected to have a negative test for either anti-SARS-CoV-2 IgM or IgG (Figure 1). However, following exposure to the infection, SARS-CoV-2 now induces a humoral immune response, which commences with the development of IgM, indicating an acute or ongoing infection from the third day of the first week of infection, as reflected by a positive outcome in either the IgM or IgM/IgG serological test.15 The level of IgM in an individual with the activated humoral immune response against SARS-CoV-2 continues to rise until it peaks during the third week following infection.15 By the end of the third week, IgM levels decrease with a concomitant elevation in the level of IgG from the third to the seventh week post symptom onset (PSO), which is revealed by a positive outcome in either the IgG or IgM/IgG serological test (Figure 2).15
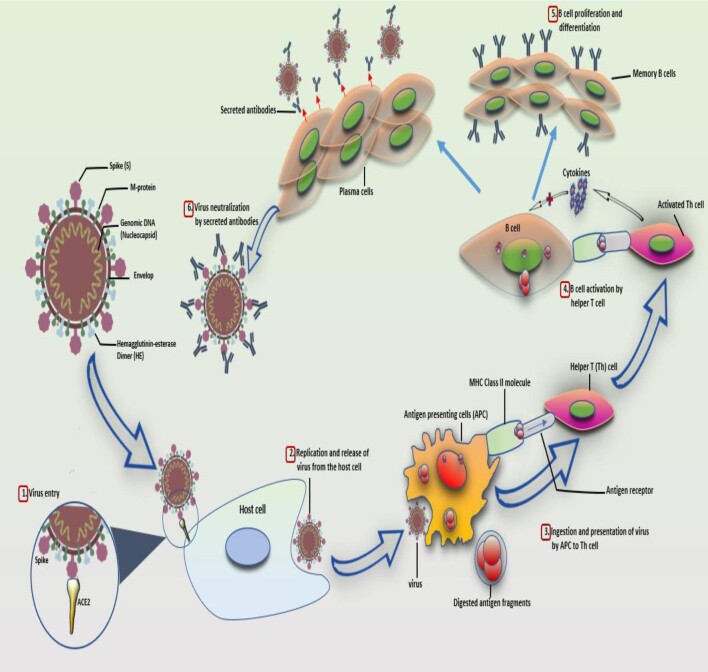
Figure 1.
Kinetics of antibody response in SARS-CoV-2 infection. The entry of the SARS-CoV-2 virus into the host cell through interaction and binding between the host's angiotensin-converting enzyme 2 (ACE2) proteins (receptor) and the viral spike (S) protein (ligand) (1). Following replication and release from the host cells (2), antigen-presenting cells (APCs) like macrophages and dendritic cells engulf some of the viruses, digest and present the digested antigen fragments on their class II MHC molecules to the helper T (Th) cells (3). Th cells, in turn, activate B cells (4), activated B cells proliferate and differentiate into memory B cells or plasma cells with high affinity to the SARS-CoV-2 antigens (5). Plasma cells release SARS-CoV-2-specific antibodies (IgM, IgG or IgA) that bind and neutralize the viruses, thus preventing the viral entry into the host cell (6).
Figure 2.
Timeline of IgM, IgG and total antibody kinetics during SARS-CoV-2 infection. The level of IgM in an individual with the activated humoral immune response against SARS-CoV-2 continues to rise until it peaks at the third week following infection. By the end of the third week, IgM levels decrease with a concomitant elevation in the level of IgG from the third to the seventh week postonset symptom (POS). For the total antibody, it peaks at the middle of the second week and reaches a plateau in the middle of the third week. The blood concentration persists for several weeks and months postinfection (image made with Biorender.com).
The median period for the development of all the classes of immunoglobulins following the activation of humoral immune response is 13 d.16 Individually, IgM, IgG and total immunoglobulins have an average duration of 11, 12 and 14 d, respectively.16 These immunoglobulins can be measured and monitored by a diverse range of antibody-based serological testing techniques, which include rapid diagnostic assay (e.g. lateral flow immunoassay [LFIA] with colloidal gold], CLIA, ELISA and neutralization assay with various diagnostic performance ratings (e.g. sensitivity, specificity, accuracy, PPV and NPV), sampling methods (e.g. finger prick, venipuncture), turnaround time and setting.16 Previous studies have demonstrated the diagnostic roles of these antibody-based serological testing techniques based on their performance. Zhao et al.17 demonstrated that within the first 7 d PSO of COVID-19 infection, the sensitivity of total antibody, IgM and IgG were 38.3%, 28.7% and 19.1%, respectively, which was lower compared with the RNA-based test of 66.7% sensitivity. As the duration of PSO increased, the sensitivity of the RNA-based test decreased by 21.2%, while those of the total antibody, IgM and IgG increased by 61.7%, 65.6% and 60.78%, respectively, within the 15th to 39th day PSO. When the RNA- and antibody-based tests were combined, sensitivity significantly improved to 78.7%, 97.0% and 100% within 1–7, 8–14 and 15–39 d PSO, respectively. The implication of this study indicates the unreliability and unsuitability of serology within the window period of infection, but also reveals an impressive sensitivity for total antibody-based assay in detection of SARS-CoV-2 as the PSO period progresses.
The study further revealed that the percentage of patients with undetectable RNA but with detectable immunoglobulin increased from 28.7% within the first 3 d to 100% within 15–39 d of PSO. This is where the total Ab (which is better than testing IgM and IgG individually) comes in, to rule out people with undetectable RNA.17 The same study recommended the combination of both RNA- and antibody-based tests to scale up the sensitivity of RNA during the course of the infection. This combined approach was observed to attain timely diagnosis of SARS-CoV-2 infection, prevent multiple sampling several days for infection status confirmation, and enhance the ability to prioritize relevant treatments and isolation management.18–20
The changes of the antibody response against SARS-CoV-2 are presently under study, as antibodies may be regarded as potent diagnostic tools to complement RT-PCR-based findings. The SARS-CoV-triggered humoral S- and N-specific IgM response reached a climax within 4 wk and was no more detectable at 3 mo PSO; the switch to IgG often occurred about day 14 and IgG were demonstrated up to 36 mo.21,22
In another study, the authors demonstrated that in 34 SARS-CoV-2 laboratories established, the cases studied were positive for IgM and IgG at week 3 PSO.15 Therefore, in the majority of those patients, the acute phase of infection persisted for >30 d. In an inverse relation, as IgM levels decrease, IgG levels rise gradually from the third to the seventh week, signifying the activation of the humoral immune response against the virus.15 Thus, the humoral response activated by SARS-CoV-2 may be similar to that elicited by SARS-CoV.15,16
In an immunodynamics study reported by Zhao et al.,17 it was observed that the antibody profile in COVID-19 patients showed that seroconversion sequentially appeared for total antibodies, IgM and IgG with a median time of 11, 12 and 14 d, respectively. Full concentrations of SARS-CoV-2 antibodies were detected by double recombinant antigen sandwich immunoassay, which utilized the receptor-binding domain (RBD) of S1 protein and the horse raddish peroxidase-conjugated antigen; IgM μ-chain capture immunoassay was used for anti-SARS-CoV-2 IgM detection. On the other hand, an indirect ELISA kit based on recombinant NP antigen was used for anti-SARS-CoV-2 IgG detection.17 The seroconversion rates recorded were 93.1%, 82.7% and 64.7% for total antibodies, IgM and IgG, respectively, and no significant difference was observed between severely and mildly affected COVID-19 patients.
The sensitivity of serum anti-SARS-CoV-2 detection was lower than the RT-PCR RNA assay within 7 d from the onset of illness (38.3% vs 66.7% for serological vs RT-PCR). However, the sensitivity increased steadily from the eighth to the 39th day PSO and overtook that of the RT-PCR test.17 More significantly, detectable and measurable levels of total anti-SARS-CoV-2 in the sera were found in COVID-19 patients with undetectable SARS-CoV-2 RNA in their respiratory tract samples.17 These results highlighted the importance of combining molecular and serological tests for the correct diagnosis of COVID-19 patients at different stages of the disease. In agreement with these reports, Jin et al. recorded the specificity of serum anti-SARS-CoV IgM and IgG as 90% compared with that of the RT-PCR test.23
In a study by Guo et al., which was carried out on two cohorts of SARS-CoV-2-infected patients, the early antibody response to NP protein was evaluated. Of 208 patients, 90.4% and 93.3% harbored plasma IgM and IgA, respectively. Also, 77.9% of plasma samples were IgG positive, and the median time for both IgM and IgA detection was on day 5 PSO (IQR 3–6) and day 14 PSO (IQR 10–18) for IgG.24 The authors observed that swift and unanticipated IgA seroconversion might be an upshot of the cytokine storm promoting the germline transcription of α and μ genes of the heavy chain constant.
Furthermore, it has been reported and established that T-cell-independent antibody responses can cause excitation of a specialized B cell subset to produce both IgA and IgM throughout the infection of some pathogens.25 Although T-cell-independent antibody response against viruses is still controversial, some viruses can act in vivo as T-cell-independent antigens and therefore cause eliciting protective isotype-switched antibodies in the non-appearance of conventional T-cell help. Inactivated virus or virus-like particles can also elicit IgM response, but factors induced when an active virus infection is ongoing seem very important and are required before there can be induction of the isotype switch and then IgG or IgA responses.26
In another study of 214 COVID-19 patients, 68.2% and 70.1% were positive for rN-specific IgM and IgG, respectively; and 77.1% and 74.3% were positive for rS-specific IgM and IgG, respectively.27 These findings indicated that the detection of rS-specific IgM was more sensitive compared with that of rN-specific IgM, which may be because of the lower immunogenicity of the N protein compared with that of the S protein. A bioinformatics study reported a lower number of B cell epitopes in the NP protein of SARS-CoV-2 than in the S protein, especially as the positive rates of IgM and IgG were low during the early stages of the disease (0–10 days post-disease onset (DPO)). On the other hand, IgM and/or IgG specific for rN and rS reached a climax at 11–15 DPO.27
The sensitivity of the tests and the epitope on which the test is based are significant factors for the well-organized detection of specific SARS-CoV-2 antibodies and timing of the humoral response. Consequently, several tests are rapidly being developed in many laboratories. For example, Li et al. developed a point-of-care LFIA test based on the RBD antigen of the SARS-CoV-2 S1 protein that can help in the concomitant detection of IgM and IgG in human blood within 15 min, with higher sensitivity than the individual IgG and IgM tests; however, the detection limit of the test was not determined.19 Also, Amanat et al. developed sensitive and specific ELISA assays based on the recombinant full-length S protein and RBD epitope, permitting the screening and detection of seroconversion upon SARS-CoV-2 infection 3 d PSO.28 Of note, no cross-reactivity from other human coronaviruses was noted, in agreement with another study highlighting that S1 is a specific antigen for SARS-CoV-2 diagnosis, as cross-reactive antibodies against the S protein of Middle East respiratory syndrome-related coronavirus (MERS-CoV) were not detected in a COVID-19 patient.29 Additionally, strong IgA and IgM responses were discovered and the IgG3 response was stronger than that of IgG1.28 The sensitivity of the test may create challenges for the early detection of IgM. Several patients were more positive for IgG than IgM during the time of hospital stay and 5 d later; likewise, they had an earlier IgG than IgM seroconversion.30
Furthermore, SARS-CoV-2-specific antibodies were detected in the sera of six infants born to mothers with COVID-19. Five of the six infants and their mothers had elevated levels of IgG and two of them also had elevated levels of anti-SARS-CoV-2 IgM. Three of the six infants who had elevated levels of IgG also had normal levels of IgM. However, two of their mothers displayed elevated levels of IgM. How the newborns that developed IgM require additional investigation. Undeniably, due to its large magnitude, IgM is not typically transferred through the placenta; however, it is affected by some pathology that compromises its configuration. The newborn might be in contact with the virus if the latter crosses the placenta, although no virus was detected from RT-PCR analysis.31
Currently, several studies are investigating the connection between antigen-specific antibodies and the clinical characteristics of COVID-19 patients, but interestingly, among people with comorbidities, lesser anti-RBD IgG, but not anti-NP IgM or IgG, have been reported, although the difference was not significant when compared with people without comorbidities.
COVID-19 serological assays
The recent pandemic outbreak of the SARS-CoV-2 virus and its rapid spread poses an urgent need for both diagnostic and therapeutic interventions to manage the infection and the outcome of the disease. The diminishment or absence of IgG and persistence of IgM are considered biomarkers for recent infection. As the epidemic progresses more individuals could get infected. The measurement of these antibodies is a good differential that helps to distinguish between recent and older infections.17 The detection of IgM (from days 1 to 7) in the absence of IgG represent an acute/recent infection, whereas the simultaneous detection of IgM and IgG could represent acute reinfection.18 On the other hand, the detection of IgG in the absence of IgM denotes a past infection.18
The increasing number of confirmed COVID-19 cases has resulted in an unprecedented rise in demand for antibody-based tests from researchers and healthcare policymakers. Recently, a list of >200 serological products was released by the Foundation for Innovative New Diagnostics (FIND); these products, which are predominately from China, are currently either available for use or are in industrial development and evaluation. However, only 12 have received emergency use authorization from the FDA. Serological products from a host of other countries, including South Korea, Germany, the USA and the UK, were also present on the FIND list.
Some commercially available serology-based tests have been considered to be inadequate for COVID-19 diagnosis if used alone, due to their low degrees of sensitivity and specificity. For instance, anti-SARS-CoV-2 IgG takes a relatively long period (not yet reported) for quantification.18 More details regarding the limitations of COVD-19 serological assay follow later in this article.
