Abstract
Antiviral therapies targeting the pandemic coronavirus disease 2019 (COVID-19) are urgently required. We studied an already-approved botanical drug cepharanthine (CEP) in a cell culture model of GX_P2V, a severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-related virus. RNA-sequencing results showed the virus perturbed the expression of multiple genes including those associated with cellular stress responses such as endoplasmic reticulum (ER) stress and heat shock factor 1 (HSF1)-mediated heat shock response, of which heat shock response-related genes and pathways were at the core. CEP was potent to reverse most dysregulated genes and pathways in infected cells including ER stress/unfolded protein response and HSF1-mediated heat shock response. Additionally, single-cell transcriptomes also confirmed that genes of cellular stress responses and autophagy pathways were enriched in several peripheral blood mononuclear cells populations from COVID-19 patients. In summary, this study uncovered the transcriptome of a SARS-CoV-2-related coronavirus infection model and anti-viral activities of CEP, providing evidence for CEP as a promising therapeutic option for SARS-CoV-2 infection.
Keywords: SARS-CoV-2, COVID-19, GX_P2V, cellular stress, cepharanthine, transcriptome
Introduction
The coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is a global public health problem that is impacting social and economic damage all over the world. As of 6 November 2020, 48,534,508 confirmed cases with 1,231,017 deaths were reported worldwide [1]. COVID-19 was characterized as a pandemic by the World Health Organization; however, there is no approved specific cure. Several medicines have been evaluated in COVID-19 patients in clinical trials, including interferon [2], lopinavir–ritonavir [2, 3], chloroquine [4], hydroxychloroquine [3, 5, 6], favipiravir [7] and remdesivir [8, 9], but their clinical efficacies and safety remain controversial. Therefore, additional anti-viral treatment options are urgently needed.
Cepharanthine (CEP) is naturally occurring alkaloid extracted from the plant Stephania cepharantha Hayata. It has been used in Japan since the 1950s to treat a number of acute and chronic diseases, including leukopenia, xerostomia, alopecia, and snake bites [10]. Our previous work found that CEP shows significant anti-viral functions for pangolin coronavirus GX_P2V, a SARS-CoV-2-related coronavirus, at both entry and post-entry stages [11]. GX_P2V is a kind of coronavirus isolated from Malayan pangolins with close genome sequence to SARS-CoV-2 [12]. Sharing 92.2% amino acid identity on spike protein of SARS-CoV-2 and using ACE2 as the receptor for infection, GX_P2V is an ideal alternative model for SARS-CoV-2 research [11, 12]. Another study confirmed that CEP inhibits SARS-CoV-2’s attachment and entry into cells [13]. However, the underlying mechanisms of the anti-viral activity of CEP remain unclear.
Cell responds to stress in a variety of ways ranging from activation of pathways promoting survival to elicitation of programmed cell death eliminating damaged cells. Many viruses such as varicella–zoster, SARS-CoV and hepatitis C induce various cellular stress responses, including heat shock response, endoplasmic reticulum (ER) stress, unfolded protein response and autophagy [14–16]. It is reported that Spike protein of SARS-CoV could modulate the unfolded protein response to facilitate viral replication [15]. Like other coronaviruses, SARS-CoV-2 is an enveloped positive-stranded RNA virus that has a large genome of ~30 kb [17], and its Spike protein shares 75.96% amino acid sequence identity with that of SARS-CoV [18]. SARS-CoV-2 was reported to reshape multiple cellular pathways such as translation, splicing, carbon metabolism, protein homeostasis and nucleic acid metabolism in host cells while targeting these pathways prevented viral replication [19]. However, cellular stress response induced by SARS-CoV-2 is not clear.
In the present study, we explored the transcriptome of virus infection and the transcriptional reprograming of CEP by bulk RNA-sequencing (RNA-seq) in a GX_P2V cell culture model and the single-cell RNA-seq of COVID-19 patients. We found cellular stress responses were in the pathophysiologic mechanism of GX_P2V infection, and HSF-1-mediated heat shock response was the core functional elements. CEP was potent to inhibit the virus by inferring with a majority of virus-perturbed genes and pathways, such as heat shock response, ER stress/unfolded protein response and autophagy, adding evidence for possible application of CEP in treatment of COVID-19. Additionally, we propose that modulating cellular stress is promising for treating SARS-CoV-2 infection.
Methods
Cell lines, coronavirus, key reagents and RNA-seq library preparation
Vero-E6 cells (derived from the kidney of an African green monkey kidney, American Type Culture Collection, Manassas, VA, USA) were plated in T75 culture flasks (1.5 × 107 cells/flask) for 24 h and treated with GX_P2V (MOI = 0.01), CEP (6.25 μM, Aladdin, China, Shanghai) or CEP and GX_P2V, and then cultured for another 72 h. Cells were harvested and mRNA was isolated using TRIzol (Invitrogen). The rRNA was depleted by QIAseq FastSelect-rRNA HMR Kit (Qiagen). The RNA-seq libraries were constructed on Vero-E6 cells using the NEBNext Ultra™ RNA Library Prep Kit for Illumina (NEB) and sequenced on Hiseq 2500 sequencing system (Illumina).
Western blotting and mass spectrum
Treatment of Vero-E6 cells with CEP was performed as described previously [11]. Briefly, Vero-E6 cells (5 × 104/well) were plated in 12-well plates and treated with CEP at different stages of viral infection (full time, entry stage and post-entry stage). The plates were washed three times with phosphate-buffered saline (PBS) and lysed with radioimmunoprecipitation (RIPA) buffer in the presence of a cocktail of proteinase inhibitors (ThermoFisher, USA). The samples were loaded on a 12% SDS-PAGE gel and transferred to a polyvinylidene fluoride membrane. Antibodies against nucleocapsid protein of GX_P2V (GenScript, USA) and GAPDH (Proteintech, USA) were used at 1:1000 and 1:20,000 dilutions, respectively. The second antibody of HRP-conjugated AffiniPure goat anti-mouse IgG (H + L) was diluted at 1:10,000. SuperSignal® West Femto Maximum Sensitivity Chemiluminescent Substrate (Thermo Scientific, USA) was used for imaging. For mass spectrum, equal amount of each protein sample was enzymatically lysed, mixed with five times of the sample volume and precipitated with pre-cooled acetone at −20°C for 2 h. The protein precipitate was sonicated in buffer containing 200 mM Triethylammonium bicarbonate. The digestion was then incubated with trypsin at a ratio of 1: 50 (enzyme: protein, m/m) for overnight. Finally, the samples were desalted according to the C18 ZipTips instructions, and vacuum freeze-dried for HPLC analysis.
