Abstract
Segmentation of head and neck cancer (HNC) primary tumors on medical images is an essential, yet labor-intensive, aspect of radiotherapy. PET/CT imaging offers a unique ability to capture metabolic and anatomic information, which is invaluable for tumor detection and border definition. An automatic segmentation tool that could leverage the dual streams of information from PET and CT imaging simultaneously, could substantially propel HNC radiotherapy workflows forward. Herein, we leverage a multi-institutional PET/CT dataset of 201 HNC patients, as part of the MICCAI segmentation challenge, to develop novel deep learning architectures for primary tumor auto-segmentation for HNC patients. We preprocess PET/CT images by normalizing intensities and applying data augmentation to mitigate overfitting. Both 2D and 3D convolutional neural networks based on the U-net architecture, which were optimized with a model loss function based on a combination of dice similarity coefficient (DSC) and binary cross entropy, were implemented. The median and mean DSC values comparing the predicted tumor segmentation with the ground truth achieved by the models through 5-fold cross validation are 0.79 and 0.69 for the 3D model, respectively, and 0.79 and 0.67 for the 2D model, respectively. These promising results show potential to provide an automatic, accurate, and efficient approach for primary tumor auto-segmentation to improve the clinical practice of HNC treatment.
Keywords: PET, CT, Tumor segmentation, Head and neck cancer, Deep learning, Auto-contouring
1. Introduction
Head and neck cancer (HNC) affects over 50,000 individuals and has a mortality rate of over 10,000 annually [1]. A vast majority of HNC patients receive radiotherapy, which targets the tumor tissue with focused radiation beams from different directions, while trying to spare the surrounding tissues as much as possible [2]. Performed by the radiation oncologist, definite primary and lymph node tumor delineation dictates subsequent radiation dose optimization. The high prescribed dose is delivered to the segmented tumor, while limiting the dose directly surrounding the segmentation. Inadequate tumor definition can therefore directly lead to under-dosage of the tumor, increasing treatment failure risk, or, in contrast, administering too much dose to the surrounding normal tissues. Adequate manual tumor segmentation is labor-intensive and subject to inter-observer variation [3-8]. Since, at present, CT tissue density information is needed for dose calculation, contours are defined on the CT, and often secondarily by 18F-FDG Positron Emission Tomography (PET), providing additional information on the tissue’s metabolic activity. Automatic segmentation of the primary tumor effectively utilizing the synergistic information from the PET and CT together is an unmet need to decrease the work-load of tumor delineation, as well as to decrease inter-variability between observers.
Deep learning (DL), an artificial intelligence subtype, is a strong tool for segmentation problems [9, 10]. DL techniques for segmentation applications on medical images for HNC radiotherapy purposes is a relative novel, yet emerging field [11]. An array of studies have peered into the difficult task of primary tumor segmentation with DL in single modality images, predominantly CT [12]. DL studies utilizing dual modalities, such as PET/CT [13-23], demonstrate the potential to outperform DL networks based on single image modalities [13, 21, 22, 24]. Likely due to the complex regional head and neck anatomy, PET/CT DL for HNC auto-contouring showed variable success, with dice similarity coefficients (DSC) ranging from 0.61 to 0.785 [13, 20-22]. These studies are often limited by small numbers of patients in the training and test datasets. The DL architectures for these studies vary, with 2D image (i.e. predictions made on a slice by slice basis) or 3D image approaches (i.e. predictions made by inputting the entire image volume) predominating.
The aim of this study was to develop and validate primary tumor auto-contouring with 2D/3D DL approaches that utilize PET and CT images simultaneously based on multi-institutional HNC data, as part of the MICCAI 2020: HECKTOR challenge.
2. Methods
We developed a deep learning model (Sect. 2.3) for auto-segmentation of primary tumors of HNC patients using co-registered 18F-FDG PET and CT imaging data (Sect. 2.1). The ground truth manual segmentation of the tumors and the normalized imaging data (Sect. 2.2) were used to train the model (Sect. 2.4). The performance of the trained model for auto-segmentation was validated using a 5-fold cross validation approach (Sect. 2.5).
