Abstract
Viruses are responsible for causing various epidemics and pandemics with a high mortality rate e.g. ongoing SARS-CoronaVirus-2 crisis. The discovery of novel antivirals remains a challenge but drug repurposing is emerging as a potential solution to develop antivirals in a cost-effective manner. In this regard, we collated the information of repurposed drugs tested for antiviral activity from literature and presented it in the form of a user-friendly web server named ‘DrugRepV’. The database contains 8485 entries (3448 unique) with biological, chemical, clinical and structural information of 23 viruses responsible to cause epidemics/pandemics. The database harbors browse and search options to explore the repurposed drug entries. The data can be explored by some important fields like drugs, viruses, drug targets, clinical trials, assays, etc. For summarizing the data, we provide overall statistics of the repurposed candidates. To make the database more informative, it is hyperlinked to various external repositories like DrugBank, PubChem, NCBI-Taxonomy, Clinicaltrials.gov, World Health Organization and many more. ‘DrugRepV’ database (https://bioinfo.imtech.res.in/manojk/drugrepv/) would be highly useful to the research community working to develop antivirals.
Keywords: viruses, drug repurposing, pandemics/epidemics, web server, SARS-CoV-2
Introduction
Identification of the promising antiviral agents always remains a challenge. The development of antivirals started more than 50 years ago [1]. In 1967, the description of DNA dependent RNA polymerase for pox virus was the first viral enzyme to be discovered, which led to the discovery of antiviral drugs [2]. The major concerns in the development of a novel drug are high costs, time, mutation rates, etc. [3]. However, the periodic emergence of viral epidemics and pandemics, poses serious challenges for the speedy development of novel and effective antivirals [4]. Thus, there is a need for parallel drug discovery rate as per the emergence and reemergence of viral infections [5]. Since the last decade, the drug repurposing approach shows promising results in the identification of antivirals.
Drug repurposing (drug repositioning or retasking or reprofiling) is the investigation of existing approved or investigational drugs for new medical indications [6]. The drug repurposing strategy is highly advantageous, as it has lower failure rate because it has been already tested safe in the preclinical trials; reduced time frame as most of the preclinical trials and safety assessment were already tested; and economical due to skipped clinical trial phases [7]. The first successful repurposed antiviral drug is zidovudine for human immunodeficiency virus (HIV) whose original indication was anticancer [8]. Numerous drugs like hydroxychloroquine, remdesivir, dexamethasone, etc. have been tested as potential candidates to fight COVID-19 [9, 10]. For Zika virus (ZIKV), the drugs like ribavirin and sofosbuvir were previously used to treat Hepatitis C virus (HCV) infections [11]. The Coronaviridae family viruses like severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) have been treated with Chlorpromazine, an antipsychotic drug [12]. For the dengue virus (DENV), the prochlorperazine (a previously used antiemetic drug) has been successfully repurposed [13]. Thus, drug repurposing emerged as a fast and successful approach, as a solution to target various viral diseases.
Till date, no comprehensive manually curated database dedicated for the viruses has not been reported in the literature. However, few databases are available consisting of other information for general repurposed drugs. The RepoDrug database provides information about the repurposing of only approved drugs from only Clinicaltrial.gov and DrugCentral [14]. The PROMISCUOUS database consisted of the drug-protein interaction of repurposing drug data [15]. However, we have developed a ‘DrugRepV’ database with various characteristics, which make it unique from the rest of the databases. Thus, in the current scenario of the virus pandemic, this database would be very helpful for the researchers to speed up the research to identify promising repurposed drug candidates.
The computational interventions can further reduce the cost and time of the drug repurposing that facilitates researchers to monitor drug candidates and test for the variety of diseases within a short period of time [16–18]. In this endeavor, we have developed ‘DrugRepV’, the first dedicated one-stop solution for the repurposed drugs against epidemics and pandemics viruses like SARS Coronavirus-2 (SARS-CoV-2), ZIKV, Ebola (EBV), DENV, Nipah (NiV), influenza (IFV), chikungunya (CHIKV), Lassa virus (LASV), Crimean-Congo hemorrhagic fever virus (CCHV), enterovirus (EV), HIV, SARS, MERS, Hendra virus (HeV), Japanese encephalitis virus (JEV), Marburg virus (MARV), Measles virus (MV), polio virus (PV), Rift Valley fever virus (RVFV), Vaccinia virus (VACV), Variola virus (VARV), West Nile virus (WNV) and Yellow fever virus (YFV). DrugRepV is a complete package of repurposed drugs against viruses. It contains information of biological, chemical, clinical and structural details of each repurposed drug.
Material and methods
Data collation
We build the search query ‘(((virus) OR viral)) AND (((repurpos*) OR reposition*))’ in the advanced search option of the PubMed database and got 1210 papers. Then, we filter out papers for only those viruses that are listed as epidemic/pandemic diseases by World Health Organization (WHO) (https://www.who.int/emergencies/diseases/en/). These 23 viruses are SARS-CoV-2, ZIKV, EBV, DENV, NiV, IFV, CHIKV, HIV, SARS, MERS, CCHV, EV, HeV, JEV, LASV, MARV, MV, PV, RVFV, VACV, VARV, WNV and YFV. This leads to the shortlisting of 986 articles. Further, we started scanning the 986 articles for relevant data but still there are articles, which do not have the required information as per our database instead describing other aspects like a drug synthesis, in silico studies, etc. In addition, we also excluded reviews, non-journal papers, books, documents, etc. Finally, we extracted 8485 entries for 23 viruses from the 351 research articles having the relevant data for the development of the DrugRepV resource.
