Abstract
Detection of SARS-CoV-2 RNA in wastewater is a promising tool for informing public health decisions during the COVID-19 pandemic. However, approaches for its analysis by use of reverse transcription quantitative polymerase chain reaction (RT-qPCR) are still far from standardized globally. To characterize inter- and intra-laboratory variability among results when using various methods deployed across Canada, aliquots from a real wastewater sample were spiked with surrogates of SARS-CoV-2 (gamma-radiation inactivated SARS-CoV-2 and human coronavirus strain 229E [HCoV-229E]) at low and high levels then provided “blind” to eight laboratories. Concentration estimates reported by individual laboratories were consistently within a 1.0-log10 range for aliquots of the same spiked condition. All laboratories distinguished between low- and high-spikes for both surrogates. As expected, greater variability was observed in the results amongst laboratories than within individual laboratories, but SARS-CoV-2 RNA concentration estimates for each spiked condition remained mostly within 1.0-log10 ranges. The no-spike wastewater aliquots provided yielded non-detects or trace levels (<20 gene copies/mL) of SARS-CoV-2 RNA. Detections appear linked to methods that included or focused on the solids fraction of the wastewater matrix and might represent in-situ SARS-CoV-2 to the wastewater sample. HCoV-229E RNA was not detected in the no-spike aliquots. Overall, all methods yielded comparable results at the conditions tested. Partitioning behavior of SARS-CoV-2 and spiked surrogates in wastewater should be considered to evaluate method effectiveness. A consistent method and laboratory to explore wastewater SARS-CoV-2 temporal trends for a given system, with appropriate quality control protocols and documented in adequate detail should succeed.
Keywords: COVID-19, Wastewater surveillance, Public health, Quality assurance, Quality control
Graphical abstract
Introduction
Monitoring of SARS-CoV-2 RNA in wastewater has been shown to be a promising, rapid tool that complements other indicators of COVID-19 disease prevalence in a community. Although the analytical techniques to quantify the genetic signal cannot be used to infer the viability or infectivity of viruses, sufficient levels of the genetic signal have been found to persist for its detection and quantification in wastewater using such methods (Bivins et al., 2020; Foladori et al., 2020; La Rosa et al., 2020; Rimoldi et al., 2020). As wastewater is not subject to the limitations of individual case level clinical diagnostic testing, it is a potentially useful aggregate indicator of a community's SARS-CoV-2 infections across a gradient of severity including asymptomatic/pre-symptomatic/mild to severe disease (Lorenzo and Picó, 2019). Since its first reported use in March 2020 at the outset of this pandemic, monitoring of SARS-CoV-2 RNA in wastewater has indicated possible early warning of increased COVID-19 case loads, facilitated investigations of spatial and temporal trends, as well as informed public-health management responses such as application of more intensive clinical testing (e.g., Ahmed et al., 2020a; Betancourt et al., 2020; Daughton, 2020; Hrudey et al., 2020; Kumar et al., 2020; Medema et al., 2020a, 2020b; Randazzo et al., 2020; Street et al., 2020).
Despite the acceleration of research and trials involving detection of SARS-CoV-2 RNA in wastewater since the initial trials (Medema et al., 2020a), methods for concentration of SARS-CoV-2 viruses, extraction of viral RNA and subsequent quantification are not standardized. However, it is not yet clear what variations among results are due to differences between methodologies, and whether inter- and intra-laboratory variability would compromise use of these methods for reliably tracking temporal trends in SARS-CoV-2 occurrence and/or evaluating the degree of disease prevalence in a community.
In wastewater and other environmental samples, SARS-CoV-2 RNA signals observed from a shared sample might be subject to additional variability attributable to sample preparation step(s) (i.e., concentration of virus from wastewater matrices) that are not applicable to the processing of clinical samples (e.g., nasopharyngeal swabs or saliva tests) (Kitajima et al., 2020; Lu et al., 2020). Accordingly, the purpose of this study was to characterize inter- and intra-laboratory variability of results that can be expected when using various methods currently deployed across Canada for quantification of SARS-CoV-2 virus in a common spiked wastewater sample. The common sample was provided blind by the coordinating laboratory and distributed to all participants. A cross-section of eight laboratories across Canada that had already demonstrated experience and capacity to analyze SARS-CoV-2 RNA in wastewater by RT-qPCR participated in this study. This approach was designed to capture the collective experience, capacity, and expertise of individual laboratories, and to allow for broader inferences to be drawn related to differences in data generation and handling approaches. Imposing a common analytical methodology on this study—or considering an even broader suite of sample preparation methods for that matter—was not feasible for a variety of practical considerations. These considerations include availability of analytical equipment in the participating laboratories as well as supply chain limitations on sample processing materials and reagents.
1. Study design
While composite wastewater (Medema et al., 2020a) or samples of sludge from primary clarifiers (D'Aoust et al., 2020; Graham et al., 2020; Peccia et al., 2020) collected over the course of a day might provide a better capture of the variation of in situ SARS-CoV-2 concentrations in wastewater, a composite sample was not needed for the purpose of the spike-and-recovery study intended to evaluate inter- and intra- laboratory variability of sample processing and analysis. A common raw wastewater grab sample, post-grit, was obtained from the Winnipeg Wastewater Treatment Plant on August 31, 2020 (Table 1 ). At the time the sample was collected, approximately 85 reported COVID-19 cases remained active in Winnipeg (population ~750,000).