2.1. Imaging Data
The data set used in this paper, which was released by AIcrowd [25] for the HECKTOR challenge at MICCAI 2020 [26], consists of co-registered 18F-FDG PET and CT scans for 201 HNC patients, of which the majority were oropharyngeal cancer patients. All imaging data was paired with manual segmentations of the HN primary tumors, i.e. primary gross tumor volume (GTVp), which were considered as the ground truth, in Neuroimaging Informatics Technology Initiative (NIfTI) format.
2.2. Image Processing
To mitigate the variable resolution and size of the PET and CT image per patient, all images (i.e., PET, CT, and GTVp masks) were cropped to fixed bounding box volumes of size 144 × 144 × 144 mm3 in the x, y and z dimensions. These bounding boxes were provided with the imaging data (Sect. 2.1) by [25]. Then, the cropped images were resembled to a fixed image size of 144 × 144 × 96 voxels. These specified number of voxels were chosen to match the maximum number of voxels found in the cropped CT images in the x, y, and z dimensions in all patients. The CT intensities were truncated in the range of [−200, 200] Hounsfield Units (HU) to increase soft tissue contrast. The intensities of the truncated CT images were then rescaled to a [−1, 1] range. The intensities of PET images were truncated between the 10th and 99th percentile to improve the images’ contrast, and subsequently with z-normalization ([intensity-mean]/standard_deviation), resulting in a mean of zero and standard deviation of one for the entire cohort.
2.3. Segmentation Model Architecture
We developed 2D and 3D fully convolutional neural network (CNN) models based on the U-net architecture [27] and our previous 2D U-net model [28], using 4 convolution blocks in the encoding and decoding branches of the U-net. For each block, we used one convolution layer. The down sampling in the encoding branch was performed using a stride 2 convolution instead of max pooling layers to improve the model expressive ability through learning pooling operations compared to fixed pooling operations [29]. The up-sampling in the decoding branch was performed using convolution transpose layers which have been shown to be effective in previous studies [30-33]. Each convolution layer was directly followed by a batch normalization and a Leaky Relu activation layer; a Leaky Relu was chosen instead of Relu to mitigate the effect of improper model weight initialization and data normalization on the model training performance due to the “dying Relu” problem [34, 35]. The encoding and decoding blocks were linked using concatenation layers. Finally, the last layer was a Sigmoid activation layer. Figure 1 shows an illustration of the 3D U-net architecture proposed in this work. A similar architecture, but substituting the 3D with 2D convolution layers, was used to build the 2D U-net model. The batch normalization and Leaky Relu activation layers after each convolution layers were omitted from Fig. 1 for clarity. The number of filters used for the 4 convolution blocks were 16, 32, 48, 64, and 80 (Fig. 1). We maximized the number of filters (16 filters) in the first convolution block such that the data could be fit in GPU memory used for the model training, while an increment of 16 filters were used for the other convolution blocks. The total number of trainable parameters were 1,351,537 for the 3D model, and 452,113 for the 2D model.
Fig. 1.
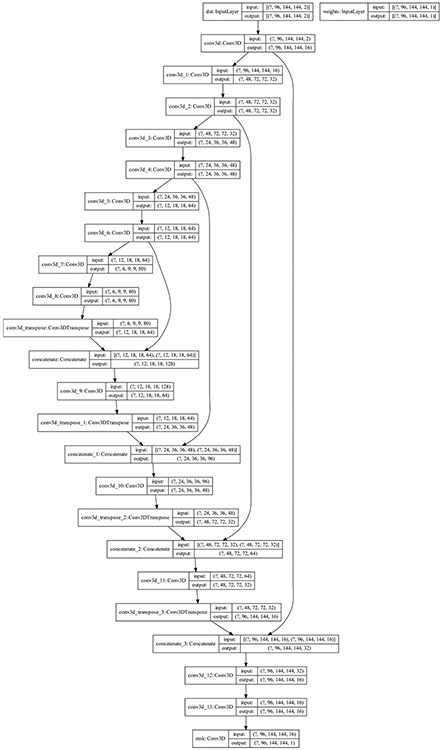
An illustration of the 3D U-net model architecture.
