ABSTRACT
Severe acute respiratory syndrome, coronavirus 2 (SARS-CoV-2), remains to be a threat across the globe. SARS-CoV-2 entry into the host is mediated by binding of viral spike protein to the Human angiotensin-converting enzyme 2 (ACE2) receptor. ACE2 is an essential member of the Renin–Angiotensin system (RAS) involved in maintaining the blood pressure and vascular remodelling. Although ACE2 receptor is the entry point to the host, recent studies show activation of ACE2 to modulate the host to develop a suitable environment for its replication. However, the ACE2 activating the immune signals on SARS-CoV-2 attachment is still under investigation. We have used systems biological approach to construct the host regulatory network upon SARS-CoV-2 attachment to the ACE2 receptor. Since lungs are the primary infection site, we integrate human lung gene expression profile along with the host regulatory network to demonstrate the altered host signalling mechanism in viral infection. Further, the network was functionally enriched to determine immune modulation in the network. We also used the proteomic database to assess the occurrence of similar signalling events in other human tissues that exhibit lineage of infection across different organs. The constructed network contains 133 host proteins with 298 interactions that directly or indirectly connect to the ACE2 receptor. Among 133 proteins, 29 were found to be differentially regulated in the host lungs on SARS-CoV-2 infection. Altered proteins connect multiple proteins in a network that modulates kinase, carboxypeptidase and cytokine activity, leading to changes in the host immune system, cell cycle and signal transduction mechanisms. Further investigation showed the presence of similar signalling events in the kidneys, placenta, pancreas, testis, small intestine and adrenal gland as well. Overall, our results will help in understanding the immune molecular regulatory networks influenced by the ACE2 mediated interaction in other body tissues, which may aid in identifying the secondary health complications associated with SARS-CoV-2 infection.
Keywords: COVID-19, SARS-CoV-2, ACE2/AGT-axis, immune response
The authors expect their results will help advance understanding of the immune molecular regulatory networks influenced by the ACE2 receptor-mediated interaction in other body tissues, which could help find the secondary health complications associated with SARS-CoV-2 infection.
INTRODUCTION
Recent pandemic of SARS-CoV-2 coronavirus causes an acute respiratory infection that leads to pneumonia and multi-organ failure. SARS-CoV-2 transmits from human-to-human that increases its prevalence day by day worldwide. To date, there is no definitive medicine or recommended drug for the treatment for this life-threatening infection. According to the World Health Organization (WHO) report on January 05, 2021, there are 8,39,10386 confirmed cases of COVID-19, including 18,39,660 deaths reported from 216 countries, areas or territories around the world (WHO 2021). Therefore, SARS-CoV-2 continues to be a threat to the world population (Cascella et al. 2020). To mitigate the exponential spread of SARS-CoV-2, infected patients are isolated and treated, later quarantined; these protocols have significantly increased the public's social and financial burden.
SARS-CoV-2 is a positive-sense, single-stranded RNA virus that belongs to the Betacoronavirus lineage (Fehr and Perlman 2015). SARS-CoV-2 encodes 24 proteins and is reported to hijack the human host machinery for its genome synthesis and replication (Viswanathan et al. 2020). The presence of spike protein projecting from the SARS-CoV-2 surface plays a vital role in the interaction with the human host, host specificity, cellular attachment and penetration leading to infection (Shang et al. 2020; Walls et al. 2020). In a comparative study, the spike protein of the SARS-CoV-2 virus showed significant homogeneity with spike proteins of other coronaviruses, suggesting similar or unified fusion events, which takes place in the process of host–pathogen interaction among the coronaviruses (Shang et al. 2020a). For instance, the spike protein of HCoV-NL63 and SARS-CoV binds to ACE2 receptor to enter human cells (Li et al. 2007). ACE2 gene is localized on chromosome X, spanning 39.98 kb in human genomic DNA comprising 18 exons (Samavati and Uhal 2020; Devaux, Rolain and Raoult 2020). ACE2 encodes a glycoprotein comprised of 805 amino acids, which functionally regulates the RAS (Riordan 2003; Kuba et al., 2010), a control machinery involved in blood pressure homeostasis and electrolyte balance (Zaman et al. 2002). Modulation of the RAS is expected on binding of the SARS-CoV-2 virus with the ACE2 receptor. In this context, clinical studies report the high mortality and severity in COVID-19 patients with hypertension (Yang et al. 2020). Also, studies from the past indicate the association of ACE2/Ang1–7 and ACE2/Ang/1–9/Mas axis control on the regulation of expression of pro-inflammatory cytokines like interleukin (IL)-1b, IL-6, TNF-alpha, chemo-attractant protein-1, monocyte and transforming growth factor-b (TGF-β). These pro-inflammatory cytokines are responsible for the processes of vascular remodelling, cardiac, lung fibrosis and pulmonary hypertension (Iwata et al. 2005; Ferreira et al. 2009; Zeng et al. 2009; Sriramula et al. 2011). Recent study by our group reported that the binding of SARS-CoV-2 to the ACE2 receptor modulates the host to develop a suitable environment for the replicating viral genome (Ahmed et al. 2020). The altered host regulatory signal and the mechanism mediating the immune process on SARS-CoV-2 attachment with the ACE2 receptor is still under investigation.
