Abstract
A chimeric antigen receptor-modified T-cell therapy recipient developed severe coronavirus disease 2019, intractable RNAemia, and viral replication lasting >2 months. Premortem endotracheal aspirate contained >2 × 1010 severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA copies/mL and infectious virus. Deep sequencing revealed multiple sequence variants consistent with intrahost virus evolution. SARS-CoV-2 humoral and cell-mediated immunity were minimal. Prolonged transmission from immunosuppressed patients is possible.
Keywords: COVID-19, SARS-CoV-2 immune responses, SARS-CoV-2 RNAemia, SARS-CoV-2 intrahost variation, SARS-CoV-2 infectivity, severe acute respiratory syndrome coronavirus 2
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection in patients with hematologic malignancies results in poor coronavirus disease 2019 (COVID-19)–related outcomes [1]. Recipients of chimeric antigen–receptor-modified T-cell therapy (CAR T-cell) are at risk for severe COVID-19 because of chronic B-cell aplasia and hypogammaglobulinemia due to “on-target/off-tumor” effects of CAR T cells [2], which occur when CAR T cells kill normal B cells that express the CAR T-cell target antigen. Conditioning regimens for CAR T-cell therapy can also cause lymphopenia and diminish B- and T-cell function. Although persistence of SARS-CoV-2 RNA in respiratory specimens is not thought to represent infectiousness [3, 4], it is possible that CAR T-cell recipients can persistently shed infectious SARS-CoV-2. Similarly, SARS-CoV-2 RNAemia, which correlates with disease severity [5], is thought to be short-lived [6] but may be more protracted in immunosuppressed hosts.
We present a case of prolonged SARS-CoV-2 infection in a patient with multiple myeloma who received CAR T cells targeting the B-cell maturation antigen (BCMA), which is universally expressed on malignant plasma cells and on some normal plasma cells and mature B cells and is involved in plasma cell survival and B-cell differentiation into plasma cells [7–10]. We characterize viral persistence, intrahost viral evolution, and immune profiles from longitudinal samples and demonstrate that the patient experienced high-level SARS-CoV-2 RNAemia and viral replication for >2 months, viral diversification, and massive lung infection before dying from the infection.
CASE DETAILS
The patient was a 73-year-old man with treatment-refractory multiple myeloma. He had undergone an autologous hematopoietic cell transplant 2 years prior but developed recurrent disease and therefore underwent anti-BCMA CAR T-cell therapy after fludarabine/cyclophosphamide lymphodepletion. Nasopharyngeal (NP) swab reverse-transcription polymerase chain reaction (RT-PCR) testing for SARS-CoV-2 was negative 17 and 2 days before CAR T-cell therapy. He received tocilizumab for cytokine release syndrome, a known toxicity of CAR T-cell therapy [2], but not corticosteroids. He was discharged home in stable condition. Twelve days after discharge (and 25 days after the CAR T-cell infusion), he was admitted to the intensive care unit (ICU) with 2 days of a productive cough, dyspnea, anorexia, and lightheadedness. Temperature was 37.1°C; oxygen saturation was 86% on room air. Laboratory evaluation showed a white blood cell (WBC) count of 3.5 × 109 cells/L (normal range, 3.8 × 109–10.6 × 109), with an absolute lymphocyte count (ALC) of 0.7 × 109 cells/L (normal range, 0.8 × 109–3.65 × 109) and an absolute neutrophil count (ANC) of 2.6 × 109 cells/L (normal range, 2.24 × 109–7.68 × 109). NP swab RT-PCR testing was positive for SARS-CoV-2 RNA (day 0); cycle thresholds were 20.1 and 21.5 for the nucleoprotein 1 (N1) and envelope (E) genes, respectively (Figure 1-I). Chest radiography revealed bibasilar and midzone opacities. He received convalescent plasma (day 2) and remdesivir (days 5–10). During this hospitalization, he had escalating oxygen requirements, necessitating the use of noninvasive positive pressure ventilation on day 5. He never required intubation, and his oxygen requirements gradually improved over the following week. On day 14, he no longer needed supplemental oxygen and was discharged in stable condition on day 17, with minimal residual dyspnea. Follow-up SARS-CoV-2 NP RT-PCR testing remained positive (days 15, 26, and 37). Cycle threshold values were unavailable.