RNA-seq data analysis
The FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) tools and fastx_trimmer wrapped in FASTX toolkit were used to trim low-quality bases (Q < 20) and adapter for raw sequences. Cleaned RNA-seq reads were mapped to the reference chlorocebus sabaeus genome ChlSab1.1 (GCA_000409795.2) by HISAT2 (v2.1.0) [20]. Duplicated reads for pair-end data were removed by SAMtools (v1.5) [21]. Counts for each gene were obtained by using HTseq. DESeq2 was used to identify differentially expressed genes between experimental groups [22]. False discovery rate (FDR q value) was calculated by adjusting P values with the Benjamini–Hochberg method. Genes with FDR q value <0.05 and |Log2 (fold change)| > 1 were considered as differential expressed genes. The volcano plot was drawn using the ggplot2 package by R.
Gene set enrichment analysis and selected genes representation
The gct format files including Vero versus Vero + Virus, Vero + Virus versus Vero + Virus + CEP were used as the reference files. Gene sets including (i) regulation of heat shock factor 1 (HSF1)-mediated heat shock response, regulation of cellular response to heat, HSF1 dependent transactivation, HYPOXIA, defense response to virus, response to virus, HIF1 targets, fat cell differentiation and autophagy were download from the MSigDB, KEGG and Reactome database and (ii) gene sets of up/downregulated genes of virus were the differentially expressed genes of Virus versus Vero RNA-seq group described above with the threshold of FDR q value <0.05 and |Log2 (fold change)| > 1. Gene set enrichment P values, normalized enrichment score (NES) values and FDR values reported throughout were calculated with 1000 permutations with GSEA4.0.3 (https://www.gsea-msigdb.org/gsea/index.jsp) [23, 24], ran in Signal2Noise mode.
Heatmap visualization was performed by an R package of GENE-E. Genes selected to presented in the Heatmap as pathway mode were also obtained from MSigDB, KEGG and Reactome database.
Gene ontology and network analysis
Gene ontology analysis was performed with a web-based tool of Metascape (https://metascape.org) [25] with differentially expressed genes obtained as described above. Pathway with P value <0.05 was considered as significantly enriched pathway. Top enriched pathways were shown in bubble plot created by R package ggplot2. The interaction network for each significant enriched pathway and the protein–protein interaction (PPI) network were drawn by Cytoscape [26], which is also wrapped in Metascape website. In detail, for each given gene list, PPI enrichment analysis has been carried out with the following databases: BioGrid [27] and OmniPath [28]. To further capture the relationships between the terms, a subset of enriched terms has been selected and rendered as a network plot, where terms with a similarity >0.3 are connected by edges. We select the terms with the best P values from each of the 20 clusters, with the constraint that there are no more than 15 terms per cluster and no more than 250 terms in total. The network is visualized using Cytoscape, where each node represents an enriched term and is colored first by its cluster ID. Nodes that share the same cluster ID are typically close to each other. The resultant network contains the subset of proteins that form physical interactions with at least one other member in the differentially expressed gene list. If the network contains between 3 and 500 proteins, the molecular complex detection (MCODE) algorithm has been applied to identify densely connected network components. Pathway and process enrichment analysis has been applied to each MCODE component independently, and the three best-scoring terms by P value have been retained as the functional description of the corresponding components, as shown in Figure 2G.
Figure 2.
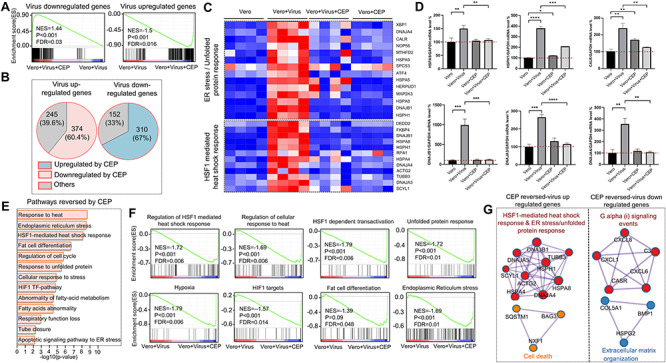
CEP reprograms the transcriptional profile of SARS-CoV-2-related coronavirus in monkey kidney epithelial cells. (A) GSEA of expression profiles on virus up/downregulated genes from virus-infected-Vero-E6 cell with or without CEP treatment. (B) Venn diagrams showed percentages of virus up/downregulated genes reversed by CEP treatment. (C) Heatmap showed genes of unfolded protein response pathway and HSF-1-mediated heat shock response pathway in Vero-E6 cells, virus-infected Vero-E6, virus-infected Vero-E6 treated with CEP and CEP treated Vero-E6 cells. Red cells represented upregulated genes while blue cells represented downregulated genes. (D) RT-qPCR showed expression of HSPA8, HSPH1, CALR, DNAJB1, DNAJA3 and DNAJA4 genes were significantly higher in virus-infected Vero-E6 cells, compared to Vero-E6 cells, Vero-E6 cells with CEP, virus-infected Vero-E6 cells with CEP treatment. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. (E) Bar diagram showed the top CEP-reversed pathways in virus-infected cells. (F) GSEA of expression profile from the CEP treated-virus-infected-Vero-E6 cell and virus-infected-Vero-E6 cell with signatures downloaded from MSigDB, KEGG and Reactome databases. (G) Gene component underneath the pathway showed in (F) with MCODE algorithm, colors indicate different core nodes.