Cases of poor performance characteristics of some serological kits/devices underscore the need for re-evaluation and validation before being made available to end-users. This is to prevent clinicians and healthcare professionals from using these serological kits/strips off the shelf for clinical purposes. Furthermore, despite kits’ satisfactory diagnostic performance, it is important to include internal quality control and external quality assurance measures in all tests run on human samples to ensure accuracy, precision and reproducibility of test results.
Challenges of SARS-CoV-2 serological testing
Serological tests rely on the detection of specific anti-viral antibodies (IgM, IgA, IgG or total antibody) in patient sera, plasma or whole blood.32 Determining the optimal antigenic epitopes to maximize sensitivity, but minimize cross-reactivity, particularly against other human coronaviruses, has meant that the development of high-quality serological testing has been slower than molecular-based diagnostics.17,32 Initial candidate epitopes have largely focused on the immunogenic viral structural proteins which include nucleocapsid (N) and spike (S) protein, particularly the S1 subunit and the RBD.32 To date, a range of serological tests for COVID-19 have been developed, each with particular test characteristics. Broadly, these serological tests can be divided into tests that (1) can be performed at the point of care; (2) can be performed in routine diagnostic laboratories; and (3) can only be performed in specialized reference laboratories.
Initial studies have reported that most patients with COVID-19 seroconvert by day 10–14 (approximately 80%), with almost 100% seroconversion by day 20.6,7 However, comparisons across published studies are challenging due to (1) different antigens used in assays; (2) differences in the complexity of patient populations; and (3) variations in the RT-PCR assays used as the gold standard for determining the sensitivity of serological assays. Further, it is not clear whether the type and number of antibodies correlate with the severity of COVID-19, or more importantly, with immune protection from reinfection.
At present, the most widely available (and most publicized) serological tests are POCT, which involves the detection of anti-SARS-CoV-2 antibodies through binding to immobilized antigen, generally bound to colloidal gold on a test strip. The relatively cheap and simple nature of lateral flow assays means that production is suited to scaling up for increased testing capacity. However, there are limited published data on the performance characteristics of serological POCT, and high-quality data are urgently needed to guide laboratories, public health agencies and governments in the appropriate and responsible deployment of POCT, and serological assays more broadly. Currently, the WHO recommends the use of POCT immunodiagnostic assays in research settings only, and not for clinical decision-making until further evidence is available.13 Ideally, validation of serological assays, including POCT, should be performed against a serum panel that includes samples from (1) patients at acute and convalescent stages of infection (to assess sensitivity) and (2) patients with other human coronavirus infections (to assess specificity).
Also, serological tests are relevant to fully characterize the SARS-CoV-2-specific antibody response. Differences in the profile of the antibody response across patients might reveal important aspects of the pathogenesis of COVID-19, explaining the great differences observed in the general population. Indeed, the correlation with disease severity and clinical characteristics is poorly understood. Old age and comorbidities seem to increase the risk for a poor outcome of the disease; however, increasing cases of young people who experience severe illness, requiring hospitalization for assistance by mechanical ventilation, may pose questions about the leading factors of disease progression.32
Some challenges are posed by the potential cross-reactivity with other human coronaviruses, due to their high homology at the genetic level. The evidence related to this aspect are still controversial; however, SARS-CoV-specific antibodies are undetectable in the sera of patients 6 y after infection. This observation excludes the presence of cross-reactivity in the sera of COVID-19 patients and might make researchers confident about the specificity of these antibodies.32 Moreover, it would be interesting to understand whether the differences in the progression of the disease might be related to the level of the immune response.
Accuracy and applications of COVID-19 serological assays
Although various reports of reputable serology assays are incredibly encouraging, product end-users must be pragmatic regarding their accuracy and applicability for COVID-19 clinical and epidemiological use. The unsustainability of RT-PCR tests for the COVID-19 laboratory response in some countries has necessitated a search for alternative assays with high sensitivity and specificity with a short turnaround time from preanalytical (sample collection) to postanalytical phases (availability of test results),32 thus enabling prompt and large-scale testing for COVID-19. While none of the antibody-based serological assays have been approved by the WHO, a number of them have been approved for clinical and epidemiological use in some countries.32 Antibody testing might have a useful role in clinically diagnosing COVID-19 patients with late presentations, prolonged symptoms and those with negative results from RT-PCR tests. Furthermore, these tests could be used to monitor the quality and duration of humoral immune response in COVID-19 patients and vaccination. Epidemiologically, SARS-CoV-2 antibody tests can be used for seroprevalence studies in public health research and to inform decisions about returning to work following asymptomatic SARS-CoV-2 infection.
This could offer an opportunity for clinical diagnosis and interruption of transmission through targeted isolation of the most infectious cases and their close contacts.32 SARS-CoV-2 antibody testing has been shown to have good clinical applications, given the varied symptoms of COVID-19 and reported cases of false-negative results of RT-PCR tests when respiratory swabs are collected ≥5 d PSO as their sensitivity begins to decline.32
Considering this, many researchers are now conducting an independent performance evaluation of these antibody-based assays. For instance, a study referred to as the ‘COVID-19 Testing Project’ was conducted by the University of California, Massachusetts General Hospital, the Chan Zuckerberg Biohub and the University of California.33 This study evaluated 10 lateral flow assays and 2 ELISAs to assess performance characteristics for anti-SARS-CoV-233 on plasma/serum of 80 symptomatic COVID-19 patients with RT-PCR positive results, 52 non-SARS-COV-2 patients’ respiratory viral infections (SARS-CoV-2 RT-PCR negative) and 108 archived sera of blood donors collected in 2018 or earlier.33
The assessment found that the assays of the products had varying sensitivities that increased over time, increasing from about 81% to 100% at ˃20 d PSO.33 Based on this, it was inferred that anti-SARS-CoV-2 tests were important for longitudinal studies because a negative result may indicate an actively infected person who has not developed a detectable level of antibodies to the virus. Conversely, the proportion of false-positive samples reported from the non-COVID-19 group ranged from 0% to 16%. The detection agreement indices of the lateral flow assays and ELISAs ranged from 75% to 94%.33
In another evaluation study of SARS-CoV-2 antibody-based tests by the Chinese company Innovita, anti-SARS-CoV-2 antibodies were found in 83% of COVID-19-confirmed patients with an assay specificity of 96%.34 After FDA authorization, these tests were anticipated for at-home use.34 Despite the merits of serological devices, limitations abound due to issues of misdiagnosis following indications of significant false-negative and false-positive results observed during the evaluation of these kits and devices during quality checks.
These rapid test kits have been observed to be unsuitable for testing patients with ≤14 d PSO. To augment these lapses, several studies recommend combining both the serological and RT-PCR-based protocols to provide a more accurate diagnosis of COVID-19 instead of only using the molecular testing approach, which introduces a myriad of strenuous demands on diagnostic and healthcare delivery establishments and regulatory bodies, as well as material, financial and human resources meant to sustain testing capacity.17,19,20,35 Also, there are several studies that have been conducted by diagnostic industries and independent researchers aiming to evaluate the performance characteristics of various anti-SARS-CoV-2 test protocols, some of which have reported promising results.35–42 It is worth noting that the clinical use of SARS-CoV-2 antibody tests should be on products that evaluated and reported the performance characteristics (especially sensitivity and specificity) during the acute phase of COVID-19.
Performance characteristics of COVID-19 serological assays
The most promising (best) LFA on total SARS-CoV-2 antibody test had a sensitivity, specificity, PPV and NPV of 100%, 100%, 100% and 100% at days 4, 5, 4 and 5, respectively, while the worst had a sensitivity, specificity, PPV and NPV of 35.95%, 63.6%, 33.3% and 26.2% at days 1, 3, 1 and 2, respectively. The most promising LFA with the best anti-SARS-CoV-2 IgM test had a sensitivity, specificity, PPV and NPV of 95.8%, 100%, 100% and 98.4% at days 5, 6, 6 and 5, respectively, while the least had a sensitivity, specificity, PPV and NPV of 15.7%, 36.4%, 43.2% and 17.4% at days 1, 3, 1 and 3, respectively. The most promising (best) LFA on anti-SARS-CoV-2 IgG test had a sensitivity, specificity, PPV and NPV of 100%, 100%, 100% and 100% at days 8, 9, 8 and 10, respectively, while the least had a sensitivity, specificity, PPV and NPV of 13.2%, 59.9%, 65.1% and 25.0% at days 1, 2, 3 and 2, respectively (Table 1).
Table 1.
Diagnostic performance of point-of-care test serological protocol from published data
| Citation | Product name/source | Type | Sample size | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Limitation of study |
|---|---|---|---|---|---|---|---|---|