2.4. Model Implementation
The processed PET and CT images (Sect. 2.2) were used as two inputs channels to the segmentation model (Sect. 2.3), resulting in an input layer size of [96, 144, 144, 2] for the 3D model and [144, 144, 2] for the 2D model which represent [z, y, x, channels] and [y, x, channels], respectively. The processed manual segmentation GTVp masks were used as the ground truth target to train the segmentation model. The processed images and masks were split into a training, validation, and test dataset (Sect. 2.5) and then used to train, validate, and test the segmentation model, accordingly. The optimizer used was ‘Adam’ with a learning rate of 5 * 10−5. The batch size was 1 for the 3D model and 96 for the 2D model. To minimize risk of over-fitting, data augmentation of the processed linked PET, CT, and mask images was implemented using a rotation range of 5°, image scaling (i.e. zoom), intensity range shifting of 5%, and horizontal-flipping of images. The same random transformations were applied to the whole PET/CT/masks images for the 3D model per patient, while for the 2D model, each single image has different random transformations. The model performance metrics were the dice similarity coefficient (DSC), the recall or sensitivity, and precision or positive predictive value [28]. We note there is a class imbalance of tumor representation compared to normal tissue (i.e., the number of images that contain GTVp’s is less than the number of images without GTVp’s – i.e. normal tissue). This problem can lead to a low sensitivity in tumor identification by the model and lower the model performance for tumor segmentation. Therefore, to reduce the class imbalance effect, the model was trained using a loss function given as the summation of the loss function of DSC and a weighted Binary Cross Entropy (BCE) loss function as shown in Eqs. (1), (2), and (3).
| (1) |
| (2) |
| (3) |
where MGT and MPred are the ground truth and predicted tumor masks, respectively, and W is the sample-weight used to scale the loss for each image. The sample-weight is a function of the number of pixels in the provided ground truth manual segmentation mask as show in previous work [28]. The weight of the loss function that corresponds to tumor with larger cross-sectional area will be larger than that with smaller areas as well as normal tissue image. Figure 2 show an example of the sample-weight used to scale the loss of each image based on the size of the tumor of the image. The use of the weight-loss biases the model to focus on reducing the loss function more on images with larger tumor size compared to those with lower tumor size and normal tissue images and therefore improves the model sensitivity and overall model performance for tumor segmentation. The sample-weight is provided to the model as a second model input and it has the same size as the target ground truth tumor masks as shown in Fig. 1.
Fig. 2.
An illustration of the sample-weight used to scale the BCE loss function for each image per patient based on the cross-sectional area of the tumor. The small squares show overlays of the tumor ground truth contours (red) and the cross-sectional images. Scale of the background grayscale color is the BCE weights.
2.5. Model Training, Optimization and Validation
There is no available separate data that can be used to evaluate and validate the performance of the segmentation model. Therefore, we used a 5-fold (80% training and 20% validation) cross-validation approach where the 201 patients’ imaging data and the corresponding ground truth tumor masks (Sect. 2.1) were split into 5 sets (Set 1 to Set 5). Each set contains imaging data of 40 patients randomly selected from the 201 patient dataset. The random split did not take into consideration the institutional sources of the data since the number of contributing patients varies significantly between these institutional centers (201 patients distributed as 72, 55, 18, and 56 from 4 different institutions); i.e. 4-fold cross validation based on patients from different institutions would provide significantly un-balanced train-validations sets and could lead to inaccurate estimation of the model performance. For each iteration of cross validation, each set of 40 patients serves as test data for the segmentation model trained using imaging data from the remaining 4 sets (i.e., 161 patients). Using this approach, the segmentation model was trained and tested 5 times. To estimate the number of epochs that should be used for training, the model was trained using 161 patients for training and 40 patients for validation, randomly selected from the 201 patients. The calculated loss using the validation data was used to obtain the maximum number of epochs before the model starts to overfit. In other words, when there is no further improvement of the loss evaluated in the optimization data. Using this approach, 50 epochs was estimated to be used for the 2D and 3D models training. Then, the segmentation model was trained for 50 epochs using 161 patients’ data – 80% and tested using 40 patients’ data – 20% 5 times. The overall DSC, recall, and precision values were obtained using the average of the mean DSC, recall, and precision values generated for the individual test data sets using the corresponding trained segmentation models. Subsequently, the model was trained one additional time, using 50 epochs, on the entire dataset (i.e., 201 patients’ data) to generate the final model for the use of predicting the tumor masks the MICCAI challenge test set, i.e. a representation of other unseen datasets.
