Abstract
Background
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) immunoglobulin G (IgG) antibody measurements can be used to estimate the proportion of a population exposed or infected and may be informative about the risk of future infection. Previous estimates of the duration of antibody responses vary.
Methods
We present 6 months of data from a longitudinal seroprevalence study of 3276 UK healthcare workers (HCWs). Serial measurements of SARS-CoV-2 anti-nucleocapsid and anti-spike IgG were obtained. Interval censored survival analysis was used to investigate the duration of detectable responses. Additionally, Bayesian mixed linear models were used to investigate anti-nucleocapsid waning.
Results
Anti-spike IgG levels remained stably detected after a positive result, for example, in 94% (95% credibility interval [CrI] 91–96%) of HCWs at 180 days. Anti-nucleocapsid IgG levels rose to a peak at 24 (95% CrI 19–31) days post first polymerase chain reaction (PCR)-positive test, before beginning to fall. Considering 452 anti-nucleocapsid seropositive HCWs over a median of 121 days from their maximum positive IgG titer, the mean estimated antibody half-life was 85 (95% CrI 81–90) days. Higher maximum observed anti-nucleocapsid titers were associated with longer estimated antibody half-lives. Increasing age, Asian ethnicity, and prior self-reported symptoms were independently associated with higher maximum anti-nucleocapsid levels and increasing age and a positive PCR test undertaken for symptoms with longer anti-nucleocapsid half-lives.
Conclusions
SARS-CoV-2 anti-nucleocapsid antibodies wane within months and fall faster in younger adults and those without symptoms. However, anti-spike IgG remains stably detected. Ongoing longitudinal studies are required to track the long-term duration of antibody levels and their association with immunity to SARS-CoV-2 reinfection.
Keywords: SARS-CoV-2, COVID-19, serology, antibody, longitudinal
Serially-measured severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) anti-nucleocapsid immunoglobulin G (IgG) titres from 452 seropositive healthcare workers demonstrate levels fall by half in 85 days. Anti-spike IgG remains elevated in most seropositive individuals up to 6 months.
Measurable immunoglobulin G (IgG) antibodies to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antigens develop after many, but not all, SARS-CoV-2 infections [1–4]. Serological responses are typically detectable within 1–3 weeks [5–8]. This allows antibody assays to be used to estimate the proportion of a population exposed or infected. Additionally, although the extent of immunity associated with different antibody titers and other immune responses is yet to be fully determined, it is probable that antibody levels will provide some information about the risk and/or severity of future infection.
However, SARS-CoV-2 IgG antibody levels are dynamic over time [9]. This has implications for epidemiological studies; for example, if IgG levels fall below detection thresholds before they are measured, past infections may be underascertained. Similarly, it has implications for estimating population protection if antibodies are a marker for protective immunity.
Contrasting data have been made available on the longitudinal trajectory and longevity of antibodies induced by SARS-CoV-2 infection. For example, a US study showed IgG antibody levels to trimerized spike were relatively stable in 121 individuals around 110 days post symptom onset [10]. Similarly, data from 1215 individuals in Iceland suggest that IgG responses to nucleocapsid and the S1 component of spike were sustained for 100–125 days [11]. However, others have noted declines in neutralizing antibodies over similar time periods [12–14].
We have recently undertaken baseline serological testing in a cohort of >10 000 healthcare workers (HCWs) [15]. We now describe serial SARS-CoV-2 antibody measurements, demonstrating quantitative anti-nucleocapsid responses fall over time and vary with age, ethnicity, and previous symptoms, but anti-spike levels antibodies remain stably detected in most individuals.
METHODS
Setting and Participants
Oxford University Hospitals (OUH) offers both symptomatic and asymptomatic SARS-CoV-2 testing programs to staff at its 4 teaching hospitals in Oxfordshire, UK. In total, 12 411 HCWs have undergone serological testing to date; data on HCWs who attended more than once for antibody testing are presented.
SARS-CoV-2 polymerase chain reaction (PCR) testing of nasal and oropharyngeal swabs for all symptomatic (new persistent cough, fever ≥37.8°C, anosmia/ageusia) staff was offered from 27 March 2020 onward. Asymptomatic HCWs were invited to participate in voluntary staff testing for SARS-CoV-2 by nasal and oropharyngeal swab PCR and serological testing from 23 April 2020 onward. The cohort, associated methods, and findings from the first test per individual have been previously described [15]. Following initial PCR and antibody testing, asymptomatic HCWs were invited to optionally attend for serological testing up to once every 2 months, with some offered more frequent screening as part of related studies. Asymptomatic staff were also offered optional SARS-CoV-2 PCR tests every 2 weeks.
Laboratory Assays
Serology for SARS-CoV-2 IgG to nucleocapsid protein was performed using the Abbott Architect i2000 chemiluminescent microparticle immunoassay (CMIA; Abbott, Maidenhead, UK). Antibody levels ≥1.40 manufacturer’s arbitrary units were considered positive, 0.50–1.39 equivocal (following Abbott Diagnostics Product Information Letter PI1060–2020) and <0.5 negative. Anti-trimeric-spike IgG levels were measured using an enzyme-linked immunosorbent assay (ELISA) developed by the University of Oxford [2], using net-normalized signal cutoff of ≥8 million units to determine antibody presence and defining 4.0–7.9 million units as equivocal [16]. Details on PCR assays are provided in the Supplementary Data.
Statistical Methods
For anti-nucleocapsid antibodies, individuals with ≥1 positive antibody result (titer ≥1.40) and ≥2 antibody results were classified as showing rising titers only, falling or stable titers only, or both. Those with only one measurement could not be classified and were excluded. In those with falling/stable titers we estimated the duration of antibody responses following the maximum observed result using Bayesian linear mixed models and their association with age, gender, ethnicity, previous self-reported symptoms, and PCR results (allowing correlated random intercept and slope terms, see Supplementary Data for details and Supplementary Table 1). We assumed antibody levels fell exponentially and so modeled log2 transformed antibody levels over time (observed data and fitted models demonstrated close congruence, Supplementary Data). The incidence of coronavirus disease 2019 (COVID-19) in our hospital fell after a peak in March and April 2020 [15], such that reexposure of HCWs during the study was uncommon; we therefore had insufficient data to study boosting of antibody responses.
We additionally modeled the antibody trajectory from a first positive PCR test using a similar approach but allowing for nonlinear effects of time rather than assuming an exponential decline.
It was not possible to model anti-spike IgG titers over time in those with a positive result as most positive readings were above the upper limit of quantification of the assay (9 million units). Therefore, we considered changes in binary results for both anti-spike and anti-nucleocapsid using Bayesian interval censored regression to estimate the proportion of individuals remaining antibody positive (as opposed to equivocal or negative) at varying times following their maximum antibody result (Supplementary Data).