| GeurtsvanKessel et al.b 43 | Rapid SARS -CoV-2 antibody (IgM/IgG) test from InTec (Test of lot S2020021505) | Total | 93 | 96.55 (28/29) | 73.44 (47/64) | 62.22 (28/45) | 97.92 (47/48) | a. No additional validation in participants with mild symptoms; this is required to rule out any possible underevaluated patient |
| IgG | 93 | 96.55 (28/29) | 76.56 (49/64) | 65.12 (28/43) | 98.0 (49/50) | b. There is a risk of interpreting a positive outcome as a measure of protection against the virus | ||
| IgM | 93 | 89.66 (26/29) | 73.44 (47/64) | 60.47 (26/43) | 94.0 (47/50) | c. Sensitivity in the early phase of infection was poor | ||
| qSARS-CoV-2 IgG/IgM cassette rapid test (GICA) from Cellex Inc. (test lot 0200311WI5513C-3) | Total | 93 | 87.76 (43/49) | 84.09 (37/44) | 86.0 (43/50) | 86.05 (37/43) | d. Relatively small size used for determining kit specificity for orient gene RDT | |
| IgG | 93 | 83.67 (41/49) | 84.09 (37/44) | 85.42 (41/48) | 82.22 (37/45) | |||
| IgM | 93 | 87.76 (43/49) | 81.82 (36/44) | 84.31 (43/51) | 85.71 (36/42) | |||
| COVID-19 IgG/IgM rapid test cassette (whole blood/serum/plasma) from orient gene/Healgen (test lot 2003260) | Total | 90 | 91.36 (74/81) | 100.0 (9/9) | 100 (74/74) | 56.25 (9/16) | ||
| IgG | 90 | 91.36 (74/81) | 100.0 (9/9) | 100 (74/74) | 56.25 (9/16) | |||
| IgM | 90 | 88.89 (72/81) | 100.0 (9/9) | 100.0 (72/72) | 50.0 (9/18) | |||
| Li et al.a 19 | Goat anti‐human IgG and IgM antibodies, rabbit IgG and goat anti‐rabbit IgG antibodies were obtained from Sigma‐Aldrich | Total | 525 | 88.66 (352/397) | 90.63 (116/128) | 96.7 (352/364) | 72.05 (116/161) | a. Inability to confirm the presence of the SARS-CoV-2 |
| IgG | 525 | 70.53 (280/397) | 98.44 (126/128) | 99.29 (280/282) | 53.09 (126/243) | b. Cross-reactivity studies with other coronaviruses and flu viruses were not performed | ||
| IgM | 525 | 82.62 (328/397) | 91.41 (117/128) | 96.76 (328/339) | 62.90 (117/186) | c. The level of changes in immunoglobulins was not compared with the various stages of SARS-CoV-2 infection | ||
| Cassaniti et al.a 44 | VivaDiagTM COVID-19 IgM/IgG rapid test | Total | 50 | 18.42 (7/38) | 91.67 (11/12) | 87.5 (7/8) | 26.19 (11/42) | a. Small sample size |
| IgG | 50 | 13.16 (5/38) | 100 (12/12) | 100 (5/5) | 26.67 (12/45) | b. Poor sensitivity | ||
| IgM | 50 | 15.79 (6/38) | 91.67 (11/12) | 85.71 (6/7) | 25.58 (11/43) | c. High false-negative value, which can lead to misdiagnosis | ||
| Porte et al.a 45 | Fluorescence immunochromatographic SARS-CoV-2 antigen test (Bioeasy Biotechnology Co., Shenzhen, China) | Total | 127 | 93.9 (77/82) | 100 (45/45) | 100 (77/77) | 90.0 (45/50) | a. Use of samples not specifically permitted by the manufacturer of the kit |
| IgG | 127 | NAa | NAa | NAa | NAa | b. Retrospective use of clinical data | ||
| IgM | 127 | NAa | NAa | NAa | NAa | |||
| Pan et al.a 46 | Colloidal gold-based immunochromatographic strip (Zhuhai Livzon Diagnositic Inc.) | Total | 108 | 68.6 (59/86) | 63.64 (14/22) | 88.06 (59/67) | 34.15 (14/41) | a. Intensity of color bands formed do not correlate with the abundance of immunoglobulin |
| IgG | 108 | 54.65 (47/86) | 59.09 (13/22) | 83.93 (47/56) | 25.0 (13/52) | b. Very low probability of having negative outcomes without the infection | ||
| IgM | 108 | 55.81 (48/86) | 36.36 (8/22) | 77.42 (48/62) | 17.39 (8/46) | |||
| Lassaunière et al.a 35 | 2019-nCOV IgG/IgM rapid test (Dynamiker Biotechnology, Tianjin, China Cat # DNK-1419–1) | Total | 62 | 90 (27/30) | 100 (32/32) | 100 (27/27) | 89 (32/35) | a. Small sample size |
| IgG | 62 | NAb | NAb | NAb | NAb | b. All kits were not tested uniformly with the same number of control sera | ||
| IgM | 62 | NAb | NAb | NAb | NAb | c. Acro Biotech and Alltest Biotech had comparatively poor test performances, which led to the suspension of further testing | ||
| OnSiteTM COVID-19 IgG/IgM rapid test (CTK Biotech, Poway, CA, USA; cat. # R0180C) | Total | 62 | 90 (27/30) | 100 (32/32) | 100 (27/27) | 89 (32/35) | c. Acro Biotech test had a cross-reaction with a control serum of a patient infected with human coronavirus HKU1 | |
| IgG | 62 | NAb | NAb | NAb | NAb | d. The indications of the presence of SARS-CoV-2 has no correlation with immunity against SARS-CoV-2 infection | ||
| IgM | 62 | NAb | NAb | NAb | NAb | e. Sample size used was small | ||
| Anti-SARS-CoV-2 rapid test (AutoBio Diagnostics, Zhengzhou, China; cat. # RTA0204) | Total | 62 | 93 (28/30) | 100 (32/32) | 100 (28/28) | 94.1 (32/34) | ||
| IgG | 62 | NAb | NAb | NAb | NAb | |||
| IgM | 62 | NAb | NAb | NAb | NAb | |||
| Coronavirus diseases 2019 (COVID-19) IgM/IgG antibody test (Artron Laboratories, Burnaby, Canada; cat. # A03–51-322) | Total | 47 | 83 (25/30) | 100 (17/17) | 100 (25/25) | 74 (17/22) | ||
| IgG | 47 | NAb | NAb | NAb | NAb | |||
| IgM | 47 | NAb | NAb | NAb | NAb | |||
| 2019-nCoV IgG/IgM rapid test cassette (Acro Biotech, Rancho Cucamonga, CA, USA; cat. # INCP-402) | Total | 20 | 80 (4/5) | 80 (12/15) | 57.1 (4/7) | 92.3 (12/13) | ||
| IgG | 20 | NAb | NAb | NAb | NAb | |||
| IgM | 20 | NAb | NAb | NAb | NAb | |||
| 2019-nCoV IgG/IgM rapid test cassette (Hangzhou Alltest Biotech, Hangzhou, China; cat. # INCP-402) | Total | 16 | 100 (1/1) | 86.7 (13/15) | 33.3 (1/3) | NAb | ||
| IgG | 16 | NAb | NAb | NAb | NAb | |||
| IgM | 16 | NAb | NAb | NAb | NAb | |||
| Hoffman et al.a 20 | COVID-19 IgG/IgM rapid test cassette (Zhejiang Orient Gene Biotech Co Ltd, Huzhou, Zhejiang, China; product/model: GCCOV-402a, Lot: 2003242) | Total | 153 | 93.1 (27/29) | 100 (124/124) | 100 (27/27) | 98.4 (124/126) | a. Inadequate comparison with clinical symptoms of positive cases |
| IgG | 153 | 68.97 (20/29) | 100 (124/124) | 100 (20/20) | 93.23 (124/133) | |||
| IgM | 153 | 93.1 (27/29) | 99.19 (123/124) | 96.43 (27/28) | 98.4 (123/125) | |||
| Pallet et al.a 47 | COVID-19 IgG/IgM rapid test cassettes (OrientGene) | Total | 200 | 82.67 (124/150) | 96.0 (48/50) | 98.48 (130/132) | 70.59 (48/68) | a Poor performance in patients with ≤14 d POS |
| IgG | 200 | 82.67 (124/150) | 96.0 (48/50) | 98.41 (124/126) | 64.86 (48/74) | |||
| IgM | 200 | 83.33 (125/150) | 100 (50/50) | 100 (125/125) | 66.67 (50/75) | |||
| Spicuzza et al.a 48 | 2019-nCoV IgG/IgM antibody rapid test kit (Beijing Diagreat Biotechnologies Co., Ltd) | Total | 37 | 82.61 (19/23) | 92.86 (13/14) | 95.0 (19/20) | 76.47 (13/17) | a. Small sample size |
| IgG | 37 | NAb | NAb | NAb | NAb | b. Not reliable for patients with symptoms within the early days of infection | ||
| IgM | 37 | NAb | NAb | NAb | NAb | |||
| Wu et al.a 49 | ALLTEST 2019-nCoV IgG/IgM rapid test (Hangzhou ALLTEST Biotech Co., Ltd. [China]) | Total | 122 | 100 (22/22) | 98 (98/100) | 91.67 (22/24) | 100 (98/98) | a. Single-center study |
| IgG | 122 | 100 (22/22) | 98 (98/100) | 91.67 (22/24) | 100 (98/98) | b. Inadequate number of cases, which could not reveal the statistical difference in the performance characteristics for the various POCT | ||
| IgM | 122 | 90.91 (20/22) | 96 (96/100) | 83.33 (20/24) | 97.96 (96/98) | c. Laboratory investigation for cross-reactivity studies were inadequate | ||
| Dynamiker 2019-nCoV IgG/IgM rapid test (Dynamiker Biotechnology [Tianjin] Co., Ltd. [China]) | Total | 462 | 89.51 (145/162) | 96.33 (289/300) | 92.95 (145/156) | 94.44 (289/306) | d. Possible misclassification of COVID-19 pneumonia patient with patients having subclinical pulmonary infiltration | |
| IgG | 462 | 89.51 (145/162) | 96.33 (289/300) | 92.95 (145/156) | 94.44 (289/306) | |||
| IgM | 462 | 87.65 (142/162) | 95.33 (286/300) | 91.03 (142/156) | 93.46 (286/306) | |||
| Wondfo SARS-CoV-2 antibody test (Guangzhou Wondfo Biotech Co., Ltd [China]) | Total | 596 | 86.43 (312/361) | 82.11 (234/285) | 99.68 (312/313) | 82.69 (234/283) | ||
| IgG | 596 | NAb | NAb | NAb | NAb | |||
| IgM | 596 | NAb | NAb | NAb | NAb | |||
| Green et al.a 50 | COVID-19 IgM-IgG Rapid Test (BioMedomics, BD, USA) | Total | 525 | 88.66 (352/397) | 90.63 (116/128) | 96.7 (352/364) | 72.05 (116/161) | a. No detail on the diagnostic performance of both IgG and IgM for most POC diagnostic devices that were evaluated |
| IgG | 525 | NAb | NAb | NAb | NAb | |||
| IgM | 525 | NAb | NAb | NAb | NAb | |||
| Xpert SARS- CoV-2 (Cepheid [USA/worldwide distribution]) | Total | 65 | 100 (30/30) | 100 (35/35) | 100 (30/30) | 100 (35/35) | ||
| IgG | 65 | NAb | NAb | NAb | NAb | |||
| IgM | 65 | NAb | NAb | NAb | NAb | |||
| VitaPCR COVID-19 assay (Credo [Singapore]) | Total | 180 | 100 (120/120) | 100 (60/60) | 100 (120/120) | 100 (60/60) | ||
| IgG | 180 | NAb | NAb | NAb | NAb | |||
| IgM | 180 | NAb | NAb | NAb | NAb | |||
| Accula SARS- CoV-2 (Mesa Biotech [USA]) | Total | 80 | 100 (50/50) | 100 (30/30) | 100 (50/50) | 100 (30/30) | ||
| IgG | 80 | NAb | NAb | NAb | NAb | |||
| IgM | 80 | NAb | NAb | NAb | NAb | |||
| ID NOW COVID-19 (Abbott Diagnostics [worldwide]) | Total | 60 | 100 (30/30) | 100 (30/30) | 100 (30/30) | 100 (30/30) | ||
| IgG | 60 | NAb | NAb | NAb | NAb | |||
| IgM | 30 | NAb | NAb | NAb | NAb | |||
| GT-100 SARS-CoV-2 IgG/IgM kit (Goldsite Diagnostics Inc. [China]) | Total | 70 | 100 (20/20) | 98 (49/50) | 90.9 (20/22) | 100 (49/49) | ||
| IgG | 70 | 100 (20/20) | 98 (49/50) | 90.9 (20/22) | 100 (49/49) | |||
| IgM | 70 | 85 (17/20) | 96 (48/50) | 89.47 (17/19) | 94.12 (48/51) | |||
| Mlcochova et al.a 51 | SAMBA II SARS-CoV-2 point of care testing | Total | 45 | 79.17 (19/24) | 100 (21/21) | 100 (19/19) | 80.77 (21/26) | a. Small sample size |
| IgG | 45 | 50.0 (12/24) | 100 (21/21) | 100 (12/12) | 63.62 (21/33) | b. Recommendation of combined rapid testing protocol with PCR in order to ensure: | ||
| IgM | 45 | 87.5 (21/24) | 100 (21/21) | 100 (21/21) | 87.5 (21/24) | i. Expansive testing in areas where diagnostic centers are sparse, and transmission is rapid | ||
| COVIDIX 2019 SARS-CoV-2 IgG/IgM test (COVIDIX Healthcare, Cambridge, UK) | Total | 45 | 95.83 (23/24) | 85.71 (18/21) | 88.46 (23/26) | 94.74 (18/19) | ii. That repeated sampling is avoided, which can generate aerosols and encourage transmission | |
| IgG | 45 | 100 (24/24) | 80.95 (17/21) | 85.71 (24/28) | 100 (17/17) | iii. That patients are safely and quickly recruited for treatment | ||
| IgM | 45 | 95.83 (23/24) | 90.48 (19/21) | 92.0 (23/25) | 95.0 (19/20) | |||
| Van Elslande et al.a 52 | Clungene COVID-19 IgG/IgM rapid test | Total | 256 | 35.95 (55/153) | 99.03 (102/103) | 98.21 (55/56) | 51.0 (102/200) | a. Control samples were limited in number from patients with frequent respiratory disorders |