Table 1.
Summary of Winnipeg, Manitoba wastewater characteristics.
| Parameter | Value |
|---|---|
| Average daily flow (m3/sec) | 2.09 |
| Maximum daily flow (m3/sec) | 3.73 |
| Total solids (mg/L) | 1010 |
| Total suspended solids (mg/L) | 254 |
| BOD5 (mg/L) | 194 |
| NH4-N (mg/L) | 32.0 |
| Total phosphorus (mg/L) | 5.79 |
| Total nitrogen (mg/L) | 52.8 |
| Total organic carbon (mg/L) | 128.7 |
Aliquots of the well-mixed raw wastewater sample were individually spiked with either a low or a high concentration of SARS-CoV-2 surrogates (Table 2 ). No-spike wastewater aliquots were also provided as blanks. Each laboratory received three aliquots of 100 mL for each spike condition. Samples were chilled (4°C) during transport from the wastewater treatment plant, prior to assembly of shipping boxes, and during transport to all laboratories. To minimize possible bias due to differences in storage of samples, they were kept cold (4°C) and processed to concentrate the virus within 48 hr of spiking for this study. After obtaining the viral concentrate, some laboratories froze the concentrate before further processing to extract RNA and perform quantitative polymerase chain reactions (qPCR). Methods deployed as well as quality assurance/quality controls performed by each laboratory has been summarized anonymously ( Appendix A ). Methods deployed by the laboratories in this study generally offered a turn-around time of 24–48 hr, although steps to improve the timeliness of the methods (e.g., reducing time for polyethylene glycol (PEG) precipitation) since the completion of this study have been investigated by individual laboratories to optimize their work flow while maintaining adequate data quality.
Table 2.
Surrogates deployed in the inter-laboratory study.
| Surrogate | Description | Spike condition |
||
|---|---|---|---|---|
| No-spike (WW-N) | Low-spike (WW-A) | High-spike (WW-B) | ||
| Gamma-irradiated inactivated SARS-CoV-2 | classified as Risk Group 2; a member of the subgenus Sarbecovirus (Betacoronavirus lineage B); positive-sense single-stranded RNA (+ssRNA) virus, with a single linear RNA segment | N/A | 18 ± 2 gene copies/mL* | 1800 ± 200 gene copies/mL* |
| Human coronavirus (HCoV) strain 229E | classified as Risk Group 2; a member of the genus Alphacoronavirus and subgenus Duvinacovirus; enveloped, positive-sense, single-stranded RNA virus which enters its host cell by binding to the APN receptor; infects humans & bats | N/A | 10 infectious units/mL | 1000 infectious units/mL |
Values based on quantification of gamma-irradiated inactivated SARS-CoV-2 viral concentrate used to prepare wastewater conditions. Quantification was performed using the Bio-Rad SARS-CoV-2 triplex assay based on the N1 and N2 viral gene targets. Error has been expressed as the standard deviation of technical triplicates.
Gamma inactivated SARS-CoV-2 stocks were quantified using the Bio-Rad QX200 droplet-digital PCR platform (Bio‐Rad, USA). Briefly, 10 µL of SARS-CoV-2 purified culture was mixed with 130 µL of PBS and RNA was extracted using a QiaAMP Viral RNA mini kit (Qiagen, USA). RNA was amplified using the Bio-Rad COVID-19 Triplex assay using C1000 Touch thermocycler (Bio-Rad) as per manufacturer's instructions, using primers and probes targeting the SARS-CoV-2 N1 and N2 genes. Thermocycling conditions were as follows: 60 min reverse transcription at 50°C (1 cycle), 10 min enzyme activation at 95°C (1 cycle), 30‐sec denaturation at 94°C (40 cycles), 1 min annealing/extension cycle at 55°C (40 cycles; ramp rate of ∼2–3°C/sec), 10 min enzyme deactivation at 98°C (1 cycle). 20 µL of the RT-qPCR reaction was used for droplet generation using a Bio-Rad automated droplet generator. The concentration was determined in technical triplicates and the error is expressed in standard deviations. Data were analyzed using QuantaSoft version 1.7.4.0917 (Bio-Rad).
