Abstract
Motivation
Ligand–receptor (L–R) interactions mediate cell adhesion, recognition and communication and play essential roles in physiological and pathological signaling. With the rapid development of single-cell RNA sequencing (scRNA-seq) technologies, systematically decoding the intercellular communication network involving L–R interactions has become a focus of research. Therefore, construction of a comprehensive, high-confidence and well-organized resource to retrieve L–R interactions in order to study the functional effects of cell–cell communications would be of great value.
Results
In this study, we developed Cellinker, a platform of literature-supported L–R interactions that play roles in cell–cell communication. We aimed to provide a useful platform for studies on cell–cell communication mediated by L–R interactions. The current version of Cellinker documents over 3700 human and 3200 mouse L–R protein–protein interactions (PPIs) and embeds a practical and convenient webserver with which researchers can decode intercellular communications based on scRNA-seq data. And over 400 endogenous small molecule (sMOL) related L–R interactions were collected as well. Moreover, to help with research on coronavirus (CoV) infection, Cellinker collects information on 16L–R PPIs involved in CoV–human interactions (including 12L–R PPIs involved in SARS-CoV-2 infection). In summary, Cellinker provides a user-friendly interface for querying, browsing and visualizing L–R interactions as well as a practical and convenient web tool for inferring intercellular communications based on scRNA-seq data. We believe this platform could promote intercellular communication research and accelerate the development of related algorithms for scRNA-seq studies.
Availability and implementation
Cellinker is available at http://www.rna-society.org/cellinker/
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
It is estimated that ∼20% and 10% of human genes encode cell-surface proteins and secreted proteins, respectively (Fonseca et al., 2016; Wood and Wright, 2019). These proteins and the ligand–receptor (L–R) interactions they involve are crucial parts of the intercellular communication network (Boisset et al., 2018; Honig and Shapiro, 2020; Wood and Wright, 2019). However, compare to the intracellular protein–protein interaction networks (PPINs), the comprehensive human cell-surface interactome is still lacking (Özkan et al., 2013; Wojtowicz et al., 2020). Several recent studies have begun to screen L–R interactions via high-throughput experimental methods, providing valuable resources for research on intercellular communication (Gil et al., 2020; Husain et al., 2019; Verschueren et al., 2020; Wojtowicz et al., 2020).
In multicellular organisms, cell–cell communication mediated by L–R interactions is an essential regulatory event coordinating various biological processes, such as the immune response, neural transmission and pathogen invasion of host cells (Douam et al., 2015; Li et al., 2015; Scadden, 2014), and aberrant loss or gain of extracellular recognition functions can contribute to multiple diseases (Massagué and Obenauf, 2016; Ning et al., 2020). For instance, the immune checkpoint protein PD1 and its ligand PDL1 have been found to act as accomplices that help tumors resist immunity-induced apoptosis and to promote tumor progression, and anti-PD1/PDL1 therapy has achieved great success in the past decade (Lei et al., 2020; Seliger, 2019). A recent study has identified 11 new protein receptors of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) capsid spike protein in addition to ACE2, providing new insight into critical virus–host interactions, tropisms and SARS-CoV-2 pathogenesis (Gu et al., 2020). Moreover, due to their accessibility to systematically delivered therapeutics, L–R interactions are tractable drug and vaccine targets (Uhlén et al., 2015; Wishart, 2006).
Recently, with the rapid development of single-cell RNA sequencing (scRNA-seq) technologies, systematically decoding the intercellular communication network mediated by L–R protein–protein interactions (PPIs) has become a focus of research (Popescu et al., 2019; Sharma et al., 2020; Vento-Tormo et al., 2018; Wang et al., 2018; Zhang et al., 2019). Several algorithms have been developed to decipher cell–cell communications and facilitate intercellular communication analysis, which has become an essential bioinformatics pipeline for scRNA-seq processing (Browaeys et al., 2020; Cabello-Aguilar et al., 2020; Efremova et al., 2020). For example, Zhang et al. implicated the regulation of lymphocytes by LAMP3+ dendritic cells in the immune microenvironment of hepatocellular carcinoma via intercellular communication analysis (Zhang et al., 2019). Some regulatory interactions between NK cells and other cells at the maternal-fetal interface have been identified that can prevent harmful innate or adaptive immune responses (Vento-Tormo et al., 2018). Hepatocyte-derived VEGFA can activate PLVAP in tumor endothelial cells and likely promote onco-fetal reprogramming of the tumor immune microenvironment (Sharma et al., 2020). Given the above findings, construction of a comprehensive, high-confidence and well-organized resource of L–R interactions for research on the functional effects of cell–cell communications and acceleration of the development of related algorithms would be of great value.