| IgG | 256 | 62.09 (95/153) | 98.06 (101/103) | 97.94 (95/97) | 63.52 (101/159) | b. Antibody response studies in asymptomatic or mild individuals were not performed | ||
| IgM | 256 | 39.22 (60/153) | 91.26 (94/103) | 86.96 (60/69) | 50.27 (94/187) | c. Participants were not tested daily to accurately determine the true period of seroconversion | ||
| OrientGene COVID-19 IgG/IgM rapid test | Total | 256 | 64.05 (98/153) | 97.09 (100/103) | 97.03 (98/101) | 64.52 (100/155) | ||
| IgG | 256 | 67.97 (104/153) | 93.2 (96/103) | 93.69 (104/111) | 66.21 (96/145) | |||
| IgM | 256 | 72.55 (111/153) | 95.15 (98/103) | 95.69 (111/116) | 70.0 (98/140) | |||
| VivaDiag COVID-19 IgG/IgM rapid test | Total | 256 | 62.75 (96/153) | 100 (103/103) | 100 (96/96) | 64.38 (103/160) | ||
| IgG | 256 | 62.75 (96/153) | 99.03 (102/103) | 98.97 (96/97) | 64.15 (102/159) | |||
| IgM | 256 | 65.36 (100/153) | 100 (103/103) | 100 (100/100) | 66.03 (103/156) | |||
| StrongStrep COVID-19 IgG/IgM rapid test | Total | 256 | 30.07 (46/153) | 100 (103/103) | 100 (46/46) | 49.05 (103/210) | ||
| IgG | 256 | 64.71 (99/153) | 99.03 (102/103) | 99.0 (99/100) | 65.38 (102/156) | |||
| IgM | 256 | 32.03 (49/153) | 99.03 (102/103) | 98.0 (49/50) | 49.51 (102/206) | |||
| Dynammiker COVID-19 IgG/IgM rapid test | Total | 256 | 61.44 (94/153) | 99.03 (102/103) | 98.94 (94/95) | 63.35 (102/161) | ||
| IgG | 256 | 61.44 (94/153) | 99.03 (102/103) | 98.94 (94/95) | 63.35 (102/161) | |||
| IgM | 256 | 69.28 (106/153) | 95.15 (98/103) | 95.5 (106/111) | 67.59 (98/145) | |||
| Multi-G COVID-19 IgG/IgM rapid test | Total | 256 | 37.25 (57/153) | 100 (103/103) | 100 (57/57) | 51.76 (103/199) | ||
| IgG | 256 | 64.71 (99/153) | 97.09 (100/103) | 97.06 (99/102) | 64.94 (100/154) | |||
| IgM | 256 | 43.79 (67/153) | 91.26 (94/103) | 88.16 (67/76) | 52.22 (94/180) | |||
| Prima COVID-19 IgG/IgM rapid test | Total | 256 | 48.37 (74/153) | 98.06 (101/103) | 97.37 (74/76) | 56.11 (101/180) | ||
| IgG | 256 | 71.24 (109/153) | 90.29 (93/103) | 91.6 (109/119) | 67.88 (93/137) | |||
| IgM | 256 | 56.21 (86/153) | 93.2 (96/103) | 68.25 (86/120) | 71.67 (86/120) | |||
| Jääskeläinen et al.b 53 | 2019-nCoV IgG/IgM rapid test cassette (Acro Biotech, California, USA) | Total | 123 | 56.1 (23/41) | 74.39 (61/82) | 52.27 (23/44) | 77.22 (61/79) | a. Low PPVs for Acro Biotech IgG/IgM rapid test due to low SARS-CoV-2 seroprevalence |
| IgG | 123 | 56.1 (23/41) | 74.39 (61/82) | 52.27 (23/44) | 77.22 (61/79) | |||
| IgM | 123 | 46.34 (19/41) | 69.51 (57/82) | 43.18 (19/44) | 72.15 (57/79) | |||
| SARS-CoV-2 IgG/IgM rapid test (Xiamen Biotime, Fujian, China) | Total | 112 | 81.25 (26/32) | 97.5 (78/80) | 92.86 (26/28) | 92.86 (78/84) | ||
| IgG | 112 | 71.88 (23/32) | 97.5 (78/80) | 92 (23/25) | 89.66 (78/87) | |||
| IgM | 112 | 81.25 (26/32) | 88.75 (71/80) | 81.25 (26/35) | 92.21 (71/77) | |||
| Kohmer et al.c 54 | FasStep (COVID-19 IgG/IgM) rapid test cassettes (COV-W32M, Assure Tech (Hangzhou) Co., Ltd, China) | Total | 29 | 93.75 (15/16) | 100.0 (13/13) | 100.0 (15/15) | 92.86 (13/14) | a. Small sample size |
| IgG | 29 | 93.75 (15/16) | 100.0 (13/13) | 100.0 (15/15) | 92.86 (13/14) | |||
| IgM | 29 | 62.5 (10/16) | 100.0 (13/13) | 100.0 (10/10) | 68.42 (13/19) | |||
| Montesinos et al.a 5 | 2019-n-CoV IgG/IgM rapid test cassette (LabOn Time) (LabOn Time, Bio Marketing Diagnostics, or Akiva, Israel) | Total | 200 | 71.88 (92/128) | 100.0 (72/72) | 100.0 (92/92) | 66.67 (72/108) | a. The reference standard used for the comparative study of the serological kits |
| IgG | 200 | 67.19 (86/128) | 100.0 (72/72) | 100.0 (86/86) | 63.16 (72/114) | b. Poor diagnostic performance based on the sensitivity of IgM and IgG for LabOn and Quickzen, respectively | ||
| IgM | 200 | 48.44 (62/128) | 100.0 (72/72) | 100.0 (62/62) | 52.17 (72/138) | |||
| Novel coronavirus (2019-n-CoV) antibody IgG/IgM assay (colloidal gold) (Avioq, Biotech, Shandong, China) | Total | 200 | 68.75 (88/128) | 95.83 (69/72) | 96.7 (88/91) | 63.3 (69/109) | ||
| IgG | 200 | 68.75 (88/128) | 95.83 (69/72) | 96.7 (88/91) | 63.3 (69/109) | |||
| IgM | 200 | 68.75 (88/128) | 95.83 (69/72) | 96.7 (88/91) | 63.3 (69/109) | |||
| QuickZen COVID-19 IgM/IgG kit (QuickZen) (ZenTech, Angleur, Belgium) | Total | 200 | 71.09 (91/128) | 100.0 (72/72) | 100.0 (91/91) | 66.06 (72/109) | ||
| IgG | 200 | 49.22 (63/128) | 100.0 (72/72) | 100.0 (63/63) | 52.55 (72/137) | |||
| IgM | 200 | 68.75 (88/128) | 100.0 (72/72) | 100.0 (88/88) | 64.29 (72/112) | |||
| Adams et al.a 56 | RDT 1 | Total | 93 | 54.55 (18/33) | 100.0 (60/60) | 100.0 (18/18) | 80.0 (60/75) | a. Presence of false-positives due to cross-reactivity of non-specific immunoglobulins, which reflects past exposure to other seasonal viral infections of the coronavirus group |
| IgG | 93 | NAb | NAb | NAb | NAb | b. Small sample size, which did not encourage strong confidence intervals around the diagnostic performance of the LFIA kits | ||
| IgM | 93 | NAb | NAb | NAb | NAb | c. The kits could not distinguish the immunoglobulins | ||
| RDT 2 | Total | 129 | 60.53 (23/38) | 98.9 (90/91) | 95.83 (23/24) | 85.71 (90/105) | ||
| IgG | 129 | NAb | NAb | NAb | NAb | |||
| IgM | 129 | NAb | NAb | NAb | NAb | |||
| RDT 3 | Total | 93 | 63.64 (21/33) | 96.67 (58/60) | 91.3 (21/23) | 82.86 (58/70) | ||
| IgG | 93 | NAb | NAb | NAb | NAb | |||
| IgM | 93 | NAb | NAb | NAb | NAb | |||
| RDT 4 | Total | 98 | 65.79 (25/38) | 98.33 (59/60) | 96.15 (25/26) | 81.94 (59/72) | ||
| IgG | 98 | NAb | NAb | NAb | NAb | |||
| IgM | 98 | NAb | NAb | NAb | NAb | |||
| RDT 5 | Total | 91 | 61.29 (19/31) | 96.67 (58/60) | 90.48 (19/21) | 82.86 (58/70) | ||
| IgG | 91 | NAb | NAb | NAb | NAb | |||
| IgM | 91 | NAb | NAb | NAb | NAb | |||
| RDT 6 | Total | 91 | 64.52 (20/31) | 98.33 (59/60) | 95.24 (20/21) | 84.29 (59/70) | ||
| IgG | 91 | NAb | NAb | NAb | NAb | |||
| IgM | 91 | NAb | NAb | NAb | NAb | |||
| RDT 7 | Total | 93 | 69.70 (23/33) | 95.0 (57/60) | 88.46 (23/26) | 85.07 (57/67) | ||
| IgG | 93 | NAb | NAb | NAb | NAb | |||
| IgM | 93 | NAb | NAb | NAb | NAb | |||
| RDT 8 | Total | 92 | 56.25 (18/32) | 100.0 (60/60) | 100.0 (18/18) | 81.08 (60/74) | ||
| IgG | 92 | NAb | NAb | NAb | NAb | |||
| IgM | 92 | NAb | NAb | NAb | NAb | |||
| RDT 9 | Total | 182 | 55.0 (22/40) | 97.18 (138/142) | 84.62 (22/26) | 88.46 (138/156) | ||
| IgG | 182 | NAb | NAb | NAb | NAb | |||
| IgM | 182 | NAb | NAb | NAb | NAb | |||
| Nuccetelli et al.a 57 | SARS-CoV-2 immunochromatographic CARD 1 | Total | 83 | 83.72 (36/43) | 100.0 (40/40) | 100.0 (36/36) | 85.11 (40/47) | a. Because the performance of these kits is based on the PCR-reference standard, the determination of the actual prevalence of the viral infection is limited and cannot reveal the actual status of participants with viral load values that are below the PCR detection limit |
| IgG | 83 | 83.72 (36/43) | 100.0 (40/40) | 100.0 (36/36) | 85.11 (40/47) | |||
| IgM | 83 | 60.47(26/43) | 100.0 (40/40) | 100.0 (26/26) | 70.18 (40/57) | |||
| SARS-CoV-2 immunochromatographic CARD 2 | Total | 83 | 90.70 (39/43) | 100.0 (40/40) | 100 (39/39) | 90.91 (40/44) | ||
| IgG | 83 | 90.70 (39/43) | 100.0 (40/40) | 100 (39/39) | 90.91 (40/44) | |||
| IgM | 83 | 88.37 (38/43) | 100.0 (40/40) | 100.0 (38/38) | 88.89 (40/45) | |||
| SARS-CoV-2 immunofluorescence CARD 3 | Total | 83 | 93.02 (40/43) | 100.0 (40/40) | 100.0 (40/40) | 93.02 (40/43) | ||
| IgG | 83 | 93.02 (40/43) | 100.0 (40/40) | 100.0 (40/40) | 93.02 (40/43) | |||
| IgM | 83 | 83.72 (36/43) | 100.0 (40/40) | 100.0 (36/36) | 85.11 (40/47) | |||
| Pérez-García et al.a 58 | AllTest COV-19 IgG/IgM kit (AllTest Biotech, Hangzhou, China) | Total | 190 | 64.44 (58/90) | 100.0 (100/100) | 100.0 (58/58) | 75.76 (100/132) | a. Study location was restricted to a healthcare center, which produced data that needs to be reinforced using a multicenter study |
| IgG | 190 | 60.0 (54/90) | 100.0 (100/100) | 100.0 (54/54) | 73.53 (100/136) | b. No consideration of the study participants with a range of clinical manifestations so as to generate non-biased data | ||
| IgM | 190 | 27.78 (25/90) | 100.0 (100/100) | 100.0 (25/25) | 60.61 (100/165) | c. Validation of just a kit | ||
| d. Poor diagnostic performance for IgM based on sensitivity |
Abbreviations: CEFA, cyclic enhanced fluorescence assay; CLIA, chemiluminescence immunoassay; LFIA, lateral flow immunoassay; MNT, microneutralization test; NAa, not applicable; NAb, not available; NPV, negative predictive value; POCT, point of care test; POS, postonset of symptoms; PPV, positive predictive value; PRNT, plaque reduction neutralization test.
a diagnostic performance performed with reference to RT-PCR.
b diagnostic performance performed with reference to a microneutralization test (MNT).
c Diagnostic performance performed with reference to plaque-reduction neutralization test (PRNT).
Note:
1. All computed values were PSO.
2. All products with ≥95% each for sensitivity, specificity, PPV and NPV may be used for epidemiological purposes. Furthermore, performance characteristics ≥95% values reported from acute COVID-19 samples could be considered for clinical use (in conjunction with clinical presentations of patients).
The most promising (best) ELISA on total SARS-CoV-2 antibody test had a sensitivity, specificity, PPV and NPV of 93.9%, 100%, 100% and 100% at days 3, 5, 4 and 3, respectively, while the least had a sensitivity, specificity, PPV and NPV of 46.1%, 86.6%, 76.6% and 55.3% at days 1, 3, 2 and 1, respectively. The most promising ELISA with best anti-SARS-CoV-2 IgM test had a sensitivity, specificity, PPV and NPV of 89.5%, 100%, 100% and 95.7% at days 4, 6, 4 and 5, respectively, while the least had a sensitivity, specificity, PPV and NPV of 64.9%, 88.1%, 70.6% and 80.0% at days 1, 3, 2 and 3, respectively. The most promising (best) ELISA on anti-SARS-CoV-2 IgG test had a sensitivity, specificity, PPV and NPV of 100%, 100%, 100% and 100% at days 8, 10, 8 and 9, respectively, while the least had a sensitivity, specificity, PPV and NPV of 46.1%, 86.6%, 72.5% and 56.2% at days 5, 7, 6 and 7, respectively. The most promising (best) ELISA on anti-SARS-CoV-2 IgA test had a sensitivity, specificity, PPV and NPV of 97.4%, 100%, 100% and 98.0% at days 4, 5, 6 and 5, respectively, while the least had a sensitivity, specificity, PPV and NPV of 46.1%, 68.3%, 58.1% and 53.3% at days 14, 13, 14 and 13, respectively (Table 2).
Table 2.