2. Results
2.1. Highly reproducible results at the scale of individual laboratories
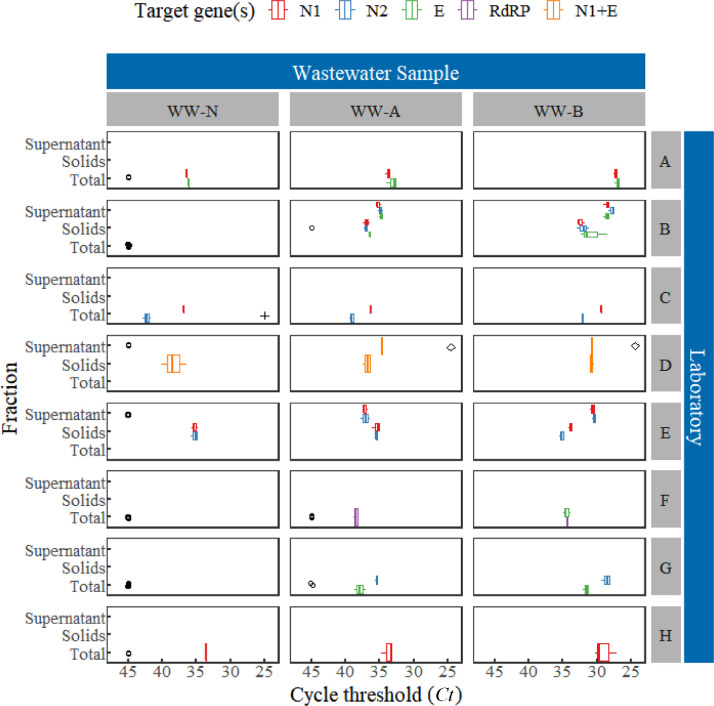
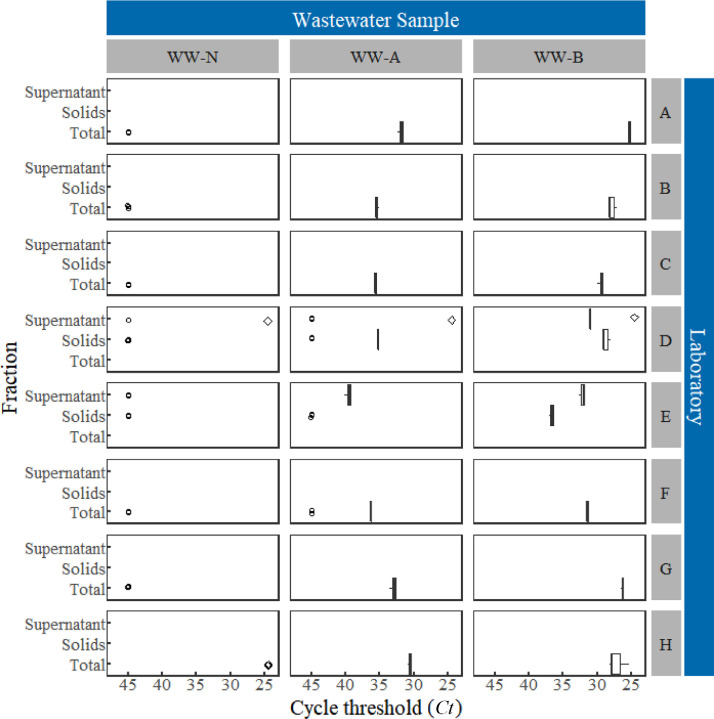
Overall, the results indicated a high level of reproducibility using the methods employed by each of the laboratories to estimate SARS-CoV-2 RNA concentrations (Figs. 1 and 2 ). Within each laboratory, aliquots from the same spiked condition consistently yielded estimated concentrations (reported as gene copies per mL) of the same order of magnitude (i.e., <1-log10). The greatest variation observed in the total SARS-CoV-2 RNA concentration estimate reported by a laboratory for either spiked condition was 0.81-log10 (coefficient of variation=153%, Table 3 ), which suggested reproducibility at the scale of individual laboratories. Statistics associated with the log10-SARS-CoV-2 concentration estimates (Canchola et al., 2017) for each spiked condition and gene targeted are summarized in Table 3. As expected, less relative variation was generally observed in the high-spike condition compared to that of the low-spike condition. Since not all laboratories had access to standards for quantification of HCoV-229E, only cycle threshold values (Ct) were reported (Fig. 3 ). Ct values corresponding to HCoV-229E quantification similarly exhibited a high degree of precision.
Fig. 1.
Estimates of concentrations of SARS-CoV-2 RNA in three wastewater samples with no-spike (WW-N), low-spike (WW-A, 18 ± 2 gene copies/mL) and high-spike (WW-B, 1800 ± 200 gene copies/mL). Concentrations of spikes are denoted by the dashed lines. Non-detects (o), detected but not quantifiable (+), as well as samples not analyzed (◊) are also shown. Laboratories have been anonymized; laboratory A was the coordinating laboratory that was responsible for the preparation and dissemination of the spiked wastewater samples.
Fig. 2.
Ct values of SARS-CoV-2 RNA reported for three wastewater samples with no-spike (WW-N), low-spike (WW-A, 18±2 gene copies/mL) and high-spike (WW-B, 1800±200 gene copies/mL). Non-detects (o), detected but not quantifiable (+), as well as samples not analyzed (◊) are also shown.
Table 3.