Ethics
Deidentified data from staff testing were obtained from the Infections in Oxfordshire Research Database (IORD), which has generic Research Ethics Committee, Health Research Authority, and Confidentiality Advisory Group approvals (19/SC/0403, 19/CAG/0144).
RESULTS
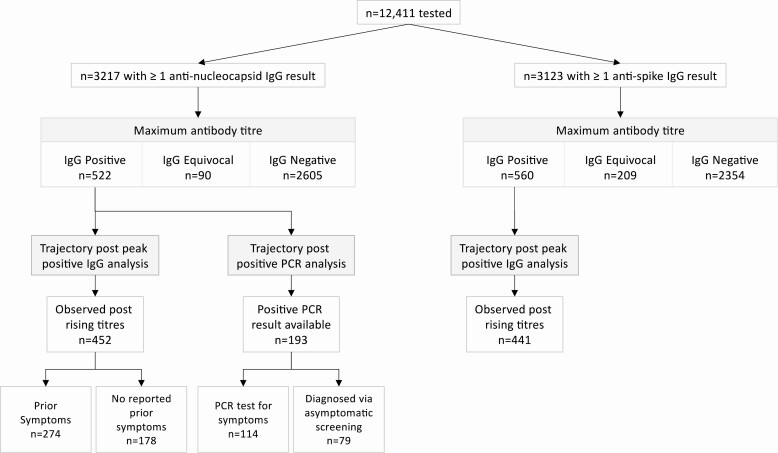
A total of 3276 HCWs provided ≥2 samples for serological testing between 23 April and 20 October 2020 (3217 had anti-nucleocapsid results [Table 1], 3123 anti-spike results, 3064 had both) (Figure 1, Supplementary Table 2). For both assays the median (interquartile range [IQR]) [range] number of samples tested was 2 (2–2) [2–8], and time from the first to last sample was 124 (95–144) [3–174] days.
Table 1.
Baseline Cohort Demographics for 3217 HCWs and 452 HCWs With ≥1 Positive SARS-CoV-2 Anti-Nucleocapsid IgG Result and ≥1 Subsequent Follow-up Sample
| Characteristic | Whole Cohort, n (%) or Median (IQR) [Range] | 452 HCWs With a Positive Antibody Result and ≥1 Subsequent Sample, n (%) or Median (IQR) [Range] |
|---|---|---|
| Age | ||
| Age, y | 39 (29–50) [16–76] | 41 (29–50) [17–69] |
| Gender | ||
| Female | 2542 (79) | 340 (75) |
| Male | 673 (21) | 112 (25) |
| Not disclosed | 2 (<1) | |
| Self-reported ethnicity | ||
| White | 2473 (77) | 302 (67) |
| Black | 90 (3) | 25 (6) |
| Asian | 440 (14) | 89 (20) |
| Other | 214 (7) | 36 (8) |
| COVID-19-like symptoms between 01 February 2020 and testing | ||
| Yes | 898 (28) | 274 (61) |
| No | 2319 (72) | 178 (39) |
| Previous positive SARS-CoV-2 PCR | ||
| Symptomatic | 128 (4) | 95 (21) |
| Asymptomatic | 117 (4) | 59 (13) |
| None | 2972 (92) | 298 (66) |
Abbreviations: COVID-19, coronavirus disease 2019; HCW, healthcare worker; IgG; immunoglobulin G; IQR, interquartile range; PCR, polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Figure 1.
SARS-CoV-2 antibody trajectory cohorts. Abbreviations: IgG, immunoglobulin G; PCR, polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
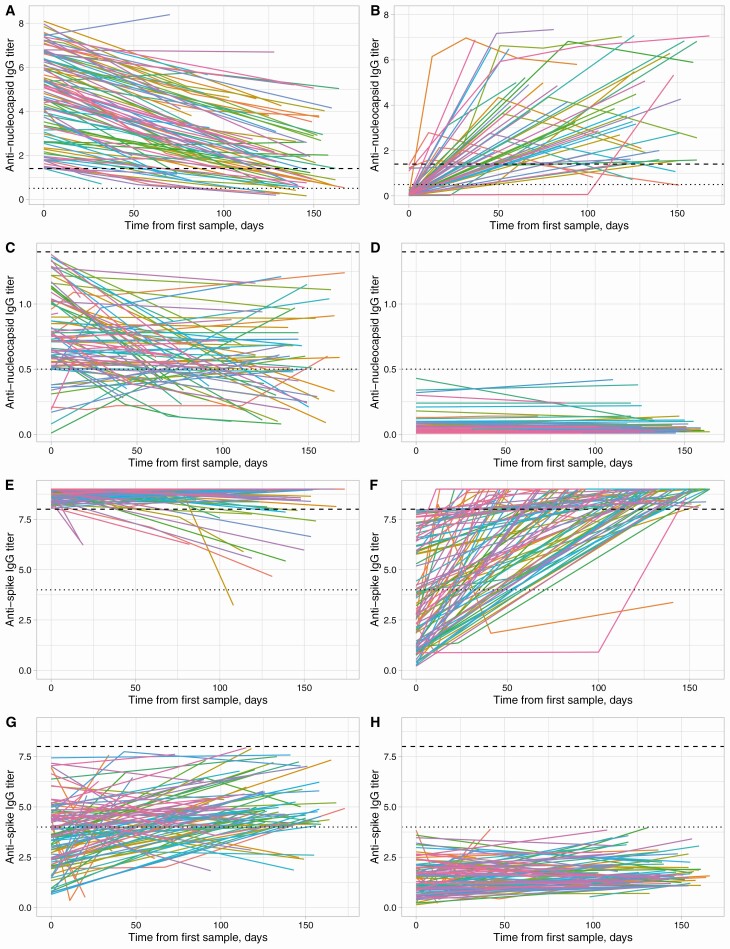
Observed IgG antibody trajectories are shown in Figure 2. In sum, 522/3217 (16%) HCWs had ≥1 sample with detected anti-nucleocapsid antibodies (Figure 2A, 2B), and another 90 (3%) had ≥1 sample with an equivocal titer (Figure 2C). Antibody titers in those with consistently negative results were broadly stable (Figure 2D), whereas falls were observed in 438/466 (94%) initially anti-nucleocapsid antibody positive individuals and in 61/83 (73%) with an initial equivocal titer (Figure 2A, 2C). Also, 560/3123 (18%) HCWs had a positive anti-spike IgG (Figure 2E, 2F) and 209 (7%) an equivocal result. Among 457 HCWs with an initially positive anti-spike titer, 362 (79%) had a final titer that remained above the upper limit of quantification, and 49 (11%) had a fall in titer (Figure 2E).
Figure 2.
