Abstract
Reported outcomes, such as incidence rates of mortality and intensive care unit admission, vary widely across epidemiological coronavirus disease 2019 (COVID-19) studies, including in the nephrology field. This variation can in part be explained by differences in patient characteristics, but also methodological aspects must be considered. In this review, we reflect on the methodological factors that contribute to the observed variation in COVID-19-related outcomes and their risk factors that are identified in the various studies. We focus on issues that arose during the design and analysis phase of the European Renal Association COVID-19 Database (ERACODA), and use examples from recently published reports on COVID-19 to illustrate these issues.
Keywords: COVID-19, epidemiology, kidney failure, mortality, outcomes, statistics
INTRODUCTION
In the past months, an impressive number of studies has been published on the epidemiology of coronavirus disease 2019 (COVID-19). Many of these studies tried to identify risk factors that predispose to developing severe COVID-19 leading to hospitalization, intensive care unit (ICU) admission and death. This plethora is not surprising since there is a great deal of uncertainty surrounding this new disease. Understanding which individuals are at risk for severe COVID-19 is critical in managing the present pandemic. For instance, current management strategies are to inform high-risk individuals—including patients with chronic kidney disease (CKD)—to take strict precautions and avoid getting infected, and for healthcare authorities to design preventive strategies and weigh them against their effects on individual freedom and social-economic consequences. In addition, information on high-risk groups is essential for designing adequate vaccination strategies.
The results of epidemiological COVID-19 studies vary widely in the field of nephrology, but also in other disease areas. This includes the point estimates of the strength of individual risk factors associated with the development of severe COVID-19, but also the incidence rates of endpoints under study, including death. This variation can in part be explained by differences in patient characteristics, such as age distribution of the studied populations [1]. However, this is usually not the only reason for diverging study results. Methodological aspects also play an important role and must be taken into account [2].
Readers of nephrological journals may not always be aware of the impact of the choice of a specific study design, outcome definition and applied method when they interpret study results. The aim of this review is therefore to increase awareness of these methodological issues. To this end, we will reflect on existing methodological factors that are commonly applied and that may contribute to the observed variation across studies in COVID-19-related outcomes and risk factors. We will focus on issues that arose during the design and analysis of the European Renal Association COVID-19 Database (ERACODA), and we illustrate this with examples from recently published reports on COVID-19.
ERACODA
ERACODA was established in March 2020, soon after the COVID-19 pandemic reached Europe, with the aim to gain insight into the consequences of this disease for patients with kidney failure treated with kidney replacement therapy (KRT) in Europe. In this cohort study, the clinical course and outcomes—including hospital admission, ICU admission and mortality—of KRT patients with COVID-19 is investigated. In addition, detailed information on patient, disease and treatment factors is gathered [3].
Data are collected from adult patients who are treated with maintenance dialysis or living with a functioning kidney allograft and who are diagnosed with COVID-19. The COVID-19 diagnosis needs to be based on a positive result on a real-time polymerase chain reaction (PCR) assay of nasal or pharyngeal swab specimens, and/or compatible findings on a computed tomography (CT) scan or chest X-ray of the lungs. Outpatients, as well as hospitalized patients, are included and the primary outcome of the study is vital status at Day 28 after diagnosis. Physicians responsible for the care of these patients are asked to enter data of all KRT patients from their centre with COVID-19 into the central ERACODA database, which is hosted at the University Medical Center Groningen, The Netherlands. By 1 December 2020, almost 2400 records had been entered into this database by 208 physicians from 128 centres in 30 countries.