SARS-CoV-2 concentration estimates observed in spiked samples of Winnipeg wastewater.
| Laboratory | Target gene(s) | WW-A Low-spike condition |
WW-B High-spike condition |
||||
|---|---|---|---|---|---|---|---|
| Mean | SD | COV (%) | Mean | SD | COV (%) | ||
| A | E | 0.47 | 0.26 | 64.9 | 2.37 | 0.15 | 35.4 |
| N1 | 0.55 | 0.15 | 36.2 | 2.60 | 0.14 | 32.8 | |
| B | E | 0.81 | 0.08 | 18.0 | 2.54 | 0.09 | 19.8 |
| N1 | 0.69 | 0.11 | 24.8 | 2.47 | 0.13 | 30.0 | |
| N2 | 0.77 | 0.19 | 47.0 | 2.63 | 0.13 | 30.0 | |
| C | N1 | 0.77 | 0.48 | 152.7 | 2.48 | 0.01 | 3.3 |
| N2 | – | – | – | 2.28 | 0.04 | 9.1 | |
| D | N1+E | −0.18 | 0.32 | 85.4 | 1.50 | 0.04 | 9.6 |
| E | N1 | 0.37 | 0.20 | 49.7 | 1.70 | 0.13 | 31.5 |
| N2 | 0.33 | 0.18 | 42.9 | 1.51 | 0.06 | 14.8 | |
| F | E | – | – | – | 1.74 | 0.25 | 63.4 |
| RdRP | – | – | – | 1.75 | 0.05 | 12.1 | |
| G | E | – | – | – | 1.92 | 0.19 | 47.1 |
| N2 | – | – | – | 2.14 | 0.26 | 64.2 | |
| H | N1 | 1.28 | 0.17 | 41.8 | 2.84 | 0.40 | 114.3 |
Note: WW-A and WW-B represent the low-spike (WW-A) and high-spike (WW-B) conditions, respectively. Mean, standard deviation (SD) and coefficient of variation (COV; Canchola et al., 2017) of the log10-transformed concentration estimates (log10-gene copies/mL) statistics were only calculated where all three aliquots of each condition yielded quantifiable values.
Fig. 3.
Ct values reported for HCoV-229E RNA in three wastewater samples with no-spike (WW-N), low-spike (WW-A, 10 infectious units/mL) and high-spike (WW-B, 1000 infectious units/mL). Non-detects (o), detected, but not quantifiable (+), as well as samples not analyzed (◊) are also shown.
2.2. Low and high spikes consistently distinguished by all laboratories
Among laboratories, total concentration estimates of SARS-CoV-2 in gene copies per mL—accounting for both supernatant and solids fractions analyzed where applicable—were generally consistent and mostly within an order of magnitude for a given spike condition, with no clear advantage based on the type of sample preparation employed. Although there was greater variability among than within laboratories, all laboratories successfully distinguished between the low- and high-spikes of SARS-CoV-2 inactivated by gamma-irradiation. The 10th and 90th percentile concentration estimates for the low- and high-spikes across all results excluding non-detects were 1.2 and 10.3 gene copies/mL (10–90 percentile range: 0.92-log10) and 34.5 and 446.8 gene copies/mL (10–90 percentile range: 1.1-log10), respectively. Ct values associated with HCoV-229E also denoted a similar ability to distinguish between the low- and high-spikes.
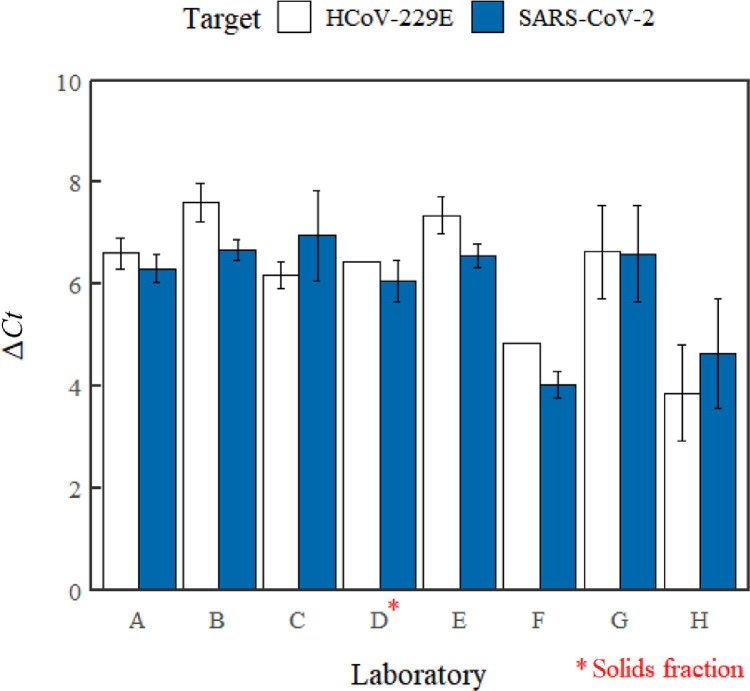
Most laboratories were able to observe approximately the intended 100-fold difference (ΔCt ≈ 6.6) in concentration between low- and high-spikes for both surrogates (Fig. 4 ). However, it was recognized that the Ct values observed for the low-spike was not likely in the linear range of PCR amplification and approached the sensitivity limit attainable by most RT-qPCR methods. Therefore, the Ct value obtained from analysis of the surrogates in the low-spike sample and the ΔCt calculated therefrom might not provide an accurate evaluation of variation in analyses during this study. Differences in reporting thresholds among laboratories is also recognized. Collectively, these results demonstrate the flexibility of sample preparation methods to yield comparable results given a common wastewater matrix and supports the notion that currently deployed methods can reliably distinguish low from high levels of viral surrogates at the conditions investigated. Given an intended purpose of detecting temporal trends of SARS-CoV-2, the ability to distinguish low from high concentrations consistently and accurately is necessary.
