Abstract
Background
Resolving the coronavirus disease 2019 (COVID-19) pandemic requires diagnostic testing to determine which individuals are infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The current gold standard is to perform reverse-transcription polymerase chain reaction (PCR) on nasopharyngeal samples. Best-in-class assays demonstrate a limit of detection (LoD) of approximately 100 copies of viral RNA per milliliter of transport media. However, LoDs of currently approved assays vary over 10,000-fold. Assays with higher LoDs will miss infected patients. However, the relative clinical sensitivity of these assays remains unknown.
Methods
Here we model the clinical sensitivities of assays based on their LoD. Cycle threshold (Ct) values were obtained from 4700 first-time positive patients using the Abbott RealTime SARS-CoV-2 Emergency Use Authorization test. We derived viral loads from Ct based on PCR principles and empiric analysis. A sliding scale relationship for predicting clinical sensitivity was developed from analysis of viral load distribution relative to assay LoD.
Results
Ct values were reliably repeatable over short time testing windows, providing support for use as a tool to estimate viral load. Viral load was found to be relatively evenly distributed across log10 bins of incremental viral load. Based on these data, each 10-fold increase in LoD is expected to lower assay sensitivity by approximately 13%.
Conclusions
The assay LoD meaningfully impacts clinical performance of SARS-CoV-2 tests. The highest LoDs on the market will miss a majority of infected patients. Assays should therefore be benchmarked against a universal standard to allow cross-comparison of SARS-CoV-2 detection methods.
Keywords: SARS-CoV-2, viral load, limit of detection, cycle threshold, antigen detection
Reverse-transcription polymerase chain reaction assays for severe acute respiratory syndrome coronavirus 2 provide quantitative data. Patient results are relatively evenly distributed across log10 viral load bins. Each 10-fold increase in limit of detection corresponds to an approximate 13% loss in sensitivity.
In response to the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic being declared a public health emergency, clinical and commercial laboratories as well as test kit manufacturers have been submitting diagnostic devices and assays for expedited Emergency Use Authorization (EUA) by the United States Food and Drug Administration (FDA). As of 1 June 2020, there were >85 such EUA issuances for coronavirus disease 2019 (COVID-19) diagnostics (https://www.fda.gov/medical-devices/emergency-situations-medical-devices/emergency-use-authorizations). However, optimal use of these assays requires consideration of several issues.
First, nasopharyngeal swabs are generally considered to provide optimal detection early in disease. However, even for this sample type, there is currently no ideal reference standard to establish clinical sensitivities of the available EUA SARS-CoV-2 diagnostic assays [1]. Second, details about assay limit of detection (LoD) are often not provided with sufficient detail and transparency to allow facile comparisons. For molecular diagnostic assays, the LoD is generally considered the lowest concentration of target that can be detected in ≥95% of repeat measurements. Of importance, the LoD is a measure of analytic sensitivity, as opposed to clinical sensitivity, which measures the fraction of infected people detected by a given test. LoDs are reported in several formats, (viral genomic RNA per milliliter or microliter of transport media, copies per reaction volume, Median Tissue Culture Infectious Dose [TCID50], or molarity of assay target), and are often based on testing of different analytical standards, making ready comparison of assay performance less than straightforward. Fourth, although reverse-transcription polymerase chain reaction (RT-PCR) tests are inherently quantitative, existing SARS-CoV-2 EUA tests only report qualitative results (ie, positive or negative), even though viral load may provide both clinically and epidemiologically important information.
Two barriers to quantitative reporting are demonstration that quantitative PCR cycle threshold (Ct) values are repeatable with acceptably low variance and a reliable means of converting from Ct value to viral load. The latter is complicated by a traditional requirement for a standard curve that must span a range of viral loads at least as large as what is observed in the patient population, which can be expensive and time-consuming, especially in a pandemic where the limits of this range are unknown; however, there have been reports demonstrating how appropriate measurements, based on the principles of RT-PCR, can be used as an alternative for reliable conversion of Ct values to viral loads [2, 3].