Database enrichment
The database was made highly informative by cross-linking each entry with their respective biological, chemical, structural and clinical information from various repositories.
Biological information
It includes the information manually extracted from the articles like primary indication, secondary indication, assay used, the concentration of the antiviral agent, type of inhibition, cytotoxicity and many more.
Chemical information
The details like IUPAC, canonical and isomeric SMILES, molecular weight, molecular formula, InChl, common name and synonyms were extracted from various sources like PubChem, ChEMBL, DrugBank and ChemSpider. However, various chemicals were drawn using Chemicalize or Marvin Sketch from scratch followed by the extraction of all other information. All the entries are hyperlinked to PubChem, ChEMBL and DrugBank for making the database more informative [19, 20].
Structural information
The structural details are an important component of the chemical agents. Thus, we calculated the 2D and 3D sdf format for every agent. Further, both the 2D and 3D structures were displayed on the web server using mol2ps (http://merian.pch.univie.ac.at/~nhaider/cheminf/mol2ps.html) and Jmol from JavaScript-based molecular viewer (JSmol) (http://jmol.sourceforge.net/), respectively.
Clinical information
It incorporates details like the category of drug, clinical trials phases, hyperlink to clinicaltrails.gov, WHO and Centers for Disease Control and Prevention (CDC) websites, etc.
Database statistics
We have summarized the important fields of the database in the form of various user-friendly plots. The fields like antiviral agents, viruses, drug target, disease category of antiviral agents, taxonomy and drug categories are displayed. We also provided the network-based statistics of the highly effective antivirals according to the families. The summarized plots and networks would be helpful for getting valuable insight into the repurposed drugs against viral diseases.
Web server development
The DrugRepV web server is designed in a user-friendly manner. It contains browse, search, statistics, manual, frequently asked questions and contact web pages. The manual/help page is well documented to address queries of the users and explain the usage of each component of the server. The DrugRepV web server was made functional using the LAMPP package. The front end is developed using PHP, Javascript, CSS, XML, etc., whereas the backend is comprehended using MySQL, Python, Perl, etc. [19, 21, 22].
Results
Database architecture
DrugRepV is a comprehensive repository with 8485 entries, which include multi-variant information like biological, chemical, clinical and structural. The web server includes extensive browse and search options. The overall architecture of DrugRepV is shown in Figure 1.
Figure 1.
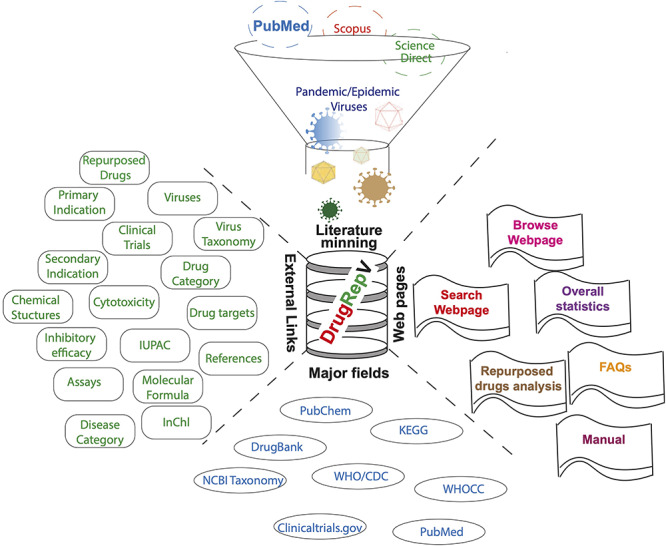
DrugRepV architecture.
Repurposed drugs analysis
We have compiled the antiviral drugs of 23 viruses responsible to cause pandemic/epidemic from literature. Further, we analyzed the data to check the trend of the research done in antiviral discovery for viruses. We scanned the important fields like antiviral drugs, viruses, drug targets, drug categories and taxonomy. Further, we also perform the network-based analysis for the virus families.
Repurposed drugs
Among all the experimentally validated antiviral drugs we found that ribavirin utilized in majorly and consisted of 102 entries. Further, other drugs like chloroquine, favipiravir, mycophenolic acid, dolutegravir, raltegravir, elvitegravir and bictegravir have been checked in 92, 90, 89, 60, 60, 59 and 58 cases, respectively (Figure 2A). However, we also provided the summarized tabulated view of the repurposed drugs from the DrugRepV database. The table includes the information of the repurposed drug, primary indication, secondary indication, inhibition efficiency and references (Table 1).
Figure 2.