Diagnostic performance of ELISA and ELFA protocol from published data
| Citation | Product name/source | Type | Sample size | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Limitation of study |
|---|---|---|---|---|---|---|---|---|
| Van Elslande et al.a 52 | Euroimmun | Total | 256 | NAb | NAb | NAb | NAb | a. Samples used to determine both specificity and sensitivity were challenging |
| IgG | 256 | 55.56 (85/153) | 96.12 (99/103) | 95.51 (85/89) | 56.28 (99/167) | b. Diagnostic performance data both for total antibody and IgM were not made available | ||
| IgM | 256 | NAb | NAb | NAb | NAb | |||
| Zhao et al.a 17 | COVID-19 ELISA kit (Beijing Wantai Biological Pharmacy Enterprise Co. Ltd) | Total | 386 | 93.06 (161/173) | 99.06 (211/213) | 98.77 (161/163) | 94.62 (211/223) | a. Sampling was for upper respiratory tract instead of lower respiratory tract with higher sensitivity for RNA tests |
| IgG | 386 | 64.74 (112/173) | 98.98 (195/197) | 98.25 (112/114) | 76.17 (195/256) | b. No evaluation of the persistence of antibodies as sampling was performed during the acute phase of the participants | ||
| IgM | 370 | 82.66 (143/173) | 98.59 (210/213) | 97.95 (143/146) | 87.5 (210/240) | c. Cross-reactivity studies were not performed for the serological kits | ||
| Xiang et al.a 59 | Sandwich ELISA kit (Livzon Inc, Zhuhai, China, lot numbers 20200308 [IgM] and 20200308 [IgG]) | Total | 126 | 83.33 (55/66) | 100 (60/60) | 100 (55/55) | 84.51 (60/71) | a. Small sample sizes were used to determine the seropositive rate of IgG |
| IgG | 126 | 83.33 (55/66) | 95.0 (57/60) | 94.83 (55/58) | 83.82 (57/68) | b. Unreliable for testing within the window period of infection due to misdiagnosis; retesting was recommended for those with early seronegative immunoglobulins | ||
| IgM | 126 | 77.27 (51/66) | 100 (60/60) | 100 (51/51) | 80.0 (60/75) | |||
| Jääskeläinen et al.b 53 | Anti-SARS-CoV-2 IgA and IgG EIA (Euroimmun, Lübeck, Germany) | Total | 123 | 87.8 (36/41) | 86.59 (71/82) | 76.6 (36/47) | 93.42 (71/76) | a. No extensive investigation on prozone phenomenon capable of causing false-negative results |
| IgG | 123 | 70.73 (29/41) | 86.59 (71/82) | 72.5 (29/40) | 85.54 (71/83) | b. IgA detection is not useful for screening purposes but can only be applied for follow-up investigations in patients with proven COVID-19 infections | ||
| IgA | 123 | 87.8 (36/41) | 68.29 (56/82) | 58.06 (36/62) | 91.8 (56/61) | |||
| Jääskeläinen et al.a 60 | Anti-SARS-CoV-2 IgA and IgG EIA (Euroimmun, Lübeck, Germany) | Total | 40 | 92.86 (13/14) | 92.31 (24/26) | 86.67 (13/15) | 96.0 (24/25) | a. Small sample size |
| IgG | 40 | 92.86 (13/14) | 92.31 (24/26) | 86.67 (13/15) | 96.0 (24/25) | |||
| IgA | 40 | 78.57 (11/14) | 73.08 (19/26) | 61.11 (11/18) | 86.36 (19/22) | |||
| Geurtsvan Kessel et al.c 43 | Wantai SARS-CoV-2 total Ig and IgM ELISA (Beijing Wantai Biological Pharmacy Enterprise Co., Ltd) | Total | 226 | 98.68 (75/76) | 99.33 (149/150) | 98.68 (75/76) | 99.33 (149/150) | a. Not entirely adequate for population screening during an early phase of the pandemic |
| IgG | 226 | NAb | NAb | NAb | NAb | |||
| IgM | 226 | 89.47 (68/76) | 98.67 (148/150) | 97.14 (68/70) | 94.87 (148/156) | |||
| Anti-SARS-CoV-2 IgG and IgA ELISA assay (EUROIMMUN Medizinische Labordiagnostika AG) | Total | 237 | 97.37 (74/76) | 99.38 (160/161) | 98.67 (74/75) | 98.77 (160/162) | ||
| IgG | 237 | 81.58 (62/76) | 99.38 (160/161) | 98.41 (62/63) | 91.95 (160/174) | |||
| IgA | 237 | 97.37 (74/76) | 93.79 (151/161) | 88.10 (74/84) | 98.69 (151/153) | |||
| Müller et al.d 61 | EUROIMMUN anti-SARS-CoV-2 IgA and IgG ELISA test | Total | 42 | 46.15 (12/26) | 100.0 (16/16) | 100.0 (12/12) | 53.33 (16/30) | a. Diagnostic performance based on sensitivity was very poor |
| IgG | 42 | 46.15 (12/26) | 100.0 (16/16) | 100.0 (12/12) | 53.33 (16/30) | |||
| IgA | 42 | 46.15 (12/26) | 100.0 (16/16) | 100.0 (12/12) | 53.33 (16/30) | |||
| Kohmer et al.c 54 | Euroimmun SARS-CoV-2 IgG ELISA (Euroimmun, Lübeck, Germany) | Total | 40 | 93.75 (15/16) | 95.65 (22/23) | 93.75 (15/16) | 95.65 (22/23) | a. Small sample size |
| IgG | 40 | 93.75 (15/16) | 95.65 (22/23) | 93.75 (15/16) | 95.65 (22/23) | |||
| IgA | 40 | 58.82 (10/17) | 95.65 (22/23) | 90.91 (10/11) | 75.86 (22/29) | |||
| Vircell COVID-19 ELISA IgG (Vircell Spain S.L.U., Granada, Spain) | Total | 38 | 100.0 (16/16) | 95.24 (20/21) | 94.12 (16/17) | 100.0 (20/20) | ||
| IgG | 38 | 100.0 (16/16) | 95.24 (20/21) | 94.12 (16/17) | 100.0 (20/20) | |||
| IgA | 38 | 70.59 (12/17) | 95.24 (20/21) | 92.31 (12/13) | 80.0 (20/25) | |||
| Kohmer et al.c 62 | Anti-SARS-CoV-2 ELISA IgG (S1 protein-based) (Euroimmun, Lübeck, Germany) | Total | 65 | NAb | NAb | NAb | NAb | a. Both ELISA assays could not detect immunoglobulins in samples of participants with mild form of COVID-19 |
| IgG | 65 | 71.11 (32/45) | 100.0 (20/20) | 100.0 (32/32) | 60.61 (20/33) | b. The study on the protective mechanism and the duration of immune response, which was not performed in detail, was further proposed | ||
| IgA | 65 | NAb | NAb | NAb | NAb | |||
| Virotech SARS-CoV-2 IgG ELISA (N protein-based) (Virotech Diagnostics GmbH Riisseisheim, Germany) | Total | 80 | NAb | NAb | NAb | NAb | ||
| IgG | 80 | 66.67 (30/45) | 100.0 (35/35) | 100.0 (30/30) | 70.0 (35/50) | |||
| IgA | 80 | NAb | NAb | NAb | NAb | |||
| Wolff et al.a 63 | Euroimmun anti-SARS CoV-2 ELISA IgG and IgA assays (Euroimmun, Luebeck, Germany) | Total | 207 | 82.88 (92/111) | 95.83 (92/96) | 95.83 (92/96) | 82.88 (92/111) | a. Prolonged average sampling collection period was 12 d, which could influence the diagnostic performance of assays |
| IgG | 207 | 75.68 (84/111) | 95.83 (92/96) | 95.45 (84/88) | 77.31 (92/119) | b. Based on the use of qRT-PCR as reference protocol for the study, there is a possibility of missing positive cases whose respiratory viral load is lower than the detection limit for PCR | ||
| IgA | 207 | 82.88 (92/111) | 95.83 (92/96) | 95.83 (92/96) | 82.88 (92/111) | |||
| VIDAS anti-SARS CoV-2 (ELFA) (BioMérieux, Marcy-l'Etoile, France) | Total | 207 | 72.97 (81/111) | 100.0 (96/96) | 100.0 (81/81) | 76.19 (96/126) | ||
| IgG | 207 | 72.97 (81/111) | 100.0 (96/96) | 100.0 (81/81) | 76.19 (96/126) | |||
| IgM | 207 | 64.86 (72/111) | 100.0 (96/96) | 100.0 (72/72) | 71.11 (96/135) | |||
| Francesca et al.e 64 | Anti-SARS-CoV-2 IgG, IgM and IgA ELISA tests (ENZY-WELL SARS-CoV-2 ELISA, DIESSE Diagnostica Senese S.p.a.) | Total | 468 | 93.91 (108/115) | 98.02 (346/353) | 93.91 (108/115) | 98.02 (346/353) | a. All assays indicated moderate cross-reactivity with samples from participants for other communicable and non-communicable disorders |
| IgG | 468 | 92.17 (106/115) | 91.78 (324/353) | 78.52 (106/135) | 97.30 (324/333) | |||
| IgM | 468 | 87.83 (101/115) | 88.10 (311/353) | 70.63 (101/143) | 95.69 (311/325) | |||
| IgA | 468 | 93.91 (108/115) | 98.02 (346/353) | 93.91 (108/115) | 98.02 (346/353) | |||
| Montesinos et al.a 55 | Euroimmun anti-SARS-CoV-2 ELISA IgG and IgA assays (Euroimmun, Luebeck, Germany) | Total | 200 | 84.38 (108/128) | 87.5 (63/72) | 92.31 (108/117) | 75.9 (63/83) | a. The retrospective nature of the study, which involved no fresh samples, could adversely affect the accuracy of results |
| IgG | 200 | 61.72 (79/128) | 98.61 (71/72) | 98.75 (79/80) | 59.17 (71/120) | |||
| IgA | 200 | 83.59 (107/128) | 86.11 (62/72) | 91.45 (107/117) | 74.7 (62/83) | |||
| Adams et al.a 56 | In-house ELISA recombinant SARS-CoV-2 trimeric spike protein | Total | 90 | NAb | NAb | NAb | NAb | a. No detailed data to investigate immunoglobulin-positivity as a correlate of protective immunity. |
| IgG | 90 | 85.0 (34/40) | 100.0 (50/50) | 100.0 (34/34) | 89.29 (50/56) | b. No further studies to confirm the lack of evidence to establish the relationship between severity of the disorder and antibody titers | ||
| IgM | 90 | NAb | NAb | NAb | NAb | |||
| Beavis et al.a 65 | EUROIMMUN anti-SARS-CoV-2 assay | Total | 168 | NAb | NAb | NAb | NAb | a. Small sample size |
| IgG | 168 | 67.07 (55/82) | 97.67 (84/86) | 96.49 (55/57) | 75.68 (84/111) | b. Prolonged average sampling collection period, which could affect the diagnostic performance of the kit | ||
| IgA | 168 | 82.93 (68/82) | 88.37 (76/86) | 87.18 (68/78) | 84.44 (76/90) |
Abbreviations: ELFA, enzyme linked fluorescence assay; IFA, immunofluorescence assay; IFT, immunofluorescence test; MNT, microneutralization assay; NAb, not available; NPV, negative predictive value; NT, neutralization test; POS, postonset of symptoms; PPV, positive predictive value; PRNT, plaque-reduction neutralization assay.
a Diagnostic performance performed with reference to RT-PCR.
b Diagnostic performance performed with reference to microneutralization test (MNT).
c Diagnostic performance performed with reference to plaque-reduction neutralization test (PRNT).
d Diagnostic performance performed with reference to NT and IFT.
e Diagnostic performance performed with reference to IFA.
Note:
1. All computed values were POS.
2. All products with ≥95% each for sensitivity, specificity, PPV and NPV may be used for epidemiological purposes. Furthermore, performance characteristics ≥95% values reported from acute COVID-19 samples could be considered for clinical use (in conjunction with clinical presentations of patients).
The most promising (best) CLIA on total SARS-CoV-2 antibody test had a sensitivity, specificity, PPV and NPV of 100%, 100%, 100% and 100% at days 2, 3, 2 and 3, respectively, while the least had a sensitivity, specificity, PPV and NPV of 58.7%, 92.3%, 81.6% and 61.5% at days 1, 2, 1 and 1, respectively. The most promising (best) CLIA on anti-SARS-CoV-2 IgM test had a sensitivity, specificity, PPV and NPV of 96.8%, 100%, 100% and 98.5% at days 1, 3, 3 and 2, respectively, while the least had a sensitivity, specificity, PPV and NPV of 63.1%, 90.5%, 84.9% and 58.1% at days 12, 10, 9 and 11, respectively. The most promising (test) CLIA on anti-SARS-CoV-2 IgG test had a sensitivity, specificity, PPV and NPV of 95.7%, 100%, 100% and 98.7% at days 7, 9, 8 and 7, respectively, while the least had a sensitivity, specificity, PPV and NPV of 43.8.1%, 68.7%, 76.1% and 54.9% at days 1, 3, 2 and 1, respectively (Table 3).
Table 3.
Diagnostic performance of chemiluminescence immunoassay (CLIA), electro-chemiluminescence (ECLIA) and chemiluminescent microparticle immunoassay (CMIA) protocol from published data
| Citation | Product name/source | Type | Sample Size | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Limitation of study |
|---|---|---|---|---|---|---|---|---|
| Jin et al.a 23 | CLIA test kit Shenzhen YHLO Biotech Co., Ltd (China) | Total | 76 | 88.37 (38/43) | 100 (33/33) | 100 (38/38) | 86.84 (33/38) | a. Sample size used was small as just 43 lab-confirmed COVID-19 participants and 33 apparently healthy participants |
| IgG | 76 | 88.37 (38/43) | 90.91 (30/33) | 92.68 (38/41) | 85.71 (30/35) | b. The period to viral molecular detection and to serological investigation was not constant and was based on clinical judgment | ||
| IgM | 76 | 48.84 (21/43) | 100 (33/33) | 100 (38/38) | 86.84 (33/38) | c. The value of serological investigation in participants with severe cases requires assessment as those enrolled into the study had mild to moderate COVID-19 cases | ||
| d. The average period from clinical hallmark onset to serological investigation was long due to the late availability of testing kits | ||||||||