Here we report on (1) the reproducibility of Ct values from the Abbott SARS-CoV-2 EUA [4] obtained by sampling the same patients within 6 and 12 hours; (2) conversion from Ct to viral load; (3) the distribution of viral loads in sampling of 4700 first-time positive patients; and (4) based on these findings, an estimate of clinical sensitivity of testing in relation to assay LoD. These findings have clear implications for patient care, epidemiology, and the social and economic management of the ongoing pandemic.
MATERIALS AND METHODS
Setting and Time Period
All SARS-CoV-2 testing data from the Beth Israel Lahey Health Network from 26 March to 2 May 2020 were included in our analysis. Testing was performed on the basis of clinical suspicion. The study was deemed exempt by our hospital institutional review board.
Testing
Tests were performed using the Abbott RealTime SARS-CoV-2 assay, a real-time RT-PCR test for qualitative detection of SARS-CoV-2 in nasopharyngeal and oropharyngeal swabs [5]. The dual target assay detects both the SARS-CoV-2 RdRp and N genes with a reported LoD of 100 copies/mL. The assay also includes an internal control. Results are reported as positive if the Ct value is ≤31.5, based upon the signal threshold determined by the manufacturer. Ct values for all first-time positive test results were analyzed. Repeat tests were excluded to estimate the range of Ct values of the infected population upon presentation at our medical center. In our internal validation, we determined that the detection rate was 100% for the Abbott m2000 platform at 100 copies/mL (n = 80), with Ct mean and standard deviation at this LoD of 26.06 ± 1.03 [4], including 20 replicates each on 4 separate M2000 platforms on different dates. The genome copy number was based on the reference standard produced by SeraCare (AccuPlex SARS-CoV-2 Reference Material Kit, catalog number 0505-0126). This control material consists of replication-incompetent, enveloped, positive-sense, single-stranded RNA Sindbid virus into which both SARS-CoV-2 PCR targets detected by the Abbott PCR assay are cloned. The control material was quantified by the manufacturer using digital droplet PCR, and diluted into viral transport medium for analysis. Using the SeraCare standard, the coefficient of variation of the cycle threshold was 4.0% at 100 copies/mL and 2.31% at 1000 copies/mL (20 replicates total run on 4 analyzers over 3 days), suggesting high methodological reproducibility and precision. To determine the LoD, we tested a fine dilution series in replicates of 10 at several levels below 100 copies/mL. By simple logistic regression (Logit), the LoD (95% detection rate) was approximately 50 copies/mL (data not shown).
Note that the Ct determination on Abbott M2000rt platform is alternatively called the fractional cycle number (FCN) and is specifically one way of determining the cycle number at the maximum amplification efficiency inflection point (ie, the maxRatio) of each amplification curve [6]. The FCN has been reported to be a more robust measure for Ct determination than a fixed fluorescence threshold.
Statistical Analysis
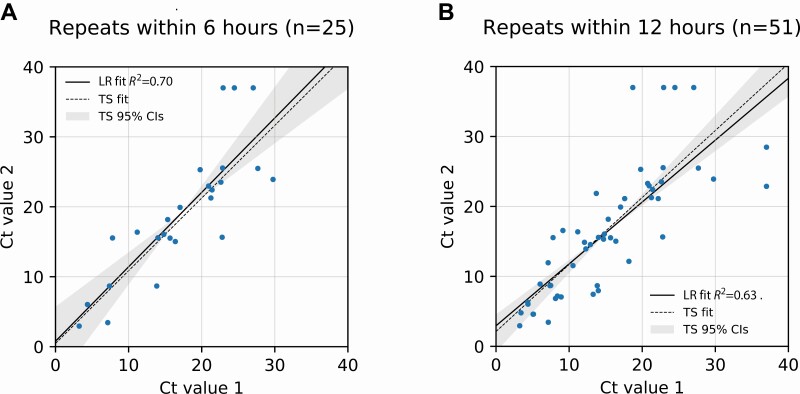
Variance was estimated by R2 of Ct values for repeat tests obtained within 6 hours (n = 25 patients, excluding one obvious outlier that by itself accounted for half the total variance: initial Ct 4.4, but repeat negative and attributed to preanalytic or analytic technical error) and 12 hours (n = 51 patients, excluding the same outlier). The conversion from Ct value to viral load was performed using the definition of exponential growth with variable efficiency [2, 3]. Efficiency was measured from plots of fluorescence intensity vs cycle number for 50 positive samples chosen at random, yielding an expression for viral load in copies/mL as a function of Ct (see Eq. 6 in the Supplementary Methods). Per this expression, the expected negative cutoff corresponds to 9.14 copies per mL or approximately 2 virions per RT-PCR reaction volume, supporting the validity of our parameter estimation.