SELECTION OF STUDY POPULATIONS
Bias introduced by selection
The first methodological challenge arose already in the design phase of the study. The choice of the exact patient group to be included is of major importance in all clinical studies, because this may result in selection bias. Selection bias originates from any error in the enrolment of study participants and/or from factors affecting the study participation. As a result, the relationship between exposure and outcome may differ between individuals who are included in the study and those who were potentially eligible for the study, but were not included [4]. The problem of selection bias can be illustrated by a hypothetical example. Suppose an investigator is interested in the recovery of CKD patients with COVID-19 who were admitted to a hospital. The investigator sends these patients a request to complete an extensive survey on how they are feeling 1 month after their COVID-19 diagnosis. One can imagine that those patients who recovered well will respond to the survey, while patients who are still feeling ill or lack energy, will not respond. Hence, this method of selecting study participants will lead to overrepresentation of relatively healthy study participants and consequently to an underestimation of the effect COVID-19 on patient well-being.
Even when there is no selection bias on individual study level, like in population-based registries that do not take samples but report on the entire population, the choice of the study population has significant implications for its comparability with other studies that investigate the same outcome. For instance, some of the recent papers on COVID-19 describe outcomes in the general population [5, 6], whereas others report on disease-specific populations, such as patients with a history of cardiovascular disease or with diabetes mellitus [7, 8]. It can be expected that analyses based on disease-specific cohorts yield higher mortality rates than those based on cohorts from the general population. The same is true for studies that included only hospitalized patients versus those that included all patients with a COVID-19 diagnosis, including those who were not admitted to a hospital.
Example 1: Williamson et al. published one of the largest studies on COVID-19 in Europe, which was based on OpenSAFELY, a health analytics platform that covers ∼40% of the National Health Service population in the UK [5]. The investigators included over 17 million individuals in their recent study, of whom almost 11 000 died of COVID-19. The primary outcome of the study was death from COVID-19. In this study design, COVID-19-related mortality is not only determined by the risk of death for individual patients once diagnosed with COVID-19, but also by their risk of being infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) at all [2, 5]. This risk of getting infected may vary between subgroups in the study population. For instance, people with assumed risk factors for mortality may have shielded themselves to prevent infection. As a result, these assumed risk factors become less strongly related with mortality when compared with a situation in which people have no assumptions on what risk factors for mortality might be.
In studies that include only patients who are infected with SARS-CoV-2, the risk of getting infected does not play a role. Consequently, the mortality rates in the study by Williamson et al. were 100- to 1000-fold lower when compared with other reports that reported mortality among infected patients [2, 5]. This makes comparison of outcomes of these different types of study populations complicated.
The study design used by Williamson et al. could explain, at least in part, some of their findings. For instance, the finding that South Asian and Black people had a 43–48% higher risk of COVID-19-related death than White people even after adjustment for comorbidities or other known risk factors. This higher mortality risk in non-White ethnic groups could be interpreted as a genetic preponderance to a more severe disease course. However, it could also reflect a higher risk of infection due to differences in standards of living or occupational exposure. For instance, it may be that non-White people live with more people of more generations per household, or have a profession with more close physical contact to other people. This will increase transmission rates and therefore mortality rates when expressed as percentage of the overall population instead of when expressed as percentage of diseased persons.
Case ascertainment
A second important issue is how patients infected with SARS-CoV-2 are identified. Testing protocols and accuracy may have a large impact on the results [9].
In some studies, patients are classified as having COVID-19 based merely on clinical symptoms, while in others patients must (also) have a positive PCR assay of nasal or pharyngeal swab specimens and/or a CT scan or chest X-ray showing abnormalities compatible with the disease. For example, in ERACODA, the COVID-19 diagnosis needs to be based on a positive result on a real-time PCR assay of nasal and/or pharyngeal swab specimens, and/or compatible findings on CT scan or chest X-ray of the lungs, whereas the ERA-EDTA Registry in their recent study included also patients with a clinical diagnosis only [3, 10, 11]. This latter group may be expected to be less ill or even not to have COVID-19. This may explain at least in part the lower mortality rate in the ERA-EDTA Registry when compared with ERACODA. On the other hand, making a positive test result mandatory may also lead to bias, since only a small fraction of the population has been tested [12]. This was especially an issue in the early days of the pandemic when there was a shortage of testing capacity. Testing was therefore mostly performed in patients with severe symptoms and among those in high-risk groups. Including in a study only those patients who tested positive will therefore have led in these early studies to a higher proportion of cases with a severe course of disease when compared with later studies that also included asymptomatic patients with a positive PCR [12], and thus to higher mortality rates. Also the risk factor profile is likely to be different in patients with a clinical diagnosis in which the physician did not ask for confirmation by a PCR test, since these patients were generally younger and had fewer comorbidities [2]. The effect of this type of bias is expected to have diminished over time, because more and more testing capacity has become available.