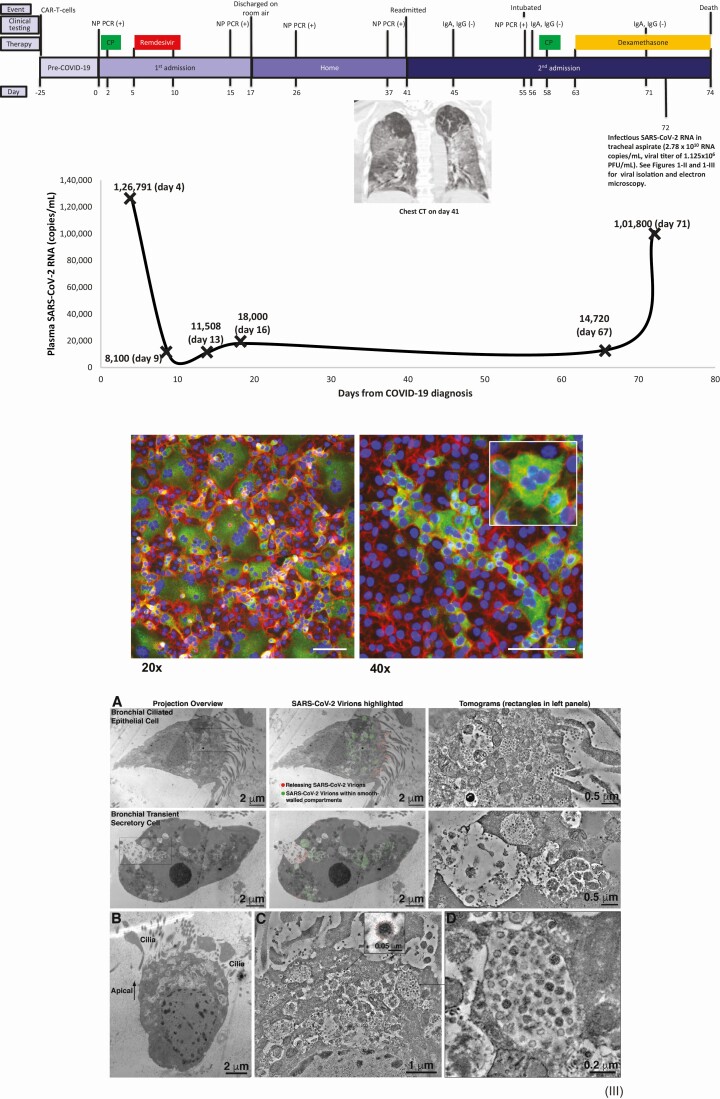
Figure 1.
I, Clinical timeline showing chimeric antigen receptor-modified T-cell infusion (days –25 to 0), first hospital admission (days 0–17), home stay (days 18–41), and second hospital admission (days 41–74). Day 0 denotes day of first positive nasopharyngeal (NP) swab polymerase chain reaction (PCR) for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA. The top panel shows the clinical course and timeline, with results of clinical SARS-CoV-2 NP swab reverse-transcription PCR (RT-PCR) testing and clinical immunoglobulin G and immunoglobulin A testing. The bottom graph shows serial plasma SARS-CoV-2 RNA quantification obtained using a research quantitative RT-PCR assay, with viral quantification from endotracheal aspirate fluid using research assays (quantitative RT-PCR and infectious viral titer). Inset shows a coronal view of chest computed tomography from day 41. Green squares: convalescent plasma; red squares: remdesivir; yellow squares: dexamethasone. II, Indirect immunofluorescence of SARS-CoV-2 (green) isolated in Vero E6 cells (red/blue) from day 72 endotracheal aspirate samples. Green: antibody directed against SARS CoV-2 spike protein (Sinobiologicals). Blue: DNA counterstained with DAPI. Red: filamentous actin counterstained with phalloidin. A syncytium is shown in the inset (right) at higher magnification. Scale bars: 100 µm. III, Electron microscopy of endotracheal aspirate sample obtained on day 72. A, Comparison of virus localization in a bronchial epithelial cell (top) and a transient secretory cell (bottom). Each image is shown as montaged 2D overviews of the whole cell in a 200-nm section (left), overlaid with colored dots to indicate positions of virions (center) and 3D tomographic reconstructions detailing virus populations within cytoplasmic compartments. B, 2D overview of SARS-CoV-2–infected ciliated epithelial cell in a 200-nm semi-thick section. The apical side of the cell is characterized by numerous membrane-bound compartments that are filled with SARS-CoV-2 virions. C, Montaged tomogram of the apical side of the cell shown in (B); hundreds of virus particles are contained within smooth-walled cytoplasmic compartments. Inset: Detail of a single virion with spikes indicated by red dots. D, Tomogram detail of a smooth-walled cytoplasmic compartment containing at least 30 SARS-CoV-2 virions within the shown 15-nm thick volume. Abbreviations: CAR, chimeric antigen receptor; COVID-19, coronavirus disease 2019; CP, convalescent plasma; CT, computed tomography; IgA, immunoglobulin A; IgG, immunoglobulin G; NP, nasopharyngeal; PCR, polymerase chain reaction; PFU, plaque-forming units; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Forty-one days after being diagnosed with COVID-19, he was readmitted to the ICU with a 1-week history of weakness and 4 days of progressively worsening dyspnea, a minimally productive cough, and diarrhea. During the 24 days between hospital discharge and readmission, he had not left his residence except for obtaining outpatient SARS-CoV-2 PCR testing. Temperature was 36.9°C. Oxygen saturation was 81% on room air, and he was started on heated high-flow supplemental oxygen with improvement in his saturation to 97%. WBC count and ANC were normal, but ALC was 0.1 × 109 cells/L. Lymphocyte subset testing showed zero CD19+ B cells and 32 CD3+ T cells/μL (normal range, 856–2669/μL). Chest computed tomography demonstrated bilateral ground glass opacities (Figure 1-I).
Since the literature at the time of the patient’s presentation suggested that prolonged PCR positivity indicated the presence of noninfectious RNA [3, 4], he initially received no specific treatments for COVID-19. Instead, an extensive evaluation was sent to identify other infections, which included blood cultures (grew Escherichia coli that rapidly cleared with piperacillin-tazobactam) and the following tests, all of which were negative: urine Legionella antigen, plasma PCR for adenovirus, cytomegalovirus, and Epstein-Barr virus, serum for Aspergillus galactomannan and β-D-glucan, non–SARS-CoV-2 respiratory virus PCR, and stool pathogen PCR. He required pressor support on day 50 for hypotension, and his respiratory status continued to worsen. He was intubated on day 55 and required mechanical ventilation with 80%–100% fraction of inspired oxygen for the remainder of his hospital stay. SARS-CoV-2 NP RT-PCR testing remained positive on day 55; cycle thresholds were 13.3 and 16 for the N1 and E genes, respectively. Bronchoalveolar lavage fluid from day 55 grew rare Klebsiella pneumoniae, which was not thought to be significantly contributing to his condition but was treated with meropenem, with no improvement in his respiratory status.
Because no etiology aside from COVID-19 could be identified, he received another course of convalescent plasma (day 58) and dexamethasone (days 63–74) (Figure 1-I). He was unable to receive additional remdesivir due to limited availability. Serum from days 45, 56, and 71 was negative for SARS-CoV-2 immunoglobulin G (IgG) and immunoglobulin A (IgA) (Euroimmun assay [11]). Despite myeloma biomarkers showing an excellent response to CAR T-cell therapy, his family ultimately decided to focus on comfort measures because of nonresolving respiratory failure. The patient passed away on day 74 after COVID-19 diagnosis.