The validity of our model was tested by establishing a calibration curve using inactivated SARS-CoV-2 strain USA-WA1/2020 A virus from the FDA Verification Panel (FDA SARS-CoV-2 Reference Panel) as further described in the Supplementary Materials.
We used Python (version 3.6) and its NumPy, SciPy, Matplotlib, and Pandas libraries to plot linear regression and Theil-Sen slopes with 95% confidence intervals on repeat positives; a normalized cumulative distribution (histogram) of positive results (with reversed x-axis for ease of interpretation); binned histogram by 0.5 log10 units, and linear regression on log10-transformed data.
RESULTS
Of the 27 098 tests performed on 20 076 patients over the testing period, 6037 tests were positive (22%), representing 4774 unique patients. Analysis of repeats within 6 or 12 hours of each other [7] demonstrated general repeatability of Ct values over these short time windows (R2 = 0.70, n = 25 and R2 = 0.63, n = 51, respectively), supporting the validity of this quantitative measure as a basis for assessment of viral load in patients (Figure 1). We used basic principles of PCR and detailed measurements of PCR efficiency on 50 randomly chosen positive samples to convert from Ct values to viral load, in units of copies of viral RNA/mL of viral transport medium. We confirmed the validity of our mathematical model by comparison with a calibration curve established through testing of serial dilutions of an inactivated SARS-CoV-2 virus reference material (see the Supplementary Materials and Supplementary Figure 3). To study the patient population upon presentation without confounding by repeat measurements on the same patients, the remainder of the analysis was on the first positive value for the above-mentioned 4774 unique patients.
Figure 1.
Cycle threshold (Ct) values are highly repeatable. Data points shown are Ct values for severe acute respiratory syndrome coronavirus 2 testing of pairs of nasopharyngeal samples obtained within either 6 hours (A) or 12 hours (B) of each other from the same patient, represented by the X and Y coordinates of each data point. Shaded areas indicate 95% confidence intervals for Theil-Sen linear regression fit. Abbreviations: CI, confidence interval; LR, linear regression fit; TS, Theil-Sen linear regression fit.
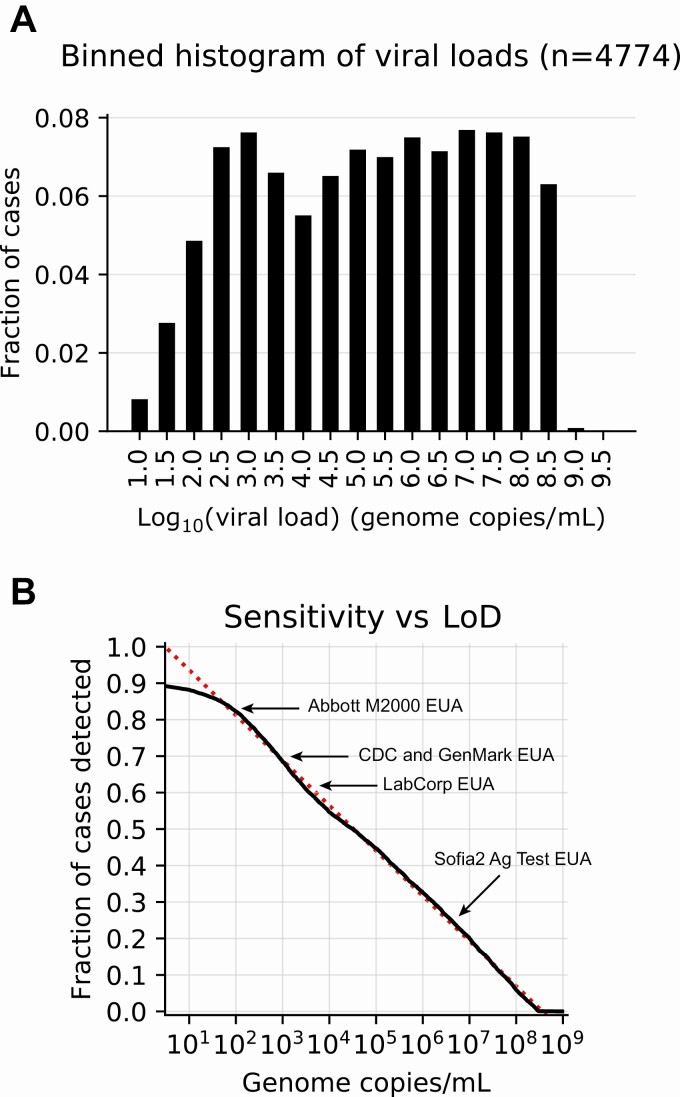
Viral loads spanned nearly 9 orders of magnitude, from 9 copies/mL to 2.5 billion copies/mL (Figure 2). Notably, patients were almost equally likely to exhibit low, medium, or high viral loads upon initial testing, with remarkable uniformity down to the LoD of 50 copies/mL (R2 = 0.99). The reason for this uniformity is unknown. Fewer patients had viral loads below the LoD, as reflected by the curve’s departure from the trend in this range. Because the LoD is a 95% confidence limit, the difference between the curve and the trend likely reflects false negatives: The lower the viral load, the greater the likelihood that infection will be missed. By definition, only 5% of patients with viral load at the LoD are expected to be missed (1 in 20 patients); this percentage grows for patients with viral loads below this threshold. Thus, extending the observed trend leftward to the assay’s positive cutoff, which corresponds to approximately 2 genome copies per reaction volume, yields an estimate of the total false-negative rate for this assay of 10%, and thus a clinical sensitivity of 90%, or 9 in 10 infected individuals.