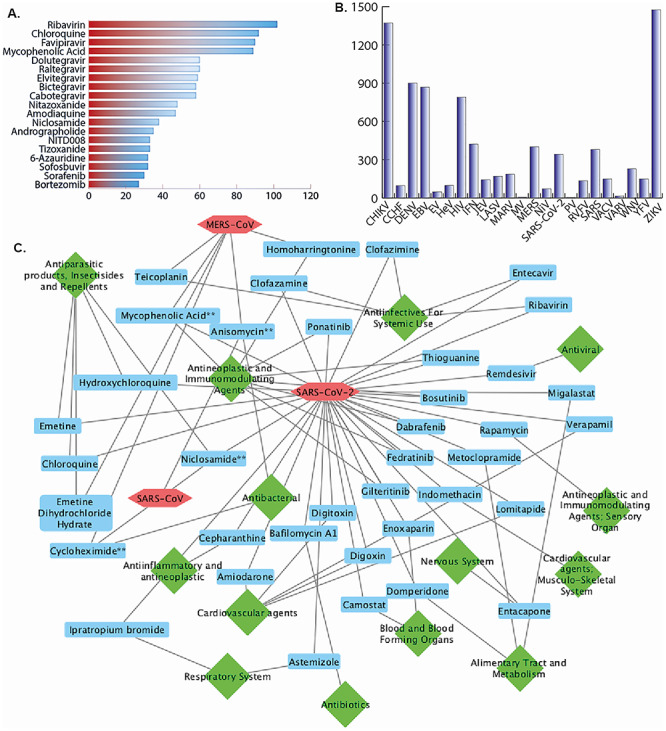
Data statistics of DrugRepV (A) repurposed drugs, (B) viruses, (C) interaction network showing the highly efficient repurposed drugs having < 1 μM IC50/EC50 among coronaviruses namely SARS, MERS and SARS-CoV-2. Hexagon shape (pastel red color) represents the virus, rectangle shape (light blue color) displays repurposed ‘drug’ and diamond shape (green color) shows drug-categories. Drugs tested for two viruses are denoted with ** sign.
Table 1.
Table showing the information of majorly used antiviral drugs with the information of primary indication, secondary indication, inhibition efficiency` and references
| Repurposed drugs | Primary indication | Repurposed indication | Inhibition efficacy | References |
|---|---|---|---|---|
| Ribavirin | HCV | CHIKV | EC50 = 2.5 ± 0.3 μM | [23] |
| CCHV | IC50 = 0.04 ± 0.01 mM | [24] | ||
| DENV | EC50 = 7.7 ± 0.6 μM | [25] | ||
| EBV | EC50 = 60.1 μM | [26] | ||
| MERS | EC50 = 9.99 ± 2.97 μM | [27] | ||
| SARS-CoV-2 | EC50 = 26.739 μg/ml | [28] | ||
| SARS | EC50= >100 μM | [29] | ||
| ZIKV | EC50 = 12 ± 6.6 μM | [30] | ||
| Chloroquine | Malaria | SARS-CoV-2 | EC50 = 5.47 μM | [31] |
| SARS | EC50 = 3.3 μM | [32] | ||
| MERS | EC50 = 3.0 μM | [32] | ||
| EBV | EC50 = 16 μM | [33] | ||
| CHIKV | EC50 = 10 ± 5 μM | [34] | ||
| Favipiravir | IFV | ZIKV | EC50 = 22 ± 15 μM | [30] |
| RVFV | EC50 = 5 μg/ml | [35] | ||
| SARS-CoV-2 | EC50 = 61.88 μM | [36] | ||
| NiV | EC50 = 14.57 μM | [37] | ||
| Mycophenolic Acid | Organ rejection (immunosuppressants) | SARS-CoV-2 | IC50 = 20 nM | [38] |
| WNV | IC50= > 100 μg/ml | [39] | ||
| MERS | EC50 = 0.17 ± 0.03 μM | [40] | ||
| EBV | IC50 = 0.96 μg/ml | [41] | ||
| LASV | IC50 = 0.34 μg/ml | [41] | ||
| Nitazoxanide | Diarrhea | IFV | EC50 = 1 μM | [42] |
| ZIKV | Log Reduction= > 100 fold | [43] | ||
| SARS-CoV-2 | EC50 = 2.12 μM | [44] | ||
| MERS | IC50 = 0.92 μg/ml | [45] | ||
| CHIKV | EC50 = 3.01 ± 0.61 μM | [46] | ||
| Amodiaquine | Malaria | SARS-CoV-2 | EC50 = 1832.7 mg/ml | [28] |
| ZIKV | EC50 = 3.07 ± 0.36 μM | [47] | ||
| MARV | IC50 = 2.3 μM | [48] | ||
| DENV | EC50 = 7.41 ± 1.09 μM | [49] |
Viruses
We have analyzed the viruses, against which the drugs have been repurposed. We found that most tested viruses are ZIKV, CHIKV, DENV, EBV, HIV, IFV, MERS, SARS and SARS-CoV-2 with 1475, 1371, 900, 868, 790, 422, 401, 380 and 342 cases (Figure 2B).