METHODS
The patient was enrolled in the University of Pittsburgh’s Acute Lung Injury Registry and Biospecimen Repository (institutional review board number STUDY19050099). SARS-CoV-2 RNA quantification was performed using a sensitive (limit of detection, 3 copies/reaction) quantitative RT-PCR (qRT-PCR) assay on plasma (days 4, 9, 13, 16, 67, and 71) and in endotracheal aspirate (ETA) fluid (from day 72). SARS-CoV-2 Spike-specific deep next-generation sequencing (NGS) was performed on plasma (days 4, 13, 67, and 72) and ETA fluid (day 72). Virus isolation from plasma (days 4, 65, and 71) and ETA fluid (day 72) was attempted on Vero E6 cells, followed by indirect immunofluorescence and dideoxy sequencing of the Spike (S) gene. Anti–SARS-CoV-2 IgG antibody titers directed against the S-protein receptor binding domain (RBD) using an indirect enzyme-linked immunosorbent assay (ELISA), competitive ELISA using human ACE2, and pseudovirus neutralization assays were performed with samples from days 4, 9, 13, 16, and 72. Plaque reduction neutralization assays on live SARS-CoV-2 (laboratory strain) were also performed (days 4, 65, and 71). To characterize T-cell and B-cell responses, single-cell RNA sequencing (scRNA-seq) was performed on peripheral blood mononuclear cells (PBMCs) isolated from blood obtained on days 4 and 9. SARS-CoV-2 S-specific T-cell responses were determined by flow cytometric quantification of T-cell frequencies with intracellular staining of interferon-γ and CD107a, following co-culture with S-protein and nucleocapsid protein peptide pools. Assay details can be found in the Supplementary Materials.
RESULTS
SARS-CoV-2 RNA Measurements
High levels of SARS-CoV-2 RNA were detected in all plasma samples (Figure 1-I). RNAemia was greatest on day 4 (126 792 copies/mL) and showed a >10-fold decrease by day 9 (8100 copies/mL), after administration of convalescent plasma and remdesivir. RNAemia remained readily detected but <100 000 copies/mL on day 13 (11 508 copies/mL), day 16 (18 000 copies/mL), and day 67 (14 720 copies/mL), then increased to 101 800 copies/mL on day 71 during administration of dexamethasone. More than 20 billion copies/mL of SARS-CoV-2 RNA (2.78 × 1010 copies/mL) were detected by qRT-PCR in a serially diluted ETA sample from day 72.
Next -Generation Sequencing of SARS-CoV-2 Spike Gene in Longitudinal Samples
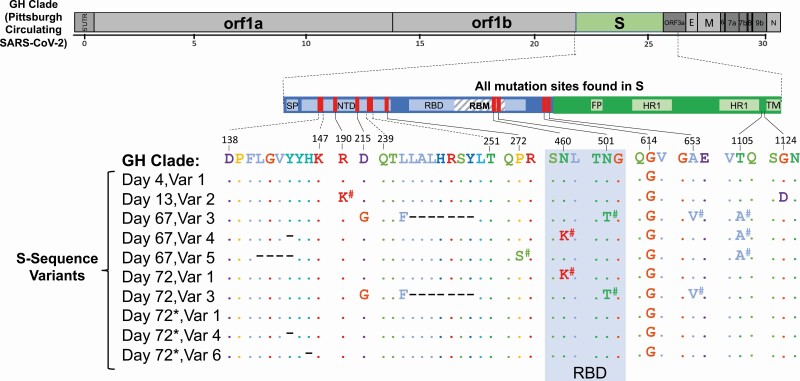
Six different SARS-CoV-2 sequence variants were identified in longitudinal plasma and ETA fluid between days 4, 13, 67, and 72 (Figure 2 and Supplementary Methods). On day 4, plasma viral S sequences matched the SARS-CoV-2 GH clade (containing D614G) that was circulating in Pittsburgh at the time. However, by day 13, while the patient was still in the hospital, additional mutations were detected, including R190K and G1124D substitutions, which were previously observed in 0.005% (n = 9/189 163) and 0% (n = 0/189 163), of GISAID SARS-CoV-2 sequences, respectively. Comparison of sequences between days 13 and 67 demonstrated the emergence of several additional mutations, such as a Y144 deletion and a D215G substitution, which were later identified in the United Kingdom (UK) and South African variants, respectively, several months after the patient’s death [12, 13]. We also identified an N501T substitution on day 67, which was recently shown to enhance binding affinity to the ACE2 receptor [14]. Interestingly, a substitution at the same location (N501Y) is currently circulating in the UK and South Africa [12, 13]. Finally, mutations continued to emerge between days 67 and 72 (eg, H146 deletion). The longitudinal emergence of multiple different and novel sequence variants is indicative of intrahost evolution of SARS-CoV-2.