Example 2: The choice to include only symptomatic or to include also asymptomatic patients who had a positive PCR test may lead to different outcomes in patients treated with dialysis and those who are living with a functioning kidney transplant (KTx). Haemodialysis (HD) units have increasingly implemented routine screening procedures, leading to identification of patients with COVID-19 who do not (yet) have any signs or symptoms of the disease. On the other hand, KTx recipients and peritoneal dialysis (PD) patients are usually only tested for COVID-19 when they present with symptoms, which is confirmed by the findings from ERACODA (Figure 1). Especially in HD patients, there will be a difference in mortality rates when only symptomatic patients or also asymptomatic patients with a positive PCR are included. The former will lead to a higher, and the latter to a lower mortality rate. How the patient population is defined will therefore affect the comparison of mortality rates in dialysis versus transplant patients. For a fair comparison of the effect of treatment modality, a comparison can therefore best be made in patients who are identified by symptoms in both groups in a similar manner.
FIGURE 1:
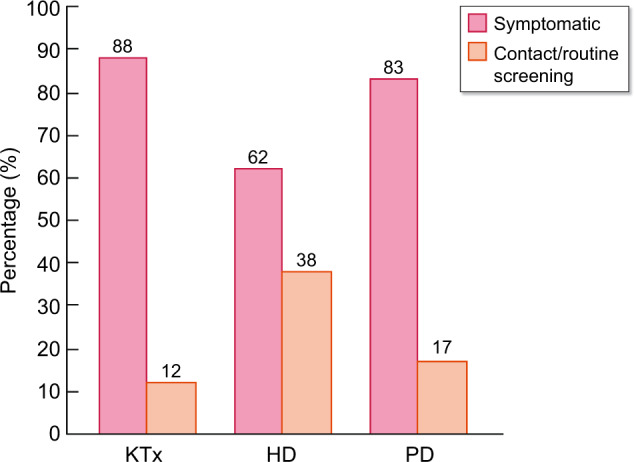
Distribution (%) of type of COVID-19 identification among KTx recipients (n = 338), HD (n = 861) and PD (n = 41) patients in ERACODA. Among symptomatic patients mortality rates were 22, 28 and 41%, respectively, for KTx, HD and PD patients, whereas mortality rates in patients identified by contact/routine screening were lower (19, 18 and 14%, respectively).
DEFINITION OF OUTCOME PARAMETERS
COVID-19-related mortality
The outcome that has most often been studied in relation to COVID-19 is, without any doubt, mortality. Mortality can be expressed in many ways, for example by mortality rates, case fatality rates and infection fatality rates. For most of the measures used to express COVID-19-related mortality one needs (i) the number of deaths from the disease and (ii) the number of cases. As was discussed in the previous section, the number of cases suffers from selective testing and from variation in testing policies across studies, across countries and over time [12]. Data on mortality are in general reliable but can still be subject to limited comparability due to a significant reporting lag (registration authorities can be slow when reporting the number of deceased subjects for a certain period), and potential under- or over-reporting [12]. Each of the ways to express mortality (Table 1) leads to different results and has a different interpretation, as is shown by Example 3.
Table 1.