In this study, we developed Cellinker (http://www.rna-society.org/cellinker/), a platform of manually curated L–R interactions for intercellular communication analysis. We aimed to provide a useful platform for studies on cell–cell communication mediated by L–R interactions. The current version of Cellinker documents more than 3700 human and 3200 mouse L–R PPIs, as well as over 400 endogenous small molecule (sMOL) related L–R interactions Cellinker also includes a webserver for intercellular communication analysis based on scRNA-seq data. Moreover, to aid research on CoV infection, Cellinker contains information on 16L–R PPIs involved in CoV–human interactions (including 12L–R PPIs involved in SARS-CoV-2 infection). In summary, Cellinker provides a user-friendly interface for querying and visualizing L–R interactions and is a practical and convenient platform with which researchers can explore intercellular communications based on scRNA-seq data.
2 Materials and methods
2.1 Data collection
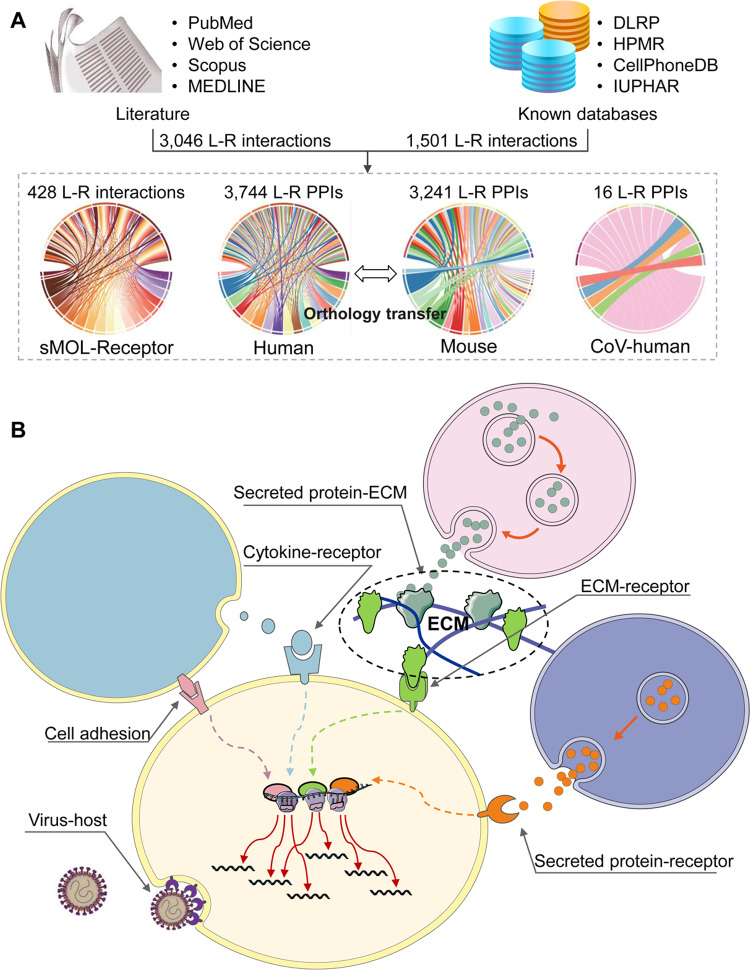
As shown in Figure 1A, the L–R interactions in Cellinker were manually curated from the literature (before July 2020) and four other known databases, including the Database of Ligand–Receptor Partners (DLRP) (Graeber and Eisenberg, 2001), the Human Plasma Membrane Receptome (HPMR) (Ben-Shlomo et al., 2003), International Union of Basic and Clinical Pharmacology (IUPHAR)/British Pharmacological Society (BPS) Guide to Pharmacology (Armstrong et al., 2020) and CellPhoneDB (Efremova et al., 2020) (only literature-supported data were collected). For curation, we retrieved literature from PubMed, Web of Science, Scopus and MEDLINE using the following keywords: ‘ligand’, ‘receptor’, ‘signal transduction’, ‘extracellular signal’, ‘cell communication’, ‘cell-surface protein’, ‘secreted proteins’, ‘extracellular matrix’, and ‘ligand–receptor interaction’. A total of 3046 experimental supported L–R interactions were retrieved from 1376 publications. We also obtained 1501L–R interactions from known databases, including 359 from the DLRP, 713 from the HPMR, 744 from the IUPHAR/BPS Guide to Pharmacology database (including 316 PPIs and 428 sMOL related L–R interactions), 349 from CellPhoneDB. The endogenous sMOLs were divided into five types: inorganic, metabolite, natural product, peptide and synthetic organic. Moreover, to aid research on CoV infection, information on 16L–R interactions involved in CoV–human interactions was collected.