Drug targets
All the drug entries recorded in the literature were used to extract the targets. Topmost targets are P35367 (histamine H1 receptor); P35348 (alpha-1A adrenergic receptor); Q7ZJM1 (integrase); P14416 (D(2) dopamine receptor); P20839 (inosine-5′-monophosphate dehydrogenase 1); P12268 (inosine-5-monophosphate dehydrogenase 2); P28223 (5-hydroxytryptamine receptor 2A) and P35368 (slpha-1B adrenergic receptor) in 269; 251; 237; 228; 225; 217; 211 and 209 cases, respectively. Diagram showing the frequently used drug targets in repurposed antiviral drugs is shown in Supplementary Figure S1B.
Drug category
The drugs which have been entered in our database were grouped into several drug categories. The majority of the drugs were categorized as ‘antiinfectives for systemic use’ in 884 entries. However, other drugs fall in categories like ‘antineoplastic and immunomodulating agents’; ‘antiparasitic products, insecticides and repellents’; ‘nervous system’; ‘cardiovascular system’ and ‘antiviral’ with 688; 465; 461; 366 and 317, respectively. Frequently used drug categories are depicted as pie charts in Supplementary Figure S1C.
Taxonomy
The 23 viruses in the database were also explored on the basis of the taxonomy. Majorly explored viruses are from the Flaviviridae family with 2895 entries. Further, the viruses from Togaviridae, Coronaviridae, Filoviridae, Reteroviridae and Orthomyxoviridae family have 1371, 1123, 1054, 790 and 422 entries, respectively. The distribution of taxonomy is shown in Supplementary Figure S1A.
Network-based analysis
We constructed the network of the highly efficient drugs (IC50/EC50 < 1 μM) from the families of the 23 viruses. The network-based analysis of the Coronaviridae family viruses like SARS-CoV-2, SARS and MERS is shown in Figure 2C. To build a network, we have used drug, virus and drug categories as input in the Cytoscape software. We have used different shapes and colors to represent the virus (hexagon shape having Pastel Red color), drug (rectangle shape with light blue color) and drug-categories (diamond shape with green color). Further, we have used a single color for edges. We have developed graphs for those drugs whose efficacy (IC50/EC50) is less than 1 μM and excluded those whose drug category information is not given. Drugs tested for two or three viruses are denoted with ** and *** signs, respectively. For example, anisomycin and cycloheximide drugs are found in SARS and MERS. Mycophenolic acid is found in MERS and SARS-CoV-2. Niclosamide drug is found in SARS-CoV-2 and SARS as shown in Figure 2C. However, the network of the virus families like Filoviridae, Flaviviridae, Orthomyxoviridae, Retroviridae and Togaviridae are shown in Supplementary Figures S2–S6.
Web server
DrugRepV web server is the first dedicated repository for repurposed antivirals. It contains browse and search options for the exploration of data. To make the database more user-friendly, we provide help and frequently asked questions on web pages. To summarize the important fields of the data, we provide statistics web pages for the fields like drugs, drug categories, drug targets, viruses and disease categories. Freely available at: https://bioinfo.imtech.res.in/manojk/drugrepv/.
Browse
The database can be explored by the ‘browse’ option. The data can be browsed by fields like viruses, chemicals, drugs, drug targets, disease categories, clinical trials and PubMed. The ‘browse’ is displayed in two web pages. The first page provides information on a few important fields like DrugRepV IDs, drug name, drug type, primary indication, secondary indication, virus strain, pathways, assay, activity, clinical status and references. In the second page, each entry is expanded as biological, chemical, structural and clinical information. The diagrammatic representation of the ‘browse’ option is shown in Supplementary Figure S7.
The ‘biological information’ contains primary indication (disease and disease category), secondary indication (approaches, methods, model system, mode of viral infection, time and duration of drug delivery, drug concentration, inhibition type and references). The ‘chemical information’ consisted of IUPAC name, SMILES (canonical and isomeric), molecular formula, molecular weight, InChl, common names and Synonyms. The ‘structural information’ provides 2D and 3D structure of each drug. The ‘clinical information’ has clinical trial phases and links to clinicaltrials.gov.
Search
The ‘search’ page is used to explore the desired query. The entries can be searched by DrugRepV_ID, drug Name, the primary target, the secondary target, assay, diseased category and PubMed ID. The biological, chemical, structural and clinical information of each entry is being displayed for the query.
Hyperlinking with external databases
To make our database highly informative, we have hyperlinked it with various resources. Chemical information is linked to PubChem and ChemSpider, whereas the DrugBank provides the details of each drug. Further, for clinical status the entries are linked to Clinicaltrails.gov. The NCBI-Taxonomy browser was used for retrieving the taxonomy of viruses. However, WHO and CDC databases were linked to get more information about viruses. Finally, the PubMed repository was used for references.
Webpages
The DrugRepV database is made on the apache server in a Linux environment. The front end is linked to the server PHP, Javascript, Perl and HTML, whereas the back end the database was linked via MySQL (relational database).
Discussion
Viruses are responsible for various pandemics/epidemics with very high mortality rates. The designing of an effective antiviral remains a challenge for the scientific community. For the same, drug repurposing emerged as a powerful tool, which reduces both time and cost for developing successful antivirals [50, 51]. In the current study, we provide all the experimentally validated repurposed antivirals in the form of a repository named ‘DrugRepV’. On analyzing the viruses against which drugs were repurposed, we found that attempts were made almost against all the major viruses. But the repositioning seems to be utilized extensively against viruses responsible to cause epidemics and pandemics like SARS-CoV-2, SARS, MERS, ZIKV, DENV, EBV, CHIKV, IFV, etc. [50, 52–54].