In this study, we use systems biological approach to identify the host regulatory network upon SARS-CoV-2 attachment to the human ACE2 receptor. Since lungs are the primary site of SARS-CoV-2 infection, we used human lung expression profile to demonstrate the altered host mechanism in viral infection. Interestingly, we have identified the proteins mediating immune signals on SARS-CoV-2 interaction with the ACE2 receptor. Additionally, the generated ACE2 regulatory network will help in understanding the immune molecular regulatory networks influenced by the ACE2 mediated interaction in other body tissues, which may aid in identifying the secondary health complications associated with SARS-CoV-2 infection.
METHODOLOGY
Protein interaction
The proteins interacting with the ACE2 receptor were collected from the databases such as BioGRID (https://thebiogrid.org/), Intact (https://www.ebi.ac.uk/intact/), Human Proteinpedia (http://www.humanproteinpedia.org/) and HPRD (https://www.hprd.org/). All collected protein interactions were checked for their expression in lung tissue using Human Proteome Map (http://www.humanproteomemap.org/), Proteomics DB (https://www.proteomicsdb.org/), Human Protein Atlas (https://www.proteinatlas.org/) and HPRD (https://www.hprd.org/). The final lung-specific regulatory network was developed to capture the possible downstream signaling events that can be initiated upon the ACE2 mediated modulation that occurs in the human lungs.
Gene expression analysis
RNA-Seq data was searched in the NCBI, GEO database using the keyword relating SARS-CoV-2 infection and human lungs. The relevant dataset, describing the SARS-CoV-2 infection in primary human lung epithelium (NHBE; GSE147507) was identified. The raw data containing three controls (GSM4432378, GSM4432379 and GSM4432380) and three SARS-CoV-2 infected samples (GSM4432381, GSM4432382 and GSM4432383) were retrieved. Using shell commands, and scripting with the relevant tools the initial quality control and the RNA-Seq pipeline was executed by aligning the reads to hg19 genome and differential expression genes with P-value < 0.05 were identified based on DESeq. Further, the involvement of differentially expressed genes in the regulatory network was determined. Further, we searched for the presence of similar regulatory network in other tissues using Human Proteinpedia, Human Protein Atlas, Human Proteome Map, Human Proteome Browser and ProteomicsDB databases.
Molecular enrichment
To determine the molecular functions exhibited by the regulatory network, we used the BiNGO module in Cytoscape software. The significantly over represented Gene Ontology (GO) in the network was confirmed by the hyper-geometric method with a false discovery rate (FDR) corrected P-values < 0.05. Further, the molecular pathways associated with the network were identified using the Reactome database. Among the various enriched molecular pathways, the over representing pathways associated with the network was confirmed by FDR corrected P-values < 0.05.
RESULTS
ACE2 mediating protein signalling
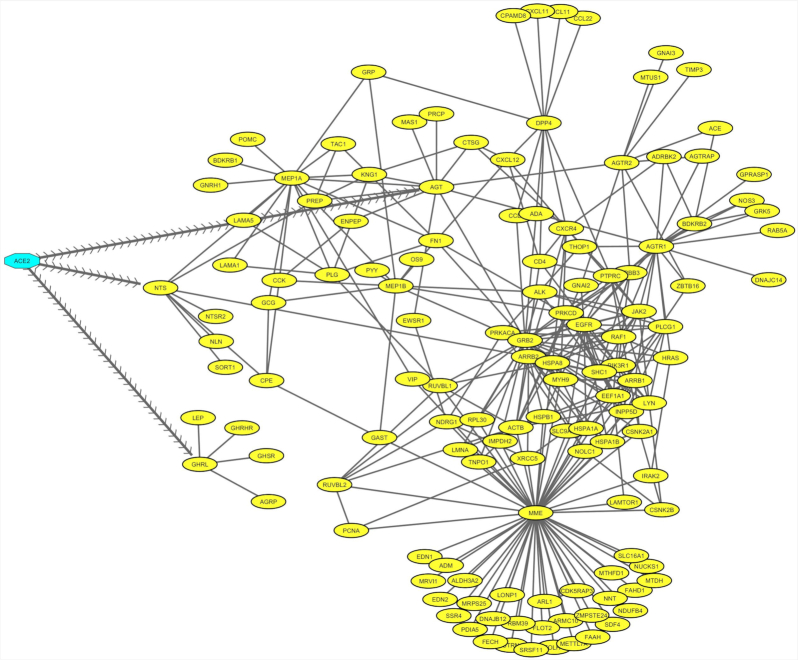
Regulatory network was constructed, highlighting the ACE2–protein interaction that represents the modulated host signalling mechanism initiated upon binding of SARS-CoV-2 with the ACE2 receptor (Fig. 1). Using a protein interaction database, several host proteins that connect directly or indirectly with the ACE2 receptor was obtained. However, to reflect the receptor signals that occurs in the lungs, all the collected network proteins were mapped with the in-house lung expression database. The network comprised of 298 interactions with 133 proteins, which provide a crucial signalling process in the lungs. Particularly, we have identified proteins such as AGT (Angiotensinogen), NTS (Large neuromedin N) and GHRL (Ghrelin and obestatin prepropeptide) that directly connect other proteins in the network, that are assumed to be activated upon viral binding to the ACE2 receptor (Fig. 1).