Fig. 1.
Collection, processing and organization of the L–R interaction data. (A) Schematic of the database architecture. (B) Schematic diagram of the categories of L–R interactions
2.2 Transfer of L–R interaction orthology information
The collected L–R PPIs were mapped to two species (human and mouse) by an orthology majority-voting scheme (Li et al., 2017). The orthology information for the interactions in Cellinker was transferred from four orthology resources, Ensembl (Yates et al., 2020), HomoloGene (Sayers et al., 2019), the KEGG (Kanehisa et al., 2017) and the eggNOG database (Huerta-Cepas et al., 2019), on the basis of the following voting scheme: orthology information were transferred if two or more databases agreed on the orthology assignment. Ultimately, 3744 human and 3241 mouse L–R PPIs were identified in Cellinker (see Fig. 1A).
2.3 Annotation and organization
To unify the ligand and receptor information from multiple resources in an authoritative reference database, the ligand/receptor proteins were mapped to the NCBI gene database (Entrez ID) (Sayers et al., 2019) and UniProt (Consortium, 2019). The sMOL ligands were mapping to NCBI PubChem database (PubChem SID and CID) (Kim et al., 2021). The subcellular localization information (cell membrane, secreted or ECM) of ligand/receptor proteins was manually curated from the literature, the KEGG pathway database and UniProt (Consortium, 2019). Human and mouse gene expression data across different tissues were collected from The Human Protein Atlas (HPA) project (62 human tissues) (Uhlén et al., 2015) and the TISSUES 2.0 database (39 mouse tissues) (Palasca et al., 2018), respectively. Moreover, we took into account ligand/receptor complexes in L–R interactions and collected information on such complexes from the KEGG pathway database and the literature for inclusion in Cellinker.
After careful consideration of common perspectives from multiple review articles (Günther et al., 2018; Guryanov et al., 2016; Long et al., 2006; Sanes and Zipursky, 2020; Wright, 2009), the known catalogue in the KEGG pathway database (category: signaling molecules and interaction) and the subcellular localization of the ligands/receptors, we divided all the L–R PPIs into five categories: ‘secreted protein-to-receptor interaction’, ‘cytokine-to-cytokine receptor interaction’, ‘ECM-receptor interaction’, ‘secreted protein-to-ECM interaction’ and ‘cell adhesion’ (see Fig. 1B).
2.4 Pipeline for intercellular communication analysis
Cellinker features a webserver for intercellular communication analysis based on the expression of ligands/receptors. However, an expression value cannot exactly represent the specificity of a ligand/receptor; in other words, a weakly expressed but specific ligand/receptor in a cell is more valuable than a strongly expressed but less specific ligand/receptor in that cell type. To resolve this inadequate representation, we used term frequency-inverse document frequency (TF-IDF) transformation to improve the expression specificity of ligands/receptors across the expression matrix (Pliner et al., 2019; Sparck Jones, 1972). For M, an m by n matrix of input scRNA-seq data, the formula is as follows:
| (1) |
where ei, j is the expression value of gene i in cell j from M.
To infer the cell–cell communications between different cell types, the LRscore is defined as the score of an L–R interaction k between cell types i and j, which is evaluated by the expression of ligand and receptor. The formula is as follows:
| (2) |
where Li, k is the expression value of the ligand in cell i and Ri, k is the expression value of the receptor in cell j. The ligand and receptor are from the L–R interaction k.
If the ligand is a complex containing n subunits, L is defined as the geometric mean of the expression value of all subunits:
| (3) |
where lg is the expression value of subunit g in the ligand complex.
Similar to the case for ligands, if a receptor is a complex containing n subunits, R is defined as the geometric mean of the expression value of all subunits:
| (4) |
where rh is the expression value of subunit h in the receptor complex.
To examine the statistical significance of the LRscore, the P value was estimated by permutation test (by permuted the cell labels). Moreover, receptors/ligands that were expressed in less than N% (default: N = 25) of the cells of a certain cell type were removed before the intercellular communication analysis.
2.5 Architecture
Cellinker is implemented using the HTML and PHP languages with the MySQL server. The interface component consists of web pages designed and implemented in HTML/CSS. It has been tested in the Google Chrome, Firefox and Internet Explorer web browsers.