Real-time polymerase chain reaction
Total RNA of Vero-E6 cells was extracted using TRIzol (Invitrogen, USA). Reverse transcription was performed with a Hifair II 1st Strand cDNA synthesis kit with gDNA digester (Yeasen Biotech, Cat:11121ES60, Shanghai, China), and real-time polymerase chain reaction (RT-qPCR) was performed using QuantStudio 1 quantitative RT-PCR detection system (Applied Biosystems, Foster City, CA, USA) with Hieff qPCR SYBR Green Master Mix (Yeasen Biotech, Cat:11202ES08, Shanghai, China). The sequence information of primers (Tsingke Biotech, Shanghai, China) is listed in Supplementary Table 1. RT-qPCR amplification of SYBR Green method was performed as follows: 95°C for 5 min followed by 40 cycles consisting of 95°C for 10 s, 55°C for 20 s and 72°C for 31 s.
Single-cell transcriptome analysis
In total, two single-cell datasets (both generated by Seq-Well platform) were used. Peripheral blood mononuclear cells (PBMC) from COVID-19 patients were downloaded from Gene Expression Omnibus (GEO) with the accession number of GSE150728 [29]. In detail, 44,721 cells, eight peripheral blood samples from seven hospitalized patients with COVID-19 (four of which had acute respiratory distress syndrome (ARDS) and need ventilation) and six healthy controls were extracted. Single cell transcriptome datasets were read and further integrated using Seurat v3.0.2. Briefly, Seurat Object of each dataset was generated by the Seurat function of ‘CreateSeuratObject’. Cells that expressed less than 500 genes were considered outliers and discarded. Raw unique molecular identifier counts were normalized to unique molecular identifier count per million total counts and log-transformed with the Seurat global-scaling normalization function of ‘NormalizedData’. In total, 2000 most variable genes were identified based on average expression and dispersion with the selection method of ‘vst’ by the Seurat function of ‘FindVariableFeatures’. FindIntegrationAnchors function was then used to find correspondences across the different studied datasets (ie., healthy, nonvent, ARDS) with the parameters of dimensionality = 1:20. IntegrateData function was used to generate the integrated Seurat object with the result output from the FindIntegrationAnchors. Next, we applied a linear transformation function called ScaleData of Seurat to shift the expression of each gene to the mean expression across cells to 0, and scales the expression of each gene to make the variance across cells to 1, this step gives equal weight in downstream analyses. Finally, the standard workflow from Seurat was used to find the relevant components with principal component analysis (PCA) (npcs = 30) and to visualize the results with uniform manifold approximation and projection (UMAP) (reduction = ‘pca’, dims = 1:20).
In order to create feature plots and violin plots for gene set with multiple genes (HSF1-mediated heat shock response, HSF1 dependent transactivation and autophagy signature), the average normalized expression of those genes was used. Score of each signature was obtained from the average expression level of each gene set by using the ‘apply’ function of R. Next, standard FeaturePlot and VlnPlot function of Seurat was used to generate the feature plots or violin plots by replacing the original single gene with gene set signature score.
For correlation analysis between gene signatures of HSF1-mediated heat shock response and HSF1 dependent transactivation with autophagy gene signature, another single-cell RNA-seq dataset with angiotensin-coverting enzyme 2 (ACE2+) cells from human lung and Macaca mulatta lung under accession number of GSE148829 were used [30]. Next, score of each signature was obtained from the average expression level of each gene set by using the ‘apply’ function of R. Then, Pearson correlation and scatterplot were performed by GraphPad version 7.0.
Results
The SARS-CoV-2-related coronavirus disturbs homeostasis in cells
To determine the transcriptional response of SARS-CoV-2-related coronavirus, we first compared the transcriptional difference between virus treated Vero-E6 cells and Vero-E6 cells. To this end, we collected poly(A) RNA from virus-infected Vero-E6 cells and Vero-E6 cells and performed RNA-seq to estimate the differentially expressed genes. A total of 1081 genes were reserved with P value <0.05 and |Log2 (Fold change) | > 1, including 619 upregulated genes and 462 downregulated genes (Figure 1A and B). Inflammation-related genes (TRAF1, TNFRSF14, DNAH3, TOX3, BAG3 and CXCL10) and heat shock response genes (HSPA4, HSPA8, HSPA5, HSPH1 and DNAJB1) were upregulated, while genes in alternative complement pathway such as C3, and genes in pathways affecting insulin-like growth factor (IGF1)-Akt signaling such as IGFBP3 were downregulated by the virus (Figure 1B). Specifically, we observed that genes associated with defense response to virus, ER stress and HSF1-mediated heat shock response were significantly upregulated along with virus infection (Figure 1C). Gene ontology pathway enrichment analysis showed that, with the coronavirus GX_P2V infection, virus-upregulated genes were predominantly enriched in tumor necrosis factor-alpha (TNF-α) signaling via NF-κB, unfolded protein responses, ER stress, heat shock response-related pathways such as regulation of HSF1-mediated heat shock response and HSF1-dependent transactivation, cell response to stress and apoptosis (Figure 1D), whereas virus-downregulated genes were mainly enriched in host cell basic functional pathways like extracellular matrix organization and post-translational protein phosphorylation (Supplementary Figure 1). Gene set enrichment analysis (GSEA) further confirmed differentially expressed genes were significantly enriched in multiple signaling pathways including regulation of HSF1-mediated heat shock response, regulation of cellular response to heat, HSF1 dependent transactivation, unfolded protein response, ER stress, hypoxia, defense response to virus and response to virus (Figure 1E). To further decipher functionally grouped gene ontology and the crosstalk of those signaling pathways, network analyses were performed with ClueGO, a Cytoscape plug-in [31]. Interestingly, among all the enriched pathways, HSF1-dependent transactivation pathway is at the core, extensively interacting with other pathways such as cellular response to stress, ER stress, TNF-α signaling, hypoxia and autophagy (Figure 1F). In addition, PPI network analysis with the MCODE algorithm has been applied to identify densely connected network component genes. Results showed that genes such as HSPA8, HSPH1, HSPA5, DNAJA4, DNAJB1, DNAJC3, AP1G1 and BAG3 were widely linked to other genes, and these genes were typically associated with heat shock response and ER stress, again indicating cellular stress response is critical in the pathogenesis of the SARS-CoV2-related coronavirus infection (Figure 1G, Supplementary Figure 2).