Measures that can be used to express COVID-19-related mortality
| Measure | Definition | Strengths and weaknesses |
|---|---|---|
| Probability of death | Percentage of deaths at a certain time point calculated using the Kaplan–Meier method for survival analysis |
Strength: observations can be censored in case of loss to follow-up/in absence of information on vital status before the end of follow-up; this allows inclusion of all available information Weakness: the number of infected cases is not taken into account |
| Mortality rate |
Number of deaths from COVID-19 in the population, scaled to the size of that population, per unit of time Typically expressed as cases per 1000 individuals per year (also person-years can be used) |
Strengths: can easily be calculated and interpreted; suitable for comparison of populations because it is scaled to the total population size Weaknesses: the number of infected cases is not taken into account; time to death (early or late) or loss to follow-up is not taken into account |
| Case fatality rate |
Proportion of deaths from COVID-19, compared with the total number of people diagnosed with the disease for a particular period Represents a measure of disease severity; most often used for diseases with limited-time courses, such as outbreaks of acute infections |
Strength: can easily be calculated and interpreted Weaknesses: asymptomatic and otherwise undiagnosed cases are not taken into account; often calculated while the individual outcome (recovery or death) is known only for a proportion of infected patients |
| Infection fatality rate |
Proportion of deaths from COVID-19 compared with the total number of infected people—including those who are asymptomatic and undiagnosed—for a particular period Similar to case fatality rate, but aims to estimate the fatality rate in both the sick (with detected disease) and healthy (with undetected disease) groups of infected people |
Strengths: includes the whole spectrum of infected people, from asymptomatic to severe; recommended as a more reliable parameter for evidence-based assessment of the SARS-CoV-2 pandemica Weakness: it may be difficult to capture asymptomatic and undiagnosed subjects |
The Centre for Evidence-Based Medicine, Global COVID-19 Case Fatality Rates, CEBM.
Example 3: Suppose that from the 20 million people in a country, 750 000 individuals were diagnosed with COVID-19 between 1 March 2020 and 1 March 2021. Within this (calendar) year, 15 000 of them died from the disease. If we would calculate the mortality rate, this would be 15 000/20 000 000 × 1000 = 0.75 per 1000 individuals per year. Alternatively, if we chose to present the case fatality rate in the year between March 2020 and March 2021, this would be 15 000/750 000 = 2.0%. In reality, more than 750 000 individuals will have been infected with the virus, since part of infected patients are known to be asymptomatic or were not tested for some other reason. Thus, the total number of infected people could for instance be around 900 000 (estimation based on [13]). As a result, the infection fatality rate is lower than the case fatality rate: 15 000/900 000 = 1.7%. Most studies, and also the popular media, use the case fatality rate to express mortality, because it is easy to obtain and understand [10, 11]. However, to compare the severity of a new viral disease with that of other longer existing viral diseases, such as influenza, it may be better to compare infection fatality rates. The latter is more difficult to calculate, but is not affected by the availability of test capacity, which especially at the start of an epidemic may be limited.
In addition to the chosen measure for mortality, the time period in which mortality is assessed is important. Most studies choose Day 28 mortality because this measure is easily understandable and also used in many clinical trials. Other studies, however, report on in-hospital mortality until discharge, which is longer than 28 days in many COVID-19 cases. In contrast, infection fatality rate is defined as fatality rate until the viral disease and its sequelae have fully recovered, which is a measure that is independent of hospital admission and of follow-up duration. Infection fatality rate will therefore be higher than in-hospital mortality, which in turn will be higher than Day 28 mortality.
When the length of follow-up time is discussed, it is also important to consider when the counting starts. Does follow-up start at the day of a positive screening test, at the first day of symptoms or at the day of a positive test when having symptoms? This choice may lead to differences in follow-up time of several days and hence to differences in mortality when 28-day mortality is used. In case of screening for COVID-19 (even in subjects without symptoms), lead-time bias can occur, meaning that the survival time of patients with a positive test falsely seems longer than that of symptomatic patients because of the earlier diagnosis. In contrast, when for symptomatic patients the day of a positive test result is used as starting point of follow-up, this leads to a relatively short survival time when compared with the situation where start of symptoms was the starting point of follow-up. Finally, bias can occur because patients are missed who have already died before a diagnosis could be made.