SARS-CoV-2 anti-nucleocapsid (A–D) and anti-spike (E–H) IgG antibody trajectories. Panels A and B show anti-nucleocapsid trajectories for HCWs with a positive result (≥1.40 arbitrary units) at some time. A, Those whose first measurement was positive (n = 466, only data from 100 randomly sampled individuals is shown to assist visualization). B, Remainder (n = 56) in whom seroconversion was observed. C, Those with a maximum titer that was equivocal (0.50–1.39, n = 90). D, Results from HCWs with a maximum titer that was negative (<0.50, n = 2605, 100 randomly sampled individuals are shown). Dashed and dotted lines indicate the thresholds for a positive and equivocal result; note the different y-axis scales in panels A and B vs panels C and D. Similarly, panels E–H show anti-spike trajectories in million net normalized units for individuals who start positive (≥8 million units, n = 457), seroconvert (n = 103), have a maximum equivocal result (4.0–7.9 million units, n = 209, 100 shown), and only negative results (<4 million units, n = 2354, 100 shown). Anti-spike assay values above the upper limited of quantification, ie, >9 million, are plotted as 9 million. Abbreviations: HCW, healthcare worker; IgG, immunoglobulin G; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Anti-Nucleocapsid IgG Trajectories After a Positive Antibody Result
Among 522 individuals with ≥1 anti-nucleocapsid IgG-positive sample, 70 (13%) seroconverted with rising titers only and so were excluded from analyses of the duration of response following a peak IgG result (39/70 had a PCR test and are included in a separate analysis below). In the remaining 452 (87%), the median (IQR) [range] number of samples tested was 2 (2–3) [2–5], and time from the first to last sample was 121 (83–143) [4–171] days. Only 3/120 (3%) of individuals with ≥3 measurements had a final titer above the minimum observed; that is, potential evidence of boosting and titer increases were all <5%. The median (IQR) age was 41 (29–50) years, and 75% of participants were female (Table 1). The most common self-reported ethnic groups were White (302, 67%) and Asian (89, 20%; predominately south Asian and Filipino). In addition, 274 (61%) recalled self-identified COVID-19-like symptoms between 1 February 2020 and testing; 95 (21%) had a positive SARS-CoV-2 PCR following symptomatic testing, and 59 (13%) a positive PCR during asymptomatic screening. It is likely that many of the remainder were infected prior to widespread availability of testing. The first positive PCR in each individual was prior to or on the day of their maximum antibody titer in all but 5/154 (3%, tested 3–17 days later).
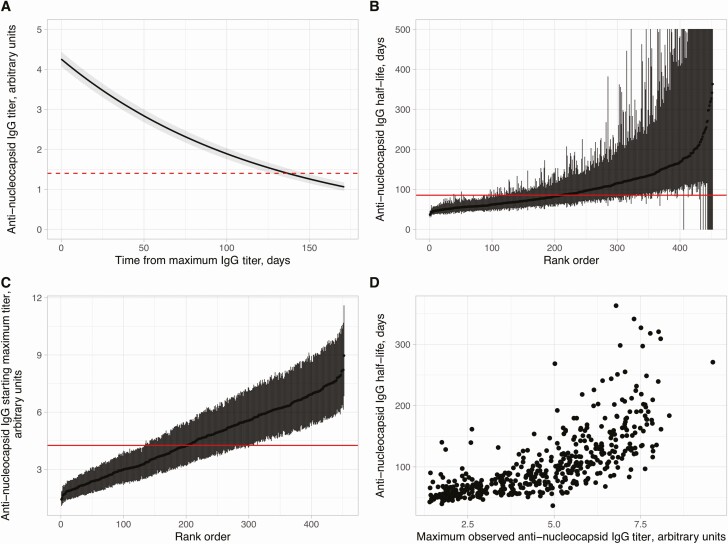
Using a Bayesian statistical model, the trajectory of anti-nucleocapsid IgG levels following the maximum measured titer in each individual is shown in Figure 3A. The estimated mean antibody half-life was 85 (95% CrI 81–90) days and estimated mean maximum antibody level 4.3 (95% CrI 4.1–4.4) arbitrary units. The mean trajectory crossed the diagnostic threshold of 1.40, switching from a positive to equivocal result at 137 (95% CrI 127–148) days. IgG half-lives and maximum titers varied between individuals (Figure 3B, 3C). Higher maximum observed anti-nucleocapsid levels correlated with longer IgG half-lives, that is, slower rates of decline over time (Figure 3D; Spearman rank R2 = .65, P < .0001). Findings were similar in a sensitivity analysis investigating the impact of starting with each individual’s maximum result on half-life estimates (see Supplementary Data).
Figure 3.
SARS-CoV-2 anti-nucleocapsid IgG antibody trajectories in 452 SARS-CoV-2 seropositive HCWs. A, Overall mean trajectory of anti-nucleocapsid IgG antibody levels from the maximum observed level (ie, model fixed effect). Posterior mean and 95% CrI are shown as a solid line and shaded area. Dashed line represents the diagnostic threshold of 1.40 arbitrary units. B, Estimated anti-nucleocapsid IgG half-life with 95% CrI by days for all participants, ranked by its value. Solid horizontal line indicates the overall mean. CrIs exceeding 500 days are truncated at 500 days. C, Estimated maximum anti-nucleocapsid IgG antibody level with 95% CrI for all participants, ranked by its value. Solid horizontal line indicates the overall mean. D, Comparison of maximum observed anti-nucleocapsid IgG antibody level and the estimated anti-nucleocapsid IgG half-life per individual. Abbreviations: CrI, credibility interval; HCW, healthcare worker; IgG, immunoglobulin G; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Effect of Demographics and Other Covariates on Anti-Nucleocapsid Trajectories
Within this cohort of HCWs of working age, age, self-reported ethnicity, prior symptoms compatible with COVID-19, and a positive SARS-CoV-2 PCR were independently associated with changes in anti-nucleocapsid trajectories (Table 2, Supplementary Figure 1–5, Supplementary Table 3). Increasing age was independently associated with higher maximum anti-nucleocapsid levels and a longer half-life, .17 (95% CrI .07–.25) arbitrary units and 3.40 (1.79–4.62) days per 10 years, respectively (Table 2, Supplementary Figure 5). HCWs of Asian ethnicity had higher maximum anti-nucleocapsid levels with adjusted increases of 0.54 (0.18–0.95) arbitrary units compared to White HCWs, with marginal evidence for longer half-lives, by 6.28 (−0.44 to 14.8) days. Within the limits of the power of the study, there was no strong statistical evidence that antibody trajectories varied in other ethnic groups.
Table 2.