Figure 1.
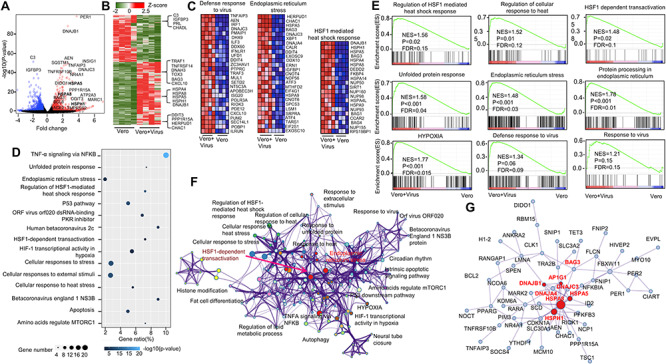
Host transcriptional response to SARS-CoV-2-related coronavirus in monkey kidney epithelial cells. (A and B) Volcano plot (A) and heatmap (B) of total 1081 differential expression genes with P value <0.05. Red color represented upregulated genes in virus-infected cell with Log2(fold change) > 1 and blue represented downregulated genes with Log2(fold change) < −1, relative expression in z-score mode was showed in heatmap. Red cells represented upregulated genes; green cells represented downregulated genes. (C) Heatmap of pathways of defense response to virus, ER stress and HSF1-mediated heat shock response. Red cells represented upregulated genes; blue cells represented downregulated genes. (D) The bubble plot of top upregulated pathways enriched from 619 upregulated genes in virus-infected cells. The bubble color scaled the enrichment score. Light color means more significant enrichment. The size of the bubble scaled the count of the enriched genes. X-axis is equal to gene ratio, which means the percentage of enriched target genes among total 619 genes. Y-axis is the name of the Metascape enriched pathway. (E) GSEA of expression profile from the virus-infected Vero-E6 cell and uninfected Vero-E6 cells with signatures downloaded from MSigDB, KEGG and Reactome databases. (F) Network visualization of pathways enriched by 619 upregulated genes in virus-infected cells performed by ClueGO plug-in of Cytoscape, edges width indicates the number of overlapped genes between individual terms, color indicate different GO term groups. (G) Protein–protein network visualization of gene component underneath the pathway networks showed in (F) with MCODE algorithm, red nodes indicate the core nodes of the network. Red font indicates genes on ER stress and HSF-1-mediated heat shock response pathways.
CEP was potent to reverse virus-perturbed gene expression and pathways
Our previous work showed that CEP has significant anti-viral power against the SARS-CoV-2-related coronavirus, GX_P2V [11]. Moreover, the anti-viral effect of CEP mainly takes effect at post-entry stage (Supplementary Figure 3); however, the exact mechanisms remain unclear. Therefore, transcriptome analyses were performed and we found that CEP demonstrates strong power to reverse virus-perturbed gene expression and pathways (Figure 2A and B, Supplementary Figure 4). In general, most viral downregulated genes were upregulated by CEP (NES = 1.44, P < 0.001, FDR = 0.03), and viral upregulated genes were downregulated by CEP (NES = −1.5, P < 0.001, FDR = 0.016) (Figure 2A). Specifically, among 619 upregulated genes by the virus, 374 genes (60.4%, only significant core enrichment were included here) were downregulated by CEP, whereas 310 genes (67%) among 462 downregulated genes were upregulated by CEP (Figure 2B). Among them, gene signatures of unfolded protein response and HSF1-mediated heat shock response were significantly upregulated in virus-infected Vero-E6 cells, but their expression was comparable in CEP treated virus-infected Vero-E6 cells and CEP treated Vero-E6 cells compared to the control (Figure 2C). For example, the expression of HSPH1 gene and CALR gene were upregulated in virus-infected cells but were brought down by CEP treatment (Supplementary Figures 4 and 5). In consistent with this, RT-qPCR showed that expression of HSPA8, HSPH1, CALR, DNAJB1, DNAJA3 and DNAJA4 in virus-infected Vero-E6 cells were significantly higher than in Vero-E6 cells, Vero-E6 cells with CEP, and infected Vero-E6 cells with CEP, whereas the expression of these genes was similar in Vero-E6 cells and Vero-E6 cells with CEP except CALR (Figure 2D). Furthermore, protein-level of selected genes from related pathways showed the same trend (Supplementary Figure 6). The reversed genes by CEP were predominantly enriched in heat shock response-related pathways (such as response to heat, HSF1-mediated heat shock response), ER stress/response to unfolded protein and hypoxia pathways (Figure 2E and F). Interestingly, fat metabolism pathways such as fat cell differentiation, abnormality of fatty acid metabolism, fatty acids abnormality were also affected by the virus but were reversed by CEP (Figure 2E and F). This is consistent with the clinical observation that statin use is associated with lower risk of mortality in COVID-19 patients [32], indicating lipid metabolism may take a part in the pathogenesis of SARS-CoV-2 infection and can be a therapeutic target. To identify densely connected network component genes located at the core of the CEP reversed genes, MCODE were applied. Consistently, we identified four clusters of genes, which were components of HSF1-mediated heat shock response and unfolded protein response, cell death, G alpha signaling events and extracellular matrix organization pathway, respectively (Figure 2G, Supplementary Figure 7). The former two pathways were essential for cell response to stress, while the latter two pathways were essential for cell to execute the basic biological functions.