3. Results
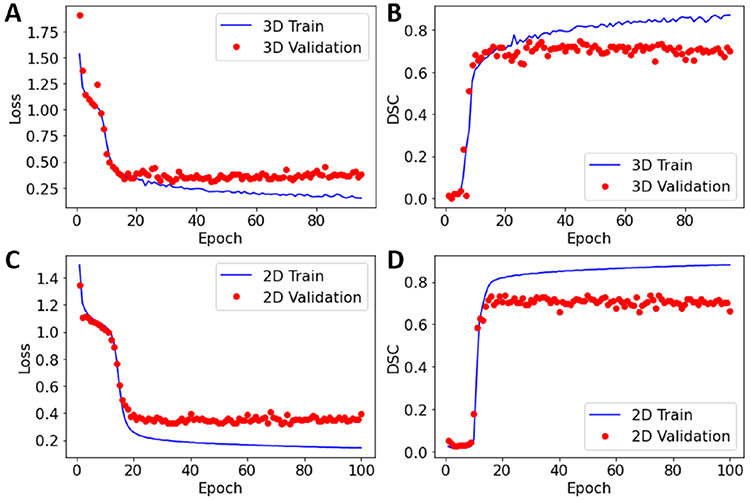
The training performance of the model is illustrated in Fig. 3. The validation loss and DCS values do not show further improvements after epoch 45 for the 3D model and epoch 50 for the 2D model, consequently further model training led to model overfitting.
Fig. 3.
The loss and DSC values as a function of epochs obtained during the 3D (A) and (B) and the 2D (C) and (D) model training.
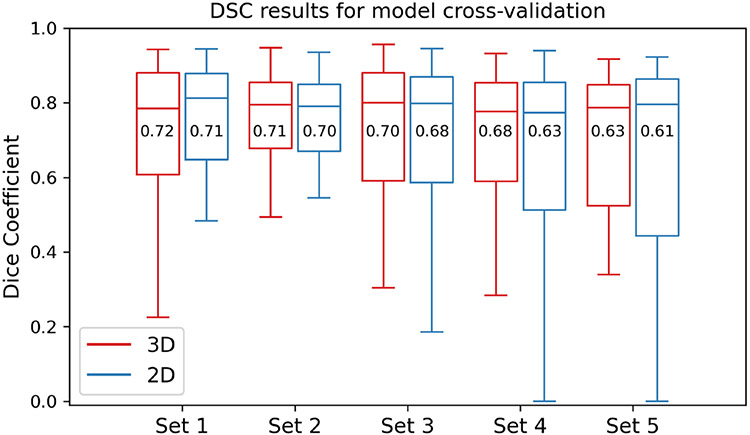
The DSC values’ distributions obtained by the 3D and 2D segmentation models for the 5 test data sets using 50 epochs are illustrated in Fig. 4. The DSC median and mean values for the 3D model for Set 1 to Set 5 are 0.79, 0.79, 0.80, 0.78, and 0.79, respectively, and 0.72, 0.71, 0.70, 0.68, and 0.63, respectively. The DSC median and mean values for the 2D model for Set 1 to Set 5 are 0.81, 0.79, 0.80, 0.77, and 0.80, respectively, and 0.71, 0.70, 0.68, 0.63, and 0.61, respectively. The overall average (mean) values for the DSC, recall, and precision using all test data sets are presented in Table 1.
Fig. 4.
Boxplots of the DSC distribution for the 5 test data sets (Set 1 to Set 5) used for the 3D and 2D segmentation model cross validation. The DSC mean values are given in the boxes and the lines inside the box refer to the DSC median values.
Table 1.
3D and 2D model performance metrics.