Fig. 4.
Differences between Ct values for low-spike and high-spike (i.e. ΔCt) observed for SARS-CoV-2 and HCoV-229E by each laboratory. Error bars represent the standard deviation of the mean ΔCt; the absence of error bars denote cases where no replicates were available. The ΔCt values represent those observed in the supernatant fraction or from the processing of both fractions unless otherwise indicated.
2.3. Surrogate spikes may partition differently in wastewater than authentic in-situ SARS-CoV-2
Where the method used by a given laboratory allowed for the distinction of the supernatant and particle-associated fractions of the wastewater matrix, more of the spiked viral surrogates, SARS-CoV-2 inactivated by gamma radiation or HCoV-229E, were generally detected in supernatants rather than solids phases (Figs. 1, 2, and 3). Given the spike preparation method and the relatively short time between sample spiking and sample processing in this study, it was recognized that spike-and-recovery approaches might not adequately represent the recovery of authentic in-situ SARS-CoV-2 that are likely to be transported predominantly via fecal matter in the wastewater matrix. For this study, the demonstrated effective recovery of SARS-CoV-2 from the supernatant phase does not guarantee efficient recovery of authentic, in situ SARS-CoV-2 from wastewater samples, in which the virus might predominate in solids phases. This underscores the fact that partitioning of the target virus among phases can substantially influence apparent effectiveness of the methods used in this study.
2.4. Trace amounts of SARS-CoV-2 RNA detected in “no-spike” wastewater samples
No-spike wastewater aliquots were also provided to and evaluated by participating laboratories. As expected, none of the laboratories detected HCoV-229E RNA in those aliquots (Fig. 3) because it was not inoculated as a spike and has been previously documented to be absent in stool samples (Esper et al., 2010). In contrast, the same samples yielded either non-detects or trace levels (<20 gene copies/mL) of SARS-CoV-2 RNA (Figs. 1 and 2; laboratories A, C, D, E, and H). Considering that approximately 85 active cases of COVID-19 in Winnipeg were reported at the time of wastewater sample collection—which suggests that a larger number of asymptomatic, pre-symptomatic, and recovering cases of COVID-19 are likely to exist in the community that can contribute to SARS-CoV-2 concentrations in Winnipeg wastewater—the trace amounts observed might represent authentic in-situ SARS-CoV-2 RNA recovered from the wastewater. Notably, detections appear linked to methods that included or focused on the solids fraction of the wastewater matrix rather than the supernatant (laboratories C, D, and E). This is in apparent contrast to the partitioning exhibited by spiked viral surrogates in this study to the supernatant phase as noted above.
3. Discussion
3.1. Consistent results and inferences drawn from different methods support the use of these methods for temporal-trend detection
Overall, the results are consistent with the view that there is flexibility of sample preparation methods to yield reproducible and comparable results given a split wastewater sample. The notion that currently deployed methods can reliably distinguish lesser from greater amounts of viral surrogates at the conditions investigated is also supported. These inferences are largely consistent with other international studies performed in parallel (Pecson et al., 2020; Pocock et al., 2020). Both of these parallel studies concluded that sample preparation methods did not have a clear, systematic impact on results (Pecson et al., 2020; Pocock et al., 2020). Remarkably, there is substantially less variability in the results amongst the methods deployed in this study than that observed in the parallel study of 36 methods (Pecson et al., 2020). This observation might be linked to 1) the limited range of methods captured in this study (i.e., primarily concentration methods involving PEG precipitation and ultrafiltration) amongst various other types of sample preparation methods that exist (e.g., aluminum salt coagulation, skim milk flocculation, charged membrane filtration) and/or 2) the use of known, quantified SARS-CoV-2 surrogate spikes in this work rather than the quantification of in-situ SARS-CoV-2 in the parallel study. Given that the low-spike level used in our study was at/near the level of sensitivity typically attainable using RT-qPCR methods, it was expected that the two viral surrogates might not be consistently detected within all three aliquots of the lesser amounts spiked into samples. Consequently, the ability for all laboratories to consistently observe the 100-fold difference between the low- and high-spike conditions might have been impacted. In the future, a regression design using a gradient of spike concentrations would allow for a more rigorous examination of method sensitivity at the lower end of spike concentrations. This would be particularly useful for understanding limitations of the method for monitoring in areas where active COVID-19 cases are less prevalent.
Overall, the findings of the study, results of which are reported here, support that the cross-section of methods investigated in this study can be used to explore temporal trends of SARS-CoV-2 RNA in wastewater for a specific community. For this purpose, it is recommended that a consistent method and laboratory for each system/community be used to prevent avoidable bias. However, the minimization of methodological biases does not imply that the resulting SARS-CoV-2 RNA status trend for a given community/system are directly comparable to others. The data generated using these methods—and the status trends generated therefrom—must be explored considering sampling program design (e.g., composite vs. grab samples) and specific conditions (e.g., location within a sewershed, community water use patterns, and hydraulic conditions) that are unique for every system.