Figure 1.
ACE2 interacting protein network in the lungs. Network describing the attachment of SARS-CoV-2 with ACE2 receptor (highlighted in blue), that modulate the signalling of 133 proteins (highlighted in yellow), that form 298 interaction (black edges) in the lungs. The arrow edges indicate direct interacting proteins with the ACE2 receptor.
Dysregulated genes
All proteins in the regulatory network were evaluated to determine their changes in the SARS-CoV-2 infection. The analysis of RNA-Seq data showed, 2126 genes were found to be significantly (p < 0.05) altered in the lungs on SARS-CoV-2 infection. Among which, 133 genes were found to be associated with regulatory network, in that 29 were differentially expressed. This highlights the hijacking of the host regulatory network by the SARS-CoV-2 for their downstream process (Table 1). Among the differentially expressed genes, in particular, AGTR2, PIK3CB and MME are predicted to be important genes regulated in the host, considering their established role in the viral replication machinery and immune response.
Table 1.
Differentially expressed genes in BAL cells infected with SARS-CoV-2, which are predicted to interact with ACE2 receptor.
| S. No. | Gene | Differentially expressed | p-value |
|---|---|---|---|
| 1 | Membrane Metallo-Endopeptidase | Down-regulated | 0.004 |
| 2 | Prolylcarboxypeptidase (angiotensinase c) | Down-regulated | 0.002 |
| 3 | Phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | Down-regulated | 0.005 |
| 4 | Fibronectin 1 | Down-regulated | 0.004 |
| 5 | Eukaryotic translation elongation factor 1 alpha 1 | Down-regulated | 0.046 |
| 6 | Shc (src homology 2 domain containing) transforming protein 1 | Down-regulated | 0.027 |
| 7 | Proliferating cell nuclear antigen | Down-regulated | 0.018 |
| 8 | Lyn proto-oncogene, src family tyrosine kinase | Down-regulated | 0.002 |
| 9 | Ruvb-like aaa atpase 2 | Down-regulated | 0.021 |
| 10 | Actin, beta | Down-regulated | 0.033 |
| 11 | Methylenetetrahydrofolate dehydrogenase (nadp + dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase | Down-regulated | 0.054 |
| 12 | Aldehyde dehydrogenase 3 family, member a2 | Down-regulated | 0.004 |
| 13 | Nicotinamide nucleotide transhydrogenase | Down-regulated | 0.044 |
| 14 | Heat shock 27kda protein 1 | Down-regulated | 0.047 |
| 15 | Fatty acid amide hydrolase | Down-regulated | 0.025 |
| 16 | Protein disulfide isomerase family a, member 5 | Down-regulated | 0.004 |
| 17 | Methyltransferase like 7a | Down-regulated | 0.000 |
| 18 | Endothelin 1 | Up-regulated | 0.000 |
| 19 | Heat shock 70kda protein 8 | Down-regulated | 0.043 |
| 20 | ATPase, h+/k + transporting, nongastric, alpha polypeptide | Down-regulated | 0.020 |
| 21 | Interleukin-1 receptor-associated kinase 2 | Up-regulated | 0.000 |
| 22 | Utrophin | Down-regulated | 0.033 |
| 23 | Solute carrier family 9, subfamily a (nhe3, cation proton antiporter 3), member 3 regulator 1 | Down-regulated | 0.029 |
| 24 | DNAJ (hsp40) homolog, subfamily b, member 12 | Down-regulated | 0.035 |