3 Results
3.1 Statistics of L–R interactions
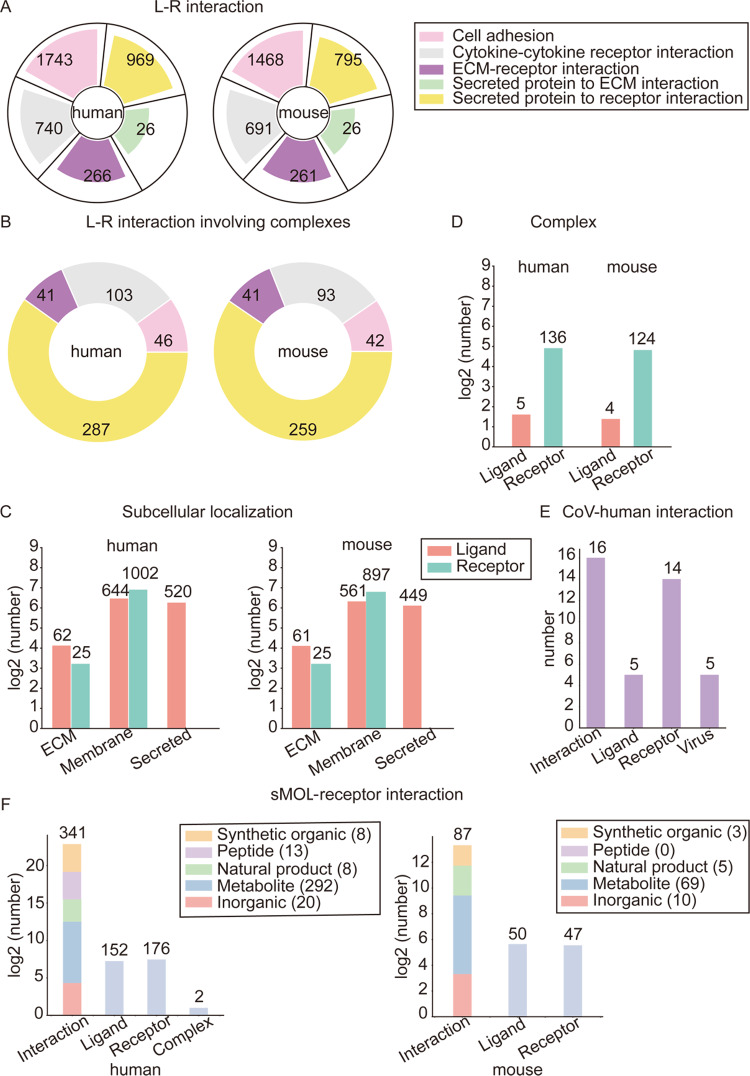
Cellinker documents 3744 human L–R PPIs (including 477 ligand/receptor complex-related interactions) and 3241 mouse L–R PPIs (including 435 ligand/receptor complex-related interactions). In addition, 1139 human and 987 mouse L–R PPIs can be annotated into specific KEGG pathways. The category distributions of the L–R PPIs are shown in Figure 2A and B. For humans, there are 1743 cell adhesion interactions (including 46 ligand/receptor complex-related interactions), 969 secreted protein-to-receptor interactions (including 287 ligand/receptor complex-related interactions), 740 cytokine-to-cytokine receptor interactions (including 103 ligand/receptor complex-related interactions), 266 ECM-to-receptor interactions (including 41 ligand/receptor complex-related interactions) and 26 secreted protein-to-ECM interactions (with no ligand/receptor complex-related interactions). For mice, there are 1468 cell adhesion interactions (including 42 ligand/receptor complex-related interactions), 795 secreted protein-to-receptor interactions (including 259 ligand/receptor complex-related interactions), 691 cytokine-to-cytokine receptor interactions (including 93 ligand/receptor complex-related interactions), 261 ECM-to-receptor interactions (including 41 ligand/receptor complex-related interactions) and 26 secreted protein-to-ECM interactions (with no ligand/receptor complex-related interactions). The subcellular localization distributions of the ligand/receptor proteins are shown in Figure 2C. Most of the ligand proteins are membrane proteins (human: 644, mouse: 5561) or secreted proteins (human: 520, mouse: 449), and most of the receptor proteins are membrane proteins (human: 1002, mouse: 897). In addition, 141 complexes (including 5 ligand complexes and 136 receptor complexes) are involved in human L–R PPIs, and 128 complexes (including 4 ligand complexes and 124 receptor complexes) are involved in mouse L–R PPIs (see Fig. 2D). Meanwhile, Cellinker documents 16 CoV–human interactions involving 5 ligands, 14 receptors (four of which are complexes) and 5 CoVs (see Fig. 2E). Moreover, Cellinker collects 341 human sMOL related L–R interactions (refers to 152 sMOL ligands and 176 receptors) and 87 mouse sMOL related L–R interactions (refers to 50 sMOL ligands and 47 receptors) (see Fig. 2F).
