Abstract
Background
COVID-19 is infrequently complicated by bacterial co-infection, but antibiotic prescriptions are common. We used community-acquired pneumonia (CAP) as a benchmark to define the processes that occur in bacterial pulmonary infections, testing the hypothesis that baseline inflammatory markers and their response to antibiotic therapy could distinguish bacterial co-infection from COVID-19.
Methods
Retrospective cohort study of CAP (lobar consolidation on chest radiograph) and COVID-19 (PCR detection of SARS-CoV-2) patients admitted to Royal Free Hospital (RFH) and Barnet Hospital (BH), serving as independent discovery and validation cohorts. All CAP and >90% COVID-19 patients received antibiotics on hospital admission.
Results
We identified 106 CAP and 619 COVID-19 patients at RFH. Compared with COVID-19, CAP was characterized by elevated baseline white cell count (WCC) [median 12.48 (IQR 8.2–15.3) versus 6.78 (IQR 5.2–9.5) ×106 cells/mL, P < 0.0001], C-reactive protein (CRP) [median 133.5 (IQR 65–221) versus 86.0 (IQR 42–160) mg/L, P < 0.0001], and greater reduction in CRP 48–72 h into admission [median ΔCRP −33 (IQR −112 to +3.5) versus +14 (IQR −15.5 to +70.5) mg/L, P < 0.0001]. These observations were recapitulated in the independent validation cohort at BH (169 CAP and 181 COVID-19 patients). A multivariate logistic regression model incorporating WCC and ΔCRP discriminated CAP from COVID-19 with AUC 0.88 (95% CI 0.83–0.94). Baseline WCC >8.2 × 106 cells/mL or falling CRP identified 94% of CAP cases, and excluded bacterial co-infection in 46% of COVID-19 patients.
Conclusions
We propose that in COVID-19, absence of both elevated baseline WCC and antibiotic-related decrease in CRP can exclude bacterial co-infection and facilitate antibiotic stewardship efforts.
Introduction
The COVID-19 pandemic caused by the novel beta coronavirus SARS-CoV-2 has caused >65 million infections and >1.5 million deaths worldwide.1 The drivers of pathology remain to be elucidated, but a hyperinflammatory response is associated with worse case fatality.2 Other viral respiratory tract infections, best characterized by influenza, can be complicated by bacterial co-infections that also raise inflammatory markers and are associated with high mortality,3,4 but distinguishing severe viral pneumonia from bacterial co-infection is challenging.5 In COVID-19, several studies have found bacterial co-infection to be rare, as determined by identification of causative pathogens.6–10 However, routine microbiological culture takes several days, lacks sensitivity,11 and does not readily distinguish bacterial colonization from infection. Moreover, microbiological respiratory tract sampling is not performed routinely in patients admitted with COVID-19.9 Therefore, despite guidance aimed at rationalizing antibiotic use,12 it is unsurprising that diverse and elevated rates of antibiotic prescriptions have been reported in patients admitted for COVID-19 infection.8,9
It is likely that many COVID-19-associated antibiotic prescriptions are given in the absence of bacterial co-infection, hampering antimicrobial stewardship efforts and potentially increasing antimicrobial resistance.13–16 Many studies have focused on clinical and laboratory features that risk-stratify outcome in COVID-19,17–20 but currently infections caused by virus alone cannot be readily distinguished from those with a bacterial component. C-reactive protein (CRP), white cell count (WCC) and procalcitonin (PCT) have been used to distinguish between influenza and bacterial pneumonia, allowing antibiotic treatment to be omitted or stopped.21–24 Serial measurements of inflammatory markers may also assist in distinguishing bacterial from viral infections.25,26 A small retrospective study comparing COVID-19 with community-acquired pneumonia (CAP) patients identified differences in admission neutrophil counts, D-dimers and CRP, but did not provide a rigorous definition for the pneumonia cases, or explore changes in these markers over time.27
In this study, we aimed to identify features that discriminated viral COVID-19 infections from those complicated by bacterial co-infection. We used CAP as a benchmark to define the processes that occur in bacterial pulmonary infections and tested the hypothesis that baseline inflammatory markers and their response to antibiotics could distinguish CAP from most COVID-19 infections. To address this research question, we performed a retrospective, cohort study from a large split-site academic hospital in the UK. We used the independent nature of the two sites to discover and validate our findings, extending their generalizability.
Methods
Data extraction and ethics
Anonymized demographics, antimicrobial prescriptions, haematological and biochemical investigations were extracted from the Clinical Practice Group analysis team, Cerner Electronic Patient Records and the electronic Clinical Infection Database (elCID), and microbiological investigations from WinPath at Royal Free London (RFL) NHS Trust.28 The study was approved by the Research and Innovation Group at RFL NHS Trust, which stated that confidential patient information could be used under the COVID-19 COPI notice issued by the UK Department of Health and Social Care, and that as this was a retrospective review of routine clinical data, formal ethics approval was not required.
Patient selection
We identified patients from two hospital sites of RFL NHS Trust in London, UK: Royal Free Hospital (RFH) and Barnet Hospital (BH). These hospitals are separated by 11 km and patient care is delivered by non-overlapping clinical staff, using non-identical clinical care bundles and antibiotic policies. We included patients aged >18 years old admitted to hospital, of which a subset was admitted for >48 h (Table 1), excluding patients with haematological malignancies. We defined COVID-19 by RT-PCR detection of SARS-CoV-2 from nasopharyngeal swabs, identifying patients between 1 March and 31 May 2020. These criteria yielded 619 and 181 COVID-19 patients from RFH and BH, respectively. CAP was defined by a clinical diagnosis of CAP made between 1 January and 31 May 2019 with focal consolidation on chest radiograph reported by consultant radiologists (106 patients at RFH and 169 at BH). We used RFH patients as a discovery cohort in our analyses to build a model and cut-off parameters to discriminate between CAP and COVID-19, and BH patients were used as an independent cohort to validate the findings from RFH.