| 25 | Nuclear casein kinase and cyclin-dependent kinase substrate 1 | Down-regulated | 0.049 |
| 26 | Timp metallopeptidase inhibitor 3 | Down-regulated | 0.001 |
| 27 | Microtubule associated tumor suppressor 1 | Down-regulated | 0.001 |
| 28 | Laminin, alpha 1 | Down-regulated | 0.022 |
| 29 | Chemokine (c-x-c motif) ligand 10 | Up-regulated | 0.020 |
Functional enrichment
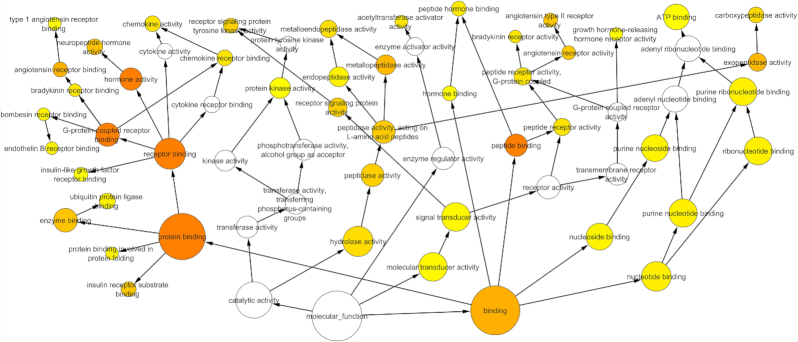
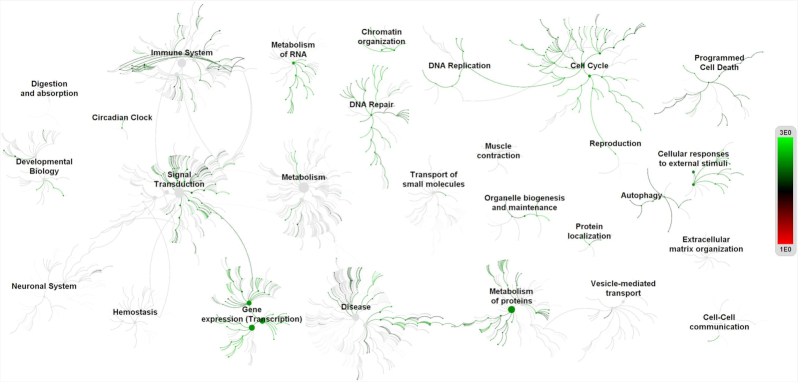
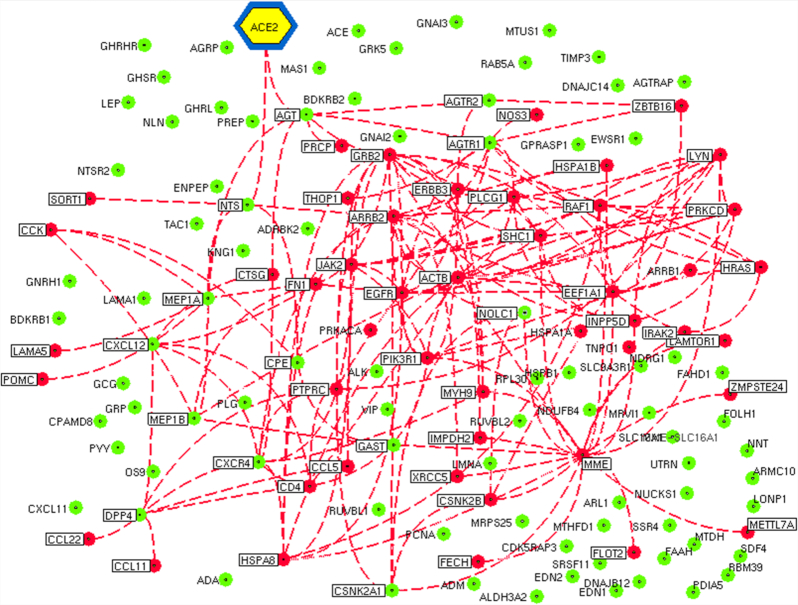
The regulatory network was further enriched by gene ontology using the BiNGO module in Cytoscape. The Gene Ontology (GO) based on the molecular function confirmed multiple host processes ( p < 0.001) related to hydrolase, protein kinase, carboxypeptidase, metallopeptidase and cytokine activity (Fig. 2). Besides this, molecular function such as type 1 angiotensin receptor binding (AGTR1), G-protein coupled receptor, insulin receptor, ribonucleotide, chemokine and cytokine receptor binding were observed to be altered by the modulation in the regulatory network. The pathway analysis using the Reactome database revealed the association of regulatory network with several biological pathways (adjusted p < 0.05) responsible for the disease, immune system, autophagy, cell cycle, programmed cell death, transcription, DNA repair and signal transduction (Fig. 3). We captured the immune signals in the network influenced by the ACE2 receptor. In the regulatory network, interestingly there were around 47 proteins, identified to be linked with immune signalling. For instance, upon receptor modulation, the ACE2 was found to directly influence the AGT, AGTR2 and AGRT1 through which the immune proteins such as THOP1, GRB2, NOS3 and PRCP were signalled (Fig. 4). THOP1, GRB2, NOS3 and PRCP proteins play a vital role in antigen presentation, activation of cytokine production and inflammatory response. We have also identified the presence of regulatory network across tissues using the Human Protein Atlas, Human Proteome Browser, Human Proteome Map, Proteomics and Human Proteinpedia databases. Of the analysed tissues, the kindney, placenta, pancreas, testis, small intestine and adrenal gland showed immune signalling proteins that are found to be deregulated on binding of the SARS-CoV-2 with the ACE2 receptor; however lung tissue harbored more immune molecules than other tissues (Fig. 5).
Figure 2.
Molecular enrichment of the ACE2 network. Functional Enrichment analysis of 133 proteins in the lung protein network showed the molecular functions related to hydrolase, protein kinase, carboxypeptidase, metallopeptidase and cytokine activity. Found to modulate type 1 angiotensin receptor binding (AGTR1), G-protein coupled receptor, insulin receptor, ribonucleotide, chemokine and cytokine receptor binding. Circumference of the circle represents number proteins associated with molecular function. An increase in color intensity and circumference of the circle in the network represents the number of proteins involved in the respective molecular function. Arrow represents the functional connectivity between the functional nodes.
Figure 3.