Figure 2.
Mutations and deletions in the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Spike gene identified in the patient’s samples compared to the SARS-CoV-2 GISAID GH Clade circulating in Pittsburgh. The full genome at the top shows the GH clade sequence. The enlarged S gene shows all of the mutations identified in the patient’s samples compared with the GH clade. The sequence alignments in S compared to the GH clade are shown for each of the multiple (6) sequence variants (var) identified by deep next-generation sequencing (Illumina). All of the sequence variants were detected in plasma. Day 72 (*) shows the matching sequence variants identified in the endotracheal aspirate sample. The D614G substitution was found in all samples. #Mutations detected as mixed populations <100% but >20%. Abbreviations: E, envelope; FP, fusion peptide; HR 1, heptad repeat 1; M, membrane; N, nucleocapsid; NTD, N-terminal domain; ORF, open reading frame; RBD, receptor binding domain; RBM, receptor binding motif; SP, signal peptide; TM, transmembrane domain; Var, sequence variant.
Viral Isolation and Dideoxy Sequencing
Infectious SARS-CoV-2 was recovered from the day 72 ETA sample (Figure 1-II) with a titer of 1.125 × 106 plaque-forming units (PFU)/mL. Numerous SARS-CoV-2 virions were detected by electron microscopy in the ETA sample (Figure 1-III). Dideoxy sequencing of the S-gene of cultured virus showed mutations consistent with those identified by NGS (eg, D614G substitution, Y144 deletion, and H146 deletion). Viral isolation from plasma was unsuccessful despite the presence of SARS-CoV-2 RNA.
Immune Responses
Analysis of PBMCs (days 4 and 9) by immunophenotyping and scRNA-seq demonstrated absence of B cells and near total depletion of T cells, consistent with lymphodepleting chemotherapy with fludarabine and cyclophosphamide (Supplementary Figure 2B). The scRNA-seq profiling showed interferon-stimulated genes in the monocyte lineage suggestive of a viral infection. We evaluated SARS-CoV-2–specific T-cell responses from days 4 and 67. The day 4 sample contained too few live T cells (<1%) to assess. The day 67 PBMC sample showed somewhat higher levels of live T cells (9.2%), with 1.72% and 0.79% of CD8+ T cells expressing the cytotoxicity marker CD107a following stimulation with S protein and nucleocapsid peptide pools, respectively, suggesting that a small fraction of the T cells were able to react to SARS-CoV-2 antigens ex vivo (Supplementary Figure 3). However, the low number of events (<5000 per sample) acquired by flow cytometry prevents any conclusions from being drawn from these analyses.
Despite the administration of convalescent plasma on days 2 and 58, no IgG antibody targeting the S-protein RBD was detected at any time point, nor was there any antibody that competed with human ACE2 binding to RBD, possibly because the convalescent plasma used had a low titer of SARS-CoV-2 antibody, or there was rapid clearance of antibody bound to very higher numbers of virions (Supplementary Table 2). Additionally, SARS-CoV-2 plaque reduction neutralization assays showed no plaque reduction in the day 4 sample. However, pseudovirus assays showed approximately 50% inhibition at a 1:10 dilution in the day 13 sample (Supplementary Figure 1). After the second dose of convalescent plasma on day 58, samples from days 65 and 71 showed ≥50% plaque reduction at a 1:16 dilution. Since IgG antibody against the S-protein RBD was not detected at any time point (days 4, 9, 13, 16, 45, 56, 71, and 72), the observed plaque reduction suggests the presence of low titers of neutralizing antibodies or other antibodies directed to non-RBD regions of the S-protein in the patient’s plasma (Supplementary Table 2).