Other outcomes related to COVID-19
Other frequently studied clinical outcomes in relation to COVID-19 are hospitalization and ICU admission. It is important to note that these outcomes may not necessarily reflect the severity of the disease. In particular, the outcome of ICU admission has important caveats. In ERACODA, we observed that mortality in dialysis patients was higher than in transplant recipients, whereas dialysis patients were less often admitted to the ICU [11]. This may suggest that treatment limitations played a role in dialysis patients. Such limitations could be based on an assumption of futility of ICU admission, justified or not justified, but also on a lack of ICU capacity, which occurs if the healthcare system is flooded.
STATISTICAL ANALYSIS
Sample size
Most observational studies start without an a priori sample size calculation and without definition of a clinically meaningful effect size [14]. Consequently, studies that are similar in their design and research question, but have different sample sizes, may obtain similar point estimates for the association of a risk factor with the outcome, but lead to different conclusions. In larger studies, a narrower confidence interval (CI) will be found then in smaller studies, resulting in a higher chance of the effect of the same risk factor to be found statistically significant, whereas the clinical relevance of such a risk factor can sometimes be questioned.
Missing data
Any dataset may suffer from missing data, but this is especially a problem in studies that collect a large set of variables. The problem is often ignored when researchers perform a so-called complete case analysis, in which only subjects with complete data for all covariates used in the analysis are included. This may not be a problem when the data are missing completely at random, which means that the value of the variable that is missing is not related to the reason that data is missing. Unfortunately, this is often not the case and not all studies are large studies. Missing data therefore can introduce bias. A recent and reasonably sophisticated method for dealing with missing data, which may help to overcome some of this bias, is multiple imputations [15]. This method predicts missing values using information that is available for the same patient. The advantage of the method is that it gives unbiased results when data are missing at random, meaning that the probability of missing data is unrelated to the actual missing value.
Although it is advised that manuscripts mention specifically which proportion of study data was missing and how missing data were dealt with, this is unfortunately often not reported. Moreover, strategies to solve the problem of missing data, including multiple imputations, are not always applied (and/or described).
Adjustment for confounders
An important part of statistical analyses is how investigators deal with (potential) confounding. Confounding is the ‘mixing’ or ‘blurring’ of effects that happens when an investigator tries to assess the effect of an exposure on an outcome, but then actually measures the effect of a factor other than the exposure [16]. Such a factor is called a confounder when the following three criteria apply: (i) the variable is causally associated with the outcome or, in other words, it is a risk factor for the outcome; (ii) the variable is associated with the exposure; and (iii) the variable is not an effect of the exposure and is not a factor in the causal pathway of the disease [17]. Figure 2 illustrates an example of confounding in the association between COVID-19 and death, which is applicable to patients with kidney failure, but also to individuals from the general population.
FIGURE 2:
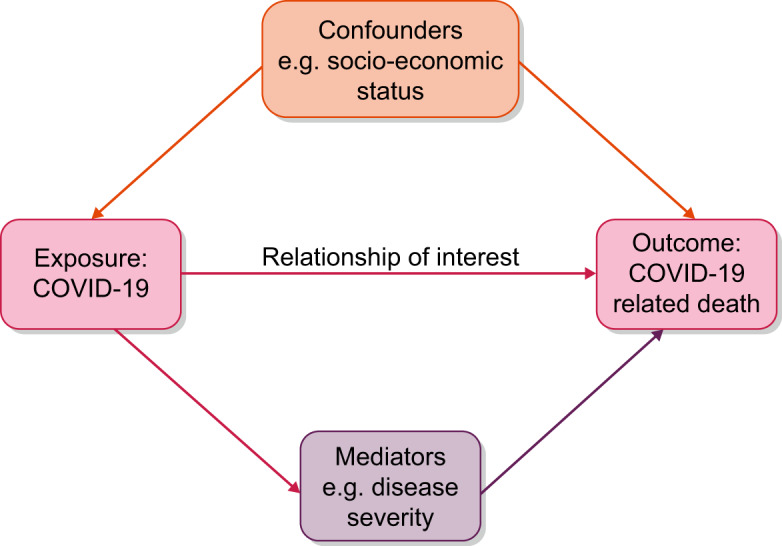
Example of confounding in the association between COVID-19 (exposure) and death (outcome).