3.2. Results of this study are a function of method of spiking and partitioning behavior of the surrogates used
The need to better understand and characterize partitioning of SARS-CoV-2 and various surrogates or standards in various wastewater matrices is underscored by the results of this study. Given the method of spiking and the relatively short duration between spiking and processing of samples during this study, it was recognized that spike-and-recovery approaches might not adequately represent the recovery of authentic in-situ SARS-CoV-2 that are likely to be transported predominantly via fecal matter particles in wastewater matrices. Indeed, the trace levels of SARS-CoV-2 RNA detected by participating laboratories that included or focused on the analysis of the solids fraction during this study supports other emerging reports of solids-associated behavior exhibited by in-situ SARS-CoV-2 in wastewater samples (D'Aoust et al., 2020; Graham et al., 2020; Peccia et al., 2020). Phase partitioning of the target virus(es) can therefore influence apparent effectiveness of methods applied in this study. For example, preferential partitioning of SARS-CoV-2 viruses to solid phases would be a disadvantage for methods that strictly rely on analysis of the supernatant unless additional step(s) are implemented to encourage virus mobilization into the supernatant.
The implications of the foregoing observation are threefold. First, the importance of processing the entire wastewater sample, including both solids-associated and supernatant fractions to assess RNA presence, is underscored. Second, the choice of surrogates and methods used to assess recovery should be verified (additional details elaborated upon in the recommendations on process controls below). Third, the benefits and weaknesses inherent to the design of these inter-laboratory comparisons are highlighted: spike-and-recovery approaches (such as the one applied herein this study) might provide greater confidence of intended spiking levels but may not be truly representative of authentic in-situ SARS-CoV-2 behavior, while inter-laboratory studies that estimate in-situ SARS-CoV-2 to wastewater samples (e.g., Pecson et al., 2020) might be limited by inevitable uncertainty arising from not being able to know the true concentration, but may better reflect the ability to detect authentic in-situ SARS-CoV-2 in wastewater.
3.3. Inter-laboratory comparisons require a focus on how standard curves for quantification are generated
Despite the differences between methods employed during this study, all approaches yielded comparable results. This underscores the value and need for standardizing quality assurance and quality control (QA/QC) protocols and reporting. Standardizing analytical methods themselves is currently not practical for the diverse range of laboratories seeking to respond to the growing need for analytical services. However, doing so for QA/QC is achievable and has been shown to be effective (Ahmed et al., 2020c). Indeed, discrepancies between initial values reported by laboratories arose due to differences of standards and protocols used to generate standard curves for quantification from Ct values. Specifically, some laboratories that originally used plasmid standards reported SARS-CoV-2 RNA concentration estimates approximately an order of magnitude greater than those laboratories which utilized linear DNA/RNA standards, such as Integrated DNA Technologies gBlocks gene fragments or Twist Synthetic SARS-CoV-2 RNA controls. Although the supercoiled structure of plasmids affords these biomolecules relative stability for their storage and use as standards, this property can also result in delayed Ct values in developing the standard curve against which samples are evaluated (Chen et al., 2007; Hou et al., 2010). The undetected lower efficiency of plasmid amplification in the early stages of PCR when the supercoiled plasmid is the dominant template has been suggested to result in substantial overestimation of nucleic acid abundance (Hou et al., 2010). The degree to which results are biased may differ across methods/laboratories. An extreme example of how the use of SARS-CoV-2 plasmid standards without linearization resulted in overestimation of results by over two orders of magnitude in this study is shown in Fig. 5 . Accordingly, linearized DNA, RNA or PCR amplicons might be preferable for quantification of SARS-CoV-2 RNA. Regardless, the impact of this feature on quantification is substantial.
Fig. 5.
An example of standard curves generated from a plasmid DNA standard and an RNA standard. In this extreme case, SARS-CoV-2 RNA concentration estimates derived from the use of plasmid DNA standards yielded results that were two orders of magnitude higher than those generated using RNA standards.
Protocols for standard curve preparation and instrument threshold settings can also be coordinated to reduce systematic variations of standard curves and cycle thresholds. Discrepancies in preliminary results reported were also noted when standard curves against which samples are evaluated were developed from the preparation of serial dilution of the positive control standards in a matrix other than distilled water. For instance, a standard curve prepared using spikes of gamma-irradiated inactivated SARS-CoV-2 into a wastewater matrix can result in biased SARS-CoV-2 RNA concentration estimates. Differences in instrument threshold settings and cut-off thresholds/protocols for reporting were also noted as potential contributors to inter-laboratory variability. Collectively, reference materials for quantification, protocols for standard curve preparation, as well as instrument and reporting thresholds should be coordinated among laboratories to better facilitate direct inter-laboratory comparisons of these results.