Univariable and Multivariable Models of Determinants of SARS-CoV-2 Anti-Nucleocapsid Antibody Trajectories
| Univariable Model | Multivariable Model | ||||||
|---|---|---|---|---|---|---|---|
| Posterior Mean | 95% CrI | Posterior Mean | 95% CrI | ||||
| Baseline model | Maximum antibody level (intercept) | 4.26 | 4.07 | 4.44 | 3.33 | 3.02 | 3.67 |
| Antibody level half-life | 85.38 | 81.17 | 90.01 | 69.94 | 63.74 | 77.19 | |
| Gender model | Maximum antibody level (intercept): Female | 4.24 | 4.04 | 4.46 | |||
| Antibody level half-life: Female | 84.88 | 80.14 | 90.25 | ||||
| Change in intercept: Male | .03 | −.40 | .47 | −.05 | −.34 | .25 | |
| Change in half-life: Male | 1.95 | −8.26 | 13.12 | −.40 | −5.66 | 5.30 | |
| Age model | Maximum antibody level (intercept): 41 years (median) | 3.82 | 3.56 | 4.10 | |||
| Antibody level half-life: 41 years (median) | 75.27 | 69.71 | 81.46 | ||||
| Change in intercept: per 10-year older | .21 | .10 | .30 | .17 | .07 | .25 | |
| Change in half-life: per 10-year older | 4.07 | 2.52 | 5.17 | 3.40 | 1.79 | 4.62 | |
| Ethnicity model | Maximum antibody level (intercept): White | 4.03 | 3.82 | 4.24 | |||
| Antibody level half-life: White | 81.48 | 76.69 | 86.66 | ||||
| Change in intercept: Black | .48 | −.30 | 1.37 | .37 | −.21 | 1.03 | |
| Change in intercept: Asian | .83 | .32 | 1.34 | .54 | .18 | .95 | |
| Change in intercept: Other | .59 | −.10 | 1.37 | .44 | −.07 | 1.00 | |
| Change in half-life: Black | 7.03 | −11.41 | 31.13 | 2.63 | −6.92 | 15.03 | |
| Change in half-life: Asian | 14.64 | 1.54 | 29.29 | 6.28 | −.44 | 14.76 | |
| Change in half-life: Other | 14.85 | −3.12 | 38.26 | 8.24 | −1.13 | 20.55 | |
| Prior symptom model | Maximum antibody level (intercept): No | 3.90 | 3.64 | 4.16 | |||
| Antibody level half-life: No | 83.57 | 77.38 | 90.61 | ||||
| Change in intercept: Yes | .60 | .24 | .96 | .40 | .12 | .69 | |
| Change in half-life: Yes | 2.85 | −5.99 | 11.59 | .26 | −4.52 | 5.19 | |
| PCR model | Maximum antibody level (intercept): No | 4.09 | 3.87 | 4.32 | |||
| Antibody level half-life: No | 80.23 | 75.48 | 85.28 | ||||
| Change in intercept: Positive (symptomatic) | .65 | .17 | 1.16 | .30 | −.04 | .67 | |
| Change in intercept: Positive (asymptomatic) | .23 | −.31 | .81 | .14 | −.24 | .55 | |
| Change in half-life: Positive (symptomatic) | 19.60 | 7.22 | 35.66 | 9.56 | 2.35 | 19.09 | |
| Change in half-life: Positive (asymptomatic) | 13.49 | −.31 | 30.37 | 6.45 | −.86 | 15.85 | |
Posterior mean and 95% credibility intervals for the maximum antibody level (model intercept) and antibody half-life (model slope) are shown. See Supplementary Table 3 for other model parameters and statistical analysis quality metrics.
Abbreviations: CrI, credibility interval; PCR, polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Prior self-reported symptoms were associated with a higher starting maximum anti-nucleocapsid levels (adjusted increase .40 [95% CrI .12–.69]) but not changes in half-lives. There was moderate evidence that a positive PCR result undertaken for symptoms, independently increased the starting maximum level by .30 (−.04 to .67) arbitrary units, and half-life by 9.56 (2.35–19.09) days compared to those with no positive PCR. We observed no effect of gender on either maximum level or antibody half-life.
Anti-Nucleocapsid Trajectories Following a Positive PCR Test
In total, 245 of the 3217 HCWs with ≥2 anti-nucleocapsid samples had a positive PCR test. And 114/128 (89%) symptomatic HCW seroconverted (maximum IgG titer ≥1.40), including 11/12 (92%) who required hospital treatment, all other infections were mild. 79/117 (68%) identified through asymptomatic screening seroconverted. In the 52 individuals not showing evidence of seroconversion, all but 1 (98%) had ≥1 antibody test ≥14 days after their PCR-positive test, and in 29 (56%) this was before 90 days. PCR cycle threshold values were lower in individuals who seroconverted (Supplementary Table 4).
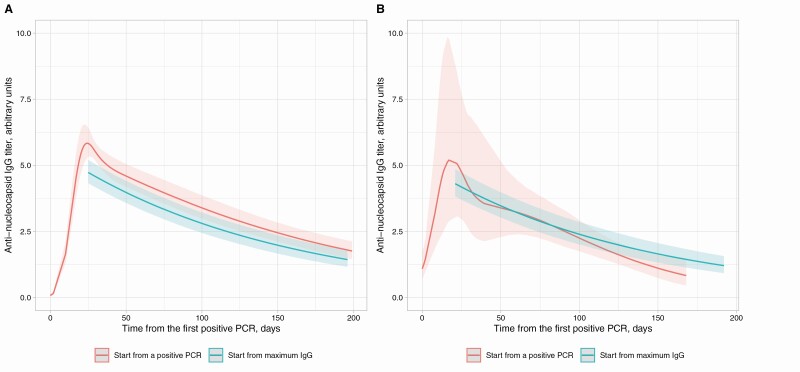
Data from PCR-positive individuals who seroconverted were used to model antibody trajectories relative to a first positive PCR test. Antibody levels rose to a peak at 24 (95% CrI 19–31) days post-first positive PCR test, before beginning to fall (Supplementary Figure 6). Comparing with the antibody trajectory estimated in the main analysis, the estimated rates of waning were consistent between the 2 models. Antibody trajectories were similar in those being tested following symptoms or during asymptomatic screening (Figure 4).
Figure 4.