Pathway analysis of ACE2 network. The pathway analysis based on Reactom database suggests most of the proteins in the ACE2 network were found to be associated with functions such as the disease, immune system, autophagy, cell cycle, programmed cell death, transcription, DNA repair and signal transduction.
Figure 4.
Immuno-modulation influenced by the ACE2 network in the SARS-CoV-2 infection. The network visualizes the immune signals that are modulated by the binding of the SARS-CoV-2 with the ACE2 receptor. The dotted lines highlight the interaction of immune molecules influenced by the ACE2 receptor. Red nodes represent immune molecules altered in SARS-CoV-2 infection, whereas the unaltered immune molecules are represented by the green nodes.
Figure 5.
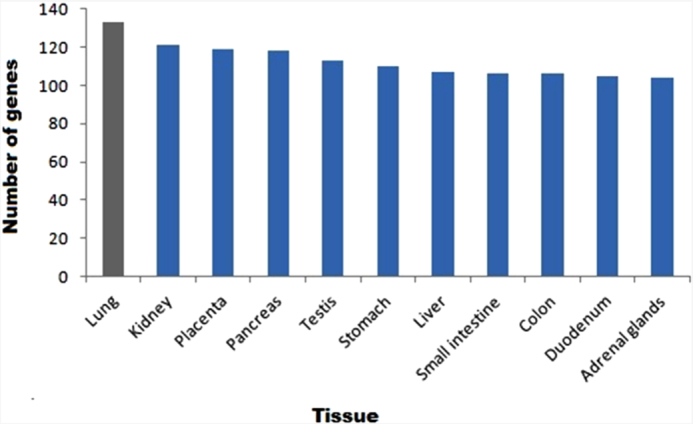
ACE2 interacting immune molecules in human tissues. The bar represents the number of genes associated with ACE2 mediated immune response in tissues.
DISCUSSION
A recent study using single-cell RNA-sequencing of humans' nasal epithelial cells revealed the expression of SARS-CoV-2 entry factors together with innate immune genes (Sungnak et al. 2020). Thus, suggesting for a potential tropism involved between the viral entry factors and associated genes involved in innate immunity, highlighting potential role in modulation of the host immune environment for the SARS-CoV-2 infection and spread. Interactome data from the current study reveals that ACE2 is found to interact with AGT directly mediated by AGTR2, NTS and GHRL. These proteins form secondary interaction with PI3K, PIK3R1 and MME. AGTR2 is implicated in the immune-mediated response leading to inflammation and fibrogenesis in cardiovascular system and liver (Okada et al. 2006). Thus, suggesting that the role of AGTR2 not only restricted to the hemodynamic regulation, but also render proinflammatory and fibrogenic functions. Even though the key role of AGTR2 is associated with vascular tone and blood pressure regulation, it was also being reported with function associated with cellular immune responses through a calcineurin-dependent pathway (Nataraj et al. 1999). NTS has been reported to involve in the regulation of immunity and inflammation. NTS and its receptors are expressed by macrophages, T lymphocytes and dendritic cells. Besides, NTS also regulates neutrophil chemotaxis, cytokine synthesis, mast cell activation and nitric oxide (NO) generation (Otten et al. 1994; Vega et al. 2003). Apart from the well-studied role of ghrelin in energy balance and metabolic functions, studies investigated and reported the functional association of ghrelin on multiple organ systems such as central nervous, cardiovascular and immune systems (Dixit and Taub 2005). Both in vitro and in vivo experiments describe ghrelin as an anti-inflammatory agent and immunoregulatory hormone or cytokine (Baatar, Patel and Taub 2011). MME has been reported with functions such as inflammatory response and insulin signalling (Ramirez et al. 2019). Patients with immune cell defects have studied to contain loss-of-function mutations in PIK3R1 gene (Conley et al. 2012). From the knowledge which is gathered from the available literature, it can be conceived that the functional hubs involved in ACE2/AGT-axis not only modulated host environment for viral entry and replication, but also secondarily can lead to inadvertent signalling leading to exaggerated immune response. Further studies have to be undertaken to decipher the role of ACE2/Ang-axis involvement with the immune system in detail. Based on the annotation of genes interacting with ACE2 receptor from our current study, it can be observed that ACE2 interacting genes are essential for host-virus interaction and essential for replication, survival and spread of the virus mediated through the deregulated host immune signalling.