| Model | DSC | Recall | Precision |
|---|---|---|---|
| 3D | 0.69 ± 0.03 | 0.75 ± 0.07 | 0.72 ± 0.03 |
| 2D | 0.67 ± 0.04 | 0.71 ± 0.06 | 0.71 ± 0.03 |
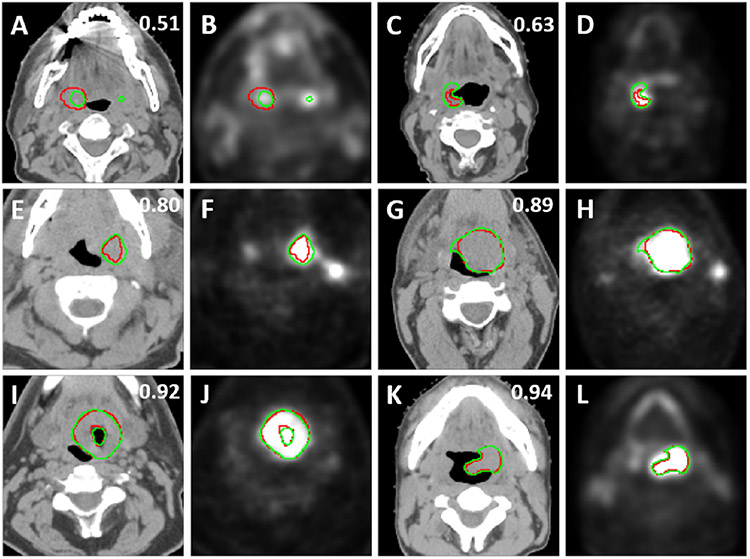
To illustrate the performance of the segmentation model, samples of overlays of CT and PET images with the outlines of tumor masks using ground truth (red) and model segmentations (green) from the test data sets are shown in Fig. 5. The figure shows representative segmentation results for DSC values 0.51, 0.63, 0.80, 0.89, 0.92, and 0.94 which are below, comparable, and above the segmentation model’s median DSC value of 0.79.
Fig. 5.
2D axial examples of overlays of the ground truth segmentations (red) and predicted segmentations (green) and CT images (first and third columns) and PET images (second and forth columns) with different 3D volumetric DSC values given at the right top.
4. Discussion
As shown in Fig. 4 and Table 1, the 3D model outperforms the 2D model for all performance metrics. Specifically, the 3D model performance was superior to that of the 2D model for the mean DCS values for each test set (Set 1 to Set 5) (Fig. 4). Moreover, the mean DCS, recall, and precision values for all test sets (Table 1) are higher for the 3D model compared to the 2D model. As shown in Table 1, the mean DSC value of the 3D model (0.69) is larger than that of the 2D model (0.67). We performed a paired t-test on the 200 DSC values obtained from the 3D and 2D models (combining the DSC values of the 5 validation sets), resulting in a significant p-value of 0.043.
The image size of 144 × 144 used in the model training is relatively small compared to 512 × 512 and 256 × 256 usually used in the training of the standard U-net. Therefore, in the current 2D and 3D U-net models, to overcome overfitting, we used a small number of filters and 1 convolution layer for each convolution block which leads to total numbers of trainable parameters of 1,351,537 for the 3D model, and 452,113 for the 2D model compared to 7,759,521 for the standard U-net used in our previous model [28]. In addition, we used data augmentation to further mitigate the overfitting to improve the model performance. However, as seen in Fig. 3, the model starts to overfit after epoch 30 for the 3D model when trained on 161 patients. This indicates that the size of the data set used to train the model needs to be increased to improve the model performance and to mitigate the overfitting problem.
There are some limitations in the current approach. The proposed model has not been evaluated using an independent data set, instead, we performed a five-fold cross-validation approach (80% training and 20% validation) to estimate the expected model performance when applied to unseen test data. Therefore, it was assumed that the training data used by the model have a similar statistical representation of true unseen data which may not be accurate, especially if the unseen data has different image resolutions than the ones used to train the model. The model has been trained using images with 144 × 144 × 96 voxels which are the maximum number of voxels found in the CT scans of the 201 patients within the provided bounding boxes of 144 × 144 × 144 mm3. Therefore, all images were up-sampled to that number of voxels. Training the model with a larger size (i.e., 144 × 144 × 144) gives a lower model performance and increases overfitting as increasing the image size does not add additional useful information to the model describing the tumor. To show this, we trained the 3D model for 50 epochs using image size of 144 × 144 × 144 voxels; the mean DSC values obtained from the 5 validation sets was lowered to 0.66 ± 0.04 compared to 0.69 ± 0.03 using images with size of 144 × 144 × 96 voxels. Therefore, the model performance may be degraded when used to predict tumor masks using images with voxel size larger than 144 × 144 × 96 as the input images will need to be down-sampled to a smaller size 144 × 144 × 96 for masks’ prediction.
The proposed models combine several novel features to improve performance such as a reduced U-net size, data augmentation, and a novel loss function for improving model sensitivity. For the 3D model, these features aid in achieving overall average median and mean DSC values of 0.79 and 0.69 respectively, comparable to a mean DSC between radiation oncologists for HNC GTV delineation using PET/CT images (0.69) [36]. These features can be implemented in several other network architectures proposed for tumor segmentations such as ResNet [37], Inception [38], and DenseNet [39], which are worth investigating for future model improvement.