DISCUSSION
We report here sustained SARS-CoV-2 RNAemia with viral replication for >2 months and intrahost viral evolution in a patient with COVID-19 and both B- and T-cell depletion as a result of the therapies he had received for multiple myeloma. The initial clinical improvement and >10-fold reduction in plasma RNAemia (days 4–9) suggests a treatment response following convalescent plasma (day 2) and remdesivir (days 5–10). RNAemia was never completely suppressed, however, and we hypothesize that the absence of anti–SARS-CoV-2 humoral responses and the paucity of T-cell responses related to prior chemotherapy and anti-BCMA CAR T cells resulted in uncontrolled viral replication and overwhelming SARS-CoV-2 pneumonia (1.125 × 106 PFU/mL and 2.78 × 1010 RNA copies/mL in ETA) that likely contributed to death. The detection of a distinct SARS-CoV-2 sequence variant on day 13 of the first hospitalization with 3 coding mutations (Figure 2) compared to the initial sequence variant detected on day 4 that matched the circulating GH clade argues for viral evolution within the host and against superinfection or reinfection. Several additional sequence variants, including those with coding mutations and deletions in the S gene that have yet to be detected in SARS-CoV-2 in Pittsburgh as of January 2021, were identified on day 67. Furthermore, additional sequence variants rapidly developed in the span of 5 days between days 67 and 72 while the patient was still in the hospital (Figure 2), further supporting continued intrahost viral evolution, since reinfection with 5 distinct variants is highly improbable. Taken together, these findings provide insights into the potential duration of continued SARS-CoV-2 replication and the plasticity of the SARS-CoV-2 S gene, as others have noted [15].
RNAemia has been described in immunocompromised patients with non–SARS-CoV-2 respiratory viruses. Respiratory syncytial virus (RSV) RNAemia developed in 30% of hematopoietic cell transplant recipients 2 days after onset of RSV pneumonia and was a predictor of mortality [16]. Emerging data suggest that SARS-CoV-2 RNAemia may be a marker of severity of COVID-19 pneumonia [5, 17], but sustained RNAemia for >2 months has not yet been described. We were unable to isolate replication-competent virus from plasma, which may be due to technical issues, or may suggest that RNAemia is not caused by the presence of virions in the plasma, but rather due to “spillover” of infected cells with SARS-CoV-2 RNA from the lung. The reduction in plasma RNAemia between days 4 and 9 (after administration of convalescent plasma on day 2 and of remdesivir between days 5 and 10) suggests that viral replication was inhibited by the therapies given. Unfortunately, measurement of plasma RNAemia is not readily available for clinical use in the United States, and it is not currently known whether suppression of SARS-CoV-2 RNAemia can improve clinical outcomes. The rebound of viremia back to 101 800 RNA copies/mL on day 71 occurred in the setting of the administration of dexamethasone. Whether corticosteroids should be avoided in heavily immunosuppressed individuals in whom prolonged SARS-CoV-2 replication is suspected warrants further study. Ultimately, randomized trials are needed to evaluate whether measurement of RNAemia should be used to determine clinical response to SARS-CoV-2 therapies.
We isolated virus from respiratory samples collected 72 days after COVID-19 onset, demonstrating the potential for infectivity late into the clinical course. In a prior study of immunocompetent patients with mild COVID-19, virus isolation from samples obtained after 8 days was unsuccessful [3]. Our findings suggest that certain severely immunosuppressed patients with COVID-19 may require isolation longer than the 20-day period currently proposed by the Centers for Disease Control and Prevention [18]. Whether PCR cycle thresholds (which were consistently <25 in our patient) can be used to make decisions related to discontinuation of transmission-based precautions warrants further investigation. Importantly, replication-competent variants isolated in cell culture harbored deletions associated with increased transmissibility similar to what has since been identified in the UK months later [12]. These findings raise the possibility that the origin of the highly mutated UK and South African variants may have been persons with protracted infection.