Figure 2.
Viral load distribution and limit of detection (LoD). A, Fraction of positive tests binned by 0.5 log10 bins of viral load. B, Cumulative histogram distribution of viral loads showing percentage detected as a function of LoD: actual (solid line) and trend line (dotted line). For purposes of discussion, examples of LoD for several other methods, obtained from Emergency Use Authorization package inserts or the scientific literature, are overlaid. Abbreviations: Ag, antigen; CDC, Centers for Disease Control and Prevention; EUA, Emergency Use Authorization.
Notably, our results can be used to estimate the clinical sensitivity of assays with other LoDs. For example, an assay with LoD of 1000 copies/mL, such as that of the Centers for Disease Control and Prevention assay [8], or Genmark ePlex EUA, based on a study using a less labile target than used for the LoD determination in the original EUA [9], is expected to detect 77% of infected individuals. With an LoD of 6250 copies/mL, the LabCorp COVID-19 RT-PCR EUA test has an estimated clinical sensitivity of 67%. The first EUA antigen detection assay, the Quidel Sofia2 SARS Antigen FIA, has an LoD of approximately 6 million copies/mL in a contrived universal transport medium sample collection. Although the package insert indicates the LoD using TCID50 units, the BEI Resources control material referenced lists both TCID50 and genome copies/mL, allowing the calculation of the latter and an associated estimated clinical sensitivity of 31%.
DISCUSSION
The diagnostic priorities in the COVID-19 pandemic are to robustly identify 3 populations: the infected, the infectious, and the susceptible. Our study addresses the first of these. Specifically, it illustrates the clinical and epidemiologic impact of assay LoD on SAR-CoV-2 diagnosis and the challenges of interpreting and comparing molecular assay results across various platforms. First, viral loads vary widely among infected individuals, from individuals with extremely high viral loads (potential “superspreaders”) who presumably would be picked up by even the least sensitive assays, to those whose viral loads are near, at, or even below the LoD of many assays. Therefore, a substantial fraction of infected patients will be missed by less sensitive assays. Concerningly, some of these missed patients are, have been, or will become infectious, and such misses will undermine public health efforts and put patients and their contacts at risk. This must give pause in the rush to approve additional testing options and increase testing capacity, and emphasizes the importance of defining infectivity as a function of viral load and other factors (eg, time of exposure), which remains a critical unknown in this pandemic. The relative ability of different sampling techniques to obtain specimens with the highest viral loads may also substantially impact detection rate.