3.4. Monitoring and addressing PCR inhibition as a key component of QA/QC
Because PCR inhibitors are widespread in environmental samples, PCR can be partially or completely inhibited (Schrader et al., 2012) possibly leading to false-negatives (e.g., Graham et al., 2020; Kitajima et al., 2020). Consequently, quantified RT-qPCR results must not be presumed void of inhibition effects by making an inaccurate presumption that inhibition is consistently and successfully mitigated by the choice of reagent kits utilized. An appropriate reference target must be deployed as an inhibition control; substantially delayed amplification (i.e., higher Ct value) of this reference target within the sample matrix will be indicative of PCR inhibition. This should be monitored and documented along with steps taken to minimize inhibition. Dilution or use of a smaller effective sample volume of the viral concentrate are common approaches to mitigating inhibition effects; however, it can result in loss of sensitivity of methods (Eckhart et al., 2000; Monteiro et al., 1997; Schrader et al., 2012; Scipioni et al., 2008a, 2008b; Widjojoatmodjo et al., 1992) and/or random sampling error when few viruses are present (Emelko et al., 2008). Other strategies, such as heat treatment or chemical addition, for the removal of PCR inhibitors from stool and environmental matrices also exist and are summarized elsewhere (Schrader et al., 2012). Nevertheless, the strategy chosen must be tailored for the individual wastewater matrix analyzed. Ultimately, inhibition must be managed to be negligible to provide assurance that the estimated quantity of gene fragments in a sample are not negatively confounded by inefficient/unsuccessful PCR amplification.
3.5. Process controls and better understanding of surrogate behavior are needed to evaluate method recovery efficiency
Accurate estimates of target virus abundances in wastewater require an adjustment of the observed result from the RT-qPCR assay to account for losses incurred throughout the various sample processing, RNA extraction and quantification steps (Ahmed et al., 2020b; Bustin et al., 2009; Huggett et al., 2005; Rusiñol et al., 2020). To date, model viruses with similar structural and morphological characteristics that are absent from the wastewater matrix have been used as matrix spikes to estimate process recovery efficiency because of stringent biosafety requirements associated with SARS-CoV-2 (Ahmed et al., 2020b; Rusiñol et al., 2020). Recoveries of other enveloped viruses, such as murine hepatitis virus [MHV]; Ahmed et al., 2020b) and synthetic standards, such as quantified armored RNA (Hietala and Crossley, 2006), have also been used and assumed to represent efficiency of recovery of SARS-CoV-2, although often without additional experimental validation (Bustin et al., 2009).
In this study, HCoV-229E RNA was not detected by any of the participating laboratories in the no-spike wastewater samples. HCoV-229E are enveloped viruses of the same coronavirus family (i.e., Coronaviridae, subfamily Orthocoronavirinae) as SARS-CoV-2 (references in Li et al., 2020) and have been previously documented to be absent from stool samples (Esper et al., 2010). Furthermore, laboratories that measured greater amounts (i.e., lower Ct values) of the spiked HCoV-229E also recovered greater amounts of SARS-CoV-2. Accordingly, HCoV-229E might be a promising candidate as a common internal matrix spike of the spiked-in SARS-CoV-2. However, viral metagenome analysis of biosolids collected from a wastewater treatment facility has shown that HCoV-229E and other coronaviruses can be present in small quantities (Bibby et al., 2011). Therefore, use of this and other surrogates as matrix spikes requires ascertaining background amounts in wastewater, as well as further validation of its ability to represent authentic in situ recovery of SARS-CoV-2 from wastewater. Given the lack of evidence that all surrogates—including HCoV-229E that was spiked in—comparably represent the recovery of authentic in situ SARS-CoV-2 viruses from wastewater, the effort required to perform matrix spikes in every sample (that might be desirable to characterize recovery variability attributable to changing wastewater matrix conditions) is likely not warranted. In the parallel study by Pecson et al., 2020, five of six additional surrogates introduced as matrix spikes exhibited statistically different recoveries than their primary choice of spiked-in recovery surrogate (human coronavirus strain OC43). Therefore, a preferred approach might be to report quantitated SARS-CoV-2 concentrations prior to adjusting for recovery, along with an estimate of the recovery efficiency itself and the matrix spike surrogate used.
3.6. Use of fecal biomarkers/indicators to normalize results for improved temporal trend detection
Indicators of fecal contributions from wastewater have been increasingly advocated as a means to normalize SARS-CoV-2 RNA concentration estimates according to fecal loading in the wastewater sample that might enable improved detection of temporal trends within a given community. Common viral indicators used for this purpose include the Pepper Mild Mottle virus (PMMoV) (Graham et al., 2020; Jafferali et al., 2020), crAssphage (Green et al., 2020; Jennings et al., 2020; Stachler et al., 2017), or other human fecal specific bacteriophages or biomarkers. Chemical substances such as the excretory product creatinine and the calorie-free sugar substitute acesulfame (Lin et al., 2019), or physically-based wastewater volumetric flow rates might also be used to normalize SARS-CoV-2 RNA concentration estimates. There is merit and potential demonstrated for compensating for dilution of sewage with stormwater in combined sewer systems or for systems with substantial groundwater infiltration (Alpaslan Kocamemi et al., 2020; Balboa et al., 2020; Kaplan et al., 2020; Peccia et al., 2020; Wu et al., 2020). Applicability of these indicators to provide a consistent fecal signature for normalizing SARS-CoV-2 concentration estimates presumes that their persistence and variability in fecal contributions are consistent with the loading and behavior of SARS-CoV-2 in a given system.