Humoral and cell-mediated immunity after COVID-19 appear to be necessary to control SARS-CoV-2 infection [19, 20]. In our patient, PBMC analysis confirmed B-cell aplasia, which is compatible with negative assay results for IgGs against the S-protein RBD, and IgAs/IgGs using a clinically approved assay. IgGs against the S-protein RBD were absent at all time points (days 4, 9, 13, 16, 45, 56, 71, and 72) despite the fact that the patient received convalescent plasma on days 2 and 58 of illness. This finding suggests either low titer of antibodies in the convalescent plasma or rapid clearance of antibodies in the context of high viral burden. Interestingly, there was evidence of low-level viral neutralization on days 65 and 71 (after the second dose of convalescent plasma) using live virus neutralization assays, which may suggest the presence of low titers of neutralizing antibodies directed to non-RBD regions of Spike or low titers of other antibodies in the convalescent plasma samples. The scRNA-seq profiling from days 4 and 9 showed upregulation of interferon-stimulated genes in monocytes, suggesting the presence of innate antiviral immunity, which was unable to control the infection. SARS-CoV-2–specific T-cell response assays did show some evidence of T-cell activation late into the disease course, although the T-cell lymphopenia related to antecedent chemotherapy limited our ability to draw conclusions about the patient’s T-cell responses. Overall, these findings support the hypothesis that the diminished T-cell responses and nearly absent B-cell responses led to uncontrolled SARS-CoV-2 infection.
In conclusion, our case highlights that immunocompromised patients with profound lymphocyte defects such as CAR T-cell therapy recipients are at risk for prolonged SARS-CoV-2 replication. Other patients with severe lymphocyte deficiencies, such as anti-CD20 monoclonal antibody recipients, and hematopoietic cell or organ transplant recipients within the first few months of transplant, may also be at risk. Clinicians caring for such patients should be cautious not to attribute persistent detection of SARS-CoV-2 RNA from clinical samples of these patients to the presence of noninfectious virus and should consider revising their transmission-based precautions for these patients. Further work is needed to define the role of monitoring RNAemia in managing immunocompromised persons with COVID-19, the duration of infectivity in these patients, the importance of intrahost emergence of SARS-CoV-2 sequence variants in immune escape, and the immune phenotypes associated with recovery vs fatal infection. More effective antiviral agents to suppress SARS-CoV-2 replication are urgently needed, especially for immunodeficient hosts.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Author contributions. M. K. H. and W. G. B. contributed equally to this work as co-first authors. J. J. and S. N. contributed equally to this work. All authors have seen and approved the manuscript and contributed significantly to this work.
Acknowledgments. The authors thank Dr Rima Abdel-Massih, Luann Borowski, Michelle Busch, Dr Jennifer McComb, Dr Bryan McVerry, Heather Michael, Dr Stephanie Mitchell, Asma Naqvi, John Ries, Dr Charles Rinaldo, Caitlin Schaefer, Dr Michael Shurin, Dr Alan Wells, and Dr Sarah Wheeler.
Financial support. Research reported in this publication was supported by the National Center for Advancing Translational Sciences of the National Institutes of Health (NIH) (award number KL2TR001856 to G. H.); the United States Department of Veterans Affairs Biomedical Laboratory R&D Service (career development award number IK2 BX004886 to W. B.); the NIH (grant number P50 8 P50 AI150464–13 to P. J. B.); George Mason University Fast Grants (to P. J. B.); the Center for Vaccine Research/University of Pittsburgh (grant numbers P01HL114453 to P. R. and J. S. L. and R01 HL136143 to J. S. L.); and the University of Pittsburgh Medical Center P01 AI108545, P50 CA097190 & R01s CA203689, DK089125, AI129893, and AI144422 (to D. A. V.).