From the over representing pathways associated with regulatory network across different human tissues, highlighted testis to express specific immune molecules, which can interact with ACE2 receptor. Studies report high ACE2 expression levels in testicular cells such as spermatogonia, seminiferous duct, Sertoli and Leydig cells (Wang and Xu 2020). Thus, it has been suggested that it could act as an underlying factor that men are more susceptible to reproductive dysfunction when/after infected with COVID-19 (Fan et al. 2020; Shen et al. 2020). Leydig cells are located close to the testis's seminiferous tubules, responsible for the production of testosterone on induction by the luteinizing hormone (LH; Wang et al. 2017). A recent study report the low testosterone levels in the majority of men infected with COVID-19, who are admitted to an Intensive Care Unit (ICU) at the University Hospital Hamburg-Eppendorf, Germany (Schroeder et al. 2020), considering the hormonal and reproductive function of the Leydig cells in men, together with existing knowledge on viral-induced mutation and immuno-modulation in testis. Therefore, it urges the need for carrying follow up study in recovered individuals, especially in men, evaluating the hormonal and reproductive endpoints. Since the SARS-CoV-2 infection could affect both the semen integrity and likely transmission into the subsequent generation (Dejucq and Jegou 2001). Thus, the viral-induced long-term effect on male genital tract and their effect on reproductive system should be considered as the potential future health risk in COVID-19 recovered patients of active reproductive age. Based on the expression levels of ACE2 interacting immune factors in various human tissues, next to testicular cells, stomach, small intestine, colon and duodenum has shown to express ACE2 receptor inducible immune molecules in higher amount when compared to the other tissues. Recent research found that SARS-CoV-2 can infect and multiply in the cells of the intestine by utilizing cell culture models of the human intestine (Lamers et al. 2020). Apart from the symptoms of fever and respiratory illness, COVID-19 infected patients also presented with signs of diarrhoea, vomiting and abdominal pain accounting for gastrointestinal symptoms (Wong, Lui and Sung. 2020). In line with our results showing higher expression of regulatory network in stomach and intestine, therefore future should study the impact of COVID-19 infection on intestinal function and adverse effect on patients with gastrointestinal problems from an immunological perspective. In this work, we would also like to highlight few genes like insulin-like growth factor (IGF), G protein-coupled receptor kinases 2 (GRK2) and Phosphoinositide-3-Kinase Regulatory Subunit 1 (PI3KR1), which are well studied to have roles in insulin signalling mechanism, that were shown have to interact with the regulatory network. Further study has to be undertaken to dissect the functional role of ACE2/Ang-axis interaction with the insulin signalling pathway's genes.
Alteration in the expression of ACE2 receptor transcriptional activity has been reported with chronic heart failure, hypertension and lung injury (Tikellis and Thomas 2012). Modulation of lung ACE2 expression is associated with alteration in the Ang II and Ang1–7 levels, which lead to hypoxia and inflammation (Sodhi et al. 2018). Thus, ACE2 is implicated to play a protective and beneficial role against lung diseases. ACE serves as a rate-limiting factor for the production of Ang II protein, which is reported roles in vasoconstriction and mitogenic action (Lipworth and Dagg 1994). Besides, Ang II playing a causative role in the initiation of inflammatory response involving bradykinin, nitric oxide (NO), endothelin-1 (ET-1), cyclooxygenase-2 (COX-2) and epoxyeicosatrienoic acids (EETs) signal transduction pathways (Cheng, Vapaatalo and Mervaala 2005), Ang II also modulates the cellular oxidative stress, mediated by AT1 receptors which increases the reactive oxygen species (ROS). Thus, enhanced oxidative stress leads to cellular dysfunction and inflammation by activating the redox-sensitive transcription factors (NF-kappaB), leading to the production of chemokines and cytokines (Husain et al. 2015). In our study, the computational prediction of genes interacting with the ACE2 receptor comprised of signalling molecules from the NF-kappaB and NOS signal transduction pathways, mainly connected through the membrane ACE2/AGT/AGTR1/MME hub. Recently, the function of membrane MME in preadipocytes has been linked with inflammatory response and insulin signalling (Ramirez et al. 2019). Additionally, hypoxia itself demonstrated to influence the expression levels of MME in lung cancer cells (Leithner et al. 2014). However, at this moment it is imperative to extend our understanding of how COVID-19 infection could alter membrane MME levels, either through ACE-dependent mechanism, or modulated by the hypoxic condition resulting from the compromised lung function resulting from the SARS-CoV-2 infection, which warrants further clinical and experimental elucidation.
In a prospective cohort study investigating the biomarkers of host defense and RAS in bronchoalveolar lavage (BAL) fluid from neonates, children, adults and older adults who had acute respiratory distress syndrome (ARDS), highlighted that the activity of ACE and ACE2 remind unchanged. However, their expression was found to be altered between the age groups. Also, there is a significant alteration in the levels of myeloperoxidase, interleukin (IL)-6, IL-10 and p-selectin, which increased in an age dependent-manner (Schouten et al. 2019). Severe cases of COVID-19 were reported to develop ARDS, in a sub-group of patients with cytokine storm syndrome and hyper inflammation. The clinical outcome of ARDS has now well supported by the reports from patients with COVID-19 disease severity, observed with increased interleukin (IL)-2, IL-7, macrophage inflammatory protein 1-α, interferon-γ inducible protein 10, granulocyte-colony stimulating factor, monocyte chemoattractant protein 1, ferritin and tumor necrosis factor-α levels, resulting from viral induced hyperinflammation (Huang et al. 2020). In the present study protein–protein interaction network of ACE2 receptor found to influence the chemokine and cytokine activity, cytokine and chemokine receptor binding and bradykinin receptor binding mediated through a major hub involving ATR/G-protein coupled receptors (GPCR). In the over representing pathways, compared with other tissues in the human body, lungs contained the highest number of immune molecules, which can functionally interact with the ACE2 receptor. This could be an underlying reason for the SARS-CoV-2 infection to profoundly affect the lower respiratory tract over other tissues in the body. Future clinical and in vitro studies should investigate the SARS-CoV-2 induced immuno-modulatory outcomes in body tissues other than lungs, investigating the regulatory network to find the secondary complications associated with SARS-CoV-2 infection.