Single-cell transcriptome analysis revealed SARS-CoV-2 induces cellular stress responses and autophagy in PBMC from COVID-19 patients
To confirm the possible mechanism of CEP and the role of cellular stress responses in COVID-19, we also analyzed two published single-cell RNA-seq datasets [29, 30]. The first dataset includes eight peripheral blood samples from seven hospitalized patients with COVID-19 (four of which had ARDS and need ventilation) and six healthy controls. With the subject extracted from the original dataset, a cells-by-genes expression matrix was created and dimensionality reduction by UMAP and graph-based clustering were performed, identifying 20 clusters (Figure 3A and B, same with the original paper [29]). To determine the expression level of HSF1 regulated genes in PBMC, we performed GSEA on those 20 clusters. Interestingly, we found gene signatures of regulation of HSF1-mediated heat shock response, HSF1 dependent transactivation and ER stress were predominantly enriched in clusters of IgG and IgA plasmablasts (PB), and proliferative lymphocytes (Figure 3C and D), cell populations that were specifically increased in COVID-19 patients (Figure 3A and B). Furthermore, from healthy, nonventilated patients (NonVent) to ARDS patients need ventilation, cells with gene signatures of regulation of HSF1-mediated heat shock response as well as HSF1 dependent transactivation and ER stress pathways were increasingly enriched, indicating those gene signatures of cellular stress responses were correlated with severity of COVID-19 (Figure 3E–G, and Supplementary Figure 8).
Figure 3.
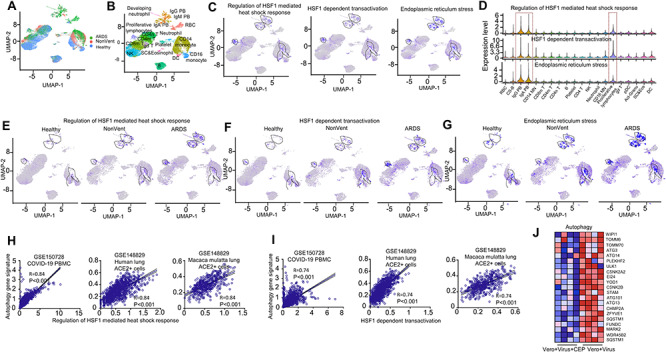
SARS-CoV-2 induces cellular stress responses in PBMC from COVID-19 patients. (A) Integrated UMAP visualization of 44,721 PBMC determined by Seurat v3. Each dot represents a single cell. Cells from different samples are color-coded (ARDS patients: green, Nonventilated patients: pink, healthy donors: light blue). (B) UMAP visualization of integrated projection PBMC that were assigned to 20 clusters and color-coded based on the clusters. (C) Enrichment of the gene signatures of regulation of HSF1-mediated heat shock response, HSF1 dependent transactivation, ER stress in PBMC from patients with COVID-19 and healthy controls, determined by multiple feature analysis of Seurat v3. Single-cell average expression levels of each gene signature were illustrated in the UMAP plots and violin plots (D). Expression levels are color-coded by gray (not expressed) and blue (expressed). Each dot corresponds to one individual cell, and the violin represents the probability density at each value. Those three gene signatures were significantly enriched in IgG/IgA PB and proliferative lymphocytes, highlighted in black circles of the UMAP (C) and red box on violins (D). (E–G) Enrichment of the individual gene signature in PBMC split by samples from healthy donors (left), Non-Vent (middle) and ARDS (right) patients were illustrated by UMAP plots. (H–I) Correlation analysis between gene signatures in regulation of HSF1-mediated heat shock response (H, X-axis), HSF1 dependent transactivation (I, X-axis) and autophagy (Y-axis) in PBMC from COVID-19 patients (left), Human lung ACE2 positive cells (middle) and M. mulatta lung ACE2 positive cells (right). (J) Heatmap showed genes of autophagy pathway in virus infected Vero-E6 cells with or without CEP treatment. Red cells represented upregulated genes while blue cells represented downregulated genes.
A recent study proposed that the heat shock protein 90 could be a target to treat COVID-19 by serving as a major component that enables viruses to hijack infected cells through the process of autophagy [33]. Our data showed the autophagy-associated genes were mainly expressed in the same PBMC populations in severe COVID-19 patients, together with red blood cells (Supplementary Figure 9). Indeed, both genes associated with the regulation of HSF1-mediated heat shock response and genes associated with HSF1 dependent transactivation were positively correlated with autophagy gene signatures in cells where they predominantly located (left panel of Figure 3H and 3I). To confirm this observation, we further analyzed another single-cell RNA-seq dataset including ACE2 positive lung epithelial cells from both human and M. mulatta lung. Both datasets showed significant correlation between HSF1 and autophagy-associated gene signatures (right panel of Figure 3H and 3I). In our bulk RNA-seq data, CEP also showed the power to reverse the expression of autophagy-associated genes in infected cells (Figure 3J). Altogether, cellular stress response and autophagy may play an important role in the pathogenesis of COVID-19.
Discussion
Here our transcriptome analysis demonstrated the induction of various cellular responses by GX_P2V infection and CEP was potent to inhibit the virus and modulate multiple pathways including cellular stress responses and autophagy, helping cell recover from the insult. Cellular stress responses were induced in PBMC, especially IgG/IgA PB and proliferative lymphocytes, in COVID-19 patients, and were associated with severity of the disease.