Table 1.
Patients identified in each diagnostic group at RFH and BH
| Royal Free Hospital |
Barnet Hospital |
|||||
|---|---|---|---|---|---|---|
| Diagnosis | CAP (n = 106) | COVID-19 (n = 619) | P value | CAP (n = 169) | COVID-19 (n = 181) | P value |
| Chest radiograph, n (%)a | ||||||
| Lobar consolidation | 106 (100) | 4 (0.6) | – | 169 (100) | 0 (0%) | – |
| CVXC0 | 62 (10.0) | 22 (12.2) | ||||
| CVCX1 | 281 (45.3) | 71 (39.2) | ||||
| CVCX2 | 136 (22.0) | 36 (19.9) | ||||
| CVCX3 | 17 (2.7) | 5 (2.8) | ||||
| Ungraded | 123 (19.9) | 47 (26.0) | ||||
| Male, n (%) | 50 (47) | 386 (62) | 0.0037 | 81 (47.9) | 104 (57.5) | 0.0865 |
| Age, years, median (range) | 72 (19–99) | 68 (18–100) | 0.1401 | 74 (18–98) | 71 (29–98) | 0.0633 |
| Ethnicity, n (%) | 0.0007 | 0.0335 | ||||
| White | 60 (57) | 250 (40) | 128 (76) | 115 (64) | ||
| Asian | 23 (22) | 83 (13) | 11 (7) | 26 (14) | ||
| Black | 6 (6) | 69 (11) | 1 (1) | 11 (6) | ||
| Mixed | 2 (2) | 7 (1) | 0 (0) | 2 (1) | ||
| Other/unknown | 15 (14) | 210 (34) | 29 (17) | 27 (15) | ||
| Charlson index co- morbidities n (%) | <0.0001 | 0.0206 | ||||
| 0 | 18 (17) | 243 (39) | 51 (30) | 81 (45) | ||
| 1 | 33 (31) | 190 (31) | 51 (30) | 58 (32) | ||
| 2 | 28 (26) | 109 (18) | 35 (21) | 23 (13) | ||
| 3+ | 27 (25) | 77 (13) | 32 (19) | 19 (10) | ||
| Patients with microbiological identification of bacteria, n (%) | 15 (14.2) | 26 (4.2) | 0.0003 | 15 (8.9) | 10 (5.5) | 0.2992 |
| Microbiology results, n (%) | ||||||
| Sputum | 6 (5.7) | 8 (1.3) | 2 (1.2) | 5 (2.8) | ||
| Blood | 2 (1.9) | 12 (1.9) | 6 (3.6) | 5 (2.8) | ||
| Urine Ag | 4 (3.8) | 3 (0.5) | 8 (4.7) | 0 (0.0) | ||
| Mycoplasma PCR | 5 (4.7) | 3 (0.5) | 0 (0.0) | 0 (0.0) | ||
| Blood samples collected 48–72 h into admission, n (%) | 53 (50.0) | 331 (53.5) | 99 (58.6) | 60 (33.1) | ||
P values represent comparisons between CAP and COVID-19 and each hospital site. Comparisons between the cohorts at each hospital site were performed by Mann–Whitney test for age, by Fisher’s exact test for gender, and by Chi-square test for ethnicity, Charlson co-morbidities and microbiological results. P values represent comparisons between CAP and COVID-19 and each hospital site.
Chest radiograph codes for COVID-19 patients based on British Society of Thoracic Imaging guidelines: CVCX0 = Normal; CVCX1 = Classic for COVID-19; CVCX2 = Indeterminate for COVID-19; CVCX3 = Non-COVID-19.
We identified 26 (4.2%) and 10 (5.5%) COVID-19 patients at RFH and BH, respectively, with microbiological evidence of bacterial co-infection. This was defined by the presence of a non-contaminant bacterial growth on blood culture, bacterial growth in sputum samples, detection of Mycoplasma pneumoniae by PCR from sputum or detection of Streptococcus pneumoniae antigen in urine (Table 1 and Table S1, available as Supplementary data at JAC Online). In addition, at RFH, 4 (0.6%) COVID-19 patients had radiological evidence of lobar pneumonia on a chest radiograph within 72 h of hospital admission. We collectively termed COVID-19 patients with microbiological or radiological evidence of bacterial co-infection ‘MR+ COVID-19’, as opposed to the remaining COVID-19 patients termed ‘MR− COVID-19’. Of the MR+ COVID-19 patients, 18 and 4 remained in hospital for ≥48 h at RFH and BH, respectively.
Statistical analysis
Baseline demographics were compared by Mann–Whitney test (age), Fisher’s exact test (gender and microbiology) or Chi-square test (ethnicity and Charlson co-morbidities). Continuous variables were expressed as median and IQR, and patient groups were compared using non-parametric two-tailed Mann–Whitney U tests. A multivariate logistic regression model was used to determine factors that discriminated between CAP and MR− COVID-19. The model’s categorical output variable was a diagnosis of CAP, and continuous dependent variables were baseline demographics and inflammatory markers. Variables were treated as interval data, with no true zero. In this way positive and negative values (i.e. the ones generated from Δ calculations) were treated equally by the model, with only the differences in their relative association between CAP and COVID-19 patients contributing to their discriminatory capacity. This analysis generated Receiver Operating Characteristic (ROC) curves and AUC as a summary statistic. For pre-determined cut-offs, we also calculated sensitivity, specificity, positive and negative predictive values, and positive and negative likelihood ratios. All analyses were performed using Microsoft Excel and GraphPad Prism.