Fig. 2.
Statistical data for Cellinker. (A) Category distributions of L–R interactions (in humans and mice). (B) Category distributions of L–R interactions involving complexes (in humans and mice). (C) Subcellular localization distributions of ligands/receptors (in humans and mice). (D) Complex distribution of ligands and receptors (in humans and mice). (E) Statistics for CoV–human interactions. (F) Statistics for human and mouse sMOL-receptor interactions
3.2 Data querying and result presentation
To make it convenient for users to query and browse data, Cellinker provides two different search methods on the Search page (see Fig. S1), including ‘Exact Search’ (which requires users to input the gene symbol/Entrez ID of the ligand/receptor) and ‘Batch Search’ (which requires users to input or upload a list of the gene symbols/Entrez IDs of ligands/receptors). A brief summary of the search results is presented in a table on the Result page. In addition, to help users interactively view L–R interactions and their associated subcellular locations, Cellinker provides an embedded Sankey plot web tool on the Result page. Users can highlight interactions of interest by moving the cursor over the diagram. Detailed information on certain L–R interactions can be viewed on the Detail page by clicking ‘more’. The Detail page presents more information (see Fig. S1), including basic information on the L–R interaction (including the interaction type, species, KEGG pathway annotation, etc.), basic information on the ligand and receptor (including the gene symbols, gene IDs, UniProt IDs and subcellular locations), expression information for the ligand and receptor across different tissues, and references for the L–R interaction (including literature and/or known databases). Moreover, if the ligand and/or receptor is a complex, information on the complex and the corresponding subunits (the gene symbols, gene IDs and UniProt ID of all subunits) is also presented on the Detail page. Moreover, Cellinker provided an independent webpage for querying, browsing and visualizing detailed information about the sMOL–receptor interactions and CoV–human interaction, respectively.
3.3 Webserver for intercellular communication analysis
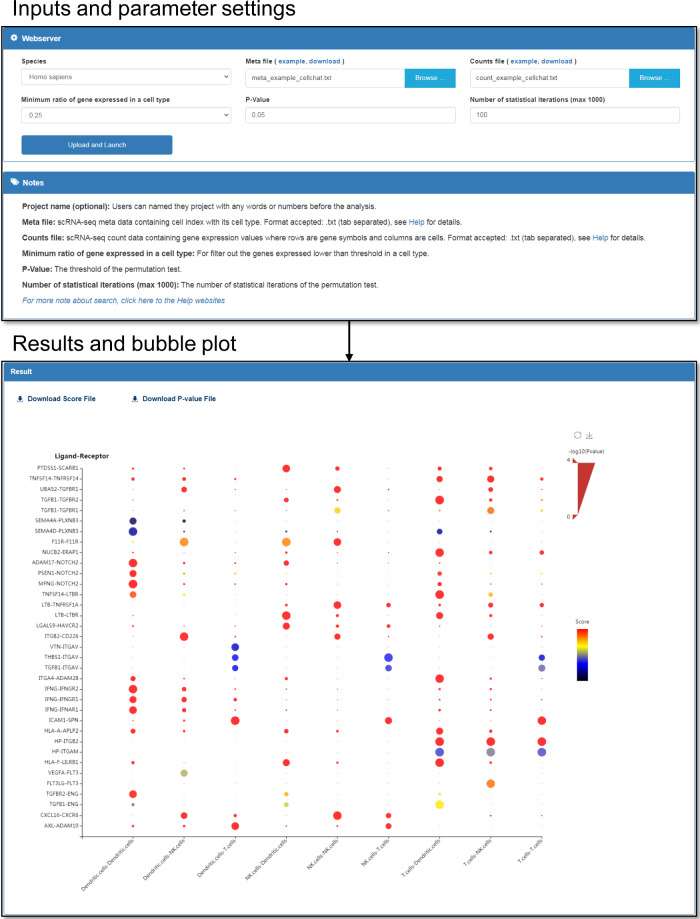
For user convenience, Cellinker launches a webserver for intercellular communication analysis based on scRNA-seq data (Fig. 3). First, users can upload scRNA-seq data in the proper format: (1) an META file containing the cell index with its cell type and (2) an expression file containing gene expression values (TPM values/counts), where the rows are the gene symbols and the columns are the cells. Then, users can specify an ‘N’ to filter out the receptors/ligands expressed in less than N% of cells of a certain cell type. Users can also determine the threshold of the p value and the number of statistical iterations for the permutation test. When the analysis is complete, the results are presented as a bubble plot on the Result page. The color of each bubble represents the LRscore, and the size of the bubble represents the significance of the LRscore. The results table containing LRscore values and p values can be downloaded from the Result page.