Disclaimer. The content of this work is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Potential conflicts of interest. G. H. and G. D. K. have received research funds from Karius, Inc. J. W. M. is a consultant to Gilead Sciences and Merck and owns shares in Co-Crystal Pharmaceuticals, Infectious Disease Connect, and Abound Bio. D. A. V. is a cofounder and stock holder of Novasenta, Tizona, and Potenza; is a stock holder of Oncorus and Werewolf; holds patents and receives royalties from Astellas and BMS; is a scientific advisory board member for Tizona, Werewolf, and F-Star; is a consultant to Astellas, BMS, Almirall, and Incyte; and receives research funding from BMS and Novasenta. All other authors report no potential conflicts of interest.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Malard F, Genthon A, Brissot E, et al. COVID-19 outcomes in patients with hematologic disease. Bone Marrow Transplant 2020; 55:2180–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Haidar G, Garner W, Hill JA. Infections after anti-CD19 chimeric antigen receptor T-cell therapy for hematologic malignancies: timeline, prevention, and uncertainties. Curr Opin Infect Dis 2020; 33:449–57. [DOI] [PubMed] [Google Scholar]
- 3. Wolfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020; 581:465–9. [DOI] [PubMed] [Google Scholar]
- 4. Xiao AT, Tong YX, Zhang S. Profile of RT-PCR for SARS-CoV-2: a preliminary study from 56 COVID-19 patients. Clin Infect Dis 2020; 71:2249–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Veyer D, Kerneis S, Poulet G, et al. Highly sensitive quantification of plasma SARS-CoV-2 RNA sheds light on its potential clinical value [manuscript published online ahead of print 17 August 2020]. Clin Infect Dis 2020. doi:10.1093/cid/ciaa1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang W, Xu Y, Gao R, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 2020; 323:1843–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ali SA, Shi V, Maric I, et al. T cells expressing an anti-B-cell maturation antigen chimeric antigen receptor cause remissions of multiple myeloma. Blood 2016; 128:1688–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carpenter RO, Evbuomwan MO, Pittaluga S, et al. B-cell maturation antigen is a promising target for adoptive T-cell therapy of multiple myeloma. Clin Cancer Res 2013; 19:2048–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. D’Agostino M, Raje N. Anti-BCMA CAR T-cell therapy in multiple myeloma: can we do better? Leukemia 2020; 34:21–34. [DOI] [PubMed] [Google Scholar]
- 10. Raje N, Berdeja J, Lin Y, et al. Anti-BCMA CAR T-cell therapy bb2121 in relapsed or refractory multiple myeloma. N Engl J Med 2019; 380:1726–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.US Food and Drug Administration. EUROIMMUN anti-SARS-CoV-2 ELISA instructions for use. Available at: https://www.fda.gov/media/137609/download. Accessed 26 October 2020.
- 12.Rambaut A, Loman N, Pybus O; on behalf of COVID-19 Genomics Consortium UK (CoG-UK). Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. Available at: https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563. Accessed 10 January 2021.
- 13. Garry RF. Mutations arising in SARS-CoV-2 spike on sustained human-to-human transmission and human-to-animal passage. Available at: https://virological.org/t/mutations-arising-in-sars-cov-2-spike-on-sustained-human-to-human-transmission-and-human-to-animal-passage/578. Accessed 10 January 2021.
- 14. Starr TN, Greaney AJ, Hilton SK, et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell 2020; 182: 1295–310.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Choi B, Choudhary MC, Regan J, et al. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N Engl J Med 2020; 383:2291–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Waghmare A, Campbell AP, Xie H, et al. Respiratory syncytial virus lower respiratory disease in hematopoietic cell transplant recipients: viral RNA detection in blood, antiviral treatment, and clinical outcomes. Clin Infect Dis 2013; 57:1731–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jacobs JL, Mellors JW. Detection of SARS-CoV-2 RNA in blood of patients with COVID-19: what does it mean? [manuscript published online ahead of print 8 September 2020]. Clin Infect Dis 2020. doi:10.1093/cid/ciaa1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Centers for Disease Control and Prevention. Duration of isolation and precautions for adults with COVID-19. Available at: https://www.cdc.gov/coronavirus/2019-ncov/hcp/duration-isolation.html. Accessed 10 January 2021.
- 19. Gudbjartsson DF, Norddahl GL, Melsted P, et al. Humoral immune response to SARS-CoV-2 in Iceland. N Engl J Med 2020; 383:1724–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sekine T, Perez-Potti A, Rivera-Ballesteros O, et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell 2020; 183: 158–68.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.