Results
Defining the discovery cohort
We identified 106 CAP and 619 COVID-19 patients at RFH. Male gender was overrepresented in COVID-19 (62% in COVID-19 versus 47% in CAP), whereas CAP patients were older (median age 72 years in CAP and 68 years in COVID-19). The proportion of Black, Asian, Mixed and Other (non-white) ethnicity patients was higher in COVID-19 compared with CAP and patients with CAP had more comorbidities and identified bacteria in routine microbiological investigations more commonly (Table 1).
Distinguishing CAP from COVID-19
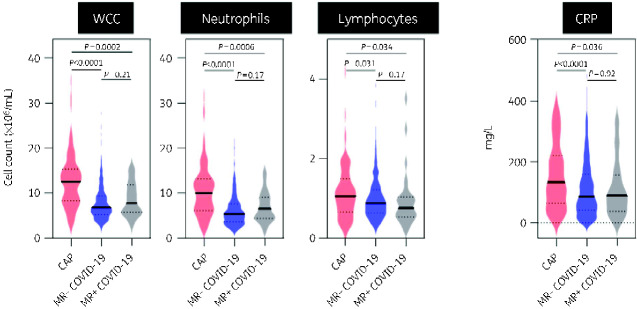
We tested the hypothesis that inflammatory markers could discriminate CAP from COVID-19 by comparing total WCC, its differential cell counts and CRP on the day of admission to hospital. We divided the COVID-19 population into 589 MR− and 30 MR+, highlighting that most COVID-19 patients did not show microbiological or radiological evidence of bacterial co-infection. Compared with CAP, COVID-19 was associated with significantly lower median WCC (12.48 versus 6.78 and 7.77 × 106/mL) and neutrophils (9.98 versus 5.36 and 6.51 × 106/mL) relative to both MR− and MR+ populations (Figure 1). Lymphocyte counts were marginally lower in COVID-19 than CAP, and CRP was significantly higher in CAP than in both COVID-19 populations (median CRP 133.5, 86.0 and 89.5 mg/L, respectively) (Figure 1). Notably, there were no differences in these markers between the COVID-19 subpopulations.
Figure 1.
Admission blood samples for all patients admitted to RFH. Violin plots represent distribution of values for CAP (n = 106), MR− COVID-19 (n = 589) and MR+ COVID-19 (n = 30) patients. Bold lines represent median values. Dotted lines represent IQR values. P values derived from two-tailed Mann–Whitney test. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
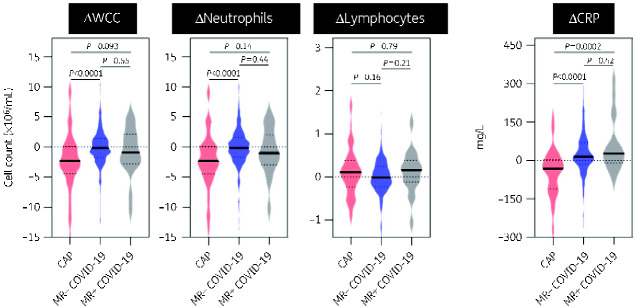
All CAP patients were prescribed antibiotics on admission and in two independent surveys of COVID-19 patients from RFH, 95/100 (95%) and 104/118 (88%) were prescribed antibiotics to treat a presumptive pulmonary bacterial co-infection. We hypothesized that CAP and COVID-19 could be further discriminated by changes in inflammatory markers following initiation of antibiotics.25 In the RFH cohort, 53 (50%) CAP, 313 (53%) MR− COVID-19 and 18 (60%) MR+ COVID-19 patients were admitted for >48 h and had a blood sample collected as part of routine clinical care 48–72 h into admission (Table 1). Differences in inflammatory markers on admission within this subset mirrored that seen in the wider cohort (Figure S1). At this later timepoint, CAP was still characterized by elevated median WCC (9.61, 7.28 and 7.41 × 106/mL, respectively), but this difference was diminished compared with admission (Figure S2). Moreover, the difference in CRP between CAP and either COVID-19 subpopulation was no longer evident (median CRP 113.0, 126.0 and 148 mg/L, respectively) (Figure S2). These changes were driven by a greater fall in WCC and CRP for CAP compared with MR− or MR+ COVID-19 (ΔWCC −2.32, −0.16, −0.94 × 106/mL and ΔCRP −33, +14, +26 mg/L respectively) (Figure 2). Similar to baseline samples, no differences were observed for changes in inflammatory markers between MR− and MR+ COVID-19 (Figure 2).
Figure 2.
