Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infected individuals that have hypertension or cardiovascular comorbidities have an elevated risk of serious coronavirus disease 2019 (COVID-19) disease and high rates of mortality but how COVID- and cardiovascular diseases interact are unclear. We therefore sought to identify novel mechanisms of interaction by identifying genes with altered expression in SARS-CoV-
and cardiovascular diseases interact are unclear. We therefore sought to identify novel mechanisms of interaction by identifying genes with altered expression in SARS-CoV-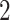 infection that are relevant to the pathogenesis of cardiovascular disease and hypertension. Some recent research shows the SARS-CoV-
infection that are relevant to the pathogenesis of cardiovascular disease and hypertension. Some recent research shows the SARS-CoV- uses the angiotensin converting enzyme-
uses the angiotensin converting enzyme- (ACE-
(ACE- ) as a receptor to infect human susceptible cells. The ACE2 gene is expressed in many human tissues, including intestine, testis, kidneys, heart and lungs. ACE2 usually converts Angiotensin I in the renin–angiotensin-aldosterone system to Angiotensin II, which affects blood pressure levels. ACE inhibitors prescribed for cardiovascular disease and hypertension may increase the levels of ACE-
) as a receptor to infect human susceptible cells. The ACE2 gene is expressed in many human tissues, including intestine, testis, kidneys, heart and lungs. ACE2 usually converts Angiotensin I in the renin–angiotensin-aldosterone system to Angiotensin II, which affects blood pressure levels. ACE inhibitors prescribed for cardiovascular disease and hypertension may increase the levels of ACE- , although there are claims that such medications actually reduce lung injury caused by COVID-
, although there are claims that such medications actually reduce lung injury caused by COVID-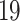 . We employed bioinformatics and systematic approaches to identify such genetic links, using messenger RNA data peripheral blood cells from COVID-
. We employed bioinformatics and systematic approaches to identify such genetic links, using messenger RNA data peripheral blood cells from COVID- patients and compared them with blood samples from patients with either chronic heart failure disease or hypertensive diseases. We have also considered the immune response genes with elevated expression in COVID-
patients and compared them with blood samples from patients with either chronic heart failure disease or hypertensive diseases. We have also considered the immune response genes with elevated expression in COVID- to those active in cardiovascular diseases and hypertension. Differentially expressed genes (DEGs) common to COVID-
to those active in cardiovascular diseases and hypertension. Differentially expressed genes (DEGs) common to COVID- and chronic heart failure, and common to COVID-
and chronic heart failure, and common to COVID- and hypertension, were identified; the involvement of these common genes in the signalling pathways and ontologies studied. COVID-
and hypertension, were identified; the involvement of these common genes in the signalling pathways and ontologies studied. COVID- does not share a large number of differentially expressed genes with the conditions under consideration. However, those that were identified included genes playing roles in T cell functions, toll-like receptor pathways, cytokines, chemokines, cell stress, type 2 diabetes and gastric cancer. We also identified protein–protein interactions, gene regulatory networks and suggested drug and chemical compound interactions using the differentially expressed genes. The result of this study may help in identifying significant targets of treatment that can combat the ongoing pandemic due to SARS-CoV-
does not share a large number of differentially expressed genes with the conditions under consideration. However, those that were identified included genes playing roles in T cell functions, toll-like receptor pathways, cytokines, chemokines, cell stress, type 2 diabetes and gastric cancer. We also identified protein–protein interactions, gene regulatory networks and suggested drug and chemical compound interactions using the differentially expressed genes. The result of this study may help in identifying significant targets of treatment that can combat the ongoing pandemic due to SARS-CoV- infection.
infection.
Keywords: SARS-CoV-2, COVID-19, coronavirus, cardiovascular, hypertension, differentially expressed gene, cell signalling pathway and ontology
1 Introduction
The present pandemic of coronavirus disease 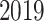 (COVID-
(COVID- ) resulting from the highly contagious severe acute respiratory syndrome coronavirus
) resulting from the highly contagious severe acute respiratory syndrome coronavirus  (SARS-CoV-
(SARS-CoV- ) is a World Health Organization (WHO) declared global public emergency. As of December
) is a World Health Organization (WHO) declared global public emergency. As of December  , almost
, almost 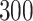 countries and territories around the world have reported more than
countries and territories around the world have reported more than 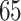 million confirmed cases resulting in over one and a half million deaths [1, 2, 6] Recent studies have found that some pre-existing conditions greatly increase the risk of severe symptoms and mortality in COVID-
million confirmed cases resulting in over one and a half million deaths [1, 2, 6] Recent studies have found that some pre-existing conditions greatly increase the risk of severe symptoms and mortality in COVID- patients. These include pulmonary diseases, cardiovascular diseases, kidney disease, type 2 diabetes and hypertension [47, 56]. Sequential Organ Failure Assessment (SOFA) scores have been reported significantly greater in SARS-CoV-2 associated deaths [49, 69]. While COVID-
patients. These include pulmonary diseases, cardiovascular diseases, kidney disease, type 2 diabetes and hypertension [47, 56]. Sequential Organ Failure Assessment (SOFA) scores have been reported significantly greater in SARS-CoV-2 associated deaths [49, 69]. While COVID- primarily affects the respiratory system in the early stages of the disease, while it also affects the cardiovascular system of patients, greatly increasing their risk of fatality [22, 28, 68]. Multiple studies reported elevated death rates in patients with SARS-CoV-
primarily affects the respiratory system in the early stages of the disease, while it also affects the cardiovascular system of patients, greatly increasing their risk of fatality [22, 28, 68]. Multiple studies reported elevated death rates in patients with SARS-CoV- infections and increased levels of markers of chronic heart failure [8, 30, 41, 69]. Similarly, studies found hypertension as comorbidity is associated with an increased risk of severe disease with SARS-CoV-
infections and increased levels of markers of chronic heart failure [8, 30, 41, 69]. Similarly, studies found hypertension as comorbidity is associated with an increased risk of severe disease with SARS-CoV- infection. Huang et al. found
infection. Huang et al. found  COVID-
COVID- patients with other health conditions, most commonly hypertension and cardiovascular disease were at risk. [17, 27]. The most frequent comorbidities in patients with severe symptomatic (such as acute respiratory distress) COVID-
patients with other health conditions, most commonly hypertension and cardiovascular disease were at risk. [17, 27]. The most frequent comorbidities in patients with severe symptomatic (such as acute respiratory distress) COVID- are hypertension
are hypertension  and cardiovascular disease
and cardiovascular disease  [62]. Since patients with COVID-
[62]. Since patients with COVID- and pre-existing cardiovascular diseases and hypertension have increased risk of severe disease and death, studies are needed to identify the interactions between these diseases and COVID-
and pre-existing cardiovascular diseases and hypertension have increased risk of severe disease and death, studies are needed to identify the interactions between these diseases and COVID- . Further the therapies under the investigation for COVID-
. Further the therapies under the investigation for COVID- may affect cardiovascular disease, hypertension or other significant comorbidities by influencing the pathways that mediate their interaction with COVID-19.
may affect cardiovascular disease, hypertension or other significant comorbidities by influencing the pathways that mediate their interaction with COVID-19.