Fig. 3.
Screenshot of webserver for intercellular communication analysis. Users could upload scRNA-seq data with proper format and parameter setting to launch the analysis. The results are presented as a bubble plot in the result page
3.4 Comparison of Cellinker with other L–R databases
We compared Cellinker with other literature-supported databases/datasets including CellPhoneDB (Efremova et al., 2020), SingleCellSignalR (Cabello-Aguilar et al., 2020), CelltalkDB (Shao et al., 2020) and L–R interaction dataset collected from Ramilowski et al. (Ramilowski et al., 2015) (see Table 1). For data collection, Cellinker collected the most L–R PPIs from literatures and databases, and only Cellinker and CelltalkDB provided mouse L–R PPIs. Meanwhile, only Cellinker and CellPhoneDB take account into the ligand/receptor complex, which should not be ignored because many ligands/receptors act as multisubunit complexes. For data annotation, Cellinker provides subcellular location and gene expression across tissues of ligands/receptors, and divided all the L–R interactions into five categories. These annotations and organization offered important references to cell–cell communication analysis. Moreover, Cellinker and CellPhoneDB provide a web tool, and SingleCellSignalR provides a R package for exploration of intercellular communications based on scRNA-seq data, respectively. Besides, Cellinker also documents 341 human and 87 mouse sMOL related L–R interactions. In summary, compared with other L–R databases, Cellinker collected the most L–R interactions for both human and mouse, and provided well-organized data with more varied and valuable annotations.
Table 1.
Comparison of Cellinker with other L–R databases
| Cellinker | CellPhoneDB | SingleCellSignalR | CelltalkDB | Ramilowski et al. | ||
|---|---|---|---|---|---|---|
| Data collection | Human L–R PPI | 3744 | 1396 | 3251 | 3398 | 1894 |
| Mouse L–R PPI | 3241 | No | No | 2021 | No | |
| Human sMOL-receptor L–R interaction | 341 | No | No | No | No | |
| Mouse sMOL-receptor L–R interaction | 87 | No | No | No | No | |
| Data source | Literatures + databases | Literatures + databases | Literatures + databases | Literatures | Databases | |
| Complexes | Yes | Yes | No | No | No | |
| Data annotation | Classification | Yes | No | No | No | No |
| Subcellular location | Yes | No | No | No | No | |
| Gene expression across tissues | Yes | No | No | No | No | |
| Tool | Inferring intercellular communication | Yes | Yes | Yes | No | No |
4 Discussion
Cell-to-cell communication in multicellular organisms plays essential roles in various biological processes that extensively rely on interactions of extracellular/surface molecules (such as proteins/peptides, RNA molecules and metabolites) between cells, such as the immune response, development and viral infection (Huang et al., 2021; Husted et al., 2017; Lin et al., 2020; Pires-daSilva and Sommer, 2003; Ramilowski et al., 2015; Zhang et al., 2019). Therefore, investigation of intercellular communications can facilitate understanding of the dynamics, mechanisms and effects of signal transmission between cells (Özkan et al., 2013). Notably, the rapid development of scRNA-seq technologies has provided an excellent foundation for systematic deciphering of intercellular communication networks based on the cell-surface protein interactome. Therefore, construction of a comprehensive and high-confidence resource of L–R interactions for research on the functional effects of cell–cell communications will be of great value. Here, we developed Cellinker, a manually curated resource of L–R interactions involved in cell–cell communication, and provide a practical and convenient platform with which researchers can explore intercellular communications based on scRNA-seq data.
First, Cellinker documents over 3700 human and 3200 mouse L–R PPIs with high-confidence. Most L–R interactions are curated from the peer-reviewed literature. And it is larger than other literature-supported databases [CellPhoneDB (Efremova et al., 2020), SingleCellSignalR (Cabello-Aguilar et al., 2020) and CelltalkDB (Shao et al., 2020)]. Moreover, Cellinker takes into account ligand/receptor complexes in L–R interactions; over 400 human and mouse ligand/receptor complex-related interactions are recorded in Cellinker. Second, for user convenience, Cellinker launches a webserver for exploration of intercellular communications based on scRNA-seq data. The algorithm is based on the expression of the ligand/receptor (also taking into account ligand/receptor complexes) combined with a statistical significance test. In addition, the TF-IDF transformation is used to improve the expression specificity of ligands/receptors across the expression matrix. Third, Cellinker also documents 341 human and 87 mouse sMOL related L–R interactions. Forth, Cellinker provides an independent webpage for querying, browsing and visualizing detailed information about the 16L–R interactions involved in CoV–human interactions (including 12 SARS-CoV-2-related L–R interactions); thus, it may be a useful resource for research on critical virus–host interactions, tropisms and viral infection pathogeneses.