For the test data of the HECKTOR challenge, our 3D model was only able to achieve a DSC of 0.637. Interestingly, while DSC performance was generally lacking when compared to the other state of the art methods in the competition, our method was among the top models in precision (0.755). However, this came at a cost of low sensitivity (0.628). This indicates that the image voxels our model classified as tumor were very likely to be tumor, however, it subsequently was unable to detect many tumor voxels. While we are currently blinded to the ground truth contours of the test data, we can make some educated guesses on why our model did not generalize well in the test dataset. The test data includes several patients with images of higher resolutions than the ones used to train the model (i.e., the cropped images within the 144 × 144 × 144 mm bounding boxes have sizes of 144 × 144 × 144 voxels). These images were down-sampled to the size of 144 × 144 × 96 voxels which is the size of the images used to train the model. Therefore, we expect a degradation of the model performance when used to predict the tumor masks of these images. The second reason for the discrepancy between the estimated DSC values using the training the test data could be due to the inaccurate estimation of the model performance on un-seen data using the proposed 5-fold cross validation the training data. Using 10-fold or larger internal validation strategies might provide a better estimation of the model performance on the test data.
5. Conclusion
This study presented a deep learning CNN model based on the U-net architecture to automatically segment primary tumors in HNC patients using co-registered FDG-PET/CT images. A combination of data normalization, dual input channel integration of PET and CT data, data augmentation, and the use of a loss function that combines contributions from the DSC while weighting BCE resulting in a promising performance of 3D tumor auto-segmentation with overall average median and mean cross-validation DSC values of 0.79 and 0.69, respectively. While our 3D model showed lower performance on a held-out test dataset, our methods are still useful for the auto-contouring community to incorporate and improve upon.
Acknowledgements.
M.A.N. is supported by a National Institutes of Health (NIH) Grant (R01 DE028290-01). K.A.W. is supported by a training fellowship from The University of Texas Health Science Center at Houston Center for Clinical and Translational Sciences TL1 Program (TL1 TR003169). C.D.F. received funding from the National Institute for Dental and Craniofacial Research Award (1R01DE025248-01/R56DE025248) and Academic-Industrial Partnership Award (R01 DE028290), the National Science Foundation (NSF), Division of Mathematical Sciences, Joint NIH/NSF Initiative on Quantitative Approaches to Biomedical Big Data (QuBBD) Grant (NSF 1557679), the NIH Big Data to Knowledge (BD2K) Program of the National Cancer Institute (NCI) Early Stage Development of Technologies in Biomedical Computing, Informatics, and Big Data Science Award (1R01CA214825), the NCI Early Phase Clinical Trials in Imaging and Image-Guided Interventions Program (1R01CA218148), the NIH/NCI Cancer Center Support Grant (CCSG) Pilot Research Program Award from the UT MD Anderson CCSG Radiation Oncology and Cancer Imaging Program (P30CA016672), the NIH/NCI Head and Neck Specialized Programs of Research Excellence (SPORE) Developmental Research Program Award (P50 CA097007) and the National Institute of Biomedical Imaging and Bioengineering (NIBIB) Research Education Program (R25EB025787). He has received direct industry grant support, speaking honoraria and travel funding from Elekta AB.