Angiotensin Converting Enzyme  (ACE
(ACE ) is known to play an important role in facilitating SARS-CoV-
) is known to play an important role in facilitating SARS-CoV- cell entry [26, 44, 46, 65]. Expression of the ACE2 gene is reported in many human tissues including intestine, testis, kidneys, heart, and lungs [36]. ACE usually converts Angiotensin I in the renin–angiotensin– aldosterone system (RAAS) of human physiology to Angiotensin II that affects human blood pressure. Further, this type
cell entry [26, 44, 46, 65]. Expression of the ACE2 gene is reported in many human tissues including intestine, testis, kidneys, heart, and lungs [36]. ACE usually converts Angiotensin I in the renin–angiotensin– aldosterone system (RAAS) of human physiology to Angiotensin II that affects human blood pressure. Further, this type  membrane protein has significant physiological functions relevant to cardiovascular diseases [70]. ACE
membrane protein has significant physiological functions relevant to cardiovascular diseases [70]. ACE interacts with SARS-CoV2 thus it may have the potential gene-pathway associations to cause vascular dysfunction that may lead to myocardial infarction [20]. ACE inhibitors and Angiotensin receptor blockers (ARBs) are used as treatments for hypertension and cardiovascular disease [14] by modulating the RAAS pathway. These drugs inhibit the RAAS and interrupt the activity of ACE and increase the level of ACE
interacts with SARS-CoV2 thus it may have the potential gene-pathway associations to cause vascular dysfunction that may lead to myocardial infarction [20]. ACE inhibitors and Angiotensin receptor blockers (ARBs) are used as treatments for hypertension and cardiovascular disease [14] by modulating the RAAS pathway. These drugs inhibit the RAAS and interrupt the activity of ACE and increase the level of ACE receptors [9, 14, 31]. It is thus possible that these drugs could increase cell entry for the virus and increase the damage to the cardiovascular and respiratory systems. However, recent studies found that increased ACE
receptors [9, 14, 31]. It is thus possible that these drugs could increase cell entry for the virus and increase the damage to the cardiovascular and respiratory systems. However, recent studies found that increased ACE and ARB levels can actually be beneficial in COVID-
and ARB levels can actually be beneficial in COVID- , a point of considerable research interest [18, 19]. This serves to illustrate the potential importance of interaction among cardiovascular diseases and hypertension with COVID-
, a point of considerable research interest [18, 19]. This serves to illustrate the potential importance of interaction among cardiovascular diseases and hypertension with COVID- .
.
In this work, we considered the interaction of COVID-19 with chronic heart failure (CHF) and three types of hypertensive diseases and idiopathic portal hypertension (IPH), pulmonary arterial hypertension (PAH), and preeclampsia. CHF is a progressive condition that reduces cardiac output and blood oxygenation insufficiency, and up to two-thirds of cases of heart failure with preserved systolic function occur in patients over  years old are results from CHF. IPH is characterised by an increased pressure gradient in the portal system without the presence of a clear cause of liver disease or cirrhosis [25, 50]. PAH is a condition of increased blood pressure within the arteries of the lungs [4]. Preeclampsia is a pregnancy complication which is characterised by hypertensive condition and in less developed countries, it remains a major cause of maternal mortality with over
years old are results from CHF. IPH is characterised by an increased pressure gradient in the portal system without the presence of a clear cause of liver disease or cirrhosis [25, 50]. PAH is a condition of increased blood pressure within the arteries of the lungs [4]. Preeclampsia is a pregnancy complication which is characterised by hypertensive condition and in less developed countries, it remains a major cause of maternal mortality with over  maternal deaths worldwide due to preeclampsia per year [67]. Several studies demonstrated that SARS-CoV-2 infection may increase the health risk to pregnant women due to physiological changes occurred during pregnancy [13, 37].
maternal deaths worldwide due to preeclampsia per year [67]. Several studies demonstrated that SARS-CoV-2 infection may increase the health risk to pregnant women due to physiological changes occurred during pregnancy [13, 37].
We have found several significant cell signalling pathways and gene networks that are commonly associated with these diseases and SARS-CoV- infection on circulating human blood cells. The pathways and gene expression ontologies were identified through examination of the common differentially expressed genes (DEGs) shared among the diseases. We have considered a COVID-
infection on circulating human blood cells. The pathways and gene expression ontologies were identified through examination of the common differentially expressed genes (DEGs) shared among the diseases. We have considered a COVID- blood sample dataset as well as a COVID-
blood sample dataset as well as a COVID- immune response dataset to determine the genes and pathways that are involved in the interaction between COVID-
immune response dataset to determine the genes and pathways that are involved in the interaction between COVID- and CHF, IPH, PAH, and PE. We have also analysed protein–protein, drug and chemical, and transcription and post-transcription expression interactions networks using the signature genes. The identified pathways and networks are associated with other diseases that might lead to improved therapeutic approaches for life threatening SARS-CoV-
and CHF, IPH, PAH, and PE. We have also analysed protein–protein, drug and chemical, and transcription and post-transcription expression interactions networks using the signature genes. The identified pathways and networks are associated with other diseases that might lead to improved therapeutic approaches for life threatening SARS-CoV- infections.
infections.
2 Results
2.1 Differentially expressed genes in whole blood reveal genetic relationships between the COVID- , chronic heart failure and hypertensive diseases
, chronic heart failure and hypertensive diseases
We identified the differentially expressed genes (DEGs) for each of these datasets using the criteria that the adjusted  -value (adj p-value) is less than
-value (adj p-value) is less than  and the absolute value of log fold-change (logFC) is not less than
and the absolute value of log fold-change (logFC) is not less than  . We identified
. We identified  DEGs from the COVID-
DEGs from the COVID- blood datasets. The number of identified DEGs for CHF, IHP, PAH and PE were
blood datasets. The number of identified DEGs for CHF, IHP, PAH and PE were  ,
,  ,
, 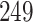 , and
, and  respectively. The four volcano plots in Figure 1 show the significant genes for CHF, IPH, PAH and PE. The red dots in the volcano plots indicate the significant genes. We have also performed a comparative analysis to identify the common DEGs between COVID-
respectively. The four volcano plots in Figure 1 show the significant genes for CHF, IPH, PAH and PE. The red dots in the volcano plots indicate the significant genes. We have also performed a comparative analysis to identify the common DEGs between COVID- and the other four diseases. The number of shared DEGs among the conditions is presented in Figure 1G. Our observation suggests that COVID-
and the other four diseases. The number of shared DEGs among the conditions is presented in Figure 1G. Our observation suggests that COVID- shares more DEGs with CHF and IHP as compared to the other two conditions. Figure 1E and 1F show heatmaps generated by the unique DEGs common to COVID-
shares more DEGs with CHF and IHP as compared to the other two conditions. Figure 1E and 1F show heatmaps generated by the unique DEGs common to COVID- and each of the other four diseases. Figure 1E shows the association between COVID-19 and other diseases from the perspective of adj P-value, while Figure 1F shows the association from the perspective of logFC. These two heat maps show that the COVID-
and each of the other four diseases. Figure 1E shows the association between COVID-19 and other diseases from the perspective of adj P-value, while Figure 1F shows the association from the perspective of logFC. These two heat maps show that the COVID- does not share a large number of DEGs with the diseases under consideration. We have found
does not share a large number of DEGs with the diseases under consideration. We have found  shared DEGs between COVID-
shared DEGs between COVID- and CHF, and the DEGs include ABCB
and CHF, and the DEGs include ABCB , CCL
, CCL , IGKC, MS
, IGKC, MS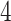 A
A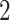 , CTSB and SAP
, CTSB and SAP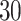 . COVID-
. COVID- has only two common DEGS with PAH including LTF and FCRL5
has only two common DEGS with PAH including LTF and FCRL5 . COVID-
. COVID- shares the highest number of DEGs with IPH compared to the other conditions. Some DEGs among these include IGKC, EZH
shares the highest number of DEGs with IPH compared to the other conditions. Some DEGs among these include IGKC, EZH , MK
, MK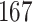 , CD
, CD , IGHA
, IGHA , IGHA
, IGHA , BTK and KNL
, BTK and KNL . We have identified
. We have identified  (NUAK
(NUAK , CCR
, CCR , RHOT
, RHOT , CNTLN and PLA
, CNTLN and PLA G
G ) DEGs common to COVID-
) DEGs common to COVID- and PE.
and PE.
Figure 1.

Volcano plots show DEGs of (A) CHF (B) IPH (C) PAH (D) PE, with genes having log fold-change of at least 1 and adjusted P value < . Heatmaps show the relationships among DEGs based on (E) adjusted
. Heatmaps show the relationships among DEGs based on (E) adjusted  value and (F) log fold-change (G) Venn diagram depicts the shared DEGs among COVID-
value and (F) log fold-change (G) Venn diagram depicts the shared DEGs among COVID- and other conditions.
and other conditions.
2.2 DEGs of COVID- immune responses in common with CHF and the hypertensive diseases
immune responses in common with CHF and the hypertensive diseases
We have identified the DEGs from each of the diseases, and identified (Figure 2C) a number of overlapping DEGs among them. We have also generated heat maps to show the association among the overlapping DEGs. The heat map presented in Figure 2A displays the association between gene and diseases from the perspective of adj P-value, while the heat map is shown in Figure 2B displays the relationship between gene and diseases in terms of the values of log fold change. Similar to the previous analysis on COVID- blood samples, COVID-
blood samples, COVID- immune response dataset shares more DEGs with CHF and IPH compared to the other two diseases (PAH and PE). The DEGs that are common between COVID-
immune response dataset shares more DEGs with CHF and IPH compared to the other two diseases (PAH and PE). The DEGs that are common between COVID- immune response and CHF include BCL
immune response and CHF include BCL , TRAF
, TRAF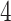 , HLA-DQB
, HLA-DQB and XCL
and XCL . The four shared DEGs with IPH include CD
. The four shared DEGs with IPH include CD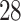 , IL
, IL RG, SLAMF
RG, SLAMF and CR
and CR . COVID-
. COVID-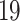 immune response share only one DEG, which is CCR
immune response share only one DEG, which is CCR (C–C chemokine receptor type
(C–C chemokine receptor type  ), with PE.