Heat shock proteins (Hsps) have crucial roles in the maintenance of the conformation, stability, activation and cellular localization of several key proteins that are involved in cell signaling, proliferation and survival, protecting cells from stressful conditions including virus infection. As expected, genes coding for Hsps was significantly upregulated in cells infected with GX_P2V (Figure 1). However, Hsps, including Hsp70 and Hsp90, can be induced by virus which in turn enhances virus replication [34]. For instance, Hsp90 is important for several virus replications and Hsp90 specific inhibitor geldanamycin blocks the replication of the coronavirus in cell culture systems. A recent drug repositioning based on datasets of SARS family of coronaviruses proposed that Hsp90 inhibitors, such as geldanamycin, could be options for COVID-19 therapy [33]. Inside a eukaryotic cell, most of the transmembrane and secreted protein are translated, modified, and folded in the ER. During the replication of virus, substantial amounts of viral proteins overload the protein processing machinery, unfolded/misfolded proteins will evoke ER stress and unfolded protein response, which result in shutdown of global translation and adjustment of biosynthetic burden [35]. A proteomics study has also shown that proteins for unfolded protein processing were increased during SARS-CoV-2 infection [19]. In consistent with this, our study revealed that GX_P2V induced significant ER stress and unfolded protein response in Vero-E6 cells. However, viruses such as SARS-CoV have evolved the capacity to overcome the protein translation shutoff to ensure the production of viral protein [15, 35]. If the damage to the ER is severe or persistent, cell death including apoptosis and autophagy will be induced.
Autophagy is often initiated to curtail infection by delivering viral particles for lysosomal degradation and inducing interferon-mediated clearance; however, some viruses have evolved anti-autophagy strategies escaping host immunity and promoting viral replication [36]. During autophagy, autophagosomes formed and can be hijacked by some viruses and used as safe sanctuary [37]. It has been concluded that coronaviruses induce autophagy but do not require the complete autophagy pathway [38]. Studies have shown SARS-CoV-2 limits autophagy flux by interfering lysosomal-autophagosomal fusion [39], whereas the cytopathic effect of SARS-CoV-2 could be blocked with autophagy inhibitors [40]. It seems conflicting that virus inhibits autophagic clearance while autophagy inhibitors suppress viral infection. Those autophagy inhibitors were speculated to interrupt early stages of viral life cycle, namely the fusion of the viral endosomes with the lysosome, reducing viral replication and protecting cells from viral induced cell death [40]. Our analysis showed that SARS-CoV-2 also induced autophagy-associated genes expression and the virus-induced heat shock response was associated with autophagy (Figure 3). Therefore, cellular stress response and autophagy could play a crucial role in pathogenesis of COVID-19.
CEP has been used to treat a diverse range of medical conditions including anemia, leukopenia, thrombocytopenia, alopecia, sarcoidosis, some cancer and even virus infection (e.g. Human immunodeficiency virus, hepatitis B virus and Ebola virus) with rare and modest toxicities [41]. The mechanism of action of CEP is multifactorial, interfering cell membranes, metabolic axes (e.g. NF-κB pathway, JAK/STAT pathway), scavenging free radicals, binding to Hsp90 [10] and inhibit autophagy/mitophagy through blockage of autophagosome-lysosome fusion [42]. We and others have shown CEP inhibits virus at entry and post-entry step [11, 13]; however, the mechanisms of CEP’s antiviral activity after virus entry into cell remains unclear. Recent researches have demonstrated CEP also strongly inhibits nonstructural protein (Nsp) 13 activity and Nsp12-Nsp8-Nsp7 complex of SARS-CoV-2, which were essential for viral replication and transcription [43, 44]. We reported the antiviral mechanism of CEP against GX_P2V appears to be associated with cell stress response-autophagy cascade, indicating that CEP exhibits broad-spectrum antiviral activity targeting multiple molecules and pathways. Moreover, NF-κB-related signaling pathway also ranked high in this GX_P2V infection model. As a hallmark of virus infections, activation of NF-κB was firstly interpreted that the host utilizes NF-κB to trigger defense mechanisms against the invader [45]. However, some viruses have evolved strategies to benefit their replication by hijacking NF-κB-driven cellular functions [45]. Bing a NF-κB suppressor, CEP was also speculated to inhibit the SARS-CoV-2-related coronavirus by inhibiting NF-κB-related pathway. Despite the close relationship of GX_P2V to SARS-CoV-2 [11], the pathogenicity of GX_P2V is much weaker than SARS-CoV-2, indicating their pathogenesis may not be exactly the same. Another limitation is that our study only used in vitro cell culture data without confirmation using in vivo infection models. Therefore, the test of efficacy of CEP in SARS-CoV-2 infection models of human cells and COVID-19 patients are urgently required.
SARS-CoV-2 infection can lead to lymphopenia [46–48], low blood lymphocyte percentage predicts severe disease of COVID-19 [48]. However, the mechanisms of lymphopenia in COVID-19 remain incompletely understood. Lymphopenia can augment T cell activation and proliferation [49]. Impressive numbers of IgG/IgA PB [29, 50] and proliferative lymphocytes [29] appeared in the peripheral blood of COVID-19 patients, especially severe individuals, which is a self-protection. During acute viral infections, abundant PB, probably derived from memory B cells, appear in the circulation transiently [51]. Rapid activation of memory B cells and their differentiation into antibody-secreting PB might provide antibodies, which can partially neutralize the virus and stop further dissemination in the host [51]. Our analyses demonstrated that both PB and proliferative lymphocytes were in apparent cell stress state, and autophagy was induced. A possible explanation is that these cells were infected by the virus directly or affected by the virus-induced hyperinflammation, resulting in cell death if the stress persists. CEP, by inhibiting viral infection and modulating cellular stress responses and autophagy, may protect lymphocytes from these insults. However, the role of activated cellular stress responses in IgG/IgA PB, and proliferative lymphocytes in SARS-CoV-2 infection warrants further study.
In summary, our study provided a transcriptome of an infection model of the SARS-CoV-2-related virus GX_P2V, and uncovered the cellular responses to the virus and anti-viral activities of CEP, providing evidence for CEP as a promising therapeutic option for COVID-19. Furthermore, the cellular stress response-stress-induced death cascade is a potential therapeutic target for SARS-CoV-2 infection.
Key Points
GX_P2V, a SARS-CoV-2-related coronavirus, disturbs homeostasis in cells.