Because confounding obscures the real effect of an exposure, it is important to eliminate it. This can be achieved using various approaches, for example by randomization, matching and stratification. Probably the most common way to control for confounding in observational studies is by adjusting for the (potential) confounding factors—that fulfil the three aforementioned criteria—in multivariable models. It is an efficient and reliable method, but it is essential to keep in mind that overadjustment can be a problem. Often researchers tend to adjust for as many factors as possible, sometimes without carefully thinking about causal aspects of the association. Consequently, they may falsely adjust for factors that are not confounders, but mediators, which may lead to unreliable effect estimates. Furthermore, with limited number of events and a long list of confounders, there may not be enough variability in data, which may result in unrealistically small or large estimates of the association between a risk factor and outcome.
The choice of the variables included in the multivariable model can have strong influence on the resulting effect estimates, i.e. hazard ratios (HRs) for Cox regression analysis and odds ratios for logistic regression analysis.
Example 4: The study based on OpenSAFELY suggested that males have a 78% higher risk of getting infected and dying from COVID-19 when only age is included in the multivariable model (HR = 1.78), and a 59% higher risk when a fully adjusted model is used (HR = 1.59) [5]. This suggests that in the general population male sex is a major risk factor for contracting the disease and subsequent mortality. In contrast, it seems that in the dialysis population male sex is a much weaker risk factor, although the HRs for (male) sex as a risk factor for mortality in studies focusing on COVID-19 in this patient group varied to some extent [10, 11].
When we further investigated these differences, it turned out that the variation could—in large part—be explained by variation in the variables included in the multivariable models. In the ERACODA and in the ERA-EDTA Registry study, the HRs for male sex were exactly equal at 1.23 when adjusted for age only [10, 11]. The strength of ERACODA is that it collects many additional variables that can be used to explore causal pathways.
Because we were interested in the cause of this difference, we performed a so-called mediation analysis. It appeared that smoking was a strong mediating factor. Males were more often smokers than women, and smoking was causally associated with COVID-19 mortality. Hence, when the factor smoking is included in the multivariable model, the effect of male sex becomes weaker, suggesting that male sex is not a strong intrinsic risk factor per se, but that the risk attributable to male sex is an indirect effect of the fact that men smoke more often.
The strength of risk factors for COVID-19-related mortality in general changes with increasing absolute risk in the population under study. In high absolute risk populations, classical risk factors may show a much weaker association compared with a relatively low-risk population. For example, risk factors lose strength of their association with outcome in older populations. We illustrate this using ERACODA data in Figure 3, where the associations of obesity, diabetes mellitus, coronary artery disease and chronic lung disease with mortality are presented for two age categories: <65 and ≥65 years. This figure shows that the HRs for the associations are in general weaker in the older than in the younger age group. When comparing the relative strength of risk factors, it is therefore important to take note of the baseline absolute risk of the populations under study.