), with PE.
Figure 2.

Heatmaps show the relationships among DEGs based on (A) adjusted P value and (B) log fold-change for the COVID- immune dataset (C) Venn diagram depicts the shared DEGs among COVID-
immune dataset (C) Venn diagram depicts the shared DEGs among COVID- immune system and other conditions.
immune system and other conditions.
2.3 Gene set enrichment analysis reveals significant shared signalling and ontology pathways
We identified the cell signalling pathways that involve the DEGs common to COVID- and each of the other diseases, then determined what other genes may play a role in those pathways. In this enrichment analysis, we have combined all the DEGs that discovered from peripheral blood cells and the immune response cells of COVID-
and each of the other diseases, then determined what other genes may play a role in those pathways. In this enrichment analysis, we have combined all the DEGs that discovered from peripheral blood cells and the immune response cells of COVID- . We identified the signalling pathways of the commonly DEGs between COVID-
. We identified the signalling pathways of the commonly DEGs between COVID- and each of the diseases using six global pathway databases include BioPlanet, BioCarta, WikiPathways, KEGG, Reactome and Panther. We have integrated the pathways from these databases, and plotted the top
and each of the diseases using six global pathway databases include BioPlanet, BioCarta, WikiPathways, KEGG, Reactome and Panther. We have integrated the pathways from these databases, and plotted the top  most significant pathways based on the adj
most significant pathways based on the adj  -value as shown in Figure 3. The pathways with higher logarithmic adj
-value as shown in Figure 3. The pathways with higher logarithmic adj  -value are highly enriched. For example, highly enriched pathways common to COVID-
-value are highly enriched. For example, highly enriched pathways common to COVID- and CHF is T cell receptor regulation of apoptosis as shown in Figure 3A and in Supplementary Table 1. The significant signalling pathways that we have identified in the relationship between COVID-
and CHF is T cell receptor regulation of apoptosis as shown in Figure 3A and in Supplementary Table 1. The significant signalling pathways that we have identified in the relationship between COVID- and PAH are mostly associated with T-cell and HIV (Figure 3D and Supplementary Table 4) such as nef and signal transduction, HIV-induced T cell apoptosis, T helper cell surface molecules, nef in HIV-
and PAH are mostly associated with T-cell and HIV (Figure 3D and Supplementary Table 4) such as nef and signal transduction, HIV-induced T cell apoptosis, T helper cell surface molecules, nef in HIV-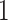 replication and disease pathogenesis and tob roles in T-cell activation.
replication and disease pathogenesis and tob roles in T-cell activation.
Figure 3.

Top  cell signalling pathways between COVID-
cell signalling pathways between COVID- and the diseases of cardiovascular and hypertension. The signalling pathways discovered using the DEGs for each of the condition and the combined genes of COVID-
and the diseases of cardiovascular and hypertension. The signalling pathways discovered using the DEGs for each of the condition and the combined genes of COVID- whole blood and immune samples. (A) shows pathways for CHF. (B) shows pathways for IPH. (C) and (D) show pathways for PAH and PE respectively.
whole blood and immune samples. (A) shows pathways for CHF. (B) shows pathways for IPH. (C) and (D) show pathways for PAH and PE respectively.
We also identified the gene ontology pathways by analysing biological process category of gene ontology. Gene ontology for each of the diseases with COVID- has been discovered. Figure 4 displays the most significant ontology pathways based on the adj
has been discovered. Figure 4 displays the most significant ontology pathways based on the adj  -value. As we see from Figure 4B, Phagocytosis is a prominent ontology pathway in the case of COVID-
-value. As we see from Figure 4B, Phagocytosis is a prominent ontology pathway in the case of COVID- and IPH (also see the Supplementary Table 6).
and IPH (also see the Supplementary Table 6).
Figure 4.

Top  gene ontology between COVID-
gene ontology between COVID- and the diseases of cardiovascular and hypertension. The ontology pathways were discovered using the DEGs for each of the condition and the combined genes of COVID-
and the diseases of cardiovascular and hypertension. The ontology pathways were discovered using the DEGs for each of the condition and the combined genes of COVID- whole blood and immune samples. (A) shows ontology for CHF. (B) shows ontology for IPH. (C) and (D) show ontology pathways for PAH and PE respectively.
whole blood and immune samples. (A) shows ontology for CHF. (B) shows ontology for IPH. (C) and (D) show ontology pathways for PAH and PE respectively.
2.4 Protein–protein interaction analysis identifies functional networks
We have constructed protein–protein interaction (PPI) network using all common DEGs among COVID-19 and the diseases of cardiovascular and hypertension. We have considered the shared DEGs discovered by analysis of gene expression profiles of blood cells and immune response cells from patients with COVID-19 and the other four diseases. The PPI network has been constructed using a web-based visualisation resource STRING [54], and the network is displayed in Figure 5 (also see the Supplementary Table 9). The figure shows the involvement and association among the signature genes in PPI network. We can also observe the relationship among the diseases from the perspective of PPI.
Figure 5.

PPI network constructed from all common DEGs.
2.5 Gene regulatory network analysis identifies DEGs–miRNA and TF–gene interactions for the shared genes
We have used the shared DEGs among COVID- and the diseases of cardiovascular and hypertension. The DEGs derived from both peripheral blood cell and immune response cells of COVID-
and the diseases of cardiovascular and hypertension. The DEGs derived from both peripheral blood cell and immune response cells of COVID- patients have been considered in this analysis. We have identified DEG–miRNA interactions by utilising TarBase and miRTarBase bases. The DEG–miRNA interactions network is displayed in Figure 6. The circles in the figure represent the DEGs whereas the squares represent the miRNAs. The colour of the circular node depends on the degree of the node. The degree of a node is the number of connections the node has with other nodes in the network. Nodes with a higher degree are considered as important hubs of the network. In addition, the size of the nodes is significant. Nodes representing genes that have strong interactions with other differentially expressed genes appear larger compared to the other nodes in the network. For example, the green colours nodes such as DLGAP5, CD28, MK167, HAVCR2, CD180 and EZH2 are more significant as we can observe from Figure 6B.
patients have been considered in this analysis. We have identified DEG–miRNA interactions by utilising TarBase and miRTarBase bases. The DEG–miRNA interactions network is displayed in Figure 6. The circles in the figure represent the DEGs whereas the squares represent the miRNAs. The colour of the circular node depends on the degree of the node. The degree of a node is the number of connections the node has with other nodes in the network. Nodes with a higher degree are considered as important hubs of the network. In addition, the size of the nodes is significant. Nodes representing genes that have strong interactions with other differentially expressed genes appear larger compared to the other nodes in the network. For example, the green colours nodes such as DLGAP5, CD28, MK167, HAVCR2, CD180 and EZH2 are more significant as we can observe from Figure 6B.
Figure 6.

Differentially expressed genes–micro RNA interaction network analysis.
We also identified the interaction network between the transcription factor (TF) and DEGs. These DEGs are common among COVID- and other conditions. Similar to the previous analysis, we have combined the DEGs from blood cells and immune response cells of COVID-
and other conditions. Similar to the previous analysis, we have combined the DEGs from blood cells and immune response cells of COVID- . To find the interactions between TF and DEG, we have used JASPER database. The TF–DEG interaction network is presented in Figure 7. The circular and squared nodes represent the DEGs and the TFs, respectively. The size of a node depends on the degree of the node. For example, PLA2G7, NUAK1, CCR1 and CNTLN, are more among more highly expressed DEGs as these genes have a higher degree in the network in the case of COVID-
. To find the interactions between TF and DEG, we have used JASPER database. The TF–DEG interaction network is presented in Figure 7. The circular and squared nodes represent the DEGs and the TFs, respectively. The size of a node depends on the degree of the node. For example, PLA2G7, NUAK1, CCR1 and CNTLN, are more among more highly expressed DEGs as these genes have a higher degree in the network in the case of COVID- and PE (Figure 7D). Transcription factors such as GATA3 and FOXC1 are more significant than others as we see in the same figure.
and PE (Figure 7D). Transcription factors such as GATA3 and FOXC1 are more significant than others as we see in the same figure.