References
- 1.Siegel RL, Miller KD, Jemal A: Cancer statistics, 2020. CA Cancer J. Clin 70, 7–30 (2020). 10.3322/caac.21590 [DOI] [PubMed] [Google Scholar]
- 2.Rosenthal DI, et al. : Beam path toxicities to non-target structures during intensity-modulated radiation therapy for head and neck cancer. Int. J. Radiat. Oncol. Biol. Phys 72, 747–755 (2008). 10.1016/j.ijrobp.2008.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vorwerk H, et al. : Protection of quality and innovation in radiation oncology: the prospective multicenter trial the German Society of Radiation Oncology (DEGRO-QUIRO study). Strahlentherapie und Onkol. 190, 433–443 (2014) [DOI] [PubMed] [Google Scholar]
- 4.Riegel AC, et al. : Variability of gross tumor volume delineation in head-and-neck cancer using CT and PET/CT fusion. Int. J. Radiat. Oncol. Biol. Phys 65, 726–732 (2006). 10.1016/j.ijrobp.2006.01.014 [DOI] [PubMed] [Google Scholar]
- 5.Rasch C, Steenbakkers R, Van Herk M: Target definition in prostate, head, and neck. Semin. Radiat. Oncol 15, 136–145 (2005). 10.1016/j.semradonc.2005.01.005 [DOI] [PubMed] [Google Scholar]
- 6.Breen SL, et al. : Intraobserver and interobserver variability in GTV delineation on FDG-PET-CT images of head and neck cancers. Int. J. Radiat. Oncol. Biol. Phys 68, 763–770 (2007). 10.1016/j.ijrobp.2006.12.039 [DOI] [PubMed] [Google Scholar]
- 7.Segedin B, Petric P: Uncertainties in target volume delineation in radiotherapy–are they relevant and what can we do about them? Radiol. Oncol 50, 254–262 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson CM, et al. : Interobserver and intermodality variability in GTV delineation on simulation CT, FDG-PET, and MR images of head and neck cancer. Jacobs J. Radiat. Oncol 1, 6 (2014) [PMC free article] [PubMed] [Google Scholar]
- 9.Guo Y, Liu Yu., Georgiou T, Lew MS: A review of semantic segmentation using deep neural networks. Int. J. Multimed. Inf. Retr 7(2), 87–93 (2017). 10.1007/s13735-017-0141-z [DOI] [Google Scholar]
- 10.Garcia-Garcia A, Orts-Escolano S, Oprea S, Villena-Martinez V, Garcia-Rodriguez J: A Review on Deep Learning Techniques Applied to Semantic Segmentation (2017) [Google Scholar]
- 11.Boldrini L, Bibault J-E, Masciocchi C, Shen Y, Bittner M-I: Deep learning: a review for the radiation oncologist. Front. Oncol 9, 977 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cardenas CE, Yang J, Anderson BM, Court LE, Brock KB: Advances in auto-segmentation. Semin. Radiat. Oncol 29, 185–197 (2019). 10.1016/j.semradonc.2019.02.001 [DOI] [PubMed] [Google Scholar]
- 13.Oreiller VAV, Vallieres M, Castelli J, Boughdad HEMJS, Adrien JOP: Automatic Segmentation of Head and Neck Tumors and Nodal Metastases in PET-CT scans (2020). http://proceedings.mlr.press/v121/andrearczyk20a.html
- 14.Li L, Zhao X, Lu W, Tan S: Deep learning for variational multimodality tumor segmentation in PET/CT. Neurocomputing 392, 277–295 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leung KH, et al. : A physics-guided modular deep-learning based automated framework for tumor segmentation in PET images. arXiv Preprint arXiv:2002.07969 (2020) [DOI] [PubMed] [Google Scholar]
- 16.Kawauchi K, et al. : A convolutional neural network-based system to classify patients using FDG PET/CT examinations. BMC Cancer 20, 1–10 (2020). 10.1186/s12885-020-6694-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhong Z, et al. : 3D fully convolutional networks for co-segmentation of tumors on PET-CT images. In: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), pp. 228–231. IEEE. (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jemaa S, Fredrickson J, Carano RAD, Nielsen T, de Crespigny A, Bengtsson T: Tumor segmentation and feature extraction from whole-body FDG-PET/CT using cascaded 2D and 3D convolutional neural networks. J. Digit. Imaging 33, 888–894 (2020). 10.1007/s10278-020-00341-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao X, Li L, Lu W, Tan S: Tumor co-segmentation in PET/CT using multi-modality fully convolutional neural network. Phys. Med. Biol 64, 15011 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang B, et al. : Fully automated delineation of gross tumor volume for head and neck cancer on PET-CT using deep learning: a dual-center study. Contrast Media Mol. Imaging 2018, 1–12 (2018). https://pubmed.ncbi.nlm.nih.gov/30473644/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moe YM, et al. : Deep learning for automatic tumour segmentation in PET/CT images of patients with head and neck cancers. arXiv Preprint arXiv:1908.00841 (2019) [Google Scholar]