Change in values between admission blood samples and those collected 48–72 h into admission at RFH. Violin plots represent distribution of difference (Δ) in investigation results between those collected on hospital admission and 48–72 h into admission in CAP (n = 53), MR− COVID-19 (n = 313) and MR+ COVID-19 (n = 18) patients. Bold lines represent median values. Dotted lines represent IQR values. P values derived from two-tailed Mann–Whitney test. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Contribution of multiple variables to discriminate CAP from COVID-19
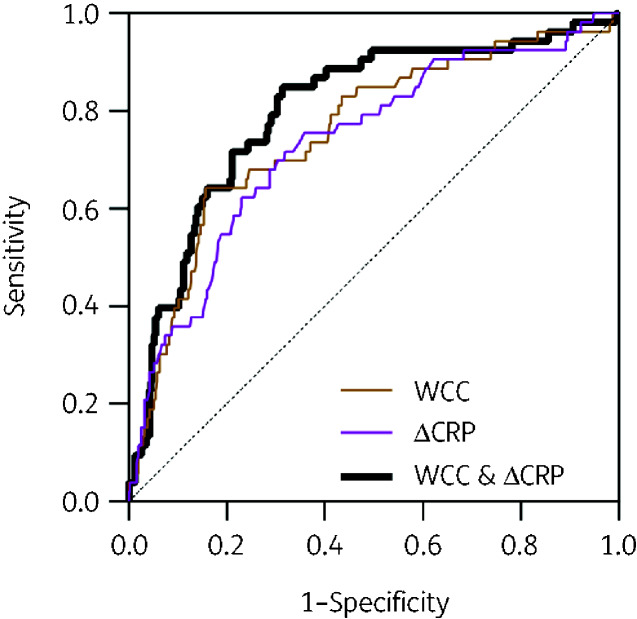
Our data suggested that elevated WCC and CRP, as well as a reduction in these parameters at 48–72 h could discriminate between COVID-19 and CAP. To test this hypothesis, we applied a logistic regression model to the data collected from the CAP and MR− COVID-19 patient groups. We used the diagnosis of CAP as the binary outcome variable and explored how baseline and changes in inflammatory markers influenced the diagnostic accuracy (Figure 3 and Table 2). The maximal AUC obtained from using each variable alone was 0.78 (95% CI 0.70–0.85) with baseline WCC. We also tested whether combining variables would improve the diagnostic accuracy of the model, and observed some improvement in discriminating CAP from COVID-19 using both baseline WCC and ΔCRP (AUC 0.81, 95% CI 0.74–0.88) (Figure 3 and Table 2).
Figure 3.
Accuracy of blood parameters to diagnose CAP in RFH cohorts of CAP and MR− COVID-19 patients. ROC curves generated from logistic regression models that incorporate combinations of WCC on admission and difference (Δ) in CRP between samples on admission and 48–72 h into admission in order to discriminate RFH patients diagnosed with CAP from those diagnosed with COVID-19. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Table 2.
Discriminatory accuracy of admission WCC, admission CRP, ΔWCC and ΔCRP for diagnosis of CAP compared with MR− COVID-19 at Royal Free Hospital (RFH) and Barnet Hospital (BH)
| Royal Free Hospital |
Barnet Hospital |
|||
|---|---|---|---|---|
| Characteristic | AUC | 95% CI | AUC | 95% CI |
| WCC on admission | 0.78 | 0.70–0.85 | 0.84 | 0.78–0.90 |
| CRP on admission | 0.71 | 0.64–0.80 | 0.79 | 0.71–0.86 |
| WCC and CRP | 0.78 | 0.71–0.85 | 0.86 | 0.80–0.92 |
| ΔWCC | 0.72 | 0.64–0.80 | 0.79 | 0.72–0.87 |
| ΔCRP | 0.77 | 0.70–0.84 | 0.82 | 0.76–0.89 |
| WCC and ΔWCC | 0.78 | 0.70–0.85 | 0.84 | 0.77–0.90 |
| WCC and ΔCRP | 0.81 | 0.74–0.88 | 0.88 | 0.83–0.94 |
Populations included were all patients admitted >48 h: n = 53 for CAP and n = 313 for MR-COVID 19 at RFH and n = 99 for CAP and n = 56 for MR− COVID-19 at BH. AUC, area under the curve; CI, confidence interval.
We performed sensitivity analyses to explore the role of possible confounders. Excluding baseline demographics from the model only minimally reduced AUCs (Table S2), indicating that inflammatory markers were the predominant discriminatory variables. We also considered the role played by admission to ICU. At RFH, 54/313 (17.2%) of MR− COVID-19 patients included in the logistic regression model were admitted to ICU within 72 h of their admission (Table 1). Excluding this subset of patients did not affect the maximal discriminatory power of the model (AUC 0.82, 95% CI 0.75–0.89) (Table S2). We also considered whether reclassification of MR+ COVID-19 patients as CAP may further improve discrimination from the MR− COVID-19 cohort. However, this approach moderately reduced the discriminatory power of the model, consistent with the differences observed between CAP and MR+ COVID-19 patients in Figures 1 and 2. Overall, these analyses confirmed that WCC and ΔCRP are the variables that most discriminate between CAP and COVID-19 and are not confounded by patient demographics, ICU attendance or microbiological/radiological evidence of bacterial co-infection.