CEP was potent to reverse most of viral-perturbed gene expression and pathways.
Single-cell transcriptome analysis revealed SARS-CoV-2 induces cellular stress responses and autophagy in PBMC from COVID-19 patients.
Supplementary Material
Shasha Li is a post-doc at the Beijing Advanced Innovation Center for Soft Matter Science and Engineering, College of Life Science and Technology, Beijing University of Chemical Technology. She is focusing on bioinformatics pipeline developing for virology.
Wenli Liu is a PhD student at the Beijing Advanced Innovation Center for Soft Matter Science and Engineering, College of Life Science and Technology, Beijing University of Chemical Technology and focused on virology.
Yangzhen Chen is a postgraduate at the College of Life Science and Technology, Beijing University of Chemical Technology and focused on virology.
Liqin Wang is a postgraduate at the College of Life Science and Technology, Beijing University of Chemical Technology and focused on virology.
Wenlin An is a professor in Beijing Advanced Innovation Center for Soft Matter Science and Engineering, College of Life Science and Technology, Beijing University of Chemical Technology and focused on virology.
Xiaoping An is a research associate at the Beijing Advanced Innovation Center for Soft Matter Science and Engineering, College of Life Science and Technology, Beijing University of Chemical Technology and is focusing on novel pathogen screening.
Lihua Song a professor in Beijing Advanced Innovation Center for Soft Matter Science and Engineering, College of Life Science and Technology, Beijing University of Chemical Technology. He is interested in novel pathogen screening.
Yigang Tong is a professor in Beijing Advanced Innovation Center for Soft Matter Science and Engineering, College of Life Science and Technology, Beijing University of Chemical Technology. He is interested in genomics and virology.
Huahao Fan is a professor in Beijing Advanced Innovation Center for Soft Matter Science and Engineering, College of Life Science and Technology, Beijing University of Chemical Technology and focusing on drug repurposing and virology.
Chenyang Lu is a scientist in the Department of Rheumatology and Immunology, West China Hospital, Sichuan University and interested in anti-viral immunity and drug discovery.
Contributor Information
Shasha Li, Beijing Advanced Innovation Center for Soft Matter Science and Engineering, College of Life Science and Technology, Beijing University of Chemical Technology.
Wenli Liu, Beijing Advanced Innovation Center for Soft Matter Science and Engineering, College of Life Science and Technology, Beijing University of Chemical Technology.
Yangzhen Chen, College of Life Science and Technology, Beijing University of Chemical Technology.
Liqin Wang, College of Life Science and Technology, Beijing University of Chemical Technology.
Wenlin An, Beijing Advanced Innovation Center for Soft Matter Science and Engineering, College of Life Science and Technology, Beijing University of Chemical Technology.
Xiaoping An, Beijing Advanced Innovation Center for Soft Matter Science and Engineering, College of Life Science and Technology, Beijing University of Chemical Technology.
Lihua Song, Beijing Advanced Innovation Center for Soft Matter Science and Engineering, College of Life Science and Technology, Beijing University of Chemical Technology.
Yigang Tong, Beijing Advanced Innovation Center for Soft Matter Science and Engineering, College of Life Science and Technology, Beijing University of Chemical Technology.
Huahao Fan, Beijing Advanced Innovation Center for Soft Matter Science and Engineering, College of Life Science and Technology, Beijing University of Chemical Technology.
Chenyang Lu, Department of Rheumatology and Immunology, West China Hospital, Sichuan University.
Contribution
S.L., H.F., W.L., Y.C., L.W., W.A., X.A. and LS. performed the experiments and data analyses. S.L. and C.L. wrote the manuscript, and Y.T. and H.F. reviewed it. Y.T., H.F. and C.L. supervised the whole project.
Funding
This research was supported by Key Project of Beijing University of Chemical Technology (No. XK1803-06), National Key Research and Development Program of China (2018YFA0903000, 2020YFC2005405, 2020YFA0712100, 2020YFC0840805), Funds for First-class Discipline Construction (No. XK1805), Inner Mongolia Key Research and Development Program (No. 2019ZD006), National Natural Science Foundation of China (81672001, 81621005), NSFC-MFST project (China-Mongolia) (No. 31961143024), Fundamental Research Funds for Central Universities (No. BUCTRC201917, BUCTZY2022).
Data availability
All datasets underlying this article have been deposited to the GEO database under the GEO accession number of GSE158050.
Reference
- 1. WHO . Coronavirus Disease 2019 (COVID-19) Weekly Epidemiological Update and Weekly Operational Update. 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports (9 November 2020, date last accessed).