Antigen detection assays promise rapid turnaround time, point-of-care implementation, and low cost. For influenza detection, such tests have exhibited substantially lower analytical and clinical sensitivity compared with nucleic acid amplification testing (NAAT) [10]. The poor historical performance for influenza detection led to reclassification of influenza rapid antigen detection tests as class II devices with a new minimal performance standard of at least 80% sensitivity compared with NAAT [11]. Previously, clinical sensitivity of 50%–88% for the Quidel Sofia influenza test was noted in several studies in different influenza seasons compared to RT-PCR comparators [12–14]. The same trend was observed in our analysis of the single SARS-CoV-2 antigen test introduced thus far with EUA status. Tests with such performance characteristics will identify individuals with the highest viral burden. However, such a high detection threshold will be unlikely to fully meet public or individual health goals in the COVID-19 pandemic.
Our findings also suggest that Ct values and imputed viral loads have clinical utility. Real-time PCR methods in particular are inherently quantitative, and we demonstrate here that they are quite reproducible during repeated clinical sampling over a short time period, with R2 of 0.70 for repeats within 6 hours (as a proxy for immediate repeats). We note that because PCR efficiency can fall substantially with PCR cycle number, as we observed here, viral load is ideally calculated not simply as a powers-of-2 transformation of Ct value but based on the observed trend between efficiency and Ct number. This trend may differ by assay: For example, the assay used here includes an internal control whose product may contribute to polymerase inhibition. (This method can be extended to provide confidence limits that incorporate the variance in, eg, the Ct of the LoD, but this extension is beyond the scope of the current work.) As yet, it is unclear whether or how viral loads affect prognosis, but they at least suggest a measure of infectivity, as well as possibly severity of illness, and, therefore may have value for public health efforts, as we learn which cutoffs may imply minimal or inconsequential infectivity, especially during clearance of infection. We make explicit our assumption that approximately 2 virions per reaction, translating to a viral load of 9 copies/mL, reflects a 100% detection rate. With stricter cutoffs, clinical sensitivity falls slightly (eg, from 90% to 86% for an assay with an LoD of 100 copies/mL, if using a cutoff of 4 copies/mL, or a single virion per reaction, and to 79% if using a cutoff of 0.7 copies/mL, or a single virion per 3-mL transport tube). Regardless, these different assumptions have essentially no effect on the relative clinical sensitivities of different assays. While it is theoretically possible that even lower levels of infection are possible, making our estimates of clinical sensitivity upper limits, we believe potential for contagion at these levels is highly unlikely, as that would assume that breathing, a cough, or a sneeze would transmit more particles than can be obtained by dedicated and vigorous physical swabbing of the actual nasopharynx.
To control the pandemic, ultimately we will need diagnostics for all 3 populations of interest—infected, infectious, and susceptible—and for that we will need to understand whether and how viral load relates to infectiousness. As we have shown, assays with higher LoD are likely to miss nonnegligible fractions of infected individuals. However, individuals with viral burdens low enough to be missed by some assays may prove to be less infectious. In vitro, approximately only 1 of 10 000 genome copies in viral cultures may be associated with a tissue culture infectious viral particle based on standard preparations such as BEI Resources NR-52866 [15]. However, it is unclear how or whether this fraction might change with viral load for patients in vivo.
The ultimate lesson from these studies bears repetition: LoD matters and directly impacts efforts to identify, control, and contain outbreaks during this pandemic. Various assays report out LoDs in manners that are often difficult to comprehend, for example, TCID50 values that may be related to viral copy numbers in different ways depending on the viral preparation, or units of copies/µL (1 copy/µL = 1000 copies/mL) or attomolar quantities (1 attomolar = 602 copies/mL). We make explicit that cross-comparisons between assays shown in Figure 2B are not based on parallel LoD determinations performed with the same quantitative standards. Therefore, relative assay performance of the examples given may prove better or worse than found in future systematic investigation. Importantly, we suggest that viral copies/mL be used as a universal standard metric for reporting SARS-CoV-2 LoD, and that an international standard, comparable to World Health Organization international standards for other viral load measurements, be established so that cross-comparison between assay LoD can be readily and rigorously made. It is clear that viral load matters, and therefore LoD values should be readily evaluable and in the public domain.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors thank the clinical laboratory scientists and volunteers in the Beth Israel Deaconess Medical Center microbiology laboratory for generating the data used in this manuscript. Abbott Molecular Inc provided the M2000 RealTime assay reagents under a coronavirus disease 2019 diagnostics evaluation agreement that was used by the authors to establish a calibration curve with the Food and Drug Administration severe acute respiratory syndrome coronavirus 2 reference material and the LoD using the SeraCare reference standard.