CONCLUSION
Overall from present study, the interactome data revealed that AGT (Angiotensinogen), LAMAS (Laminin Subunit Alpha 1), NTS (Neurotensin) and GHRL Ghrelin and (Obestatin Prepropeptide) acting as direct mediators through which the interconnected proteins hubs, which are assumed to be activated upon viral binding of the SARS-CoV-2 with the ACE2 receptor. The pathway analysis using the Reactome database revealed the association of regulatory network with several biological pathways (adjusted p < 0.05) responsible for the disease, immune system, autophagy, cell cycle, programmed cell death, transcription, DNA repair and signal transduction. The functional enrichment analysis with tissue over representation studies revealed the presence of ACE2 inducible immune factors across different tissues types, but very profound in the lungs, compared to other tissues, thus making it more sensitive and could be a molecular underlying factor for the over active immune response leading to respiratory illness.
Contributor Information
Christy Lite, Endocrine and Exposome (EE2) Laboratory, Madras Christian College, Department of Zoology, Tamil Nadu - 600059, India.
Shiek S S J Ahmed, Drug Discovery Lab, Faculty of Allied Health Sciences, Chettinad Academy of Research and Education, Kelambakkam, Tamil Nadu - 603103, India.
Melita Juliet, Department of Oral and Maxillofacial Surgery, Meenakshi Ammal Dental College and Hospital, Maduravoyal, Chennai, Tamil Nadu - 600095, India.
A J Freddy, Endocrine and Exposome (EE2) Laboratory, Madras Christian College, Department of Zoology, Tamil Nadu - 600059, India.
ETHICAL APPROVAL
This manuscript does not report any studies with human participants or animals performed by any of the authors. Informed consent for this type of paper is not required.
FUNDING
This study did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Conflicts of Interest
None declared.
REFERENCES
- Ahmed SS, Paramasivam P, Raj Ket al. Regulatory cross talk between SARS-CoV-2 receptor binding and replication machinery in the human host. Front Physiol. 2020;11:802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baatar D, Patel K, Taub DD. The effects of ghrelin on inflammation and the immune system. Mol Cell Endocrinol. 2011;340:44–58. [DOI] [PubMed] [Google Scholar]
- Cascella M, Rajnik M, Cuomo Aet al. Features, Evaluation and Treatment Coronavirus (COVID-19). In: Statpearls [internet]. StatPearls Publishing. 2, 2020. [PubMed] [Google Scholar]
- Cheng ZJ, Vapaatalo H, Mervaala E. Angiotensin II and vascular inflammation. Med Sci Monit. 2005;11:RA194–205. [PubMed] [Google Scholar]
- Conley ME, Dobbs AK, Quintana AMet al. Agammaglobulinemia and absent B lineage cells in a patient lacking the p85α subunit of PI3KHuman p85α deficiency. J Exp Med. 2012;209:463–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejucq N, Jégou B. Viruses in the mammalian male genital tract and their effects on the reproductive system. Microbiol Mol Biol Rev. 2001;65:208–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux CA, Rolain JM, Raoult D. ACE2 receptor polymorphism: susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome. J Microbiol Immunol Infect. 2020;53:425–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit VD, Taub DD. Ghrelin and immunity: a young player in an old field. Exp Gerontol. 2005;40:900–10. [DOI] [PubMed] [Google Scholar]
- Fan C, Li K, Ding Yet al. ACE2 expression in kidney and testis may cause kidney and testis damage after 2019-nCoV infection, MedRxiv. 2020. DOI: 10.1101/2020.02.12.20022418. [DOI] [PMC free article] [PubMed]
- Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. In Coronaviruses (pp. 1–23). Humana Press, New York, NY, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira AJ, Shenoy V, Yamazato Yet al. Evidence for angiotensin-converting enzyme 2 as a therapeutic target for the prevention of pulmonary hypertension. Am J Respir Crit Care Med. 2009;179:1048–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Wang Y, Li Xet al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet North Am Ed. 2020;395:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain K, Hernandez W, Ansari RAet al. Inflammation, oxidative stress and renin angiotensin system in atherosclerosis. World J Biol Chem. 2015;6:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata M, Cowling RT, Gurantz Det al. Angiotensin-(1–7) binds to specific receptors on cardiac fibroblasts to initiate antifibrotic and antitrophic effects. Am J Physiol Heart Circul Physiol. 2005;289:H2356–63. [DOI] [PubMed] [Google Scholar]
- Kuba K, Imai Y, Ohto-Nakanishi Tet al. Trilogy of ACE2: a peptidase in the renin–angiotensin system, a SARS receptor, and a partner for amino acid transporters. Pharmacol Ther. 2010;128:119–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers MM, Beumer J, van der Vaart Jet al. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369:50–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leithner K, Wohlkoenig C, Stacher Eet al. Hypoxia increases membrane metallo-endopeptidase expression in a novel lung cancer ex vivo model–role of tumor stroma cells. BMC Cancer. 2014;14:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipworth BJ, Dagg KD. Vasoconstrictor effects of angiotensin II on the pulmonary vascular bed. Chest. 1994;105:1360–4. [DOI] [PubMed] [Google Scholar]