Existing antiviral drugs are a preferred choice for repurposing against other viruses [55]. In this endeavor, the antiviral drugs ribavirin, Favipiravir and many more were repositioned against viruses [37, 56]. Ribavirin initially used to treat HCV [57], and viral haemorrhagic fevers has now been tested for ZIKV, RVFV, CHIKV, CCHV, MARV, NiV, MERS, IFV, EBV, LASV, DENV, SARS-CoV-2, HIV and WNV. It shows the good inhibition efficacy (IC50/EC50) of 0.02 μM and 0.1 μM for DENV and SARS-CoV-2, respectively. The Favipiravir with the trade name Fabiflu was used to treat IFV [58]. Later, it is checked for antiviral activity against NiV, IFV, ZIKV, CHIKV, DENV, HeV, MARV, CCHV, EBV, SARS-CoV-2, LASV, RVFV and YFV. Among them the Favipiravir shows high inhibition efficiency (IC50/EC50) of 1.03 μM and 1.9 μM against CCHV and CHIKV correspondingly. The Sofosbuvir drug with the trade name Sovaldi was originally used as an anti-HCV drug. However, it is also showing promising results against CHIKV, YFV and ZIKV with the inhibition efficiency (IC50/EC50) of <5 μM. Furthermore, the antiparasitic drug Chloroquine (antimalarial) also proved effective against SARS-CoV-2, SARS, MERS, EBV, CHIKV, etc. [31, 32]. However, Mycophenolic acid, an immunosuppressant, shows promising results against various epidemics and pandemics causing viruses like SARS-CoV-2, SARS, MERS, etc. [38, 39].
The understanding of the drug targets is important to increase the efficiency of drug repurposing approach. Thus, we identified the target of all the drugs reported in our database. Among all the drug targets, the histamine H1 receptors, Alpha-1A adrenergic receptor and integrase have been extensively used. The histamine (H1, H2, H3 and H4) and alpha-1A adrenergic receptors are a class of GPCRs. The H1 expressed in peripheral tissues and the central nervous system, and it is responsible for modulating the allergic responses. It has also been found effective for filoviruses like EBV and Marburg [59]. Various antivirals are shown to target the alpha-1A adrenergic receptors among viruses [60]. However, the integrase is a viral enzyme, which integrates the retroviral DNA in the host genome. Therefore, it is one of the most popular targets for the promising antiviral drugs [61].
The DrugRepV database is the first manually curated repository for repurposed drugs tested for their antiviral activities. There are a few databases available in the literature describing repurposed drugs candidates e.g. ‘Drug Repurposing Hub’ and ‘RepoDB’ [62]. The Drug Repurposing Hub database comprises hand curated and annotated repurposed drugs, which are categorized as FDA-approved, clinical trials and preclinical drugs [63]. It has 6798 unique entries and 670 drug indications for various diseases like cancer, diabetes, microbial infections, etc. But it contains only 60 entries as antivirals. The RepoDB database contains 6677 approved drugs against various diseases [14]. It contains data extracted from the DrugCentral and ClinicalTrials.gov. However, the RepoDB database has only 15 entries of the drugs repurposed against viruses. Therefore, with 8485 entries DrugRepV serves as a comprehensive resource of experimentally validated drugs repurposed as antivirals.
The DrugRepV database would be of immense importance for the researchers working in the field of antiviral drug discovery. As the development of the novel drug candidate is a time-consuming process. Thus, drug repurposing proves to be a successful strategy to get effective drug candidates. For example, for SARS-CoV-2 causing the recent pandemic also our database contains about 342 repurposed drug entries tested for in vitro or in vivo antiviral activities. It has about 183 unique repurposed drugs and of which 144 have efficacy in terms of IC50/EC50 value. We searched these repurposed drugs in the ClinicalTrials.gov database to know whether these drugs with <1uM IC50/EC50 value have clinical potential. We found several repurposed drugs candidates incorporated in our database have entered various stages of clinical trials for SARS-CoV-2 e.g. ritonavir, ivermectin, nitazoxanide, baricitinib, remdesivir, favipiravir, etc. This demonstrates the utility of our ‘DrugRepV’ resource for SARS-CoV-2. We anticipate high success rates for other viruses also on further exploitation of the repurposed candidates with better IC50/EC50 efficacies. As the discovery of the novel drug candidates is a time consuming and costly process, thus, this database would provide insights to speed up the antiviral drug for COVID-19 using repurposing approaches [64].
We performed the initial network-based analysis for the viruses responsible to cause epidemics and pandemics as shown in Figure 2C and Supplementary Figures S2–S6. From the network analysis, we can opt for the shared drugs between more than one virus, as promising targets as antiviral drugs. The DrugRepV data have further implications in identifying and prioritizing the new repurposed candidates using numerous approaches. For example, we can infer new repurposed candidates through detailed network analysis combining drug and target data. Moreover, the molecular docking studies can be performed on the basis of antiviral drugs and their efficacies. Likewise, the machine learning algorithms (support vector machine, random forest, artificial neural network, deep learning, etc.) based predictive methods could be developed to identify potential repurposed drug candidates. Further, the extension of this work would be the compilation of the repurposed drugs against diseases like cancer, bacteria, diabetes and many more.