Figure 7.

Transcription factors–differentially expressed genes interaction network analysis.
2.6 Suggested drug and chemical compounds analysis identifies protein–drug and protein–chemical interactions
The shared DEGs that have been discovered among the interaction of COVID- with chronic heart failure and hypertensive diseases are used in this analysis. We have identified protein–drug and protein–chemical interactions that may influence these genes. We have combined the DEGs identified from both peripheral blood cell and immune response cell populations. The protein–drug interaction is displayed in Figure 8. The squares in the figure represent drugs that affect the expression of the gene. The information of protein and drug relationship is obtained from the DrugBank database. We did not find any protein–drug relationship with the DEGs shared by COVID-
with chronic heart failure and hypertensive diseases are used in this analysis. We have identified protein–drug and protein–chemical interactions that may influence these genes. We have combined the DEGs identified from both peripheral blood cell and immune response cell populations. The protein–drug interaction is displayed in Figure 8. The squares in the figure represent drugs that affect the expression of the gene. The information of protein and drug relationship is obtained from the DrugBank database. We did not find any protein–drug relationship with the DEGs shared by COVID- and PAH. We did however identify protein–chemical interactions with the shared DEGs. The interactions are presented in Figure 9. The circular and squared nodes represent genes and chemical compounds respectively.
and PAH. We did however identify protein–chemical interactions with the shared DEGs. The interactions are presented in Figure 9. The circular and squared nodes represent genes and chemical compounds respectively.
Figure 8.

Protein–drug interactions analysis using the common DEGs among COVID- and the diseases of cardiovascular and hypertension.
and the diseases of cardiovascular and hypertension.
Figure 9.

Protein–chemical interactions analysis using common DEGs among COVID- and the diseases of cardiovascular and hypertension.
and the diseases of cardiovascular and hypertension.
3 Discussion
This study focuses on finding possible interaction pathways between COVID- and chronic heart failure hypertension diseases. We have worked with two types of COVID-
and chronic heart failure hypertension diseases. We have worked with two types of COVID- datasets, and we have chosen four different cardiovascular or cardiovascular-related diseases. First, we identified the differentially expressed genes from each of the datasets. Next, we compared the DEGs common to the COVID-
datasets, and we have chosen four different cardiovascular or cardiovascular-related diseases. First, we identified the differentially expressed genes from each of the datasets. Next, we compared the DEGs common to the COVID- datasets and the four other individual diseases of interest. We started with the whole blood cell dataset of COVID-
datasets and the four other individual diseases of interest. We started with the whole blood cell dataset of COVID- , and identified
, and identified  differentially expressed gene, then similarly identified the DEGs for the four other conditions. The number of identified DEGs for CHF, IPH, PAH and PE are
differentially expressed gene, then similarly identified the DEGs for the four other conditions. The number of identified DEGs for CHF, IPH, PAH and PE are  ,
,  ,
, 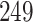 and
and  , respectively. Next, we identified the DEGs in common between COVID-
, respectively. Next, we identified the DEGs in common between COVID- and other four diseases. We found the largest number of common DEGs (
and other four diseases. We found the largest number of common DEGs ( ) are those between COVID-
) are those between COVID- and IPH, whereas in contrast only two common DEGs were identified between COVID-
and IPH, whereas in contrast only two common DEGs were identified between COVID-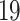 and PAH. Only one gene Immunoglobulin Kappa Constant (IGKC) has been found to be common to COVID-
and PAH. Only one gene Immunoglobulin Kappa Constant (IGKC) has been found to be common to COVID- and other two diseases, CHF and IPH. Immunoglobulin highly influences plasma cells, and plays a significant role to fight infectious pathogens in the respiratory system and digestive system at the point of pathogen entry [7]. We have also analysed COVID-
and other two diseases, CHF and IPH. Immunoglobulin highly influences plasma cells, and plays a significant role to fight infectious pathogens in the respiratory system and digestive system at the point of pathogen entry [7]. We have also analysed COVID- immune response samples with the four diseases to discover how COVID-
immune response samples with the four diseases to discover how COVID- affected immune system responses progress in people with cardiovascular and hypertension conditions. We identified
affected immune system responses progress in people with cardiovascular and hypertension conditions. We identified  DEGs from COVID-
DEGs from COVID- immune dataset. We also found a smaller number of DGEs shared by COVID-
immune dataset. We also found a smaller number of DGEs shared by COVID- immune response and other diseases. For instance, we have identified four common upregulated DEGs (CD
immune response and other diseases. For instance, we have identified four common upregulated DEGs (CD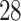 , IL
, IL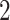 RG, SLAMF
RG, SLAMF and CR
and CR ) that are exposed in COVID-
) that are exposed in COVID- immune response having IPH condition. Most of these common DEGs show strong biological relevance with COVID-
immune response having IPH condition. Most of these common DEGs show strong biological relevance with COVID- pathology and pathogenesis. CD
pathology and pathogenesis. CD is a well known co-stimulatory receptor that provides signals for the production of classical inflammatory cytokines [32], that are involved in the cytokine storm reactions seen in SARS-CoV-
is a well known co-stimulatory receptor that provides signals for the production of classical inflammatory cytokines [32], that are involved in the cytokine storm reactions seen in SARS-CoV- infection [10]. These severe inflammatory reactions can be unpredictable, indeed there is a well known example of phase 1 clinical trial of CD
infection [10]. These severe inflammatory reactions can be unpredictable, indeed there is a well known example of phase 1 clinical trial of CD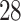 superagonists that resulted rapid and life threatening cytokine storm reactions in all the treated human participants [53] which resembled in many respects the strong inflammatory reactions of severe SARS-CoV-
superagonists that resulted rapid and life threatening cytokine storm reactions in all the treated human participants [53] which resembled in many respects the strong inflammatory reactions of severe SARS-CoV- infection [40]. IL-2RG gene express interleukin-
infection [40]. IL-2RG gene express interleukin- receptor subunit gamma chain (
receptor subunit gamma chain ( c) protein that acts as a shared receptor component for a number of cytokines, namely, IL
c) protein that acts as a shared receptor component for a number of cytokines, namely, IL , IL
, IL , IL
, IL , IL
, IL , IL
, IL and IL
and IL , which are
, which are  c family inflammatory cytokines [58]. At least two of these
c family inflammatory cytokines [58]. At least two of these  c cytokines were found elevated in plasma levels critical SARS-CoV-
c cytokines were found elevated in plasma levels critical SARS-CoV- infected patients [28]. The complement Receptor Type
infected patients [28]. The complement Receptor Type  (CR
(CR ) gene encodes a transmembrane protein on various immune cells and induces acute inflammatory immune response via complement activation [61]. The roles of soluble human CR1 (sCR1) has been manifested in Inflammatory Lung Diseases, where alveolar macrophages release sCR1 to lead acute respiratory distress syndrome (ARDS) [23]. ARDS was significantly higher in SARS-CoV-, MERS-CoV- and SARS-CoV-
) gene encodes a transmembrane protein on various immune cells and induces acute inflammatory immune response via complement activation [61]. The roles of soluble human CR1 (sCR1) has been manifested in Inflammatory Lung Diseases, where alveolar macrophages release sCR1 to lead acute respiratory distress syndrome (ARDS) [23]. ARDS was significantly higher in SARS-CoV-, MERS-CoV- and SARS-CoV- -associated deaths, where sCR1 levels were detected markedly higher in the bronchoalveolar lavage (BAL) samples [10]. Only one DEG, CCR
-associated deaths, where sCR1 levels were detected markedly higher in the bronchoalveolar lavage (BAL) samples [10]. Only one DEG, CCR , has been identified in COVID-
, has been identified in COVID- responses in the case of PE.
responses in the case of PE.