FIGURE 3:
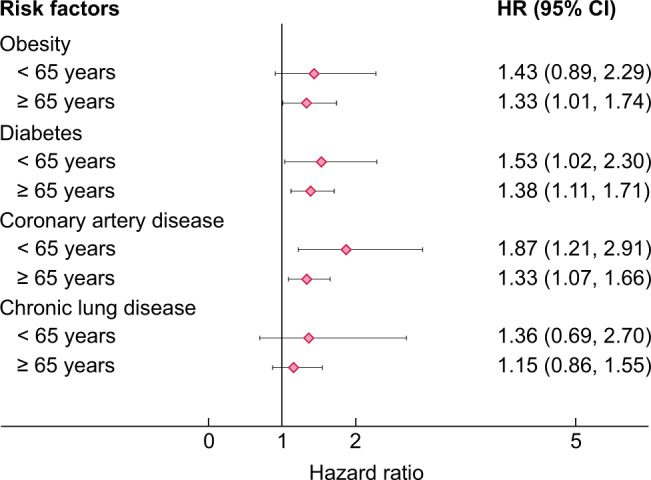
Unadjusted HRs with 95% CI for COVID-19-related mortality in subgroups of patients aged <65 and ≥65 years who are included in ERACODA.
Finally, for the interpretation of results from multivariable models, it is important to distinguish between aetiological and prognostic studies [17]. In aetiological studies, the aim is to investigate a causal relationship between risk factors and an outcome, and in this perspective confounding and knowledge of the pathophysiology of the disease process leading to outcome play a fundamental role. In contrast, prognostic research aims to predict the probability of an outcome as well as possible, and in that case the pathophysiology of the outcome is not necessarily an issue. In this review, we focused on aetiological studies and their interpretation. It is, however, important to realize that multivariable models in prognostic studies may look very similar, but have a completely different interpretation. Prognostic modelling involves predictors that should not be read and interpreted as risk factors or confounders.
IMPLICATIONS AND CONCLUSIONS
Because reported outcomes in studies on the epidemiology of COVID-19 in patients with kidney disease vary widely, careful comparison of study methods is warranted when drawing conclusions for clinical practice and designing public health policies. To be able to correctly interpret the reported findings, one should be aware of the influence of (potential) differences in study design, included study populations, outcome definitions and applied methods. In this review, we discussed the most important methodological factors (summarized in Table 2) that can play a role in COVID-19-related studies in the field of nephrology. It is essential to keep in mind that the variation in reported COVID-19-related outcomes across studies may not be true differences, but just apparent differences caused by these methodological issues.
Table 2.
Summary of the most important methodological factors that can be accountable for variation in outcome in COVID-19-related studies
| Methodological factor | Points of attention | Practical considerations |
|---|---|---|
| Choice of study population |
Specific entry criteria may lead to selection bias General population versus disease-specific population |
Selection bias may lead to the identification of spurious risk factors Disease-specific populations, e.g. only patients on dialysis, will have a higher mortality |
| Case ascertainment |
Inclusion of only symptomatic patients or also asymptomatic patients Differences in criteria for COVID-19 diagnosis, e.g. diagnosis based on positive PCR assay or merely on clinical suspicion |
Asymptomatic patients will have lower mortality Check what the definition of start of follow-up is because this may cause lead time bias |
| Definition of outcomes |
Different measures to express mortality lead to different results and interpretations Differences in time period of studies may influence results Other outcomes (hospitalization, ICU admission) do not always reflect disease severity |
See Table 1 Time period: 28-day (hospital) mortality will be lower than total in-hospital mortality, differences in start day of follow-up may cause lead-time bias Other outcomes: hospitalization, ICU admission |
| Sample size |
Too small Imprecision: wide CIs; unable to detect a clinically relevant risk factor Very large Very narrow CIs; may lead to identification of risk factors with questionable clinical relevance |
Do not confuse statistical significance and clinical relevance, and try to reason from point estimates whether associations may be clinically relevant |
| Missing data |
Incomplete information May lead to errors in statistical analysis, especially when missingness is not at random Often not reported |
Check whether missingness is at random. If not, consider resultant bias Best solution: use of multiple imputation |
ACKNOWLEDGEMENTS
We thank all clinicians that entered information in the ERACODA database for their participation and support, and all healthcare workers that have taken care of patients with COVID-19.