Comparison of SARS-CoV-2 anti-nucleocapsid IgG antibody levels following a positive PCR test and the maximum IgG level per individual in those with a positive PCR test. A, Those with a positive PCR undertaken for symptoms; B, those with a positive PCR for asymptomatic screening. The x-axis value for the model starting from the maximum IgG level is aligned to the maximum point from the model starting with a positive PCR test. Model starting from a positive PCR is fitted with a 5-knot spline (3 interior knots at t = 10, t = 30, and t = 50, locations chosen based on model fit). Abbreviations: IgG, immunoglobulin G; PCR, polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Anti-Spike Trajectories
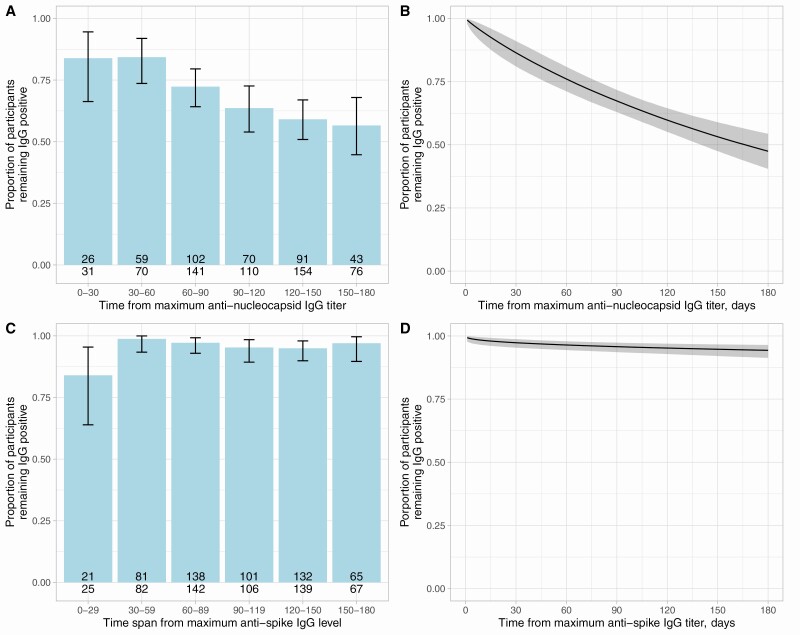
To enable comparison with anti-spike results and to facilitate comparison with studies reporting only categorical antibody results, we considered the proportion of seropositive individuals remaining antibody-positive (as opposed to equivocal or negative) when observed at varying time intervals (Figure 5A, 5C) and using an interval censored survival analysis approach (Figure 5B, 5D). Consistent with the model in Figure 4, the median time remaining anti-nucleocapsid IgG-positive was 166 (95% CrI 139–214) days. In contrast, anti-spike IgG levels remained above positive threshold in most seropositive HCWs for the duration of the study (Figure 5C, 5D), by 180 days post maximum anti-spike IgG level an estimated 94% (95% CrI 91–96%) remained positive.
Figure 5.
Proportion of HCWs remaining anti-nucleocapsid IgG (A–B) and anti-spike IgG (C–D) antibody positive by days following their maximum antibody level. Panels A and C show the observed proportion in 30-day intervals with binomial 95% confidence intervals. Number of individuals tested and the number of individuals remaining antibody positive is shown at the base of each bar. Panels B and D show the results of Bayesian interval censored survival analyses, the posterior mean and 95% credibility interval are shown. Abbreviations: HCW, healthcare worker; IgG, immunoglobulin G; PCR, polymerase chain reaction.
DISCUSSION
Most epidemiological outbreak models assume that SARS-CoV-2 infection leads to the development of postinfection immunity for a defined duration. Here we show contrasting antibody trajectories in 608 symptomatic and asymptomatic HCWs seropositive for anti-nucleocapsid and/or anti-spike antibodies followed for a median of 4 months from their maximum IgG titer. We show anti-nucleocapsid IgG levels wane with an estimated half-life of 85 (95% CrI 81–90) days. We observed variation between individuals; higher maximum observed anti-nucleocapsid titers were associated with longer half-lives. Increasing age, Asian ethnicity, and prior self-reported symptoms were independently associated with higher maximum anti-nucleocapsid levels, and increasing age and a positive PCR test undertaken for symptoms with longer antibody half-lives. In contrast, although we could not quantitively follow titers of anti-spike IgG, levels remained stably above the threshold for a positive result in 94% at 180 days post maximum titer.
IgG waning and reinfection within a year is reported for seasonal coronaviruses [17], whereas IgG remains detectable against SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV) 1–3 years later [18]. The differences we observe in SARS-CoV-2 antibody trajectories may be antigen and/or assay dependent; for example, stable anti-spike antibodies with waning of anti-nucleocapsid IgG using the same Abbott platform as in our study was also seen in an earlier smaller study, but total anti-nucleocapsid antibodies assayed using a Roche platform remained stable [14]. To some extent these findings are conditional on assay cutoffs which can be tuned to prioritise sensitivity or specificity, with inherently more specific assays having potential to also be set more sensitively, resulting in apparently longer durations of detectable antibody responses.
For anti-nucleocapsid, we observe higher IgG titers with longer durability occurring after symptomatic PCR-positive infection, consistent with data from Long et al, where 40% of asymptomatic individuals and 13% of the symptomatic group became negative for IgG in the early convalescent phase [19] and consistent with emerging coronaviruses, where antibody titers remained detectable longer after more severe illness [18], waning more rapidly after asymptomatic infection.
Relatively short-term anti-nucleocapsid IgG responses have 2 epidemiological consequences. First, antibody waning may lead to under-ascertainment of previous infections within the current pandemic, particularly in younger individuals following asymptomatic/mild infection. Additionally, IgG testing is unlikely to determine whether SARS-CoV-2 has circulated historically, for example, in a particular geographic region.
Older age (within this cohort of working age HCWs, up to 69 years) was associated with higher maximum observed anti-nucleocapsid IgG titers and longer half-lives, with similar findings associated with Asian ethnicity (many of the Asian HCWs in our study came to work in the UK healthcare system as adults). It is possible to hypothesize that this could arise from boosting of cross-reactive anti-nucleocapsid antibodies from prior exposure, for example, to a previously circulating or geographically restricted human coronavirus. However, anti-nucleocapsid cross-reactivity between endemic coronaviruses and other epidemic coronaviruses, SARS-CoV and MERS-CoV, is uncommon [18, 20].
Limitations
The assays we used could only measure quantitative trajectories for anti-nucleocapsid IgG, further studies, for example, using multiple serum dilutions, are required to quantify anti-spike IgG over time. We therefore cannot say whether the longer duration of positive anti-spike responses is due to slower waning or higher initial levels relative to assay cut-offs. Another limitation of our study was that our cohort of individuals consisted of adults of working age (17–69 years); further longitudinal studies will be required to investigate younger and older age groups. The small numbers of self-reported Black (n = 25) and other (n = 36) ethnicities reduced/limited power to detect an association between these ethnicities and antibody trajectories. We also do not account for mediators, for example, socioeconomic inequalities, that may link ethnicity to antibody responses in the absence of a direct causal relationship. Due to many of our staff developing symptoms before widespread SARS-CoV-2 PCR testing was available, only 34% of the anti-nucleocapsid-positive cohort had a documented positive PCR, and as a proportion of the cohort were asymptomatic throughout, we modeled time from maximum positive antibody test rather than time from first positive PCR or time from symptom onset in our main analysis of antibody durability. However, under an exponential assumption, half-lives can be estimated from any measurements taken after a maximum; we excluded individuals with only evidence of rising titers to avoid underestimating half-lives. Furthermore, data from those that were PCR positive were consistent with this analysis (Figure 4).
Multiple different assays are in use globally to characterize antibody responses to SARS-CoV-2. Here we study only 2; however, other antibody classes and targets, and aspects of immunity, including the innate and cellular responses, are important in conferring postinfection immunity [21]. When comparing longitudinal studies of antibody durability, care must be taken, as the various assays have not yet been cross-calibrated, and implications for protective immunity are not fully understood.