- 2. Hung IF-N, Lung K-C, Tso EY-K, et al. Triple combination of interferon beta-1b, lopinavir–ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. The Lancet 2020;395(10238):1695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tang W, Cao Z, Han M, et al. Hydroxychloroquine in patients with mainly mild to moderate coronavirus disease 2019: open label, randomised controlled trial. BMJ 2020;369:m1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Borba MGS, Val FFA, Sampaio VS, et al. Effect of high vs low doses of chloroquine diphosphate as adjunctive therapy for patients hospitalized with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. JAMA Netw Open 2020;3(4):e208857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gautret P, Lagier JC, Parola P, et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents 2020;56(1):105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6. Chen J, Liu D, Liu L, et al. A pilot study of hydroxychloroquine in treatment of patients with common coronavirus disease-19 (COVID-19). Zhejiang Da Xue Xue Bao Yi Xue Ban 2020;49(2):215–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cai Q, Yang M, Liu D, et al. Experimental treatment with favipiravir for COVID-19: an open-label control study. Engineering 2020;6(10):1192–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Grein J, Ohmagari N, Shin D, et al. Compassionate use of remdesivir for patients with severe Covid-19. N Engl J Med 2020;382(24):2327–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang Y, Zhang D, Du G, et al. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled. Multicentre Trial Lancet 2020;395(10236):1569–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bailly C. Cepharanthine: an update of its mode of action, pharmacological properties and medical applications. Phytomedicine 2019;62:152956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fan HH, Wang LQ, Liu WL, et al. Repurposing of clinically approved drugs for treatment of coronavirus disease 2019 in a 2019-novel coronavirus-related coronavirus model. Chin Med J (Engl) 2020;133(9):1051–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lam TT, Jia N, Zhang YW, et al. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature 2020;583(7815):282–5. [DOI] [PubMed] [Google Scholar]
- 13. Ohashi H, Watashi K, Saso W, et al. Multidrug treatment with nelfinavir and cepharanthine against COVID-19. bioRxiv 2020:04.14.039925. [Google Scholar]
- 14. Ke P, Chen S. Hepatitis C virus and cellular stress response: implications to molecular pathogenesis of liver diseases. Viruses 2012;4(10):2251–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chan CP, Siu KL, Chin KT, et al. Modulation of the unfolded protein response by the severe acute respiratory syndrome coronavirus spike protein. J Virol 2006;80(18):9279–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Carpenter JE, Jackson W, Benetti L, et al. Autophagosome formation during varicella-zoster virus infection following endoplasmic reticulum stress and the unfolded protein response. J Virol 2011;85:9414–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020;579(7798):270–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang B, Hu Y, Chen L, et al. Mining of epitopes on spike protein of SARS-CoV-2 from COVID-19 patients. Cell Res 2020;30:702–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bojkova D, Klann K, Koch B, et al. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature 2020;583(7816):469–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim D, Paggi JM, Park C, et al. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 2019;37(8):907–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map format and SAMtools. Bioinformatics 2009;25(16):2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102(43):15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 2003;34(3):267–73. [DOI] [PubMed] [Google Scholar]
- 25. Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 2019;10(1):1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13(11):2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stark C, Breitkreutz BJ, Reguly T, et al. BioGRID: a general repository for interaction datasets. Nucleic Acids Res 2006;34(Database issue):D535–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ceccarelli F, Turei D, Gabor A, et al. Bringing data from curated pathway resources to Cytoscape with OmniPath. Bioinformatics 2020;36(8):2632–3. [DOI] [PubMed] [Google Scholar]
- 29. Wilk AJ, Rustagi A, Zhao NQ, et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat Med 2020;26(7):1070–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ziegler CGK, Allon SJ, Nyquist SK, et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell 2020;181(5):1016, e19–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bindea G, Mlecnik B, Hackl H, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009;25(8):1091–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang XJ, Qin JJ, Cheng X, et al. In-hospital use of statins is associated with a reduced risk of mortality among individuals with COVID-19. Cell Metab 2020;32(2):176–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sultan I, Howard S, Tbakhi A. Drug repositioning suggests a role for the heat shock protein 90 inhibitor Geldanamycin in treating COVID-19 infection. 2020. doi: 10.21203/rs.3.rs-18714/v1. [DOI]
- 34. Kim MY, Oglesbee M. Virus-heat shock protein interaction and a novel axis for innate antiviral immunity. Cell 2012;1(3):646–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fung TS, Liao Y, Liu DX. Regulation of stress responses and translational control by coronavirus. Viruses 2016;8(7):184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mao J, Lin E, He L, et al. Autophagy and viral infection. Adv Exp Med Biol 2019;1209:55–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Choi Y, Bowman JW, Jung JU. Autophagy during viral infection - a double-edged sword. Nat Rev Microbiol 2018;16(6):341–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Maier HJ, Britton P. Involvement of autophagy in coronavirus replication. Viruses 2012;4(12):3440–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gassen NC, Papies J, Bajaj T, et al. Analysis of SARS-CoV-2-controlled autophagy reveals spermidine, MK-2206, and niclosamide as putative antiviral therapeutics. bioRxiv 2020:04.15.997254. [Google Scholar]
- 40. Gorshkov K, Chen CZ, Bostwick R, et al. The SARS-CoV-2 cytopathic effect is blocked with autophagy modulators. bioRxiv 2020:05.16.091520. [Google Scholar]
- 41. Rogosnitzky M, Danks R. Therapeutic potential of the biscoclaurine alkaloid, cepharanthine, for a range of clinical conditions. Pharmacol Rep 2011;63(2):337–47. [DOI] [PubMed] [Google Scholar]
- 42. Zhang Y, Jiang X, Deng Q, et al. Downregulation of MYO1C mediated by cepharanthine inhibits autophagosome-lysosome fusion through blockade of the F-actin network. J Exp Clin Cancer Res 2019;38(1):457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. White MA, Lin W, Cheng X. Discovery of COVID-19 inhibitors targeting the SARS-CoV2 Nsp13 helicase. bioRxiv 2020:08.09.243246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ruan Z, Liu C, Guo Y, et al. SARS-CoV-2 and SARS-CoV: virtual screening of potential inhibitors targeting RNA-dependent RNA polymerase activity (NSP12). J Med Virol 2020. doi: 10.1002/jmv.26222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Santoro MG, Rossi A, Amici C. NF-kappaB and virus infection: who controls whom. EMBO J 2003;22(11):2552–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chen Z, John WE. T cell responses in patients with COVID-19. Nat Rev Immunol 2020;20(9):529–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet 2020;395(10223):497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tan L, Wang Q, Zhang D, et al. Lymphopenia predicts disease severity of COVID-19: a descriptive and predictive study. Signal Transduct Target Ther 2020;5(1):33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Surh CD, Sprent J. Homeostasis of naive and memory T cells. Immunity 2008;29(6):848–62. [DOI] [PubMed] [Google Scholar]
- 50. Kuri-Cervantes L, Pampena MB, Meng W, et al. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci Immunol 2020;5(49):eabd7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fink K. Origin and function of circulating plasmablasts during acute viral infections. Front Immunol 2012;3:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets underlying this article have been deposited to the GEO database under the GEO accession number of GSE158050.