Disclaimer. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health (NIH). Abbott Molecular Inc had no role in study design, data collection/interpretation, manuscript preparation, or decision to submit the manuscript for publication.
Financial support. K. P. S. was supported by the National Institute of Allergy and Infectious Diseases, NIH (award number F32 AI124590).
Potential conflicts of interest. R. A. is principal investigator on clinical studies sponsored by Abbott Molecular, MitoLab, and E25 Bio, unrelated to the current work. All other authors report no potential conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1. Wang W, Xu Y, Gao R, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 2020; 323:1843–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lievens A, Van Aelst S, Van den Bulcke M, Goetghebeur E. Enhanced analysis of real-time PCR data by using a variable efficiency model: FPK-PCR. Nucleic Acids Res 2012; 40:e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Platts AE, Johnson GD, Linnemann AK, Krawetz SA. Real-time PCR quantification using a variable reaction efficiency model. Anal Biochem 2008; 380:315–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Smith KP, Cheng A, Chopelas A, et al. Large-scale, in-house production of viral transport media to support SARS-CoV-2 PCR testing in a multi-hospital healthcare network during the COVID-19 pandemic [manuscript published online ahead of print 23 July 2020]. J Clin Microbiol 2020. doi:10.1128/JCM.00913-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abbott Molecular Inc. Abbott RealTime SARS-CoV-2 emergency use authorization package insert, REF 09N77-095, 51-608445/R1. Des Plaines, IL: Abbott Molecular Inc, 2020. [Google Scholar]
- 6. Shain EB, Clemens JM. A new method for robust quantitative and qualitative analysis of real-time PCR. Nucleic Acids Res 2008; 36:e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Callahan CJ, Lee R, Zulauf KE, et al. Open development and clinical validation of multiple 3D-printed nasopharyngeal collection swabs: rapid resolution of a critical COVID-19 testing bottleneck. J Clin Microbiol 2020; 58:e00876-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention. 2019-novel coronavirus (2019-nCoV) real-time RT-PCR diagnostic panel emergency use authorization instructions for use, CDC-006-00019, revision: 03. Atlanta, GA: CDC,2020. [Google Scholar]
- 9. Zhen W, Smith E, Manji R, Schron D, Berry GJ. Clinical evaluation of three sample-to-answer platforms for the detection of SARS-CoV-2. J Clin Microbiol 2020; 58:e00783-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Merckx J, Wali R, Schiller I, et al. Diagnostic accuracy of novel and traditional rapid tests for influenza infection compared with reverse transcriptase polymerase chain reaction: a systematic review and meta-analysis. Ann Intern Med 2017; 167:394–409. [DOI] [PubMed] [Google Scholar]
- 11. Green DA, StGeorge K. Rapid antigen tests for influenza: rationale and significance of the FDA reclassification. J Clin Microbiol 2018; 56:e00711–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Arbefeville SS, Fickle AR, Ferrieri P. Sensitivity of the Quidel Sofia fluorescent immunoassay compared with 2 nucleic acid assays and viral culture to detect pandemic influenza A(H1N1)pdm09. Lab Med 2015; 46:230–4. [DOI] [PubMed] [Google Scholar]
- 13. Kammerer PE, Radin JM, Hawksworth AW, Myers CA, Brice GT. Performance of the Quidel Sofia rapid influenza diagnostic test during the 2012–2013 and 2013–2014 influenza seasons. Influenza Other Respir Viruses 2016; 10:220–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Selove W, Rao LV. Performance of rapid SOFIA Influenza A+B test compared to Luminex x-TAG respiratory viral panel assay in the diagnosis of influenza A, B, and subtype H3. J Investig Med 2016; 64:905–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.BEI Resources. Certificate of analysis for NR-52286. Lot numbers: 70033548 and 70034991. Manassas, VA: BEI Resources. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.