Decision-making criteria to discriminate CAP and COVID-19
We sought to convert our observations into practical decision-making criteria for clinical practice, focusing on the larger MR− COVID-19 population. We generated a series of cut-offs between CAP and COVID-19 for variables with greatest discriminatory power, admission WCC and ΔCRP, as well as baseline CRP to permit assessment at the time of admission, and explored the trade-off in sensitivity and specificity generated by these alone or in combination (Table 3). For WCC cut-offs, we used the lower quartile value of the CAP cohort (>8.2 × 106/mL) and the upper quartile value of the COVID-19 cohort (>9.4 × 106/mL). We used the same criteria for baseline CRP, yielding cut-offs of 65 and 160 mg/L, respectively. For cut-offs of ΔCRP we used the lower quartile of the COVID-19 cohort (−15 mg/L) and the upper quartile of the CAP cohort (+3.5 mg/L), rounded to 0 mg/L for simplicity. These analyses revealed that using CAP-derived quartile cut-offs for baseline WCC or CRP yielded greater sensitivity, at the expense of specificity. The lower prevalence of CAP in this cohort compared with COVID-19 offered a high negative predictive value (>90%). Requiring both a WCC > 8.2 × 106/mL and ΔCRP < 0 improved specificity, at the expense of sensitivity, but this strategy could result in many cases of CAP, and by extension pathological bacterial co-infection in COVID-19, being missed. Therefore, we explored using either parameter to define CAP, yielding a sensitivity of >90%, and although the specificity of this approach was only 43%, it was still greater than if baseline CRP had replaced the ΔCRP cut-off (Table 3). Therefore, the absence of both admission WCC > 8.2 × 106/mL and ΔCRP < 0 could exclude bacterial co-infection in 135/313 (43%) of the MR− COVID-19 cohort, in turn supporting antibiotic cessation in these patients (Table 3).
Table 3.
Discriminatory performance of WCC and ΔCRP cut-offs for diagnosis of CAP in RFH patients
| Cut-offa | Sensitivity (%) | Specificity (%) | Positive predictive value (%) | Negative predictive value (%) | Positive likelihood ratio | Negative likelihood ratio |
|---|---|---|---|---|---|---|
| WCC > 8.2 | 79.2 | 58.8 | 24.6 | 94.4 | 1.92 | 0.35 |
| WCC > 9.4 | 69.8 | 70.0 | 28.2 | 93.2 | 2.32 | 0.43 |
| CRP > 65 | 84.9 | 33.2 | 17.7 | 92.9 | 1.27 | 0.45 |
| CRP > 160 | 49.1 | 71.6 | 22.6 | 89.2 | 1.73 | 0.71 |
| WCC > 8.2 AND CRP > 65 | 69.8 | 70.3 | 28.5 | 93.2 | 2.35 | 0.43 |
| WCC > 8.2 OR CRP > 65 | 94.3 | 21.7 | 16.9 | 95.8 | 1.21 | 0.26 |
| ΔCRP<−15 | 62.3 | 75.0 | 29.7 | 92.2 | 2.50 | 0.50 |
| ΔCRP < 0 | 73.6 | 65.2 | 26.4 | 93.6 | 2.11 | 0.41 |
| WCC > 8.2 AND ΔCRP < 0 | 62.3 | 80.8 | 35.4 | 92.6 | 3.25 | 0.47 |
| WCC > 8.2 OR ΔCRP < 0 | 90.6 | 43.1 | 21.2 | 96.4 | 1.59 | 0.22 |
Populations included were all patients admitted >48 h for CAP (n = 53) and MR− COVID-19 (n = 313). aWCC values represent cell numbers ×106/mL. CRP values represent concentrations in mg/L.
Independent cohort validation
To demonstrate the reproducibility of our findings, we used independent patient cohorts from a separate hospital, BH, consisting of 169 CAP, 171 MR− COVID-19, and 10 MR+ COVID-19 patients. To ensure comparability to the RFH cohort, the patients were identified over the same time periods using identical criteria. Baseline demographic analyses were comparable to those in the RFH cohort (Table 1), and 99 (59%), 56 (31%) and 4 (2%) of CAP, MR− and MR+ COVID-19 patients respectively were admitted for >48 h and had a blood sample collected as part of routine clinical care 48–72 h into admission (Table 1).
Differences in inflammatory markers within the BH cohort reflected those observed at RFH, with admission WCC and CRP levels being higher in CAP compared with either COVID-19 population and accompanied by a reduction following 48–72 h of admission not observed in COVID-19 (Figure S3). As for RFH, we also observed no differences in the levels of these variables between MR− and MR+ COVID-19 patients (Figure S3). Applying these inflammatory marker parameters into a logistic regression model demonstrated similar findings to the RFH cohort, with optimal AUC (0.88, 95% CI 0.83–0.94) to discriminate CAP from MR− COVID-19 being derived from inclusion of both admission WCC and ΔCRP as variables in the model (Table 2). Sensitivity analyses again demonstrated that demographic differences did not play a significant role, ICU admission within 72 h of hospital admission (seen in 19/56, 34.0% COVID-19 patients) did not significantly confound the model and reassigning MR+ COVID-19 patients to the CAP cohort did not improve AUC scores (Table S3).
Next, we applied the cut-offs independently derived from the RFH cohort on BH patient data, and observed a similar trade-off between sensitivity and specificity (Table 4). High sensitivity (91.9%) with very low specificity (26.8%) was achieved when utilizing either baseline criteria (WCC > 8.2 × 106/mL or CRP > 65 mg/L) to diagnose CAP. In contrast using either admission WCC > 8.2 × 106/mL or ΔCRP < 0 mg/L yielded a sensitivity for CAP approaching 95% but with an improved specificity (46.4%), permitting exclusion of bacterial co-infection in 26/56 (46.4%) MR− COVID-19 patients that had been admitted for >48 h (Table 4).
Table 4.