DrugRepV is the first comprehensive one-stop platform for all the drugs and chemicals repurposed against viruses. This initiative would prove to be a successful step for speeding up the discovery of effective antivirals. The repository comprehends all the information from various external resources, which further make this database a complete package. The frontends of the DrugRepV are designed in a highly user-friendly manner, which can be easily accessible. We anticipate that this dedicated and comprehensive resource would help the researcher in antiviral drug development.
Key Points
‘DrugRepV’ is the first dedicated repurposed drug repository for 23 viruses causing epidemics and pandemics.
It harbors 8485 repurposed drugs with biological, chemical, structural and clinical information.
The user-friendly ‘DrugRepV’ web server can be browsed and searched in multivariate ways to get information of repurposed drugs against viruses.
The database would be really helpful to speed up the research for developing effective antivirals against various epidemics/pandemics e.g. SARS-CoV-2
Supplementary Material
Acknowledgments
We acknowledge the infrastructure support of the Department of Biotechnology, Government of India (GAP0001).
Akanksha Rajput did her PhD from CSIR-Institute of Microbial Technology, Chandigarh. She contributed to big data analysis for bacterial and viral infections.
Archit Kumar, a Project Assistant at CSIR-Institute of Microbial Technology, Chandigarh, India, has expertise and interest in molecular virology.
Kirti Megha, a Project Assistant at CSIR-Institute of Microbial Technology, Chandigarh, India, has expertise and interest in molecular biology.
Anamika Thakur is a PhD scholar at CSIR-Institute of Microbial Technology, Chandigarh, India. She is working in the area of viral bioinformatics.
Manoj Kumar is a Principal Scientist and Head of Virology Unit at CSIR-Institute of Microbial Technology, Chandigarh, India. He has significantly contributed in the field of bioinformatics and virology.
Contributor Information
Akanksha Rajput, Virology Unit and Bioinformatics Centre, Institute of Microbial Technology, Council of Scientific and Industrial Research (CSIR), Sector 39A, Chandigarh-160036, India.
Archit Kumar, Virology Unit and Bioinformatics Centre, Institute of Microbial Technology, Council of Scientific and Industrial Research (CSIR), Sector 39A, Chandigarh-160036, India.
Kirti Megha, Virology Unit and Bioinformatics Centre, Institute of Microbial Technology, Council of Scientific and Industrial Research (CSIR), Sector 39A, Chandigarh-160036, India.
Anamika Thakur, Virology Unit and Bioinformatics Centre, Institute of Microbial Technology, Council of Scientific and Industrial Research (CSIR), Sector 39A, Chandigarh-160036, India; Academy of Scientific and Innovative Research (AcSIR), Ghaziabad-201002, India.
Manoj Kumar, Virology Unit and Bioinformatics Centre, Institute of Microbial Technology, Council of Scientific and Industrial Research (CSIR), Sector 39A, Chandigarh-160036, India; Academy of Scientific and Innovative Research (AcSIR), Ghaziabad-201002, India.
Availability
The DrugRepV resource is an open access web server and available at https://bioinfo.imtech.res.in/manojk/drugrepv/. We will update the resource regularly at half yearly or when enough data are available.
Funding
This work was supported by the grants from the CSIR-Institute of Microbial Technology (OLP0501, OLP0143 and MLP0024).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- 1. Littler E, Oberg B. Achievements and challenges in antiviral drug discovery. Antivir Chem Chemother 2005;16:155–68. [DOI] [PubMed] [Google Scholar]
- 2. Kates JR, McAuslan BR. Poxvirus DNA-dependent RNA polymerase. Proc Natl Acad Sci U S A 1967;58:134–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Irwin KK, Renzette N, Kowalik TF, et al. Antiviral drug resistance as an adaptive process. Virus Evol 2016;2:vew014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. De Clercq E, Li G. Approved antiviral drugs over the past 50 years. Clin Microbiol Rev 2016;29:695–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mercorelli B, Palù G, Loregian A. Drug repurposing for viral infectious diseases: how far are we? Trends Microbiol 2018;26:865–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ashburn TT, Thor KB. Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov 2004;3:673–83. [DOI] [PubMed] [Google Scholar]
- 7. Breckenridge A, Jacob R. Overcoming the legal and regulatory barriers to drug repurposing. Nat Rev Drug Discov 2019;18:1–2. [DOI] [PubMed] [Google Scholar]
- 8. Yarchoan R, Broder S. Development of antiretroviral therapy for the acquired immunodeficiency syndrome and related disorders. A progress report. N Engl J Med 1987;316:557–64. [DOI] [PubMed] [Google Scholar]
- 9. Grein J, Ohmagari N, Shin D, et al. Compassionate use of remdesivir for patients with severe Covid-19. N Engl J Med 2020;382:2327–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Moore N. Chloroquine for COVID-19 infection. Drug Saf 2020;43:393–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bullard-Feibelman KM, Govero J, Zhu Z, et al. The FDA-approved drug sofosbuvir inhibits Zika virus infection. Antiviral Res 2017;137:134–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dyall J, Coleman CM, Hart BJ, et al. Repurposing of clinically developed drugs for treatment of Middle East respiratory syndrome coronavirus infection. Antimicrob Agents Chemother 2014;58:4885–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Simanjuntak Y, Liang J-J, Lee Y-L, et al. Repurposing of prochlorperazine for use against dengue virus infection. J Infect Dis 2015;211:394–404. [DOI] [PubMed] [Google Scholar]