| e. There was scarse follow-up data on participants who were discharged | ||||||||
| Geurtsvan Kessel et al.c 43 | DiaSorin Liaison XL | Total | 122 | 73.58 (39/53) | 98.55 (68/69) | 97.5 (39/40) | 82.93 (68/82) | a. Lack of sensitivity at the early phase of symptom onset |
| Müller et al.d 61 | LIAISON SARS-CoV-2 S1/S2 IgG CLIA test (DiaSorin S1/S2 IgG) | Total | 42 | NAb | NAb | NAb | NAb | a. Small size used for the study |
| IgG | 42 | 61.54 (16/26) | 68.75 (11/16) | 76.19 (16/21) | 52.38 (11/21) | b. All test kits missed a great proportion of neutralizing antibody | ||
| IgM | 42 | NAb | NAb | NAb | NAb | |||
| SARS-CoV-2 IgG CMIA from Abbott detecting Anti-nucleocapsid IgG antibodies (Abbott N IgG) | Total | 42 | NAb | NAb | NAb | NAb | ||
| IgG | 42 | 61.54 (16/26) | 100.0 (16/16) | 100.0 (16/16) | 61.54 (16/26) | |||
| IgM | 42 | NAb | NAb | NAb | NAb | |||
| Elecsys anti-SARS-CoV-2 ECLIA test from Roche (Roche N Ab) | Total | 42 | 65.38 (17/26) | 100.0 (16/16) | 100.0 (17/17) | 64.0 (16/25) | ||
| IgG | 42 | NAb | NAb | NAb | NAb | |||
| IgM | 42 | NAb | NAb | NAb | NAb | |||
| Kohmer et al.c 62 | SARS-CoV-2 IgG CMIA (Abbott Architect i2000 [N protein-based]; Abbott GmbH, Wiesbaden, Germany) | Total | 80 | NAb | NAb | NAb | NAb | a. These assays, especially that of elecsys anti-SARS-CoV-2, are unable to differentiate between IgA, IgM and IgG |
| IgG | 80 | 77.78 (35/45) | 100.0 (35/35) | 100.0 (35/35) | 77.78 (35/45) | b. Based on the small sample size nature of this study, it is not conclusive as to which antibodies are the most abundant and to which viral proteins (N and S) are most targeted based on the observed dissimilarities in the time frame and viral protein target of the immune response against SARS-CoV-2 | ||
| IgM | 80 | NAb | NAb | NAb | NAb | c. There is discrepancy between the diagnostic performance values of the kits determined in the current study and those generated by the manufacturer's manual and those disclosed by previous literature | ||
| Elecsys anti-SARS-CoV-2 ECLIA test (Roche cobas e 411 analyzer [N protein-based]; Roche Diagnostics International AG, Rotkreuz, Switzerland) | Total | 79 | NAb | NAb | NAb | NAb | ||
| IgG | 79 | 75.56 (34/45) | 97.06 (33/34) | 97.14 (34/35) | 75.0 (33/44) | |||
| IgM | 79 | NAb | NAb | NAb | NAb | |||
| LIAISON XL SARS-CoV-2 S1/S2 IgG CLIA test (DiaSorin S1 and S2 protein-based) (DiaSorin Deutschland GmbH, Dietzenbach, Germany) | Total | 80 | NAb | NAb | NAb | NAb | ||
| IgG | 80 | 75.56 (34/45) | 100.0 (35/35) | 100.0 (34/34) | 76.09 (35/46) | |||
| IgM | 80 | NAb | NAb | NAb | NAb | |||
| Vircell VIRCLIA automation system IgG MONOTEST (CLIA) (S1 and N protein-based) (Vircell Spain S.L.U., Granada, Spain) | Total | 76 | NAb | NAb | NAb | NAb | ||
| IgG | 76 | 88.89 (40/45) | 100.0 (31/31) | 100.0 (40/40) | 86.11 (31/36) | |||
| IgM | 76 | NAb | NAb | NAb | NAb | |||
| Jääskeläinen et al.b 53 | LIAISON SARS-CoV-2 IgG (CLIA) (DiaSorin, Saluggia, Italy) | Total | 111 | NAb | NAb | NAb | NAb | a. Poor diagnostic performance based on sensitivity; liaison rapid test kit revealed no adequacy for clinical use |
| IgG | 111 | 43.75 (14/32) | 94.94 (75/79) | 77.78 (14/18) | 80.65 (75/93) | |||
| IgM | 111 | NAb | NAb | NAb | NAb | |||
| Architect SARS-CoV-2 IgG CMIA assay (Abbott, Illinois, USA) | Total | 123 | NAb | NAb | NAb | NAb | ||
| IgG | 123 | 80.49 (33/41) | 95.12 (78/82) | 89.19 (33/37) | 90.7 (78/86) | |||
| IgM | 123 | NAb | NAb | NAb | NAb | |||
| Wolff et al.a 63 | Elecsys anti-SARS CoV-2 IgM/IgG assay (Roche Diagnostics, Vilvoorde, Belgium) | Total | 207 | 81.08 (90/111) | 100.0 (96/96) | 100.0 (90/90) | 82.05 (96/117) | a. Low detection rate at early stage of COVID-19 infection |
| IgG | 207 | NAb | NAb | NAb | NAb | |||
| IgM | 207 | NAb | NAb | NAb | NAb | |||
| Liaison SARS-CoV-2 IgG kit (CLIA) (Diasorin, Saluggia, Italy) | Total | 207 | NAb | NAb | NAb | NAb | ||
| IgG | 207 | 70.27 (78/111) | 97.92 (94/96) | 97.5 (78/80) | 74.02 (94/127) | |||
| IgM | 207 | NAb | NAb | NAb | NAb | |||
| Montesinos et al.a 55 | Maglumi 2019-n-Cov IgG and IgM (CLIA) | Total | 194 | 63.11 (77/122) | 100.0 (72/72) | 100.0 (77/77) | 61.54 (72/117) | a. The criteria for evaluating the period of illness onset were retrieved from medical archives and may include imprecisions due to subjectivity in the lack of objective determination of symptoms and periods |
| IgG | 198 | 53.17 (67/126) | 100.0 (72/72) | 100.0 (67/67) | 54.96 (72/131) | b. Low diagnostic performance based on sensitivity | ||
| IgM | 198 | 58.73 (74/126) | 100.0 (72/72) | 100.0 (74/74) | 58.06 (72/124) | |||
| Infantino et al.a 66 | SARS-CoV‐2 antibodies IgM and IgG at cuff-off values 10.0 AU/mL respectively for CLIA kits (Shenzhen YHLO Biotech Co, Ltd, China) | Total | 105 | 67.21 (47/61) | 100.0 (44/44) | 100.0 (47/47) | 75.86 (44/58) | a. Variation in the period between sampling and symptom onsets |
| IgG | 105 | 67.21 (47/61) | 100.0 (44/44) | 100.0 (47/47) | 75.86 (44/58) | b. Late-stage enrolment of study participants | ||
| IgM | 105 | 73.77 (45/61) | 93.18 (41/44) | 93.75 (45/48) | 71.93 (41/57) | c. None of the test group of participants provided a negative status sample | ||
| Nuccetelli et al.a 57 | CLIA | Total | 83 | 95.35 (41/43) | (100.0 (40/40) | 100.0 (41/41) | 95.24 (40/42) | a. No further study on the relation between antibody levels and protective immune response |
| IgG | 83 | 95.35 (41/43) | (100.0 (40/40) | 100.0 (41/41) | 95.24 (40/42) | |||
| IgM | 83 | 83.72 (36/43) | 95.0 (38/40) | 94.74 (36/38) | 84.44 (38/45) | |||
| Ma et al.a 67 | CLIA RBD-specific anti-SARS-CoV-2 IgA, IgM and IgG kit | Total | 699 | 94.44 (204/216) | 90.48 (437/483) | 81.6 (204/250) | 97.33 (437/449) | a. Irregular and prolonged average sampling duration, which could influence the accuracy of the assay |
| IgG | 699 | 96.76 (209/216) | 99.79 (482/483) | 99.52 (209/210) | 98.57 (482/489) | b. No evaluation of the relationship between immunoglobulin levels and severity of the disorder | ||
| IgM | 699 | 96.76 (209/216) | 92.34 (446/483) | 84.96 (209/246) | 98.45 (446/453) | |||
| Qian et al.a 68 | CLIA test kit Shenzhen YHLO Biotech Co., Ltd (China) | Total | 2113 | NAb | NAb | NAb | NAb | a. Insufficient data on sensitivity for convalescent samples due to limited period after the development of SARS-CoV-2 IgM/IgG assays and access to limited participant demographics |
| IgG | 2113 | 95.68 (531/555) | 98.07 (1528/1558) | 94.65 (531/561) | 98.45 (1528/1552) | |||
| IgM | 2113 | 84.68 (470/555) | 98.14 (1529/1558) | 94.19 (470/499) | 94.44 (1529/1614) | |||
| Suhandynata et al.a 69 | Diazyme DZ-LITE 2019-nCoV IgG (CLIA) Assay Kit (cat. # 130219015M)/ IgM (CLIA) assay kit (cat. # 130219016M) | Total | 289 | 100.0 (54/54) | 98.72 (232/235) | 94.74 (54/57) | 100.0 (232/232) | a. 50 of the 54 SARS-CoV-2-confirmed participants were hospitalized and were more likely have acute phase infection compared with other average participants that were infected with COVID-19 |
| b. A participant had a medical history of common variable IgG immunodeficiency, which can adversely affect the diagnostic performance of the kit based on sensitivity as the participant was wrongly categorized as false-negative for IgG despite the positive status as revealed using PCR | ||||||||
| c. Insufficient serologic data on SARS-CoV-2 participants with less severe symptoms who recovered | ||||||||
| IgG | 289 | 94.44 (51/54) | 99.14 (233/235) | 96.23 (51/53) | 98.73 (233/236) | |||
| IgM | 289 | 88.89 (48/54) | 99.57 (234/235) | 97.96 (48/49) | 97.5 (234/240) |
Abbreviations: CLIA, chemiluminescence immunoassay; IFT, immunofluorescence test; NAb, not available; NPV, negative predictive value; NT, neutralization test; POS, postonset of symptoms; PPV, positive predictive value; PRNT, plaque-reduction neutralization assay; RBD, receptor-binding domain.
a diagnostic performance performed with reference to RT-PCR.
b diagnostic performance done with reference to microneutralization assay (MNT)
c diagnostic performance performed with reference to PRNT.
d diagnostic performance performed with reference to NT and IFT.
Note:
1. All computed values were POS.
2. All products with ≥95% each for Sensitivity, Specificity, PPV and NPV may be used for epidemiological purposes. Furthermore, performance characteristics ≥95% values reported from acute COVID-19 samples could be considered for clinical use (in conjunction with clinical presentations of patients).
The detection, peak and decline periods of blood anti-SARS-CoV-2 IgM, IgG and total antibodies for POCT, ELISA and CLIA vary widely. The most promising of these assays for POCT detected anti-SARS-CoV-2 at day 3 POS in 21.1% (n=19) and peaked on the 15th day in 93.3% (n=21)58 of COVID-19 patients; ELISA products detected anti-SARS-CoV-2 IgM and IgG at days 2 and 6 in 34.1% (n=38)71 and in 46.7% (n=15)52 COVID-19 patients, respectively, and peaked on the eighth day in 92.1% (n=38)71 of COVID-19 patients. The most promising CLIA product detected anti-SARS-CoV-2 IgM and IgG at days 1 and 4 in 33.3% (n=6)23 and 60.0% (n=35),63 respectively, and peaked on the 30th day in 97.8% (n=87)73 of COVID-19 patients (Tables 4-6).
Table 4.
Diagnostic monitoring of immunoglobulins by point-of-care test serological protocol from published data
| Citation | Product name/source | Antibody assessment type | Detection time range (d) | Mean time of detection (d) | Peak period (d) | Period of decline (d) |
|---|---|---|---|---|---|---|
| Van Elslande et al.52 | Clungene COVID-19 IgG/IgM rapid test | Total | 5–6 | 5 | 17–18 | NAb |
| IgG | 5–6 | 7 | 17–18 | NAb | ||
| IgM | 5–6 | 5 | 17–18 | NAb | ||
| OrientGene COVID-19 IgG/IgM rapid test | Total | 5–6 | 5 | 17–18 | NAb | |
| IgG | 5–6 | 7 | 17–18 | NAb | ||
| IgM | 5–6 | 5 | 17–18 | NAb | ||
| VivaDiag COVID-19 IgG/IgM rapid test | Total | 5–6 | 5 | 17–18 | NAb | |
| IgG | 5–6 | 7 | 17–18 | NAb | ||
| IgM | 5–6 | 5 | 17–18 | NAb | ||
| StrongStrep COVID-19 IgG/IgM rapid test | Total | 5–6 | 5 | 17–18 | NAb | |
| IgG | 5–6 | 7 | 17–18 | NAb | ||
| IgM | 5–6 | 5 | 17–18 | NAb | ||
| Dynammiker COVID-19 IgG/IgM rapid test | Total | 5–6 | 5 | 17–18 | NAb | |
| IgG | 5–6 | 7 | 17–18 | NAb | ||
| IgM | 5–6 | 5 | 17–18 | NAb | ||
| Multi-G COVID-19 IgG/IgM rapid test | Total | 5–6 | 5 | 17–18 | NAb | |
| IgG | 5–6 | 7 | 17–18 | NAb | ||
| IgM | 5–6 | 5 | 17–18 | NAb | ||
| Prima COVID-19 IgG/IgM rapid test | Total | 5–6 | 5 | 17–18 | NAb | |
| IgG | 5–6 | 7 | 17–18 | NAb | ||
| IgM | 5–6 | 5 | 17–18 | NAb | ||
| Montesinos et al.55 | 2019-n-CoV IgG/IgM rapid test cassette (LaboOn Time) (LabOn Time, Bio Marketing Diagnostics, or Akiva, Israel) | Total | 0–7 | 4 | >15 | >15 |
| IgG | 0–7 | 6 | >15 | >15 | ||
| IgM | 0–7 | 4 | >15 | >15 | ||
| Novel coronavirus (2019-n-CoV) antibody IgG/IgM assay (colloidal gold) (Avioq, Bio-Tech, Shandong, China) | Total | 0–7 | 4 | >15 | >15 | |
| IgG | 0–7 | 6 | >15 | >15 | ||
| IgM | 0–7 | 4 | >15 | >15 | ||
| QuickZen COVID-19 IgM/IgG kit (ZenTech, Angleur, Belgium) | Total | 0–7 | 4 | >15 | >15 | |
| IgG | 0–7 | 7 | >15 | >15 | ||
| IgM | 0–7 | 4 | >15 | >15 | ||
| Pérez-García et al.58 | AllTest COV-19 IgG/IgM kit (AllTest Biotech, Hangzhou, China) | Total | NAb | NAb | NAb | NAb |
| IgG | 1–6 | 3 | 31–36 | >36 | ||
| IgM | 1–6 | 3 | 13–18 | 25–30 |
Abbreviations: NAb, not available; POS, postonset of symptoms.
Note: All computed days were POS.
Table 6.