FUNDING
ERACODA received unrestricted research grants from ERA-EDTA, the Dutch Kidney Foundation, Baxter and Sandoz. This article is part of a supplement supported by Fresenius Medical Care without any influence on its content.
CONFLICT OF INTEREST STATEMENT
None declared. The results presented in this paper have not been published previously in whole or part, except in abstract format.
REFERENCES
- 1. Sudharsanan N, Didzun O, Bärnighausen T. et al. The contribution of the age distribution of cases to COVID-19 case fatality across countries: a 9-country demographic study. Ann Intern Med 2020; 173: 714–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gansevoort RT, Hilbrands LB.. CKD is a key risk factor for COVID-19 mortality. Nat Rev Nephrol 2020; 26: 1–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Noordzij M, Duivenvoorden R, Franssen CFM. et al. ERACODA: the European database collecting information of patients on kidney replacement therapy with COVID-19. Nephrol Dial Transplant 2020; 35: 2023–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jager KJ, Tripepi G, Chesnaye NC. et al. Where to look for the most frequent biases? Nephrology (Carlton) 2020; 25: 435–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Williamson EJ, Walker AJ, Bhaskaran K. et al. Factors associated with COVID-19-related death using OpenSAFELY. Nature 2020; 584: 430–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Docherty AB, Harrison EM, Green CA. et al. Features of 20 133 UK patients in hospital with covid-19 using the ISARIC WHO clinical characterisation protocol: prospective observational cohort study. BMJ 2020; 369: m1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Clark A, Jit M, Warren-Gash C. et al. Global, regional, and national estimates of the population at increased risk of severe COVID-19 due to underlying health conditions in 2020: a modelling study. Lancet Glob Health 2020; 8: e1003–e1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Holman N, Knighton P, Kar P. et al. Risk factors for COVID-19-related mortality in people with type 1 and type 2 diabetes in England: a population-based cohort study. Lancet Diabetes Endocrinol 2020; 8: 823–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tsang TK, Wu P, Lin Y. et al. Effect of changing case definitions for COVID-19 on the epidemic curve and transmission parameters in mainland China: a modelling study. Lancet Public Health 2020; 5: e289–e296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jager KJ, Kramer A, Chesnaye NC. et al. Results from the ERA-EDTA Registry indicate a high mortality due to COVID-19 in dialysis patients and kidney transplant recipients across Europe. Kidney Int 2020; 98: 1540–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hilbrands LB, Duivenvoorden R, Vart P. et al. COVID-19-related mortality in kidney transplant and dialysis patients: results of the ERACODA collaboration. Nephrol Dial Transplant 2020; 35: 1973–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sornette D, Mearns E, Schatz M. et al. Interpreting, analysing and modelling COVID-19 mortality data. Nonlinear Dyn 2020; 101: 1751–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. https://covid19.who.int (1 December 2020, date last accessed).
- 14. Noordzij M, Tripepi G, Dekker FW. et al. Sample size calculations: basic principles and common pitfalls. Nephrol Dial Transplant 2010; 25: 1388–1393 [DOI] [PubMed] [Google Scholar]
- 15. de Goeij MCM, van Diepen M, Jager KJ. et al. Multiple imputation: dealing with missing data. Nephrol Dial Transplant 2013; 28: 2415–2420 [DOI] [PubMed] [Google Scholar]
- 16. Jager KJ, Zoccali C, MacLeod A, Dekker FW.. Confounding: what it is and how to deal with it. Kidney Int 2008; 73: 256–260 [DOI] [PubMed] [Google Scholar]
- 17. Tripepi G, Jager KJ, Zoccali C, Dekker FW.. Testing for causality and prognosis: etiological and prognostic models. Kidney Int 2008; 74: 1512–1515 [DOI] [PubMed] [Google Scholar]