- 14. Brown AS, Patel CJ. A standard database for drug repositioning. Sci Data 2017;4:170029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. von Eichborn J, Murgueitio MS, Dunkel M, et al. PROMISCUOUS: a database for network-based drug-repositioning. Nucleic Acids Res 2011;39:D1060–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cully. Anticancer drugs: advancing precision medicine in silico. Nat Rev Drug Discov 2015;14:311. [DOI] [PubMed] [Google Scholar]
- 17. Vanhaelen Q, Mamoshina P, Aliper AM, et al. Design of efficient computational workflows for in silico drug repurposing. Drug Discov Today 2017;22:210–22. [DOI] [PubMed] [Google Scholar]
- 18. Battah B, Chemi G, Butini S, et al. A repurposing approach for uncovering the anti-tubercular activity of FDA-approved drugs with potential multi-targeting profiles. Molecules 2019;24:4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rajput A, Thakur A, Sharma S, et al. aBiofilm: a resource of anti-biofilm agents and their potential implications in targeting antibiotic drug resistance. Nucleic Acids Res 2018;46:D894–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thakur N, Qureshi A, Kumar M. AVPpred: collection and prediction of highly effective antiviral peptides. Nucleic Acids Res 2012;40:W199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rajput A, Kaur K, Kumar M. SigMol: repertoire of quorum sensing signaling molecules in prokaryotes. Nucleic Acids Res 2016;44:D634–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rajput A, Gupta AK, Kumar M. Prediction and analysis of quorum sensing peptides based on sequence features. PLoS One 2015;10:e0120066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ferreira AC, Reis PA, de Freitas CS, et al. Beyond members of the family, sofosbuvir also inhibits chikungunya virus replication. Antimicrob Agents Chemother 2019;63:e01389-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Paragas J, Whitehouse CA, Endy TP, et al. A simple assay for determining antiviral activity against Crimean-Congo hemorrhagic fever virus. Antiviral Res 2004;62:21–5. [DOI] [PubMed] [Google Scholar]
- 25. Kato F, Kobayashi T, Tajima S, et al. Development of a novel Dengue-1 virus replicon system expressing secretory Gaussia luciferase for analysis of viral replication and discovery of antiviral drugs. Jpn J Infect Dis 2014;67:209–12. [DOI] [PubMed] [Google Scholar]
- 26. Mudhasani R, Kota KP, Retterer C, et al. High content image-based screening of a protease inhibitor library reveals compounds broadly active against Rift Valley fever virus and other highly pathogenic RNA viruses. PLoS Negl Trop Dis 2014;8:e3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chan JFW, Chan K-H, Kao RYT, et al. Broad-spectrum antivirals for the emerging Middle East respiratory syndrome coronavirus. J Infect 2013;67:606–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Arshad U, Pertinez H, Box H, et al. Prioritization of anti-SARS-Cov-2 drug repurposing opportunities based on plasma and target site concentrations derived from their established human pharmacokinetics. Clin Pharmacol Ther 2020;108:775–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ivens T, Van den Eynde C, Van Acker K, et al. Development of a homogeneous screening assay for automated detection of antiviral agents active against severe acute respiratory syndrome-associated coronavirus. J Virol Methods 2005;129:56–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zmurko J, Marques RE, Schols D, et al. The viral polymerase inhibitor 7-Deaza-2’-C-Methyladenosine is a potent inhibitor of in vitro Zika virus replication and delays disease progression in a robust mouse infection model. PLoS Negl Trop Dis 2016;10:e0004695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yao X, Ye F, Zhang M, et al. In vitro antiviral activity and projection of optimized dosing design of hfor the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clin Infect Dis 2020;71:732–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. de Wilde AH, Jochmans D, Posthuma CC, et al. Screening of an FDA-approved compound library identifies four small-molecule inhibitors of Middle East respiratory syndrome coronavirus replication in cell culture. Antimicrob Agents Chemother 2014;58:4875–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Madrid PB, Panchal RG, Warren TK, et al. Evaluation of Ebola virus inhibitors for drug repurposing. ACS Infect Dis 2015;1:317–26. [DOI] [PubMed] [Google Scholar]
- 34. Nothias-Scaglia L-F, Pannecouque C, Renucci F, et al. Antiviral activity of diterpene esters on chikungunya virus and HIV replication. J Nat Prod 2015;78:1277–83. [DOI] [PubMed] [Google Scholar]
- 35. Madelain V, Guedj J, Mentré F, et al. Favipiravir pharmacokinetics in nonhuman primates and insights for future efficacy studies of hemorrhagic fever viruses. Antimicrob Agents Chemother 2017;61:e01305-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Saul S, Einav S. Old drugs for a new virus: repurposed approaches for combating COVID-19. ACS Infect Dis 2020;6:2304–18. [DOI] [PubMed] [Google Scholar]