Implications
It is widely recognized that pathogen-specific IgG levels decline after the acute phase of an infection. After the initial humoral response in which short-lived plasmablasts secrete high titers of antibody, long-lived plasma cells and memory B cells then contribute to longer-term antibody-mediated protection [22]. Although declines in IgG titers are expected, understanding the assay-dependent rate of decline, whether and when titers fall below assay positive cutoffs, and how these titers relate to protection from subsequent asymptomatic and symptomatic reinfection is crucial.
Serological testing also helps quantify the extent of infection in populations, informing epidemiological models and public health strategies. However, waning antibody levels may lead to underestimated exposure due to loss of seropositivity. For example, using the anti-nucleocapsid assay, an estimated 33% of individuals seroreverted (ie, fell below the positive cutoff for the Abbott assay) within 3 months of IgG detection and an estimated 53% of individuals by 6 months (Figure 5B). Therefore, depending on the assay used, seroepidemiological surveys performed several months into this pandemic may have underestimated prior exposure, especially in younger adults who lose detectable antibody faster. Our findings contrast with the repeated cross-sectional REACT2 study [23], which reported greater reductions over time at a population level in the proportion of adults ≥65 years testing antibody-positive and more sustained responses in those 18–24 years. Nearly all HCWs in our study were <65 years; other differences may arise from study design, the assay used, and the potential for new infections predominantly in younger people [24] to replace others who had seroreverted, supporting population-level seroprevalence (in our study each individual was followed up separately).
Future Work
Antibody dynamics have significant implications for the course and management of pandemics. Durability of immunity postinfection and postvaccination will dictate the overall course of the pandemic. Further work is required to determine how prior infection and/or vaccination impacts the probability of future infection and severity of subsequent disease, determine the antibody-based correlates of this protection, and therefore the ability of serological tests to identify those who are immune. Longitudinal cohorts with baseline immunology are required to determine immune correlates of protection, to determine whether measurement of the current antibody status is enough to infer whether an individual have functional immunity or not, whether waning IgG titers are representative of waning immune protection, or whether protection remains even after an individual seroreverts.
CONCLUSION
We demonstrate that the half-life of SARS-CoV-2 anti-nucleocapsid IgG antibody responses in a cohort of adult HCWs is 85 days and varies between individuals by age, ethnicity, and prior symptom history. In contrast, anti-spike IgG responses were sustained in most HCWs up to 180 days. The extent and duration of immunity to SARS-CoV-2 infection following COVID-19 and its association with antibody titers remains a key question to be answered.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Oxford University Hospitals Staff Testing Group:
University of Oxford Medical School staff testing team (University of Oxford, Oxford, UK): Molly Abbott, Roxanna Abhari, Hibatullah Abuelgasim, Tariq Ahmed-Firani, Ahmed Alhussni, Isobel Argles, Rebecca Ashworth, Francesca Back, Susannah Black, Isabelle Brennan, Lottie Cansdale, Triya Chakravorty, Zahra Choudhury, Helen Clay, Rebecca Conway-Jones, Aisling Curtis, Joseph Cutteridge, Hannah Danbury, Hettie Davies, Lucy Denly, Julie Dequaire, Stoyan Dimitrov, Ella Dunlop, Polly Dunn, Cameron East, Ben Eastwood, Chantal Edwardes, Meirian Evans, Charlotte Fields, Sarah Flaherty, Thomas Foord, Rosie Freer, Hannah Fuchs, Alexander Grassam-Rowe, Lewis Greenan-Barrett, James Gunnell, Callum Harries, Edward Harris, Elizabeth Hatton, Conor Hennessy, Christian Holland, Ben Holloway, Gregory Howgego, Isaac Jarratt Barnham, Síle Johnson, Thomas Johnson, Daniela Johnstone, Rebecca Jurdon, Gabriella Kelly, Sven Kerneis, Beinn Khulusi, Annabel Killen, Stefan Kourdov, Mary Kumarendran, Hannah Laurenson-Schafer, Ruby Lawrence, Charlotte Lee, Angus Livingstone, Rosie Lynch, Tara Madsen, Ali Manji, James Marsh, Chris Mason, Angus Mccance, Euan Mcgivern, Ailsa Mckinlay, Alexandra Mighiu, Lancelot Millar, Yethrib Mohamed, Kenzo Motohashi, San Myat, Andrey Nezhentsev, Katherine O’Byrne, Temitope Okuwoga, Madeleine Oliver, Saxon Pattenden, Vimukthi Perera, Yva Petit, Florence Pickles, Maria Pikoula, Govind Randhawa, Krupa Ravi, Lara Reed, Caitlin Rigler, Charlotte Robinson, Evie Rothwell, Stephanie Santos-Paulo, Constantinos Savva, Alice Scharmeli, Samuel Scott, Zamin Shabir, Ishta Sharma, Remarez Sheehan, Afrah Shibu, Arun Somanathan, Samuel Sussmes, Owen Sweeney, Emily Swift, Gayatri Tadikamalla, Morwenna Tamblyn, Adan Taylor, Rebecca Te Water Naudé, Sarah Thomas, Connor Thompson, Lottie Till, Ross Toward, Charis Virgo, Imogen Vorley, Seren Waite, Harry Ward, Jocelyn Ward, Adam Watson, Lily Watson, Matthew Wedlich, Laura Wilkins, Jack Wilson, Joseph Wilson, William Wong, Rebecca Young.
University of Oxford staff testing team (University of Oxford, Oxford, UK): George Doherty, James Kavanagh, Kevin K Chau, Philip W Fowler, Jeremy Swann, Denis Volk, Fan Yang-Turner, Donal Skelly, Fredrik Karpe, Matt J Neville, Lucas Martins Ferreira, Thomas Christott, Gillian Rodger, Hayleah Pickford, Sarah Cameron, Phoebe Tamblin-Hopper.
Oxford University Hospitals staff testing team (Oxford University Hospitals NHS Foundation Trust, Oxford, UK): Ali Amini, Monique Andersson, Karen Anson, Lisa Aston, David Axten, Simran Barot, Trisha Bellinger, Adrian Bialek, Noemi Bodo, Andrew Brent, Mark Campbell, Vanessa Cox, Tammy Cronin, Timothy Davies, Alessio Deales, Louise Downs, Philip Drennan, Maria Dudareva, Laura Gates, Michelle Gates, Dominique Georgiou, Bernadett Gergely, Louise Holland, Nicola Jones, Sarah Jones, Sharon Kerr, Elaine Lawson, Stacy Looms, Michael Luciw, Diva Martins, Mary Mcknight, Alexander Mentzer, Rebecca Millard, Tracey Mitchell, Lisa Morgan, Jordan Morrow, Emma Mortimore, Harriet Mullins, Heather Nevard, Anne-Marie O’Donnell, Kaisha Patel, Leon Peto, Jane Philips, Jessica Ponting, Roshni Ray, Maria Robles, Sonam Rughani, Nellia Sande, Robert Shaw, Emma-Jane Simons, Elizabeth Sims, Kyla Smit, Lucianne Smith, Justyna Szczurkowska, Zoe Thompson, Rachel Turford, Kim Vilca, Alexander Vogt, Fiona Warren, Laura Warren, Vicki Wharton, Alison Whitty, Heather Woodley, Barbara Wozniak, Bernadette Young.