Diagnostic monitoring of immunoglobulins by CLIA test serological protocol from published data
| Citation | Product name/source | Antibody assessment type | Detection time range (d) | Mean time of detection (d) | Peak period (d) | !Period of decline (d) |
|---|---|---|---|---|---|---|
| Long et al.72 | MCLIA kits (Bioscience Co.; approved by the China National Medical Products Administration) | Total | 2–4 | 2 | 11–13 | >23 |
| IgG | 2–4 | 4 | 11–13 | >23 | ||
| IgM | 2–4 | 2 | 11–13 | >23 | ||
| Jin et al.23 | CLIA test kit Shenzhen YHLO Biotech Co., Ltd (China) | Total | 1–5 | 1 | 16–20 | >32 |
| IgG | 1–5 | 5 | 16–20 | >32 | ||
| IgM | 1–5 | 1 | 16–20 | 21–25 | ||
| Padoan et al.70 | CLIA assay (MAGLUMI 2000 Plus) | Total | NAb | NAb | NAb | NAb |
| IgG | NAb | NAb | NAb | NAb | ||
| IgM | 4 | 4 | 12 | 34 | ||
| Padoan et al.73 | MAGLUMI 2000 Plus 2019-nCov IgM and IgG assays (Snibe, Shenzhen, China) | Total | <5 | <5 | 26–30 | >30 |
| IgG | <5 | <5 | 26–30 | >30 | ||
| IgM | <5 | <5 | 12–13 | 18–19 | ||
| Wolff et al.63 | Elecsys anti-SARS CoV-2 IgM/IgG assay (Roche Diagnostics, Vilvoorde, Belgium) | Total | 4 | 4 | 11 | >24 |
| IgG | NAb | NAb | NAb | NAb | ||
| IgM | NAb | NAb | NAb | NAb | ||
| Liaison SARS-CoV-2 IgG kit (CLIA) (Diasorin, Saluggia, Italy) | Total | NAb | NAb | NAb | NAb | |
| IgG | 4 | 4 | 11–13 | >24 | ||
| IgM | NAb | NAb | NAb | NAb | ||
| Hou et al.74 | Anti-SARS-CoV-2 CLIA-YHLO kit | Total | ||||
| IgG | 3 | 3 | 48 | >48 | ||
| IgM | 3 | 3 | 30 | 48 | ||
| Montesinos et al.55 | Maglumi 2019-n-Cov IgG and IgM (CLIA) | Total | 0–7 | 4 | >15 | >15 |
| IgG | 0–7 | 7 | >15 | >15 | ||
| IgM | 0–7 | 4 | >15 | >15 | ||
| Ma et al.67 | CLIA RBD-specific anti-SARS-CoV-2 IgA, IgM, and IgG kit | Total | NAb | NAb | NAb | NAb |
| IgG | 4–10 | 10 | 16–41 | >41 | ||
| IgM | 4–10 | 7 | 11–30 | 31–41 | ||
| IgA | 4–10 | 7 | 11–20 | 21–25 | ||
| Qian et al.68 | CLIA test kit Shenzhen YHLO Biotech Co., Ltd (China) | Total | NAb | NAb | NAb | NAb |
| IgG | 6 | 6 | 20 | >35 | ||
| IgM | 6 | 6 | 20 | 35 | ||
| Suhandynata et al.69 | Diazyme DZ-LITE 2019-nCoV IgG (CLIA) assay kit (cat. # 130219015M)/IgM (CLIA) assay kit (cat. # 130219016M) | Total | NAb | NAb | NAb | NAb |
| IgG | 2–6 | 6 | 8–22 | ≥24 | ||
| IgM | 0–4 | 3 | 6–8 | 14–22 |
Abbreviations: CLIA, chemiluminescence immunoassay; NAb, not available; POS, postonset of symptoms; RBD, receptor-binding domain.
Note:
1. All computed values were POS.
Table 5.
Diagnostic monitoring of immunoglobulins by ELISA test serological protocol from published data
| Citation | Product name/source | Antibody assessment type | Detection time range (d) | Mean time of detection (d) | Peak period (d) | Period of decline (d) |
|---|---|---|---|---|---|---|
| Van Elslande et al.52 | Euroimmun | Total | NAb | NAb | NAb | NAb |
| IgG | 5–6 | 6 | 17–18 | NAb | ||
| IgM | NAb | NAb | NAb | NAb | ||
| Zhao et al.17 | COVID-19 ELISA kit (Beijing Wantai Biological Pharmacy Enterprise Co. Ltd) | Total | 7 | 4 | 14–25 | >35 |
| IgG | 14 | 14 | 25 | >35 | ||
| IgM | 7 | 4 | 14 | 21 | ||
| Xiang et al.59 | Sandwich ELISA kit (Livzon Inc., Zhuhai, China, lot numbers 20200308 [IgM] and 20200308 [IgG]) | Total | 4 | 4 | 24 | 31 |
| IgG | 4 | 4 | 24 | 28 | ||
| IgM | 4 | 4 | 18 | 28 | ||
| Padoan et al.70 | COVID-19 IgG/IgA ELISA kit (Euroimmun Medizinische Laboradiagnostika, Luebeck, Germany) | Total | NAb | NAb | NAb | NAb |
| IgG | NAb | NAb | NAb | NAb | ||
| IgA | 4 | 4 | 18 | 34 | ||
| Jääskeläinen et al.60 | Anti-SARS-CoV-2 IgA and IgG EIA (Euroimmun, Lübeck, Germany) | Total | 11 | 11 | NAb | NAb |
| IgG | 12 | 12 | NAb | NAb | ||
| IgA | 11 | 11 | NAb | NAb | ||
| Okba et al.29 | Anti-SARS-CoV-2 IgG and IgA ELISA (EUROIMMUN Medizinische Labordiagnostika AG) | Total | 5 | 5 | 13–21 | >21 |
| IgG | 5 | 5 | 13–21 | >21 | ||
| IgA | 5 | 5 | 11–15 | 20 | ||
| Montesinos et al.55 | Euroimmun anti-SARS-CoV-2 ELISA IgG and IgA assays (Euroimmun, Luebeck, Germany) | Total | 0–7 | 2 | >15 | >15 |
| IgG | 0–7 | 7 | >15 | >15 | ||
| IgA | 0–7 | 2 | >15 | >15 | ||
| Sun et al.71 | In-house ELISA produced using N protein (residue 1–419) from baculovirus insect cells (cat. # 40588-V08B, Sino biological, Beijing, China) and S protein (residue 16–685) from HEK293 cells (cat. #40591-V08H, Sino biological, Beijing, China) | Total | 0–7 | 2 | 8–15 | >22 |
| IgG | 0–7 | 7 | 8–15 | >22 | ||
| IgM | 0–7 | 2 | 8–15 | 22 | ||
| Beavis et al.65 | EUROIMMUN anti-SARS-CoV-2 assay | Total | NAb | NAb | NAb | NAb |
| IgG | 0–2 | 2 | 19–49 | ≥50 | ||
| IgA | 0–2 | 2 | 19–49 | ≥50 |
Abbreviations: NAb, not available; POS, postonset of symptoms.
Note:
1. All computed values were POS.
Conclusions
Given the varied performance characteristics of all the serological assays, there is a need to continuously improve their detection thresholds, as well as to monitor and re-evaluate their performances to ensure their significance and applicability for COVID-19 clinical and epidemiological purposes. When found satisfactory, their use will be imperative for scaling up COVID-19 testing in the face of the economic downturn in resource-limited countries. It is recommended that public institutions, private firms, researchers and healthcare policymakers consider the development and evaluation of alternative means of reducing the current PCR test turnaround time and improving the test capacity of serological tests to secure improved epidemiological data for SARS-CoV-2.
Acknowledgements
The authors greatly appreciate Gabriel Ilerioluwa Oke for proofreading and copyediting parts of the manuscript.
Contributor Information
Anthony Uchenna Emeribe, Department of Medical Laboratory Science, Faculty of Allied Medical Sciences, University of Calabar, P.M.B 1115, Calabar, Cross River State, Nigeria.
Idris Nasir Abdullahi, Department of Medical Laboratory Science, Faculty of Allied Health Sciences, College of Medical Sciences, Ahmadu Bello University, Zaria, Nigeria.
Halima Ali Shuwa, University Health Services, College of Health and Medical Sciences, Federal University, Dutse, Nigeria.
Leonard Uzairue, Department of Microbiology, Federal University of Agriculture Abeokuta, Nigeria.
Sanusi Musa, Department of Medical Laboratory Science, Faculty of Allied Health Sciences, College of Medical Sciences, Ahmadu Bello University, Zaria, Nigeria.
Abubakar Umar Anka, Department of Medical Laboratory Science, Faculty of Allied Health Sciences, College of Medical Sciences, Ahmadu Bello University, Zaria, Nigeria.
Hafeez Aderinsayo Adekola, Department of Microbiology, Olabisi Onabanjo University, Ago-Iwoye, Nigeria.
Zakariyya Muhammad Bello, Department of Medical Laboratory Science, Faculty of Allied Health Sciences, College of Medical Sciences, Ahmadu Bello University, Zaria, Nigeria.
Lawal Dahiru Rogo, Department of Medical Laboratory Science, Faculty of Allied Health Sciences, Bayero University, Kano Nigeria.
Dorcas Aliyu, Department of Medical Laboratory Science, Faculty of Allied Medical Sciences, University of Calabar, P.M.B 1115, Calabar, Cross River State, Nigeria.
Shamsuddeen Haruna, Department of Medical Laboratory Science, Faculty of Allied Health Sciences, College of Medical Sciences, Ahmadu Bello University, Zaria, Nigeria.
Yahaya Usman, Department of Medical Laboratory Science, Faculty of Allied Health Sciences, College of Medical Sciences, Ahmadu Bello University, Zaria, Nigeria.
Habiba Yahaya Muhammad, Department of Medical Laboratory Science, Faculty of Allied Health Sciences, Bayero University, Kano Nigeria.
Abubakar Muhammad Gwarzo, Department of Medical Microbiology and Parasitology, Federal University, Dutse, Nigeria.
Justin Onyebuchi Nwofe, Department of Medical Laboratory Science, University of Nigeria, Enugu, Nigeria.
Hassan Musa Chiwar, Department of Medical Laboratory Science, University of Maiduguri Maiduguri, Nigeria.
Chukwudi Crescent Okwume, Department of Medical Laboratory Services, University of Nigeria Teaching Hospital, Enugu, Nigeria.
Olawale Sunday Animasaun, Nigeria Field Epidemiology and Laboratory Training Programme, African Field Epidemiology Network, Abuja, Nigeria.
Samuel Ayobami Fasogbon, Public Health In-vitro Diagnostic Control Laboratory, Medical Laboratory Science Council of Nigeria, Lagos, Nigeria.
Lawal Olayemi, School of Medicine, Faculty of Health Sciences, National University of Samoa, Apia, Samoa.
Christopher Ogar, Department of Medical Laboratory Science, Faculty of Allied Medical Sciences, University of Calabar, P.M.B 1115, Calabar, Cross River State, Nigeria.
Chinenye Helen Emeribe, Department of Family Medicine, University of Calabar Teaching Hospital, PMB 1278 Calabar, Cross River, Nigeria.
Peter Elisha Ghamba, WHO National Polio Reference Laboratory, University of Maiduguri Teaching Hospital, Maiduguri, Nigeria.
Luqman O Awoniyi, Institute of Biomedicine, and MediCity Research Laboratories, University of Turku, 20014 Turku, Finland.
Bolanle O P Musa, Immunology Unit, Department of Medicine, Ahmadu Bello University, Zaria, Nigeria.
Authors' contributions:
INA, LDR and AUE conceptualized and planned the study. INA, AUE, HAS, LU, SM, AUA, HAA, LDR, DA, SH, YU, HYM, AMG, JON, HMC, CCO, OSA, LO, CO, CNE, PEG, LOA and BOPM conducted the literature search and compilation of data. INA, AUE, HAS, LU, SM, AUA, HAA and LDR performed the data curation and statistical analysis. All the authors participated in writing the draft, revised and final versions of the manuscript. All the authors read and approved the final version of the manuscript for intellectual content before submission. INA is responsible for the overall content as guarantor.
Funding
None received.
Competing interests
None.
Ethical approval
Not applicable (this is a review article).
Data availability
This study being a review article, the data presented in the results section and in discussing our main findings are well referenced. However, raw data will be made available on request through the corresponding author (I.N. Abdullahi).