We performed gene set enrichment analysis by discovering cell signalling pathways and biological processes of gene ontology in order to obtain the association of COVID- and other diseases from the perspective of their shared DEGs among the diseases. The DEGs identified in analysing both peripheral blood cells and immune cells (in effect a subpopulation of the peripheral blood cells) with chronic heart failure and hypertension were combined for gene set enrichment analysis. Apoptosis-related signalling pathways were mostly exposed in the analysis of common DEGs between COVID-
and other diseases from the perspective of their shared DEGs among the diseases. The DEGs identified in analysing both peripheral blood cells and immune cells (in effect a subpopulation of the peripheral blood cells) with chronic heart failure and hypertension were combined for gene set enrichment analysis. Apoptosis-related signalling pathways were mostly exposed in the analysis of common DEGs between COVID- and chronic heart failure. We observed that the family of interleukin, cancer and chemokine-related pathways were enriched. We also identified
and chronic heart failure. We observed that the family of interleukin, cancer and chemokine-related pathways were enriched. We also identified  signalling pathway, which plays a significant role in regulating proliferation of cells that have DNA damage [35, 48], although the
signalling pathway, which plays a significant role in regulating proliferation of cells that have DNA damage [35, 48], although the  pathway activation can also be induced by a number of other stress signals.
pathway activation can also be induced by a number of other stress signals.
In the case of IPH and COVID- , we have pathways involved in scavenging of heme from plasma which are highly enriched; however, the family of the fibroblast growth factor receptor (FGFR) signalling pathways were the dominant signals. FGFR signalling pathways are concerned with many aspects of vascular, connective tissue and skeletal cell functions [11]. Several T-cell, HIV and interleukin-related pathways were enriched in their interaction between COVID-
, we have pathways involved in scavenging of heme from plasma which are highly enriched; however, the family of the fibroblast growth factor receptor (FGFR) signalling pathways were the dominant signals. FGFR signalling pathways are concerned with many aspects of vascular, connective tissue and skeletal cell functions [11]. Several T-cell, HIV and interleukin-related pathways were enriched in their interaction between COVID- and PAH. The most exposed pathways discovered with the common DEGs of COVID-
and PAH. The most exposed pathways discovered with the common DEGs of COVID- and PE were G-protein-coupled receptors (GPCRs). The peptide GPCRs pathway is relevant to several physiological processes such as growth, appetite and energy metabolism, cardiac function, stress and reproductive physiology [52]. We also identified several cytokine receptors such as TSLP pathway, chemokine signalling pathway, and cytokine–cytokine receptor interactions.
and PE were G-protein-coupled receptors (GPCRs). The peptide GPCRs pathway is relevant to several physiological processes such as growth, appetite and energy metabolism, cardiac function, stress and reproductive physiology [52]. We also identified several cytokine receptors such as TSLP pathway, chemokine signalling pathway, and cytokine–cytokine receptor interactions.
The gene ontology analysis reveals B cell, T cell, cytokine, and interleukin related pathways for CHF. The biology ontological process with the common DEGs for COVID- and IPH are dominated by the families of interleukin pathways; such pathways are commonly found in disease conditions (and certainly seen in COVID-19) as interleukins are major secreted hormonal products of peripheral blood cells. Other ontology pathways observed include natural killer cell activation and several T cell-related pathways. In the case of PAH, in addition to T cell-related functions, several chemotaxis ontology terms have been observed. Our analysis reveals several calcium ion and leukocyte ontology pathways for the interaction between COVID-
and IPH are dominated by the families of interleukin pathways; such pathways are commonly found in disease conditions (and certainly seen in COVID-19) as interleukins are major secreted hormonal products of peripheral blood cells. Other ontology pathways observed include natural killer cell activation and several T cell-related pathways. In the case of PAH, in addition to T cell-related functions, several chemotaxis ontology terms have been observed. Our analysis reveals several calcium ion and leukocyte ontology pathways for the interaction between COVID- and PE. We have also observed other significant pathways include chemokine-mediated signalling pathway, cellular response to cytokine stimulus.
and PE. We have also observed other significant pathways include chemokine-mediated signalling pathway, cellular response to cytokine stimulus.
We also discovered relationships among the diseases with respect to protein–protein, gene–miRNA, tf–gene, protein–drug and protein–chemical interactions. For CHF and COVID-19, we identified PTGS2 as one of the most highly expressed genes in gene regulatory networks as well as the networks of suggested drug and chemical interactions. PTGS2 (prostaglandin-endoperoxide synthase 2) is a protein-coding gene, and the genetic variation in this gene is considered as one of the vital risk factors behind the development of cardiovascular disease [34]. PTGS2 has previously been identified as an upregulated pro-viral factor in the infection of SARS-CoV-2 [38]. Thus our finding validates previous work on cardiovascular disease and COVID-19. We have also identified B-cell lymphoma 2 (BCL2) as a highly expressed gene in CHF and COVID-19 interaction. BCL2 is another factor with known involvement in cardiovascular diseases and COVID-19 [45]. In the hypertensive conditions, we found CD28, CD180, EZH2, PRDM1, TOP2A, CENFF, LTF, FCRL5, LGALS3, PLA2G7, RHOT1, CCR1, NUAK1 and CNTLN as highly expressed. In [3], the authors found LTF as one of the potential mediators of human hypertension. Mutation in LTF highly affects the immune system [59], therefore, further study on the nature of LTF is necessary in order to combat COVID-19 since the human immune system is so greatly affected by COVID-19. Among the identified genes, PLA2G7 (phospholipase A2 group VII) is highly associated with cardiovascular, hypertension and COVID-19 complications [66]. High levels of PLA2G7 is a cause of platelet-activating factor (PAF)-related deficiency through its catalyses PAF degradation.
We have identified micro-RNA species hsa-mir-6817-3p, hsa-mir-6873-3p, hsa-mir-6833-3p, hsa-mir-3873-3p, hsa-mir-124-3p, hsa-mir-16-5p and hsa-mir-195-5p among some of the micro-RNAs shared by cardiovascular and hypertensive diseases with COVID-19. The result of this work also identifies transcription factors that are tightly coupled with cardiovascular and hypertensive diseases. For example, we have found FOXC1, GATA2, GATA3, YY1 and FOXL1 are highly expressed in the association between COVID-19 and cardiovascular and hypertensive diseases. Earlier studies found that the family of FOX and GATA are involved in hypertension and cardiovascular conditions [24]. The chemical compounds found in our analysis include nickel, valproic acid, arsenic, tretinoin, calcitriol and zinc. Valproic acid, for instance, has been considered in the drug development for cardiovascular and hypertensive diseases [39]. One recent study suggests that valproic acid can also play a significant role in the treatment of COVID-19 [51].
4 Conclusions
Recent reports suggest the co-occurence of cardiovascular and hypertension diseases on COVID- patients are high compared to other conditions, and individuals with these comorbidities have much higher rates of mortality. In this study, we have investigated how SARS-CoV-
patients are high compared to other conditions, and individuals with these comorbidities have much higher rates of mortality. In this study, we have investigated how SARS-CoV- infection might influence patients with heart failure and three types of hypertension. We identified the cell signalling and gene ontology pathways from two different scenarios of interaction between COVID-
infection might influence patients with heart failure and three types of hypertension. We identified the cell signalling and gene ontology pathways from two different scenarios of interaction between COVID- and chronic heart failure and hypertensive diseases. Our investigation revealed that although IPH, PAH and PE are all forms of hypertension, the shared DEGs between each of these diseases with COVID-
and chronic heart failure and hypertensive diseases. Our investigation revealed that although IPH, PAH and PE are all forms of hypertension, the shared DEGs between each of these diseases with COVID- differ significantly. We also considered COVID-
differ significantly. We also considered COVID- elicited immune responses and identified the pathways with heart failure and hypertensive diseases. The identified DEGs were involved in a number of important pathways. We observed that immune system-related pathways such as T cell, interleukin and toll-like receptor pathways as seen in SARS-CoV-
elicited immune responses and identified the pathways with heart failure and hypertensive diseases. The identified DEGs were involved in a number of important pathways. We observed that immune system-related pathways such as T cell, interleukin and toll-like receptor pathways as seen in SARS-CoV- infections and cardiovascular/hypertension diseases, as well as gastric cancer related pathways. In the case of PE, there are several bone and calcium ion related pathways that are are highly expressed. We also observed HIV-related pathways in the case of hypertension samples. We have identified several stress related pathways between COVID-19 and chronic heart failure samples, for example the families of FGFR and GPCR pathways are more expressed with IPH and PE conditions respectively which may indicate connective tissue problems such as fibrosis.
infections and cardiovascular/hypertension diseases, as well as gastric cancer related pathways. In the case of PE, there are several bone and calcium ion related pathways that are are highly expressed. We also observed HIV-related pathways in the case of hypertension samples. We have identified several stress related pathways between COVID-19 and chronic heart failure samples, for example the families of FGFR and GPCR pathways are more expressed with IPH and PE conditions respectively which may indicate connective tissue problems such as fibrosis.