- 22.Guo Z, Li X, Huang H, Guo N, Li Q: Deep learning-based image segmentation on multimodal medical imaging. IEEE Trans. Radiat. Plasma Med. Sci 3, 162–169 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin D, et al. : Accurate esophageal gross tumor volume segmentation in PET/CT using two-stream chained 3D deep network fusion. In: Shen D, Liu T, et al. (eds.) MICCAI 2019. LNCS, vol. 11765, pp. 182–191. Springer, Cham: (2019). 10.1007/978-3-030-32245-8_21 [DOI] [Google Scholar]
- 24.Zhou T, Ruan S, Canu S: A review: deep learning for medical image segmentation using multi-modality fusion. Array 3, 100004 (2019) [Google Scholar]
- 25.AIcrowd MICCAI 2020: HECKTOR Challenges. https://www.aicrowd.com/challenges/miccai-2020-hecktor. Accessed 07 Sept 2020
- 26.Andrearczyk V, et al. : Overview of the HECKTOR challenge at MICCAI 2020: automatic head and neck tumor segmentation in PET/CT. In: Andrearczyk V, et al. (eds.) HECKTOR 2020. LNCS, vol. 12603, pp. 1–21. Springer, Cham: (2021) [Google Scholar]
- 27.Ronneberger O, Fischer P, Brox T: U-Net: convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells WM, Frangi AF (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham: (2015). 10.1007/978-3-319-24574-4_28 [DOI] [Google Scholar]
- 28.Naser MA, Deen MJ: Brain tumor segmentation and grading of lower-grade glioma using deep learning in MRI images. Comput. Biol. Med 121, 103758 (2020). 10.1016/j.compbiomed.2020.103758 [DOI] [PubMed] [Google Scholar]
- 29.Springenberg JT, Dosovitskiy A, Brox T, Riedmiller M: Striving for simplicity: the all convolutional net. arXiv Preprint arXiv:1412.6806 (2014) [Google Scholar]
- 30.Noh H, Hong S, Han B: Learning deconvolution network for semantic segmentation. In: Proceedings of the IEEE International Conference on Computer Vision, pp. 1520–1528 (2015). 10.1109/ICCV.2015.178 [DOI] [Google Scholar]
- 31.Long J, Shelhamer E, Darrell T: Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, pp. 3431–3440 (2015). 10.1109/CVPR.2015.7298965 [DOI] [Google Scholar]
- 32.Badrinarayanan V, Kendall A, Cipolla R: SegNet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell 39, 2481–2495 (2017). 10.1109/TPAMI.2016.2644615 [DOI] [PubMed] [Google Scholar]
- 33.Krizhevsky A, Sutskever I, Hinton GE: ImageNet classification with deep convolutional neural networks. Commun. ACM 60, 84–90 (2017). 10.1145/3065386 [DOI] [Google Scholar]
- 34.Maas AL, Hannun AY, Ng AY: Rectifier nonlinearities improve neural network acoustic models. In: Proceedings of ICML, p. 3 (2013) [Google Scholar]
- 35.Xu B, Wang N, Chen T, Li M: Empirical evaluation of rectified activations in convolutional network. arXiv Preprint arXiv:1505.00853 (2015) [Google Scholar]
- 36.Gudi S, et al. : Interobserver variability in the delineation of gross tumour volume and specified organs-at-risk during IMRT for head and neck cancers and the impact of FDG-PET/CT on such variability at the primary site. J. Med. Imaging Radiat. Sci 48, 184–192 (2017). 10.1016/j.jmir.2016.11.003 [DOI] [PubMed] [Google Scholar]
- 37.Zhang Q, Cui Z, Niu X, Geng S, Qiao Y: Image segmentation with pyramid dilated convolution based on ResNet and U-Net. In: Liu D, Xie S, Li Y, Zhao D, El-Alfy ES (eds.) Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), pp. 364–372. Springer, Cham: (2017). 10.1007/978-3-319-70096-0_38 [DOI] [Google Scholar]
- 38.Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z: Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, pp. 2818–2826. IEEE Computer Society; (2016). 10.1109/CVPR.2016.308 [DOI] [Google Scholar]
- 39.Jégou S, Drozdzal M, Vazquez D, Romero A, Bengio Y: The One Hundred Layers Tiramisu: Fully Convolutional DenseNets for Semantic Segmentation (2017). https://openaccess.thecvf.com/content_cvpr_2017_workshops/w13/html/Jegou_The_One_Hundred_CVPR_2017_paper.html