Discriminatory performance of WCC and ΔCRP cut-offs for diagnosis of CAP in BH patients
| Cut-offa | Sensitivity (%) | Specificity (%) | Positive predictive value (%) | Negative predictive value (%) | Positive likelihood ratio | Negative likelihood ratio |
|---|---|---|---|---|---|---|
| WCC > 8.2 | 84.3 | 57.1 | 75.8 | 69.6 | 1.97 | 0.28 |
| WCC > 9.4 | 79.8 | 69.6 | 82.3 | 66.1 | 2.63 | 0.29 |
| CRP > 65 | 80.8 | 32.1 | 67.8 | 48.6 | 1.19 | 0.6 |
| CRP > 160 | 54.5 | 73.2 | 78.3 | 47.7 | 2.04 | 0.62 |
| WCC > 8.2 AND CRP > 65 | 68.7 | 0.75 | 82.9 | 57.5 | 2.75 | 0.42 |
| WCC > 8.2 OR CRP > 65 | 91.9 | 26.8 | 68.9 | 65.2 | 1.26 | 0.30 |
| ΔCRP<−15 | 56.2 | 80.4 | 82.0 | 53.6 | 2.86 | 0.55 |
| ΔCRP < 0 | 65.1 | 75.0 | 80.6 | 57.5 | 2.61 | 0.46 |
| WCC > 8.2 AND ΔCRP < 0 | 55.1 | 85.7 | 85.9 | 54.5 | 3.85 | 0.52 |
| WCC > 8.2 OR ΔCRP < 0 | 94.4 | 46.4 | 73.6 | 83.9 | 1.76 | 0.12 |
Populations included were all patients admitted >48 h for CAP (n = 99) and MR− COVID-19 (n = 56).
WCC values represent cell numbers ×106/mL. CRP values represent concentrations in mg/L.
Finally, we attempted to estimate the impact that the diagnostic criteria could have had on antibiotic prescribing in the MR− COVID-19 cohort at BH. We assumed antibiotic courses of 5 days duration, and thus estimated up to 855 antibiotic days in total for the 171 patients admitted in the BH cohort. The absence of both baseline WCC > 8.2 × 106/mL and CRP > 65 mg/L to exclude bacterial co-infection could have prevented 229 antibiotic days, a 27% reduction. Instead, the 56 MR− COVID-19 patients admitted for >48 h would have received up to 280 antibiotic days in total. Excluding bacterial co-infection by the absence of both WCC > 8.2 and ΔCRP < 0 has greater specificity (46.4%) but could only be applied to the final 3 days of antibiotic prescriptions. Therefore, within this cohort admitted for >48 h, we extrapolated a total saving of 78 antibiotic days, a 28% reduction in total antibiotic prescriptions.
Discussion
Elevated inflammatory responses, high case fatality and bacterial co-infections observed in influenza contribute to frequent antibiotic prescriptions in COVID-19.2,4,9,29 However, radiological findings in COVID-19 are heterogenous30 and microbiological investigations rarely identify pathogenic bacteria,6–10 precluding reliable identification of co-infection. Therefore, novel approaches to exclude bacterial co-infection in COVID-19 are a research priority to facilitate antimicrobial stewardship efforts.15,31–33
We used patients admitted with CAP prior to the COVID-19 pandemic as a benchmark to define processes that occur in bacterial pulmonary infections, including co-infection in COVID-19. This demonstrated that, at a population level, admission WCC (predominantly neutrophils) and CRP can discriminate CAP from COVID-19, and that WCC and CRP decreased following antibiotic therapy in pneumonia, but not in COVID-19. We used these observations to construct a model and decision-making criteria to assist with excluding bacterial co-infection in many cases of COVID-19. To overcome variability in individual patient responses (e.g. 26% and 35% of CAP patients at RFH and BH respectively showed a rise in CRP), we showed that using two inflammatory markers (WCC and ΔCRP) yielded the optimal sensitivity and specificity combination. We propose that absence of both admission WCC >8.2 × 106/mL and decreasing CRP could support stopping antibiotics in almost 50% of hospitalized COVID-19 patients, reducing total antibiotic prescriptions in this population by up to 25%. This approach would exceed most antimicrobial stewardship achievements,31,32 and reduce selection for antibiotic-resistant bacteria during this pandemic.13,15
The combination of selected cut-offs yielded the greatest sensitivity for CAP, ensuring ongoing antibiotic treatment where needed, but came at the expense of specificity. This was further illustrated by an alternative strategy, the use of baseline WCC and CRP, which yielded comparable sensitivity but lower specificity than assessments including the later timepoint. The absolute number of antibiotic days saved by each approach will depend on hospitals' antibiotic prescribing rates, but notably both strategies could have yielded comparable proportions of antibiotic prescribing reduction. However, we caution that predicted savings in antibiotic prescriptions dependent solely on baseline inflammatory markers are likely to be significantly over-estimated. On hospital attendance, confirmation of COVID-19 diagnosis, formal chest radiograph reporting and clinical improvement trajectory would all be lacking, supporting precautionary antibiotic prescribing in many cases. Nevertheless, for either scenario, criteria relying solely on WCC and CRP measurements remain permissive to excessive antibiotic prescribing in COVID-19, and additional markers may improve sensitivity and specificity. PCT can discriminate bacterial from some viral respiratory tract infections,23,24 but has not been systematically compared between bacterial pneumonia and COVID-19. Unfortunately, we could not investigate PCT as it was not measured routinely in our CAP cohorts. Future studies should assess the discriminatory capacity of PCT, D-dimers,9,24,34 and other novel biomarkers, such as transcriptional signatures that quantify inflammatory cytokine activity35 or those that discriminate bacterial from viral infections.36
The large, standardized populations studied, use of routinely available clinical investigations, and the reproducibility of our findings in an independent validation cohort population are key strengths of our study. We were also able to convert population level findings into practical diagnostic criteria that can be used in generalized clinical settings. The cut-offs used were derived from a separate population in which they were tested, adding scientific validity to our conclusions, but does not negate individual care providers determining institution-specific cut-offs. Moreover, all data were collected before the beneficial effects of dexamethasone were published,37 and WCC cut-offs may need revising in this context. In addition, our criteria should not be considered in isolation from clinical decision making. Clinical improvement, reduced supplemental oxygen requirement, and the absence of consolidation on chest radiograph in COVID-19 patients may all contribute to excluding bacterial co-infection and could be used alongside the WCC- and CRP-based criteria to support cessation of antibiotics.