We have combined the DEGs identified from whole peripheral blood and immune response cell samples for discovering protein–protein interactions of SARS-CoV- infection in the presence of cardiovascular and hypertension in the human body. We have also performed transcription and post-transcription analysis in order to reveal DEGs-miRNAs, TFs-DEGs, proteins-drugs and protein–chemical compound interactions. The finding of this work identifies the molecules involved in the association between COVID-19 with the diseases of cardiovascular and hypertension. These findings also validate and support the outcomes of the previous studies as we presented in the discussion section. The identified significant genes, pathways, and networks are associated with other diseases which might lead to the discovery of possible new therapies that can combat the effects of severe SARS-CoV-
infection in the presence of cardiovascular and hypertension in the human body. We have also performed transcription and post-transcription analysis in order to reveal DEGs-miRNAs, TFs-DEGs, proteins-drugs and protein–chemical compound interactions. The finding of this work identifies the molecules involved in the association between COVID-19 with the diseases of cardiovascular and hypertension. These findings also validate and support the outcomes of the previous studies as we presented in the discussion section. The identified significant genes, pathways, and networks are associated with other diseases which might lead to the discovery of possible new therapies that can combat the effects of severe SARS-CoV- infection.
infection.
5 Materials and methods
The overall approach in this work consists of seven significant phases. In the first phase, the datasets for representing each of the diseases are collected. The goal of this phase is to ensure that the samples taken to obtain the datasets were collected using the same or different experiments performed on the same tissue of the same organism. The second phase of our analytical approach deals with determining the differentially expressed genes (DEGs) from each of the selected datasets. The identification of common DEGs between COVID- and each of the other four diseases is performed in phase three. In the next phase, we have performed gene set enrichment analysis to find the biological significance among the identified DEGs. In phase five, we focused on revealing protein–protein interaction networks. We identified gene regulatory network (GRN) interactions in phase six. The last phase of our analysis identified drug–chemical compound interactions.
and each of the other four diseases is performed in phase three. In the next phase, we have performed gene set enrichment analysis to find the biological significance among the identified DEGs. In phase five, we focused on revealing protein–protein interaction networks. We identified gene regulatory network (GRN) interactions in phase six. The last phase of our analysis identified drug–chemical compound interactions.
5.1 Gene expression datasets
In this paper, we have considered two SARS-CoV- infected datasets. One of the datasets is peripheral blood sample dataset for COVID-
infected datasets. One of the datasets is peripheral blood sample dataset for COVID- is taken from [64]. The blood cell samples were collected from three SARS-CoV-
is taken from [64]. The blood cell samples were collected from three SARS-CoV- patients and three healthy individuals. The second dataset was taken from [16]. The samples of this dataset (E-MTAB-8871) were collected from whole peripheral blood cell samples from COVID-
patients and three healthy individuals. The second dataset was taken from [16]. The samples of this dataset (E-MTAB-8871) were collected from whole peripheral blood cell samples from COVID- patients and healthy individuals by analysing the immune responses using the NanoString Human Immunology Panel. The gene expression microarray datasets have been considered for the diseases of cardiovascular and hypertension. We chose four diseases that fall into the categories of cardiovascular and hypertension diseases. We collected four human gene expression raw datasets for these diseases from the Gene Expression Omnibus of the National Center for Biotechnology Information (NCBI) [5]. The samples of the selected datasets having association number GSE
patients and healthy individuals by analysing the immune responses using the NanoString Human Immunology Panel. The gene expression microarray datasets have been considered for the diseases of cardiovascular and hypertension. We chose four diseases that fall into the categories of cardiovascular and hypertension diseases. We collected four human gene expression raw datasets for these diseases from the Gene Expression Omnibus of the National Center for Biotechnology Information (NCBI) [5]. The samples of the selected datasets having association number GSE , GSE
, GSE , GSE
, GSE and GSE
and GSE were collected by analysing the peripheral blood of the patients and healthy controls of corresponding diseases. The dataset chosen in this work for chronic heart failure disease, GSE
were collected by analysing the peripheral blood of the patients and healthy controls of corresponding diseases. The dataset chosen in this work for chronic heart failure disease, GSE , is a microarray dataset derived from transcriptional profiling of peripheral blood cells. The dataset GSE69601, representing idiopathic portal hypertension, is a gene expression microarray data taken from blood samples of IPH patients and healthy individuals. The pulmonary arterial hypertension dataset, GSE
, is a microarray dataset derived from transcriptional profiling of peripheral blood cells. The dataset GSE69601, representing idiopathic portal hypertension, is a gene expression microarray data taken from blood samples of IPH patients and healthy individuals. The pulmonary arterial hypertension dataset, GSE , is obtained by gene expression profiling in blood of PAH patients and healthy volunteers. The dataset for Preeclampsia, GSE
, is obtained by gene expression profiling in blood of PAH patients and healthy volunteers. The dataset for Preeclampsia, GSE , is a microarray data obtained from the blood sample of PE patients and healthy women with normal pregnancy.
, is a microarray data obtained from the blood sample of PE patients and healthy women with normal pregnancy.
5.2 Identification of DEGs
We have identified DGEs from each of the mRNA datasets. The first step in the process of finding DEG from a dataset is to preform normalization on both disease and control state [21]. The goal is to make uniform mRNA expression in order to avoid the issue in comparing gene expression data that are derived from different platforms and experimental environment. We have normalized all sample data using Z-score transformation by following the equation presented in Equation 1.
 |
(1) |
In Equation 1, SD represents standard deviation,  represents the expression value of the gene
represents the expression value of the gene  in sample
in sample  and
and  represents the transformed value of the gene
represents the transformed value of the gene  in sample
in sample  . The outcome of this transformation allows gene expression values of different diseases at different platform comparable. After performing normalization on the dataset, the next step is to use Student’s t-test statistic in order to identify genes with altered expression association with the individual disease states. Unpaired t-test and log2 transformation operations were performed in order to determine differentially expressed genes. Finally, genes for which the adjusted
. The outcome of this transformation allows gene expression values of different diseases at different platform comparable. After performing normalization on the dataset, the next step is to use Student’s t-test statistic in order to identify genes with altered expression association with the individual disease states. Unpaired t-test and log2 transformation operations were performed in order to determine differentially expressed genes. Finally, genes for which the adjusted  -values of the t-test are less than
-values of the t-test are less than  and values of log fold change (logFC) are at least
and values of log fold change (logFC) are at least 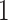 are identified as statistically significant genes.
are identified as statistically significant genes.
5.3 Gene set enrichment analysis
Gene set enrichment analysis involves signalling pathway analysis and gene ontology analysis [42]. Signalling pathway and ontology analysis are performed in order to determine the biological significance of the identified DEGs. In gene set enrichment analysis, we considered pathways for which the adjusted P-value are less than  . We have used Enrichr [33] to identify signalling pathway and ontology terms. The pathways have been considered from six databases including BioPlanet, BioCarta, WikiPathways, KEGG, Reactome and Panther.
. We have used Enrichr [33] to identify signalling pathway and ontology terms. The pathways have been considered from six databases including BioPlanet, BioCarta, WikiPathways, KEGG, Reactome and Panther.
5.4 Protein–protein interactions analysis
In order to discover associations among the diseases from the perspective of protein interactions, we have identified protein subnetworks using enriched DEGs. STRING [55]—a protein interactome database is used in this analysis. We have constructed protein–protein interaction (PPI) network using the shared DEGs among COVID- and other conditions. Gene clusters have been identified using the Markov cluster algorithm (MCL) available in STRING.
and other conditions. Gene clusters have been identified using the Markov cluster algorithm (MCL) available in STRING.
5.5 Gene regulatory networks (GRN) analysis
We have identified DEG–miRNA (microRNA) interaction networks and transcription factor(TF)–DEG interaction networks in this analysis. The networks have been discovered using Network Analyst [43, 63]. TarBase [57] and miRTarBase [29] databases are used for discovering DEG–miRNA interaction networks. In the case of TF–DEG interaction network analysis, JASPAR [15] database has been used. In GRN analysis, we have used common DEGs to reveal the transcriptional elements and miRNA that regulate DEGs at post-transcriptional level.