- Li W, Sui J, Huang ICet al. The S proteins of human coronavirus NL63 and severe acute respiratory syndrome coronavirus bind overlapping regions of ACE2. Virology. 2007;367:367–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nataraj C, Oliverio MI, Mannon RBet al. Angiotensin II regulates cellular immune responses through a calcineurin-dependent pathway. J Clin Invest. 1999;104:1693–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada H, Inoue T, Kikuta Tet al. A possible anti-inflammatory role of angiotensin II type 2 receptor in immune-mediated glomerulonephritis during type 1 receptor blockade. Am J Pathol. 2006;169:1577–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otten U, Scully JL, Ehrhard PBet al. Neurotrophins: signals between the nervous and immune systems. In Progress in Brain Research, Vol. 103, pp. 293–305.. Elsevier, 1994. [DOI] [PubMed] [Google Scholar]
- Ramirez AK, Dankel S, Cai Wet al. Membrane metallo-endopeptidase (Neprilysin) regulates inflammatory response and insulin signaling in white preadipocytes. Mol Metabol. 2019;22:21–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riordan JF. Angiotensin-I-converting enzyme and its relatives. Genome Biol. 2003;4:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samavati L, Uhal BD. ACE2, much more than just a receptor for SARS-COV-2. Front Cel Infect Microbiol. 2020;10:317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouten LR, van Kaam AH, Kohse Fet al. Age-dependent differences in pulmonary host responses in ARDS: a prospective observational cohort study. Ann Intensive Care. 2019;9:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder M, Tuku B, Jarczak Det al. The majority of male patients with COVID-19 present low testosterone levels on admission to intensive care in Hamburg, Germany: a retrospective cohort study, medRxiv. 2020. DOI: 0.1101/2020.05.07.20073817. [Google Scholar]
- Shang J, Wan Y, Luo Cet al. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci. 2020a;117:11727–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J, Ye G, Shi Ket al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Xiao X, Aierken Aet al. The ACE2 expression in Sertoli cells and germ cells may cause male reproductive disorder after SARS-CoV-2 infection. J Cell Mol Med. 2020;24:9472–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodhi CP, Wohlford-Lenane C, Yamaguchi Yet al. Attenuation of pulmonary ACE2 activity impairs inactivation of des-Arg9 bradykinin/BKB1R axis and facilitates LPS-induced neutrophil infiltration. Am J Physiol Lung Cel Mol Physiol. 2018;314:L17–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriramula S, Cardinale JP, Lazartigues Eet al. ACE2 overexpression in the paraventricular nucleus attenuates angiotensin II-induced hypertension. Cardiovasc Res. 2011;92:401–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungnak W, Huang N, Bécavin Cet al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med. 2020;26:681–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikellis C, Thomas MC. Angiotensin-converting enzyme 2 (ACE2) is a key modulator of the renin angiotensin system in health and disease. Int J Peptides. 2012;2012:256294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega JA, García-Suárez O, Hannestad Jet al. Neurotrophins and the immune system. J Anat. 2003;203:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan T, Arya S, Chan SHet al. Structural basis of RNA cap modification by SARS-CoV-2. Nat Commun. 2020;11:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls AC, Park YJ, Tortorici MAet al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chen F, Ye Let al. Steroidogenesis in Leydig cells: effects of aging and environmental factors. Reproduction. 2017;154:R111–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Xu X. scRNA-seq profiling of human testes reveals the presence of the ACE2 receptor, a target for SARS-CoV-2 infection in spermatogonia, Leydig and Sertoli cells. Cells. 2020;9:920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . Coronavirus Disease (COVID-19) – World Health Organization. 2020 [online] Available at: <https://covid19.who.int/> (05 January 2021, date last accessed).
- Wong SH, Lui RN, Sung JJ. Covid-19 and the digestive system. J Gastroenterol Hepatol. 2020;35:744–8. [DOI] [PubMed] [Google Scholar]
- Yang G, Tan Z, Zhou Let al. Effects of angiotensin II receptor blockers and ACE (angiotensin-converting enzyme) inhibitors on virus infection, inflammatory status, and clinical outcomes in patients with COVID-19 and hypertension: a single-center retrospective study. Hypertension. 2020;76:51–8. [DOI] [PubMed] [Google Scholar]
- Zaman MA, Oparil S, Calhoun DA. Drugs targeting the renin–angiotensin–aldosterone system. Nat Rev Drug Discov. 2002;1:621–36. [DOI] [PubMed] [Google Scholar]
- Zeng W, Chen W, Leng Xet al. Chronic angiotensin-(1–7) administration improves vascular remodeling after angioplasty through the regulation of the TGF-β/Smad signalling pathway in rabbits. Biochem Biophys Res Commun. 2009;389:138–44. [DOI] [PubMed] [Google Scholar]