Consistent with previous observations, we found microbiological or radiological evidence of bacterial co-infection to be rare in COVID-19.6–10 Notwithstanding the low numbers, levels of inflammatory markers in this subset, both at baseline and during admission, were strikingly almost indistinguishable from those seen in the wider COVID-19 population, and remained distinct from those in CAP patients. Furthermore, reclassifying MR+ COVID-19 patients as individuals with CAP did not improve the discriminatory capacity of our model, indicating these to be distinct populations. The vast majority of patients classified as MR+ COVID-19 showed microbiological evidence of infection and we infer that in most instances, bacterial identification, particularly from the respiratory tract, lacks sensitivity and specificity to establish causal involvement in a bacterial pulmonary infection.11,38 This is well illustrated by the low rate of microbiological identification observed in the CAP group, supporting the syndromic approach taken in our study to define populations with bacterial and viral pulmonary infections.
Our study has some notable limitations. First, the populations were identified at non-overlapping times, due to the disproportionate prevalence of COVID-19 cases in 2020. We attempted to mitigate for the enforced use of historical pneumonia comparator groups by identifying these patients over the same months of 2019 as COVID-19 cases in 2020. We also did not collect clinical severity or outcome data for the patients, and thus we cannot measure a direct impact on prognosis. Second, we used a radiological, but not microbiological, definition of pneumonia, and although standardized, it is possible that some pneumonia cases had non-bacterial aetiology. Third, we inferred that true bacterial co-infection in COVID-19 shares pathophysiology and inflammatory marker responses with CAP in the absence of COVID-19. This hypothesis remains untested and the divergence in responses between CAP and MR+ COVID-19 illustrates the difficulty of positively diagnosing COVID-19 patients with bacterial co-infection that requires antibiotic therapy. Fourth, we did not include suspected COVID-19 patients with negative SARS-CoV-2 results, therefore our findings may not be applicable to this cohort. Finally, we focused on patient assessments made within 72 h of admission, and thus our decision-making tools are not applicable to patients with prolonged hospital admissions.
In conclusion, we demonstrated that routine clinical parameters, admission WCC and changes in CRP following antibiotic administration, can be translated into a set of diagnostic criteria that can exclude bacterial co-infection in up to half of COVID-19 patients. The routine nature of the investigations required mean that, even in the context of a pandemic, this approach can form the basis of protocols to assist reductions in unnecessary antibiotic prescriptions for viral infections, minimizing drug-associated adverse effects and reducing the development of antimicrobial resistance.
Supplementary Material
Acknowledgements
We wish to thank members of the RFL Clinical Practice Group and Open Health Care UK for prompt extraction of clinical records.
Funding
This work was supported by a National Institute for Health Research (NIHR) Clinical Lectureship to G.P.
Transparency declarations
None to declare.
Supplementary data
Tables S1 to S4 and Figures S1 to S3 are available as Supplementary data at JAC Online.
References
- 1. European Centre for Disease Prevention and Control. COVID-19 pandemic: situation update. https://www.ecdc.europa.eu/en/covid-19-pandemic.
- 2. Manson JJ, Crooks C, Naja M. et al. COVID-19-associated hyperinflammation and escalation of patient care: a retrospective longitudinal cohort study. Lancet Rheumatol 2020; 2: e594–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. MacIntyre CR, Chughtai AA, Barnes M. et al. The role of pneumonia and secondary bacterial infection in fatal and serious outcomes of pandemic influenza A(H1N1)pdm09. BMC Infect Dis 2018; 18: 637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Morens DM, Taubenberger JK, Fauci AS.. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: implications for pandemic influenza preparedness. J Infect Dis 2008; 198: 962–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhou F, Yu T, Du R. et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 2020; 395: 1054–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hughes S, Troise O, Donaldson H. et al. Bacterial and fungal coinfection among hospitalized patients with COVID-19: a retrospective cohort study in a UK secondary-care setting. Clin Microbiol Infect 2020; 26: 1395–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lehmann CJ, Pho MT, Pitrak D. et al. Community Acquired Co-infection in COVID-19: a Retrospective Observational Experience. Clin Infect Dis 2020; ciaa902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rawson TM, Moore LSP, Zhu N. et al. Bacterial and fungal co-infection in individuals with coronavirus: a rapid review to support COVID-19 antimicrobial prescribing. Clin Infect Dis 2020; 71: 2459–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vaughn VM, Gandhi T, Petty LA. et al. Empiric antibacterial therapy and community-onset bacterial co-infection in patients hospitalized with COVID-19: a multi-hospital cohort study. Clin Infect Dis 2020; ciaa1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Langford BJ, So M, Raybardhan S. et al. Bacterial co-infection and secondary infection in patients with COVID-19: a living rapid review and meta-analysis. Clin Microbiol Infect 2020; 26: 1622–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. García-Vázquez E, Marcos MA, Mensa J. et al. Assessment of the usefulness of sputum culture for diagnosis of community-acquired pneumonia using the PORT predictive scoring system. Arch Intern Med 2004; 164: 1807–11. [DOI] [PubMed] [Google Scholar]
- 12. UK National Institute for Health and Care Excellence (NICE). COVID-19 rapid guideline: managing suspected or confirmed pneumonia in adults in the community. 2020. https://www.nice.org.uk/guidance/ng165/chapter/4-Managing-suspected-or-confirmed-pneumonia. [PubMed]