5.6 Suggested Drug and Chemical Compound Analysis
In this analysis, we have identified protein–drug and protein–chemical interactions using the enriched gene that COVID- shares with cardiovascular and hypertension. We have used Network Analyst [63] to identify protein–drug and protein–chemical interactions. DrugBank [60] database and Comparative Toxicogenomics Database [12] are used for protein–drug and protein–chemical compound interactions.
shares with cardiovascular and hypertension. We have used Network Analyst [63] to identify protein–drug and protein–chemical interactions. DrugBank [60] database and Comparative Toxicogenomics Database [12] are used for protein–drug and protein–chemical compound interactions.
Key Points
This work focuses on the influences of SAR-CoV-2 infection on individuals having chronic heart failure and/or hypertensive conditions.
RNA sequence data of whole blood samples were analysed to identify shared DEGs with chronic heart failure and hypertension disorders.
Gene set enrichment analysis with the identified DEGs discovered cell signalling pathways and gene ontology.
The protein–protein interaction of COVID-
 with hypertensive and heart failure condition identified significant hub genes.
with hypertensive and heart failure condition identified significant hub genes.The gene regulatory network analysis revealed the DEG–miRNA and TF–DEG interactions.
Drug and chemical compound analysis identified protein-drug and protein–chemical interactions that might help to identify the development of vaccine and medication for COVID-19.
This work may lead to the discovery of possible new therapies that can combat the effects of SARS-CoV-
 infection.
infection.
Supplementary Material
Md Asif Nashiry received his PhD degree from the University of Lethbridge, Alberta, Canada in 2018. Currently, he is working as an Associate Professor at the department of Computer Science and Engineering of the Jashore University of Science and Technology. His research areas of interest include machine learning, data science and bioinformatics.
Shauli Sarmin Sumi is currently working as an Assistant Professor at the department of Computer Science and Engineering of Jashore University of Science and Technology. She received her MSc in Computer Science from the University of Lethbridge, Canada. She works in the areas of natural language processing, data mining and bioinformatics.
Salequl Islam has completed his PhD from Japan and postdoctoral fellowship at the Johns Hopkins Bloomberg School of Public Health. His research interest is in the areas of microbiology, bioinformatics and computational biology. He is currently working as an Associate Professor (Microbiology) at Jahangirnagar University.
Julian M. W. Quinn received his doctorate from the University of Oxford, UK, in 1992, moved to Australia for postdoctoral training in bone and cancer biology at Melbourne University and St Vincent’s Institute of Medical Research later moving to Prince Henry’s Institute (as a Senior Research Fellow) and most recently to the Garvan Institute at Darlinghurst, NSW. His has strong interests in applications of biostatistics and bioinformatics.
Mohammad Ali Moni is a Research Fellow and Conjoint Lecturer at the University of New South Wales, Australia. He received his Ph.D. in clinical Bioinformatics and Machine Learning from the University of Cambridge. His research interests encompass artificial intelligence, machine learning, data science, medical image processing and clinical bioinformatics.
Contributor Information
Asif Nashiry, Department of Computer Science and Engineering, Jashore University of Science and Technology, Bangladesh.
Shauli Sarmin Sumi, Department of Computer Science and Engineering, Jashore University of Science and Technology, Bangladesh.
Salequl Islam, Department of Microbiology, Jahangirnagar University, Savar, Dhaka-1342, Bangladesh.
Julian M W Quinn, Healthy Ageing Theme, The Garvan Institute of Medical Research, Darlinghurst, NSW 2010, Australia.
Mohammad Ali Moni, Healthy Ageing Theme, The Garvan Institute of Medical Research, Darlinghurst, NSW 2010, Australia; WHO Collaborating Centre on eHealth, UNSW Digital Health, School of Public Health and Community Medicine, Faculty of Medicine, UNSW Sydney, Australia.
Data availability
We have used all the data from the publicly available; all the data links are provided to the manuscript.
References
- 1. Md MA, Aktar S, Md R-A-M, et al. A machine learning model to identify early stage symptoms of sars-cov-2 infected patients. Expert Sys Appl 2020; 160:113661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aktar S, Talukder A, Ahamad M, et al. Machine learning and meta-analysis approach to identify patient comorbidities and symptoms that increased risk of mortality in covid-19. arXiv preprint arXiv:200812683 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alexander MR, Norlander AE, Elijovich F, et al. Human monocyte transcriptional profiling identifies il-18 receptor accessory protein and lactoferrin as novel immune targets in hypertension. Br J Pharmacol 2019; 176(12): 2015–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Badesch DB, Champion HC, Sanchez MAG, et al. Diagnosis and assessment of pulmonary arterial hypertension. J Am Coll Cardiol 2009; 54(1 Supplement): S55–66. [DOI] [PubMed] [Google Scholar]
- 5. Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res 2012; 41(D1): D991–5 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Marco Cascella, Michael Rajnik, Arturo Cuomo, et al. Features, evaluation and treatment coronavirus (covid-19). In Statpearls [internet]. StatPearls Publishing, 2020. [PubMed] [Google Scholar]
- 7. Rötzschke O, Chao YX, Tan EK. The role of Iga in covid-19. Brain Behav Immun 2020; 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tao Chen, Di Wu, Huilong Chen, et al. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ, 368, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chowdhury UF, Shohan MUS, Hoque KI, et al. A computational approach to design potential sirna molecules as a prospective tool for silencing nucleocapsid phosphoprotein and surface glycoprotein gene of sars-cov-2. bioRxiv 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Coperchini F, Chiovato L, Croce L, et al. The cytokine storm in covid-19: an overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cross MJ, Claesson-Welsh L. Fgf and vegf function in angiogenesis: signalling pathways, biological responses and therapeutic inhibition. Trends Pharmacol Sci 2001; 22(4): 201–7. [DOI] [PubMed] [Google Scholar]
- 12. Johnson RJ, Sciaky D, McMorran R, et al. The comparative toxicogenomics database: update 2019. Nucleic Acids Res Sep 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Diriba K, Awulachew E, Getu E. The effect of coronavirus infection (sars-cov-2, mers-cov, and sars-cov) during pregnancy and the possibility of vertical maternal–fetal transmission: a systematic review and meta-analysis. Eur J Med Res 2020; 25(1): 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ferrario CM, Jessup J, Chappell MC, et al. Effect of angiotensin-converting enzyme inhibition and angiotensin ii receptor blockers on cardiac angiotensin-converting enzyme 2. Circulation 2005; 111(20): 2605–10. [DOI] [PubMed] [Google Scholar]
- 15. Fornes O, Castro-Mondragon JA, Khan A, et al. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res 2019; 48(D1): D87–92 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goujon M, McWilliam H, Li W, et al. A new bioinformatics analysis tools framework at embl–ebi. Nucleic Acids Res 2010; 38(suppl_2): W695–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guan W-j, Ni Z-y, Yu H, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 2020; 382(18): 1708–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guo J, Huang Z, Lin L, et al. Coronavirus disease 2019 (covid-19) and cardiovascular disease: a viewpoint on the potential influence of angiotensin-converting enzyme inhibitors/angiotensin receptor blockers on onset and severity of severe acute respiratory syndrome coronavirus 2 infection. J Am Heart Assoc 2020; 9(7):e016219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gurwitz D. Angiotensin receptor blockers as tentative sars-cov-2 therapeutics. Drug Dev Res 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guzik TJ, Mohiddin SA, Dimarco A, et al. Covid-19 and the cardiovascular system: implications for risk assessment, diagnosis, and treatment options. Cardiovasc Res 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Haidar MN, Islam MB, Chowdhury UN, et al. Network-based quantitative frameworks to identify pleotropic factors that influence for cardiomyopathy progression. In: 2019 International Conference on Computer, Communication, Chemical, Materials and Electronic Engineering (IC4ME2). IEEE, 2019, 1–5. [Google Scholar]