- 37. Dawes BE, Kalveram B, Ikegami T, et al. Favipiravir (T-705) protects against Nipah virus infection in the hamster model. Sci Rep 2018;8:7604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gordon DE, Jang GM, Bouhaddou M, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020;583:459–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Morrey JD, Smee DF, Sidwell RW, et al. Identification of active antiviral compounds against a New York isolate of West Nile virus. Antiviral Res 2002;55:107–16. [DOI] [PubMed] [Google Scholar]
- 40. Peng C, Zhou Y, Cao S, et al. Identification of vaccinia virus inhibitors and cellular functions necessary for efficient viral replication by screening bioactives and FDA-approved drugs. Vaccines (Basel) 2020;8:401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ölschläger S, Neyts J, Günther S. Depletion of GTP pool is not the predominant mechanism by which ribavirin exerts its antiviral effect on Lassa virus. Antiviral Res 2011;91:89–93. [DOI] [PubMed] [Google Scholar]
- 42. Tilmanis D, van Baalen C, Oh DY, et al. The susceptibility of circulating human influenza viruses to tizoxanide, the active metabolite of nitazoxanide. Antiviral Res 2017;147:142–8. [DOI] [PubMed] [Google Scholar]
- 43. Cao R-Y, Xu Y-F, Zhang T-H, et al. Pediatric drug nitazoxanide: a potential choice for control of Zika. Open Forum Infect Dis 2017;4:ofx009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang M, Cao R, Zhang L, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res 2020;30:269–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mahmoud DB, Shitu Z, Mostafa A. Drug repurposing of nitazoxanide: can it be an effective therapy for COVID-19? J Genet Eng Biotechnol 2020;18:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang Y-M, Lu J-W, Lin C-C, et al. Antiviral activities of niclosamide and nitazoxanide against chikungunya virus entry and transmission. Antiviral Res 2016;135:81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Han Y, Mesplède T, Xu H, et al. The antimalarial drug amodiaquine possesses anti-ZIKA virus activities. J Med Virol 2018;90:796–802. [DOI] [PubMed] [Google Scholar]
- 48. Madrid PB, Chopra S, Manger ID, et al. A systematic screen of FDA-approved drugs for inhibitors of biological threat agents. PLoS One 2013;8:e60579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Boonyasuppayakorn S, Reichert ED, Manzano M, et al. Amodiaquine, an antimalarial drug, inhibits dengue virus type 2 replication and infectivity. Antiviral Res 2014;106:125–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pizzorno A, Padey B, Terrier O, et al. Drug repurposing approaches for the treatment of influenza viral infection: reviving old drugs to fight against a long-lived enemy. Front Immunol 2019;10:531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. García-Serradilla M, Risco C, Pacheco B. Drug repurposing for new, efficient, broad spectrum antivirals. Virus Res 2019;264:22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xu M, Lee EM, Wen Z, et al. Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat Med 2016;22:1101–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Amemiya T, Gromiha MM, Horimoto K, et al. Drug repositioning for dengue haemorrhagic fever by integrating multiple omics analyses. Sci Rep 2019;9:523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Li J, Zheng S, Chen B, et al. A survey of current trends in computational drug repositioning. Brief Bioinform 2016;17:2–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sariyer IK, Gordon J, Burdo TH, et al. Suppression of Zika virus infection in the brain by the antiretroviral drug Rilpivirine. Mol Ther 2019;27:2067–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Barrows NJ, Campos RK, Powell ST, et al. A screen of FDA-approved drugs for inhibitors of Zika virus infection. Cell Host Microbe 2016;20:259–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mathur P, Kottilil S, Wilson E. Use of ribavirin for hepatitis C treatment in the modern direct-acting antiviral era. J Clin Transl Hepatol 2018;6:431–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Furuta Y, Komeno T, Nakamura T. Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proc Jpn Acad Ser B Phys Biol Sci 2017;93:449–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Schafer A, Cheng H, Xiong R, et al. Repurposing potential of 1st generation H-specific antihistamines as anti-filovirus therapeutics. Antiviral Res 2018;157:47–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Scanzano A, Cosentino M. Adrenergic regulation of innate immunity: a review. Front Pharmacol 2015;6:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Engelman AN. Multifaceted HIV integrase functionalities and therapeutic strategies for their inhibition. J Biol Chem 2019;294:15137–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tanoli Z, Seemab U, Scherer A, et al. Exploration of databases and methods supporting drug repurposing: a comprehensive survey. Brief Bioinform 2020;bbaa003:1–23. doi: 10.1093/bib/bbaa003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Corsello SM, Bittker JA, Liu Z, et al. The drug repurposing hub: a next-generation drug library and information resource. Nat Med 2017;23:405–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Han Y, Wang Z, Ren J, et al. Potential inhibitors for the novel coronavirus (SARS-CoV-2). Brief Bioinform 2020;bbaa209:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