- 13. Nieuwlaat R, Mbuagbaw L, Mertz D. et al. COVID-19 and antimicrobial resistance: parallel and interacting health emergencies. Clin Infect Dis 2020; ciaa773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rawson TM, Moore LSP, Castro-Sanchez E. et al. COVID-19 and the potential long-term impact on antimicrobial resistance. J Antimicrob Chemother 2020; 75: 1681–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rawson TM, Wilson RC, Holmes A.. Understanding the role of bacterial and fungal infection in COVID-19. Clin Microbiol Infect 2021; 27: 9–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Clancy CJ, Nguyen MH.. COVID-19, superinfections and antimicrobial development: what can we expect? Clin Infect Dis 2020; ciaa524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Knight SR, Ho A, Pius R. et al. Risk stratification of patients admitted to hospital with COVID-19 using the ISARIC WHO Clinical Characterisation Protocol: development and validation of the 4C Mortality Score. BMJ 2020; 370: m3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xiao L-S, Zhang W-F, Gong M-C. et al. Development and validation of the HNC-LL score for predicting the severity of coronavirus disease 2019. EBioMedicine 2020; 57: 102880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu F, Li L, Xu M. et al. Prognostic value of interleukin-6, C-reactive protein, and procalcitonin in patients with COVID-19. J Clin Virol 2020; 127: 104370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brill SE, Jarvis HC, Ozcan E. et al. COVID-19: a retrospective cohort study with focus on the over-80s and hospital-onset disease. BMC Med 2020; 18: 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ahn S, Kim WY, Kim S-H. et al. Role of procalcitonin and C-reactive protein in differentiation of mixed bacterial infection from 2009 H1N1 viral pneumonia. Influenza Other Respir Viruses 2011; 5: 398–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wu M-H, Lin C-C, Huang S-L. et al. Can procalcitonin tests aid in identifying bacterial infections associated with influenza pneumonia? A systematic review and meta-analysis. Influenza Other Respir Viruses 2013; 7: 349–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cuquemelle E, Soulis F, Villers D. et al. Can procalcitonin help identify associated bacterial infection in patients with severe influenza pneumonia? A multicentre study. Intensive Care Med 2011; 37: 796–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pfister R, Kochanek M, Leygeber T. et al. Procalcitonin for diagnosis of bacterial pneumonia in critically ill patients during 2009 H1N1 influenza pandemic: a prospective cohort study, systematic review and individual patient data meta-analysis. Crit Care 2014; 18: R44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Coelho LM, Salluh JIF, Soares M. et al. Patterns of c-reactive protein RATIO response in severe community-acquired pneumonia: a cohort study. Crit Care 2012; 16: R53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Coster D, Wasserman A, Fisher E. et al. Using the kinetics of C-reactive protein response to improve the differential diagnosis between acute bacterial and viral infections. Infection 2020; 48: 241–8. [DOI] [PubMed] [Google Scholar]
- 27. Yu B, Li X, Chen J. et al. Evaluation of variation in D-dimer levels among COVID-19 and bacterial pneumonia: a retrospective analysis. J Thromb Thrombolysis 2020; 50: 548–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Marks M, Pollara G, Miller D. et al. elCID: an electronic clinical infection database to support integrated clinical services and research in infectious diseases. J Infect 2015; 71: 402–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Docherty AB, Harrison EM, Green CA. et al. Features of 20133 UK patients in hospital with COVID-19 using the ISARIC WHO Clinical Characterisation Protocol: prospective observational cohort study. BMJ 2020; 369: m1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shi H, Han X, Jiang N. et al. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect Dis 2020; 20: 425–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pollara G, Bali S, Marks M. et al. Time efficiency assessment of antimicrobial stewardship strategies. Clin Infect Dis 2017; 64: 1463–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Davey P, Marwick CA, Scott CL. et al. Interventions to improve antibiotic prescribing practices for hospital inpatients. Cochrane Database Syst Rev 2017; issue 2: CD003543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sieswerda E, de Boer MGJ, Bonten MMJ. et al. Recommendations for antibacterial therapy in adults with COVID-19—an evidence based guideline. Clin Microbiol Infect 2020; doi:10.1016/j.cmi.2020.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. de Jong E, van Oers JA, Beishuizen A. et al. Efficacy and safety of procalcitonin guidance in reducing the duration of antibiotic treatment in critically ill patients: a randomised, controlled, open-label trial. Lancet Infect Dis 2016; 16: 819–27. [DOI] [PubMed] [Google Scholar]
- 35. Bell LC, Meydan C, Kim J. et al. Transcriptional response modules characterise IL-1β and IL-6 activity in COVID-19. iScience 2020; 24: 101896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sampson D, Yager TD, Fox B. et al. Blood transcriptomic discrimination of bacterial and viral infections in the emergency department: a multi-cohort observational validation study. BMC Med 2020; 18: 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. RECOVERY Collaborative Group, Horby P, WS L. et al. Dexamethasone in hospitalized patients with COVID-19—preliminary report. N Engl J Med 2020; doi:10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sanyal S, Smith PR, Saha AC. et al. Initial microbiologic studies did not affect outcome in adults hospitalized with community-acquired pneumonia. Am J Respir Crit Care Med 1999; 160: 346–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.