References
- 1. Decaro N, Lorusso A. Novel human coronavirus (SARS-CoV-2): A lesson from animal coronaviruses. Vet Microbiol. 2020;244:108693. doi: 10.1016/j.vetmic.2020.108693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Worldometers.info . Situation Update Worldwide, as of 2 July 2020. Delaware, USA: Dove, 2020.. Available at https://www.worldometers.info/coronavirus/#countries [accessed 20 July 2020]. [Google Scholar]
- 3. Padula WV. Why only test symptomatic patients? Consider random screening for COVID-19. Appl Health Econ Health Policy. 2020;18(3):333–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hellewell J, Abbott S, Gimma A et al. Feasibility of controlling COVID-19 outbreaks by isolation of cases and contacts. Lancet Global Health. 2020;8(4):e488–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ueda M, Martins R, Hendrie PC et al. Managing cancer care during the COVID-19 pandemic: agility and collaboration toward a common goal. J Natl Compr Canc Netw. 2020;18(4):366–9. [DOI] [PubMed] [Google Scholar]
- 6. Ranney ML, Griffeth V, Jha AK. Critical supply shortages — the need for ventilators and personal protective equipment during the Covid-19 pandemic. N Engl J Med. 2020;382(18):e41. [DOI] [PubMed] [Google Scholar]
- 7. Villaraza, Angangco . FDA issuances on evaluation, approval and use of COVID-19 test kits. Available at https://www.lexology.com/library/detail.aspx?g=7689832c-83ce-4683-a5ea-4630eee5788f [accessed 1 June 2020]. [Google Scholar]
- 8. Rashid ZZ, Othman SN, Samat MNA et al. ,. Diagnostic performance of COVID-19 serology assays. Malaysian J Pathol. 2020;42(1):13–21. [PubMed] [Google Scholar]
- 9. Cui J, Li F, Shi Z-L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2018;17(3):181–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chan JF-W, Kok K-H, Zhu Z et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9(1):221–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wu A, Peng Y, Huang B et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27(3):325–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ji W, Wang W, Zhao X, Zai J, Li X. Cross-species transmission of the newly identified coronavirus 2019-nCoV. J Med Virol. 2020;92(4):433–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. World Health Organization. 2020 Global Surveillance for human infection with coronavirus disease (COVID-2019), interim guidance, Geneva. Available at https://www.who.int/publicationsdetail/global-surveillance-for-humaninfection-with-novel-coronavirus-(2019-ncov) [accessed 4 April 2020]. [Google Scholar]
- 14. Happi C, Ihekweazu C, Oluniyi PE et al. SARS-CoV-2 genomes from Nigeria reveal community transmission, multiple virus lineages and spike protein mutation associated with higher transmission and pathogenicity. Virological. 2020. http://virological.org/t/sars-cov-2-genomes-from-nigeria-reveal-community-transmission-multiple-virus-lineages-and-spike-protein-mutation-associated-with-higher-transmission-and-pathogenicity/494. [accessed 2 June 2020]. [Google Scholar]
- 15. Xiao AT, Gao C, Zhang S. Profile of specific antibodies to SARS-CoV-2: The first report. J Infect. 2020;81(1):147–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Morrison A, Li Y, Loshak H. Serological tests for COVID-19. Ottawa: CADTH; 2020. (CADTH Horizon Scan; 188:13). [Google Scholar]
- 17. Zhao J, Yuan Q, Wang H et al. Antibody Responses to SARS-CoV-2 in Patients With Novel Coronavirus Disease 2019. Clin Infect Dis. 2020;71(16):2027–34. doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Long QX, Liu BZ, Deng HJ et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020;26(6):845–8. [DOI] [PubMed] [Google Scholar]
- 19. Li Z, Yi Y, Luo X et al. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J Med Virol. 2020;92(9):1518–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hoffman T, Nissen K, Krambrich J et al. Evaluation of a COVID-19 IgM and IgG rapid test; an efficient tool for assessment of past exposure to SARS-CoV-2. Infect Ecol Epidemiol. 2020;10(1):1754538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mo HY, Xu J, Ren XL et al. Evaluation by indirect immunofluorescent assay and enzyme linked immunosorbent assay of the dynamic changes of serum antibody responses against severe acute respiratory syndrome coronavirus. Chin Med J. 2005;118(6):446–50. [PubMed] [Google Scholar]
- 22. Cao W-C, Liu W, Zhang P-H, Zhang F, Richardus JH. Disappearance of antibodies to SARS-associated coronavirus after recovery. N Engl J Med. 2007;357(11):1162–3. [DOI] [PubMed] [Google Scholar]
- 23. Jin Y, Wang M, Zuo Z et al. Diagnostic value and dynamic variance of serum antibody in coronavirus disease 2019. Int J Infect Dis. 2020;94:49–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guo L, Ren L, Yang S et al. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19). Clin Infect Dis. 2020;71(15):778–85. doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cerutti A. The regulation of IgA class switching. Nat Rev Immunol. 2008;8(6):421–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Szomolanyi-Tsuda E, Welsh RM. T-cell-independent antiviral antibody responses. Curr Opin Immunol. 1998;10(4):431–5. [DOI] [PubMed] [Google Scholar]
- 27. Liu W, Liu L, Kou G et al. Evaluation of nucleocapsid and spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against SARS-CoV-2. J Clin Microbiol. 2020;58(6):e00461–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Amanat F, Stadlbauer D, Strohmeier S et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med. 2020;26(7):1033–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Okba NMA, Müller MA, Li W et al. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg Infect Dis. 2020;26(7):1478–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang W, Du R-H, Li B et al. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microb Infect. 2020;9:386–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zeng H, Xu C, Fan J et al. Antibodies in Infants Born to Mothers With COVID-19 Pneumonia. JAMA. 2020;323(18):1848–9. doi:10.1001/jama.2020.4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Weinstein MC, Freedberg KA, Hyle EP, Paltiel AD. Waiting for certainty on Covid-19 antibody tests - at what cost? N Engl J Med. 2020;383(6):e37. [DOI] [PubMed] [Google Scholar]
- 33. Whitman JD, Hiatt J, Mowery CT et al. Test performance evaluation of SARS-CoV-2 serological assays. medRxiv. [Preprint]2020 May 17. doi: 10.1101/2020.04.25.20074856. [Google Scholar]
- 34. Mandavilli A. Coronavirus Antibody Tests: Can You Trust the Results? The New York Times, 2020. Retrieved from: https://www.nytimes.com/2020/04/24/health/coronavirus-antibody-tests.html. [Google Scholar]
- 35. Lassaunière R, Frische A, Harboe ZB et al. Evaluation of nine commercial SARS-CoV-2 immunoassays. medRxiv. 2020. doi: https://doi.org/10.1101/2020.04.09.20056325. [Google Scholar]
- 36. Nicol T, Lefeuvre C, Serri O et al. Assessment of SARS-CoV-2 serological tests for the diagnosis of COVID-19 through the evaluation of three immunoassays: Two automated immunoassays (Euroimmun and Abbott) and one rapid lateral flow immunoassay (NG Biotech). J Clin Virol. 2020;129:104511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lin D, Liu L, Zhang M et al. Evaluations of the serological test in the diagnosis of 2019 novel coronavirus (SARS-CoV-2) infections during the COVID-19 outbreak. Eur J Clin Microbiol Infect Dis. 2020;39(12):2271–7. doi: 10.1007/s10096-020-03978-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chew KL, Tan SS, Saw S et al. Clinical evaluation of serological IgG antibody response on the Abbott Architect for established SARS-CoV-2 infection. Clin Microbiol Infect. 2020;26(9):1256.e9–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yang HS, Racine-Brzostek SE, Lee WT et al. SARS-CoV-2 antibody characterization in emergency department, hospitalized and convalescent patients by two semi-quantitative immunoassays. Clin Chim Acta. 2020;509:117–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nagura-Ikeda M, Imai K, Tabata S et al. Clinical Evaluation of Self-Collected Saliva by Quantitative Reverse Transcription-PCR (RT-qPCR), Direct RT-qPCR, Reverse Transcription-Loop-Mediated Isothermal Amplification, and a Rapid Antigen Test To Diagnose COVID-19. J Clin Microbiol. 2020;58(9):e01438–20. doi: 10.1128/JCM.01438-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. National SARS-CoV-2 Serology Assay Evaluation Group . Performance characteristics of five immunoassays for SARS-CoV-2: a head-to-head benchmark comparison. Lancet Infect Dis. 2020;20(12):1390–1400. doi: 10.1016/S1473-3099(20)30634-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Theel ES, Harring J, Hilgart H, Granger D. Performance characteristics of four high-throughput immunoassays for detection of IgG antibodies against SARS-CoV-2. J Clin Microb. 2020;58:e01243–20. doi: 10.1128/JCM.01243-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. GeurtsvanKessel CH, OKBA NMA, Igloi Z et al. Towards the next phase: evaluation of serological assays for diagnostics and exposure assessment. medRxiv 2020.04.23.20077156. doi: https://doi.org/10.1101/2020.04.23.20077156. [Google Scholar]
- 44. Cassaniti I, Novazzi F, Giardina F et al. Members of the San Matteo Pavia COVID-19 Task Force. Performance of VivaDiag COVID-19 IgM/IgG Rapid Test is inadequate for diagnosis of COVID-19 in acute patients referring to emergency room department. J Med Virol. 2020;92(10):1724–7. doi: 10.1002/jmv.25800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Porte L, Legarraga P, Vollrath V et al. Evaluation of novel antigen-based rapid detection test for the diagnosis of SARS-CoV-2 in respiratory samples. Int J Infect Dis. 2020;99:328–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pan Y, Li X, Yang G et al. Serological immunochromatographic approach in diagnosis with SARS-CoV-2 infected COVID-19 patients. J Infect. 2020;81(1):e28–32. doi: 10.1016/j.jinf.2020.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pallett S, Denny S, Patel A et al. Point-of-care serological assays for SARS-CoV-2 in a UK hospital population: potential for enhanced case. Research Square Preprint. 2020. doi: 10.21203/rs.3.rs-28006/v1. [Google Scholar]
- 48. Spicuzza L, Montineri A, Manuele R et al. Reliability and usefulness of a rapid IgM-IgG antibody test for the diagnosis of SARS-CoV-2 infection: A preliminary report. J Infect. 2020;81:e53–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wu J-L, Tseng W-P, Lin C-H et al. Four point-of-care lateral flow immunoassays for diagnosis of COVID-19 and for assessing dynamics of antibody responses to SARS-CoV-2. J Infect. 2020;81:435–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Green K, Graziadio S, Turner P et al. Molecular and antibody point-of-care tests to support the screening, diagnosis and monitoring of COVID-19. The Centre for Evidence-based Medicine. 2020. https://www.cebm.net/covid-19/molecular-and-antibody-point-of-care-tests-to-support-the-screening-diagnosis-and-monitoring-of-covid-19/. [Google Scholar]
- 51. Mlcochova P, Collier D, Ritchie AV et al. Combined point of care SARS-CoV-2 nucleic acid and antibody testing in suspected moderate to severe COVID-19 disease. Cell Reports Medicine. 2020;1(6):100099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Van Elslande J, Houben E, Depypere M et al. Diagnostic performance of seven rapid IgG/IgM antibody tests and the Euroimmun IgA/IgG ELISA in COVID-19 patients. Clin Microbiol Infect. 2020;26:1082–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jääskeläinen A, Kuivanen S, Kekäläinen E et al. Performance of six SARS-CoV-2 immunoassays in comparison with microneutralisation. J Clin Virol. 2020;129:104512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kohmer N, Westhaus S, Rühl C, Ciesek S, Rabenau HF. Clinical performance of different SARS-CoV-2 IgG antibody tests. J Med Virol. 2020;92(10):2243–7. doi: 10.1002/jmv.26145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Montesinos I, Gruson D, Kabamba B et al. Evaluation of two automated and three rapid lateral flow immunoassays for the detection of anti-SARS-CoV-2 antibodies. J Clin Virol. 2020;128:104413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Adams ER, Ainsworth M, Anand R et al. Antibody testing for COVID-19: a report from the national COVID scientific advisory panel. medRxiv. 2020.04.15.20066407. doi: https://doi.org/10.1101/2020.04.15.20066407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nuccetelli M, Pieri M, Grelli S et al. SARS-CoV-2 infection serology: a useful tool to overcome lockdown? Cell Death Discov. 2020;6:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pérez-García F, Pérez-Tanoira R, Romanyk J, Arroyo T, Gómez-Herruz P, Cuadros-Gonzàlez J. Alltest rapid lateral flow immunoassays is reliable in diagnosing SARS-CoV-2 infection from 14 days after symptom onset: A prospective single-center study. J Clin Virol. 2020;129:104473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Xiang F, Wang X, He X et al. Antibody detection and dynamic characteristics in patients with coronavirus disease 2019. Clini Infect Dis. 2020;71:1930–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jääskeläinen AJ, Kekäläinen E, Kallio-Kokko H et al. Evaluation of commercial and automated SARS-CoV-2 IgG and IgA ELISAs using coronavirus disease (COVID-19) patient samples. Eurosurveillance. 2020;25(18):2000603 doi: 10.2807/1560-7917.ES.2020.25.18.2000603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Müller L, Ostermann PN, Walker A et al. Sensitivity of commercial Anti-SARS-CoV-2 serological assays in a high-prevalence setting. medRxiv. 2020.06.11.20128686. doi: https://doi.org/10.1101/2020.06.11.20128686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kohmer N, Westhaus S, Rühl C, Ciesek S, Rabenau HF. Brief clinical evaluation of six high-throughput SARS-CoV-2 IgG antibody assays. J Clin Virol. 2020;129:104480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wolff F, Dahma H, Duterme C et al. Monitoring antibody response following SARS-CoV-2 infection: Diagnostic efficiency of four automated immunoassays. Diagn Microbiol Infect Dis. 2020;98:115140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Francesca C, Alessandra B, Daniele L et al. Evaluation of ELISA tests for the qualitative determination of IgG, IgM and IgA to SARSCoV-2. medRxiv preprint. 2020. doi: https://doi.org/10.1101/2020.05.24.20111682. [Google Scholar]
- 65. Beavis KG, Matushek SM, Abeleda PF et al. Evaluation of the EUROIMMUN Anti-SARS-CoV-2 ELISA Assay for detection of IgA and IgG antibodies. J Clin Virol. 2020;129:104468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Infantino M, Grossi V, Lari B et al. Diagnostic accuracy of an automated chemiluminescent immunoassay for anti-SARS-CoV-2 IgM and IgG antibodies: an Italian experience. J Med Virol. 2020;92:1671–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ma H, Zeng W, He H et al. COVID-19 diagnosis and study of serum SARS-CoV-2 specific IgA, IgM and IgG 2 by a quantitative and sensitive immunoassay. medRxiv preprint. 2020. doi: https://doi.org/10.1101/2020.04.17.20064907. [Google Scholar]
- 68. Qian C, Zhou M, Cheng F et al. Development and multicenter performance evaluation of fully automated SARS-CoV-2 IgM and IgG immunoassays. Clin Chem Lab Med. 2020;58(9):1601–7. doi: 10.1515/cclm-2020-0548. [DOI] [PubMed] [Google Scholar]
- 69. Suhandynata RT, Hoffman MA, Keiner MJ, McLawhon RW, Reed SL, Fitzgerald RL. Longitudinal monitoring of SARS-CoV-2 IgM and IgG seropositivity to detect COVID-19. J App Lab Med. 2020;5:908–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Padoan A, Sciacovelli L, Basso D et al. IgA-Ab response to spike glycoprotein of SARS-CoV-2 in patients with COVID-19: A longitudinal study. Clinica Chimica Acta. 2020;507:164–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sun B, Feng Y, Mo X et al. Kinetics of SARS-CoV-2 specific IgM and IgG responses in COVID-19 patients. Emerg Microb Infect. 2020;9(1):940–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Long Q-X, Liu B-Z, Deng H-J et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020;26(6):845–8. [DOI] [PubMed] [Google Scholar]
- 73. Padoan A, Cosma C, Sciacovelli L, Faggian D, Plebani M. Analytical performances of a chemiluminescence immunoassay for SARS-CoV-2 IgM/IgG and antibody kinetics. Clin Chem Lab Med. 2020;58(7):1081–8. [DOI] [PubMed] [Google Scholar]
- 74. Hou H, Wang T, Zhang B et al. Detection of IgM and IgG antibodies in patients with coronavirus disease 2019. Clin Transl Immunol. 2020;9(5):e1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study being a review article, the data presented in the results section and in discussing our main findings are well referenced. However, raw data will be made available on request through the corresponding author (I.N. Abdullahi).