Oxford University Hospitals microbiology laboratory (Oxford University Hospitals NHS Foundation Trust, Oxford, UK): Anita Justice, Gerald Jesuthasan, Susan Wareing, Nurul Huda Mohamad Fadzillah, Kathryn Cann, Richard Kirton.
Oxford University Hospitals Infection, Prevention and Control team (Oxford University Hospitals NHS Foundation Trust, Oxford, UK): Claire Sutton, Claudia Salvagno, Gabriella D’Amato, Gemma Pill, Lisa Butcher, Lydia Rylance-Knight, Merline Tabirao, Ruth Moroney, Sarah Wright.
Acknowledgments . The authors thank all OUH staff who participated in the staff testing program and the staff and medical students who ran the program. This work uses data provided by healthcare workers and collected by the UK’s National Health Service as part of their care and support. The authors thank all the people of Oxfordshire who contribute to the Infections in Oxfordshire Research Database. Research Database Team: L Butcher, H Boseley, C Crichton, DW Crook, DW Eyre, O Freeman, J Gearing (community), R Harrington, K Jeffery, M Landray, A Pal, TEA Peto, TP Quan, J Robinson (community), J Sellors, B Shine, AS Walker, D Waller. Patient and Public Panel: G Blower, C Mancey, P McLoughlin, B Nichols.
Financial support. This work was supported by the UK Government’s Department of Health and Social Care. This work was supported by the National Institute for Health Research Health Protection Research Unit (NIHR HPRU) in Healthcare Associated Infections and Antimicrobial Resistance at Oxford University in partnership with Public Health England (PHE) (grant number NIHR200915), the NIHR Biomedical Research Center, Oxford, and benefactions from the Huo Family Foundation and Andrew Spokes. The views expressed in this publication are those of the authors and not necessarily those of the NHS, the NIHR, or the Department of Health or Public Health England. D. W. E. is a Robertson Foundation Fellow and an NIHR Oxford BRC Senior Fellow. S. F. L. is a Wellcome Trust Clinical Research Fellow. D. I. S. is supported by the Medical Research Council (grant number MR/N00065X/1). P. C. M. holds a Wellcome Intermediate Fellowship (grant number 110 110/Z/15/Z) and is an NIHR Oxford BRC Senior Fellow. B. D. M. is supported by the structural genomics consortium (SGC), a registered charity (grant number 1097737) that receives funds from AbbVie, Bayer Pharma AG, Boehringer Ingelheim, Canada Foundation for Innovation, Eshelman Institute for Innovation, Genome Canada through Ontario Genomics Institute (grant numberOGI-055), Innovative Medicines Initiative (EU/EFPIA) (ULTRA-DD grant number 115766), Janssen, Merck KGaA, Darmstadt, Germany, MSD, Novartis Pharma AG, Pfizer, São Paulo Research Foundation-FAPESP, Takeda, and Wellcome. B. D. M. is also supported by the Kennedy Trust for Rheumatology Research. G. S. is a Wellcome Trust Senior Investigator and acknowledges funding from the Schmidt Foundation. T. M. W. is a Wellcome Trust Clinical Career Development Fellow (grant number 214560/Z/18/Z). A. S. W. is an NIHR Senior Investigator.
Potential conflicts of interest. D. W. E. reports lecture fees from Gilead, outside the submitted work. R. J. C. is a founder shareholder and consultant to MIROBio, work outside the submitted work. G. S. reports scientific advisory board fees from GSK Vaccines, outside the submitted work. M. A. reports personal fees from Prenetics, outside the submitted work. J. C. reports personal fees from Nuffield Department of Medicine, University of Oxford, outside the submitted work. T. F. reports personal fees from NHS Professionals, outside the submitted work. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Contributor Information
Oxford University Hospitals Staff Testing Group:
Molly Abbott, Roxanna Abhari, Hibatullah Abuelgasim, Tariq Ahmed-Firani, Ahmed Alhussni, Isobel Argles, Rebecca Ashworth, Francesca Back, Susannah Black, Isabelle Brennan, Lottie Cansdale, Triya Chakravorty, Zahra Choudhury, Helen Clay, Rebecca Conway-Jones, Aisling Curtis, Joseph Cutteridge, Hannah Danbury, Hettie Davies, Lucy Denly, Julie Dequaire, Stoyan Dimitrov, Ella Dunlop, Polly Dunn, Cameron East, Ben Eastwood, Chantal Edwardes, Meirian Evans, Charlotte Fields, Sarah Flaherty, Thomas Foord, Rosie Freer, Hannah Fuchs, Alexander Grassam-Rowe, Lewis Greenan-Barrett, James Gunnell, Callum Harries, Edward Harris, Elizabeth Hatton, Conor Hennessy, Christian Holland, Ben Holloway, Gregory Howgego, Isaac Jarratt Barnham, Síle Johnson, Thomas Johnson, Daniela Johnstone, Rebecca Jurdon, Gabriella Kelly, Sven Kerneis, Beinn Khulusi, Annabel Killen, Stefan Kourdov, Mary Kumarendran, Hannah Laurenson-Schafer, Ruby Lawrence, Charlotte Lee, Angus Livingstone, Rosie Lynch, Tara Madsen, Ali Manji, James Marsh, Chris Mason, Angus Mccance, Euan Mcgivern, Ailsa Mckinlay, Alexandra Mighiu, Lancelot Millar, Yethrib Mohamed, Kenzo Motohashi, San Myat, Andrey Nezhentsev, Katherine O’Byrne, Temitope Okuwoga, Madeleine Oliver, Saxon Pattenden, Vimukthi Perera, Yva Petit, Florence Pickles, Maria Pikoula, Govind Randhawa, Krupa Ravi, Lara Reed, Caitlin Rigler, Charlotte Robinson, Evie Rothwell, Stephanie Santos-Paulo, Constantinos Savva, Alice Scharmeli, Samuel Scott, Zamin Shabir, Ishta Sharma, Remarez Sheehan, Afrah Shibu, Arun Somanathan, Samuel Sussmes, Owen Sweeney, Emily Swift, Gayatri Tadikamalla, Morwenna Tamblyn, Adan Taylor, Rebecca Te Water Naudé, Sarah Thomas, Connor Thompson, Lottie Till, Ross Toward, Charis Virgo, Imogen Vorley, Seren Waite, Harry Ward, Jocelyn Ward, Adam Watson, Lily Watson, Matthew Wedlich, Laura Wilkins, Jack Wilson, Joseph Wilson, William Wong, Rebecca Young, George Doherty, James Kavanagh, Kevin K Chau, Philip W Fowler, Jeremy Swann, Denis Volk, Fan Yang-Turner, Donal Skelly, Fredrik Karpe, Matt J Neville, Lucas Martins Ferreira, Thomas Christott, Gillian Rodger, Hayleah Pickford, Sarah Cameron, Phoebe Tamblin-Hopper, Ali Amini, Monique Andersson, Karen Anson, Lisa Aston, David