5 Conclusion
Cellinker is a platform of L–R interactions involved in cell–cell communication that provides a practical and convenient webserver with which researchers can explore intercellular communications based on scRNA-seq data. Meanwhile, Cellinker documents 341 human and 87 mouse sMOL related L–R interactions. Moreover, to aid research on CoV infection, Cellinker also contains information on 16L–R interactions involved in CoV–human interactions. In summary, we believe this platform will promote intercellular communication research and accelerate the development of related algorithms for scRNA-seq data.
Supplementary Material
Acknowledgement
We would like to thank the anonymous reviewers for valuable suggestions.
Funding
This work has been supported by the National Key Research and Development Project of China [2019YFA0801800]; National Natural Science Foundation of China [82070109, 81770104, 62002153]; Basic and Applied Basic Research Fund of Guangdong Province [2019A1515010784, 2019A1515110701]; China Postdoctoral Science Foundation [2020M682623, 2020M682785]; Guangzhou science and technology project key project topic (201904020031).
Conflict of Interest: none declared.
References
- Armstrong J.F. et al. (2020) The IUPHAR/BPS guide to PHARMACOLOGY in 2020: extending immunopharmacology content and introducing the IUPHAR/MMV guide to MALARIA PHARMACOLOGY. Nucleic Acids Res., 48, D1006–D1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shlomo I. et al. (2003) Signaling receptome: a genomic and evolutionary perspective of plasma membrane receptors involved in signal transduction. Science's STKE, 2003, Re9. [DOI] [PubMed] [Google Scholar]
- Boisset J.-C. et al. (2018) Mapping the physical network of cellular interactions. Nat. Methods, 15, 547–553. [DOI] [PubMed] [Google Scholar]
- Browaeys R. et al. (2020) NicheNet: modeling intercellular communication by linking ligands to target genes. Nat. Methods, 17, 159–162. [DOI] [PubMed] [Google Scholar]
- Cabello-Aguilar S. et al. (2020) SingleCellSignalR: inference of intercellular networks from single-cell transcriptomics. Nucleic Acids Res., 48, e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium T.U. (2019) UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res., 47, D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douam F. et al. (2015) Genetic dissection of the host tropism of human-tropic pathogens. Annu. Rev. Genet., 49, 21–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efremova M. et al. (2020) CellPhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes. Nat. Protoc., 15, 1484–1506. [DOI] [PubMed] [Google Scholar]
- Fonseca A.L. et al. (2016) Bioinformatics analysis of the human surfaceome reveals new targets for a variety of tumor types. Int. J. Genomics, 2016, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil N. et al. (2020) Discovery of receptor-ligand interfaces in the immunoglobulin superfamily. Proteins 88, 135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeber T.G., Eisenberg D. (2001) Bioinformatic identification of potential autocrine signaling loops in cancers from gene expression profiles. Nat. Genet., 29, 295–300. [DOI] [PubMed] [Google Scholar]
- Gu Y. et al. (2020) Interaction network of SARS-CoV-2 with host receptome through spike protein. bioRxiv. 10.1101/2020.09.09.287508. [DOI] [Google Scholar]
- Günther T. et al. (2018) International union of basic and clinical pharmacology. CV. somatostatin receptors: structure, function, ligands, and new nomenclature. Pharmacol. Rev., 70, 763–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guryanov I. et al. (2016) Receptor-ligand interactions: advanced biomedical applications. Mater. Sci. Eng. C, 68, 890–903. [DOI] [PubMed] [Google Scholar]
- Honig B., Shapiro L. (2020) Adhesion protein structure, molecular affinities, and principles of cell-cell recognition. Cell, 181, 520–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. et al. (2021) cncRNAdb: a manually curated resource of experimentally supported RNAs with both protein-coding and noncoding function. Nucleic Acids Res., 49, D65–D70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J. et al. (2019) eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res., 47, D309–D314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain B. et al. (2019) A platform for extracellular interactome discovery identifies novel functional binding partners for the immune receptors B7-H3/CD276 and PVR/CD155. Mol. Cell. Proteomics, 18, 2310–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husted A.S. et al. (2017) GPCR-mediated signaling of metabolites. Cell Metab., 25, 777–796. [DOI] [PubMed] [Google Scholar]