- 22. Md NH, Babul Islam M, Chowdhury UN, et al. Network-based computational approach to identify genetic links between cardiomyopathy and its risk factors. IET Syst Biol 2020; 14(2): 75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hamacher J, Sadallah S, Schifferli JA, et al. Soluble complement receptor type 1 (cd35) in bronchoalveolar lavage of inflammatory lung diseases. Eur Respir J 1998; 11(1): 112–9. [DOI] [PubMed] [Google Scholar]
- 24. Hannenhalli S, Putt ME, Gilmore JM, et al. Clinical perspective. Circulation 2006; 114(12): 1269–76. [DOI] [PubMed] [Google Scholar]
- 25. Hernández-Gea V, Baiges A, Turon F, et al. Idiopathic portal hypertension. Hepatology 2018; 68(6): 2413–23. [DOI] [PubMed] [Google Scholar]
- 26. Hoffmann M, Kleine-Weber H, Schroeder S, et al. Sars-cov-2 cell entry depends on ace2 and tmprss2 and is blocked by a clinically proven protease inhibitor. Cell 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Md EH, Uddin S, Khan A, et al. A framework to understand the progression of cardiovascular disease for type 2 diabetes mellitus patients using a network approach. Int J Environ Res Public Health 2020; 17(2): 596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020; 395(10223): 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hsi-Yuan Huang, Yang-Chi-Dung Lin, Jing Li, et al. 2020: Updates to the experimentally validated microRNA–target interaction database. Nucleic Acids Res, 48(D1): D148–54, 10 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ibrahim NE, Jr JLJ. Established and emerging roles of biomarkers in heart failure. Circ Res 2018; 123(5): 614–29. [DOI] [PubMed] [Google Scholar]
- 31. Ishiyama Y, Gallagher PE, Averill DB, et al. Upregulation of angiotensin-converting enzyme 2 after myocardial infarction by blockade of angiotensin ii receptors. Hypertension 2004; 43(5): 970–6. [DOI] [PubMed] [Google Scholar]
- 32.Raymond Kaempfer, Gila Arad, Revital Levy, et al. Cd28: direct and critical receptor for superantigen toxins. Toxins, 5(9): 1531–42, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kuleshov MV, Jones MR, Rouillard AD, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 2016; 44(W1): W90–7 05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee CR, North KE, Bray MS, et al. Cyclooxygenase polymorphisms and risk of cardiovascular events: the atherosclerosis risk in communities (aric) study. Clin Pharmacol Therap 2008; 83(1): 52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Levine AJ, Hu W, Feng Z. The p53 pathway: what questions remain to be explored? Cell Death Differ 2006; 13(6): 1027–36. [DOI] [PubMed] [Google Scholar]
- 36. Li M-Y, Li L, Zhang Y, et al. Expression of the sars-cov-2 cell receptor gene ace2 in a wide variety of human tissues. Infect Dis Poverty 2020; 9:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu Y, Chen H, Tang K, et al. Clinical manifestations and outcome of sars-cov-2 infection during pregnancy. J Infect 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Loganathan T, Ramachandran S, Shankaran P, et al. Host transcriptome-guided drug repurposing for covid-19 treatment: a meta-analysis based approach. PeerJ 2020; 8:e9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. LoPinto-Khoury C, Mintzer S. Antiepileptic drugs and markers of vascular risk. Curr Treat Options Neurol 2010; 12(4): 300–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mahmudpour M, Roozbeh J, Keshavarz M, et al. Covid-19 cytokine storm: the anger of inflammation. Cytokine 2020;155151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mongirdienė A, Kuršvietienė L, Kašauskas A. The coagulation system changes in patients with chronic heart failure. Medicina 2010; 46(9): 642. [PubMed] [Google Scholar]
- 42. Moni MA, Liò P. Comor: a software for disease comorbidity risk assessment. J Clin Bioinformatics 2014; 4(1): 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Moni MA, Liò P. How to build personalized multi-omics comorbidity profiles. Front Cell Dev Biol 2015; 3(28). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nain Z, Rana HK, Liò P, et al. Pathogenetic profiling of covid-19 and sars-like viruses. Brief Bioinform 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Navratil V, Lionnard L, Longhi S, et al. The severe acute respiratory syndrome coronavirus 2 (sars-cov-2) envelope (e) protein harbors a conserved bh3-like sequence. bioRxiv 2020. [Google Scholar]
- 46. Oany AR, Mia M, Pervin T, et al. Design of novel viral attachment inhibitors of the spike glycoprotein (s) of severe acute respiratory syndrome coronavirus-2 (sars-cov-2) through virtual screening and dynamics. Int J Antimicrob Agents 2020;106177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Patel AB, Verma A. Covid-19 and angiotensin-converting enzyme inhibitors and angiotensin receptor blockers: what is the evidence? JAMA 2020; 323(18): 1769–70. [DOI] [PubMed] [Google Scholar]
- 48. Anna M. Puzio-Kuter. The role of p53 in metabolic regulation. Genes Cancer 2011; 2(4): 385–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Satu MS, Khan MI, Mahmud M, et al. Tclustvid: a novel machine learning classification model to investigate topics and sentiment in covid-19 tweets. medRxiv 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schouten JNL, Garcia-Pagan JC, Valla DC, et al. Idiopathic noncirrhotic portal hypertension. Hepatology 2011; 54(3): 1071–81. [DOI] [PubMed] [Google Scholar]
- 51. Singh S, Singh K. Valproic acid in prevention and treatment of covid-19. Authorea Preprints 2020. [Google Scholar]
- 52. Stevens RC, Cherezov V, Katritch V, et al. The gpcr network: a large-scale collaboration to determine human gpcr structure and function. Nat Rev Drug Discov 2013; 12(1): 25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Suntharalingam G, Perry MR, Ward S, et al. Cytokine storm in a phase 1 trial of the anti-cd28 monoclonal antibody tgn1412. N Engl J Med 2006; 355(10): 1018–28. [DOI] [PubMed] [Google Scholar]
- 54. Szklarczyk D, Franceschini A, Wyder S, et al. String v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015; 43(D1): D447–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2018; 47(D1): D607–13 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Taz TA, Ahmed K, Paul BK, et al. Network-based identification genetic effect of sars-cov-2 infections to idiopathic pulmonary fibrosis (ipf) patients. Brief Bioinform 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Vergoulis T, Vlachos IS, Alexiou P, et al. TarBase 6.0: capturing the exponential growth of miRNA targets with experimental support. Nucleic Acids Res 2011; 40(D1): D222–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
-
58.
Waickman AT, Park J-Y, Park J-H. The common
 -chain cytokine receptor: tricks-and-treats for t cells. Cell Mol Life Sci 2016; 73(2): 253–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
-chain cytokine receptor: tricks-and-treats for t cells. Cell Mol Life Sci 2016; 73(2): 253–69. [DOI] [PMC free article] [PubMed] [Google Scholar] - 59. Wilk AJ, Rustagi A, Zhao NQ, et al. A single-cell atlas of the peripheral immune response in patients with severe covid-19. Nat Med 2020;1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Guo AC, Lo EJ, Marcu A, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res Nov 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wong WW, Klickstein LB, Smith JA, et al. Identification of a partial cdna clone for the human receptor for complement fragments c3b/c4b. Proc Natl Acad Sci 1985; 82(22): 7711–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wu C, Chen X, Cai Y, et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gill EE, Xia J, Hancock REW. Networkanalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat Protoc 2015; 10(6): 823–44. [DOI] [PubMed] [Google Scholar]
- 64. Xiong Y, Liu Y, Cao L, et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in covid-19 patients. Emerg Microbes Infect 2020; 9(1): 761–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yan R, Zhang Y, Li Y, et al. Structural basis for the recognition of sars-cov-2 by full-length human ace2. Science 2020; 367(6485): 1444–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yang LI, Yongzhong JIANG, Yi ZHANG, et al. Abnormal upregulation of cardiovascular disease biomarker pla2g7 induced by proinflammatory macrophages in covid-19 patients. medRxiv 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Young BC, Levine RJ, Karumanchi SA. Pathogenesis of preeclampsia. Annu Rev Pathol 2010; 5:173–92. [DOI] [PubMed] [Google Scholar]
- 68. Zheng Y-Y, Ma Y-T, Zhang J-Y, et al. Covid-19 and the cardiovascular system. Nat Rev Cardiol 2020; 17(5): 259–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with covid-19 in Wuhan, China: a retrospective cohort study. Lancet 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zhou P, Yang X-L, Wang X-G, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020; 579(7798): 270–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We have used all the data from the publicly available; all the data links are provided to the manuscript.