Axten, Simran Barot, Trisha Bellinger, Adrian Bialek, Noemi Bodo, Andrew Brent, Mark Campbell, Vanessa Cox, Tammy Cronin, Timothy Davies, Alessio Deales, Louise Downs, Philip Drennan, Maria Dudareva, Laura Gates, Michelle Gates, Dominique Georgiou, Bernadett Gergely, Louise Holland, Nicola Jones, Sarah Jones, Sharon Kerr, Elaine Lawson, Stacy Looms, Michael Luciw, Diva Martins, Mary Mcknight, Alexander Mentzer, Rebecca Millard, Tracey Mitchell, Lisa Morgan, Jordan Morrow, Emma Mortimore, Harriet Mullins, Heather Nevard, Anne-Marie O’Donnell, Kaisha Patel, Leon Peto, Jane Philips, Jessica Ponting, Roshni Ray, Maria Robles, Sonam Rughani, Nellia Sande, Robert Shaw, Emma-Jane Simons, Elizabeth Sims, Kyla Smit, Lucianne Smith, Justyna Szczurkowska, Zoe Thompson, Rachel Turford, Kim Vilca, Alexander Vogt, Fiona Warren, Laura Warren, Vicki Wharton, Alison Whitty, Heather Woodley, Barbara Wozniak, Bernadette Young, Anita Justice, Gerald Jesuthasan, Susan Wareing, Nurul Huda Mohamad Fadzillah, Kathryn Cann, Richard Kirton, Claire Sutton, Claudia Salvagno, Gabriella D’Amato, Gemma Pill, Lisa Butcher, Lydia Rylance-Knight, Merline Tabirao, Ruth Moroney, and Sarah Wright
References
- 1. Deeks JJ, Dinnes J, Takwoingi Y, et al. Antibody tests for identification of current and past infection with SARS‐CoV‐2. Cochrane Db Syst Rev 2020; 6:CD013652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. The National SARS-CoV-2 Serology Assay Evaluation Group. Performance characteristics of five immunoassays for SARS-CoV-2: a head-to-head benchmark comparison. Lancet Infect Dis 2020; 20:1390–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wellinghausen N, Plonné D, Voss M, Ivanova R, Frodl R, Deininger S. SARS-CoV-2-IgG response is different in COVID-19 outpatients and asymptomatic contact persons. J Clin Virol 2020; 130:104542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fafi-Kremer S, Bruel T, Madec Y, et al. Serologic responses to SARS-CoV-2 infection among hospital staff with mild disease in eastern France. EBioMedicine 2020; 59:102915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Suhandynata RT, Hoffman MA, Kelner MJ, McLawhon RW, Reed SL, Fitzgerald RL. Longitudinal monitoring of SARS-CoV-2 IgM and IgG seropositivity to detect COVID-19. J Appl Laboratory Medicine 2020; 5:jfaa079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xiang F, Wang X, He X, et al. Antibody detection and dynamic characteristics in patients with COVID-19. Clin Infect Dis 2020; 71:1930–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. To KK-W, Tsang OT-Y, Leung W-S, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis 2020; 20:565–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Long QX, Liu BZ, Deng HJ, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med 2020; 26:845–8. [DOI] [PubMed] [Google Scholar]
- 9. Adams ER, Ainsworth M, Anand R, et al. Antibody testing for COVID-19: a report from the National COVID scientific advisory panel. Wellcome Open Res 2020; 5:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wajnberg A, Amanat F, Firpo A, et al. Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science 2020; 370:1227–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gudbjartsson DF, Norddahl GL, Melsted P, et al. Humoral immune response to SARS-CoV-2 in Iceland. N Engl J Med 2020; 383:1724–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Seow J, Graham C, Merrick B, et al. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat Microb 2020; 5:1598–607. [DOI] [PMC free article] [PubMed]
- 13. Robbiani DF, Gaebler C, Muecksch F, et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 2020; 584:437–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Muecksch F, Wise H, Batchelor B, et al. Longitudinal analysis of serology and neutralizing antibody levels in COVID19 convalescents. J Infect Dis 2020; doi.org/10.1093/infdis/jiaa659 . [Google Scholar]
- 15. Eyre DW, Lumley SF, O’Donnell D, et al. Differential occupational risks to healthcare workers from SARS-CoV-2 observed during a prospective observational study. Elife 2020; 9:e60675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Eyre DW, Lumley SF, O’Donnell D, et al. Stringent thresholds for SARS-CoV-2 IgG assays result in under-detection of cases reporting loss of taste/smell. Medrxiv 2020; 10.1101/2020.07.21.20159038. [DOI] [Google Scholar]
- 17. Edridge AWD, Kaczorowska J, Hoste ACR, et al. Seasonal coronavirus protective immunity is short-lasting. Nat Med 2020;1–3. [DOI] [PubMed] [Google Scholar]
- 18. Huang AT, Garcia-Carreras B, Hitchings MDT, et al. A systematic review of antibody mediated immunity to coronaviruses: kinetics, correlates of protection, and association with severity. Nat Commun 2020; 11:4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Long QX, Tang XJ, Shi QL, et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med 2020; 26:1200–4. [DOI] [PubMed] [Google Scholar]
- 20. Iyer AS, Jones FK, Nodoushania A, et al. Dynamics and significance of the antibody response to SARS-CoV-2 infection. Medrxiv 2020. doi: 10.1101/2020.07.18.20155374. [DOI] [Google Scholar]
- 21. Tan AT, Linster M, Tan CW, et al. Early induction of SARS-CoV-2 specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Biorxiv 2020;. doi:2020.10.15.341958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stephens DS, McElrath MJ. COVID-19 and the path to immunity. JAMA 2020; 324:1279–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ward H, Cooke G, Atchison C, et al. Declining prevalence of antibody positivity to SARS-CoV-2: a community study of 365,000 adults. Medrxiv 2020; 10.1101/2020.10.26.20219725. [DOI] [Google Scholar]
- 24. Pouwels KB, House T, Prtichard E, et al. Community prevalence of SARS-CoV-2 in England during April to September 2020: results from the ONS coronavirus infection survey. Lancet Public Health 2020; 6:E30-E38. 10.1101/2020.10.26.20219428. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.