- Kanehisa M. et al. (2017) KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res., 45, D353–D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. et al. (2021) PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res., 49, D1388–D1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Q. et al. (2020) Resistance mechanisms of anti-PD1/PDL1 therapy in solid tumors. Front. Cell Dev. Biol., 8, 672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T. et al. (2017) A scored human protein-protein interaction network to catalyze genomic interpretation. Nat. Methods, 14, 61–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. et al. (2015) ViRBase: a resource for virus-host ncRNA-associated interactions. Nucleic Acids Res., 43, D578–D582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. et al. (2020) RNAInter in 2020: RNA interactome repository with increased coverage and annotation. Nucleic Acids Res., 48, D189–D197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M. et al. (2006) Kinetics of receptor-ligand interactions in immune responses. Cell. Mol. Immunol., 3, 79–86. [PubMed] [Google Scholar]
- Massagué J., Obenauf A.C. (2016) Metastatic colonization by circulating tumour cells. Nature, 529, 298–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning L. et al. (2021) MNDR v3.0: mammal ncRNA-disease repository with increased coverage and annotation. Nucleic Acids Res., 49, D160–D164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özkan E. et al. (2013) An extracellular interactome of immunoglobulin and LRR proteins reveals receptor-ligand networks. Cell, 154, 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palasca O. et al. (2018) TISSUES 2.0: an integrative web resource on mammalian tissue expression. Database, 2018, bay003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires-daSilva A., Sommer R.J. (2003) The evolution of signalling pathways in animal development. Nat. Rev. Genet., 4, 39–49. [DOI] [PubMed] [Google Scholar]
- Pliner H.A. et al. (2019) Supervised classification enables rapid annotation of cell atlases. Nat. Methods, 16, 983–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu D.-M. et al. (2019) Decoding human fetal liver haematopoiesis. Nature, 574, 365–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramilowski J.A. et al. (2015) A draft network of ligand–receptor-mediated multicellular signalling in human. Nat. Commun., 6, 7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes J.R., Zipursky S.L. (2020) Synaptic specificity, recognition molecules, and assembly of neural circuits. Cell, 181, 536–556. [DOI] [PubMed] [Google Scholar]
- Sayers E.W. et al. (2019) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res., 47, D23–D28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scadden D.T. (2014) Nice neighborhood: emerging concepts of the stem cell niche. Cell, 157, 41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seliger B. (2019) Basis of PD1/PD-L1 therapies. J. Clin. Med., 8, 2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao X. et al. (2020) CellTalkDB: a manually curated database of ligand–receptor interactions in humans and mice. Brief. Bioinform. 10.1093/bib/bbaa269. [DOI] [PubMed] [Google Scholar]
- Sharma A. et al. (2020) Onco-fetal reprogramming of endothelial cells drives immunosuppressive macrophages in hepatocellular carcinoma. Cell, 183, 377–394.e21. [DOI] [PubMed] [Google Scholar]
- Sparck Jones K. (1972) A statistical interpretation of term specificity and its application in retrieval. J. Doc., 28, 11–21. [Google Scholar]
- Uhlén M. et al. (2015) Tissue-based map of the human proteome. Science, 347, 1260419. [DOI] [PubMed] [Google Scholar]
- Vento-Tormo R. et al. (2018) Single-cell reconstruction of the early maternal–fetal interface in humans. Nature, 563, 347–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verschueren E. et al. (2020) The immunoglobulin superfamily receptome defines cancer-relevant networks associated with clinical outcome. Cell, 182, 329–344.e319. [DOI] [PubMed] [Google Scholar]
- Wang M. et al. (2018) Single-cell RNA sequencing analysis reveals sequential cell fate transition during human spermatogenesis. Cell Stem Cell, 23, 599–614.e594. [DOI] [PubMed] [Google Scholar]
- Wishart D.S. (2006) DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res., 34, D668–D672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtowicz W.M. et al. (2020) A human IgSF cell-surface interactome reveals a complex network of protein-protein interactions. Cell, 182, 1027–1043.e1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood L., Wright G.J. (2019) Approaches to identify extracellular receptor–ligand interactions. Curr. Opin. Struct. Biol., 56, 28–36. [DOI] [PubMed] [Google Scholar]
- Wright G.J. (2009) Signal initiation in biological systems: the properties and detection of transient extracellularprotein interactions. Mol. BioSyst., 5, 1405–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates A.D. et al. (2020) Ensembl 2020. Nucleic Acids Res., 48, D682–D688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q. et al. (2019) Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell, 179, 829–845.e820. [DOI] [PubMed] [Google Scholar]
- Zhang Y. et al. (2019) RIscoper: a tool for RNA-RNA interaction extraction from the literature. Bioinformatics, 35, 3199–3202. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.