In our study, several laboratories also evaluated PMMoV endogenous to the wastewater sample. Although estimates of concentrations of PMMoV differed by an order of magnitude between laboratories, each laboratory's analysis yielded generally reproducible results (Fig. 6 ). The amount of intra-laboratory precision attained for PMMoV provides confidence and has been suggested to be useful to normalize SARS-CoV-2 RNA concentration estimates for the detection of temporal trends (e.g., D’Aoust et al., 2021). However, the use of a consistent method and laboratory for each system/community is again underscored and recommended to prevent avoidable methodological biases.
Fig. 6.
Pepper Mild Mottle Virus RNA concentration estimates observed in the Winnipeg wastewater samples, across all aliquots by three laboratories. Statistics shown are based on log10-concentration estimates. The concentration estimates reflect those observed in the supernatant fraction or from the processing of both fractions unless otherwise indicated.
4. Conclusions
The purpose of this inter-laboratory study was to characterize the inter- and intra- laboratory variability associated with results emanating from the quantification of spiked SARS-CoV-2 surrogates by use of RT-qPCR, after extraction from a common wastewater matrix. Although it was anticipated that inter-laboratory variability in results would largely be attributable to differences in preparation of the wastewater (Kitajima et al., 2020; Lu et al., 2020), the range of methods captured in this study yielded comparable results for both spiked surrogates used, which were generally within an order of magnitude of each other for the same spike condition. The methods used by each laboratory also reliably distinguished the low- from the high-spike conditions. This provides confidence that the consistent use of a particular method (and laboratory) can achieve the detection of temporal trends associated with SARS-CoV-2 RNA in wastewater for a specific community. Recognizing that the spike-and-recovery approach used in this study might not adequately represent the recovery of authentic in-situ SARS-CoV-2 present in wastewater, the findings of this study suggest that understanding partitioning of viruses among phases in the wastewater is critical for optimizing methods to improve SARS-CoV-2 RNA recovery.
Results of this inter-laboratory study further emphasized the importance of adequate QA/QC protocols, which must be in place and reported with sufficient detail. SARS-CoV-2 RNA concentration estimates from the use of circular plasmid DNA standards without first linearizing yielded concentration estimates that can be two orders of magnitude higher than other laboratories which utilized linear DNA/RNA standards. These standard materials should be coordinated to facilitate better inter-laboratory comparisons of method performance. The unadjusted, quantitated result from the sample assay should be supported with an indication that PCR inhibition controls were performed and achieved negligible inhibition, as well as an estimate of method recovery efficiency and the recovery surrogate(s) used. Community-and system-specific fecal contributions within a wastewater sample can be considered using additional fecal biomarkers (e.g., PMMoV), wastewater volumetric flow rates, or other chemical substances. Their reliable quantification might be useful to normalize SARS-CoV-2 concentration estimates to improve temporal trend detection and to facilitate comparisons of regional monitoring activities.
Statement of author contributions
Alex H.S. Chik coordinated the study, analyzed and presented the data, and prepared the manuscript; Melissa B. Glier and Mark Servos provided guidance, interpretation of the findings, processed the wastewater samples and were involved in the development of the initial manuscript; Chand S. Mangat was responsible for the collection, preparation, and distribution of the wastewater samples, processed the samples; Xiaoli (Lilly) Pang and Yuanyuan (Judy) Qiu prepared spiking materials, processed the samples; Patrick M. D'Aoust, Jean-Baptiste Burnet, Robert Delatolla, Sarah Dorner, Qiudi Geng, John P. Giesy Jr., R. Michael McKay, Natalie Prystajecky, Nivetha Srikanthan, Yuwei Xie participated in interpretation of the findings and/or processed the wastewater samples; Bernadette Conant provided leadership and oversight of the study, advised project planning; Steve E. Hrudey was the Chair of the CWN Research Advisory Group; Bernadette Conant, Steve E. Hrudey, Michael Mulvey, and James Brooks contributed to the conception and design of the project. The laboratories participating in this study (extended group author list) supported the processing of the wastewater samples. All authors reviewed the manuscript.Group author details:- Michael G. Becker, National HIV and Retrovirology Laboratory, Public Health Agency of Canada, Winnipeg,Canada - James Brooks, Centre for Communicable Disease and Infection Control, Public Health Agency of Canada, Ottawa, Canada - Jade Daigle, National Microbiology Laboratory, Public Health Agency of Canada, Winnipeg, Canada - Eyerusalem Goitom, Polytechnique Montréal, Montreal, Canada - Tyson E. Graber, Children's Hospital of Eastern Ontario Research Institute, Ottawa, Canada - Jonathon Leblanc, Great Lakes Institute for Environmental Research, University of Windsor, Windsor, Canada - Ravinder Lidder, National Microbiology Laboratory, Public Health Agency of Canada, Winnipeg, Canada - Alex E. MacKenzie, Children's Hospital of Eastern Ontario Research Institute, Ottawa, Canada - Elisabeth Mercier, University of Ottawa, Ottawa, Canada - Ziwen Ran, Microbiology and Immunology, University of British Columbia, Vancouver, Canada - David Spreitzer, National Microbiology Laboratory, Public Health Agency of Canada, Winnipeg, Canada - Yuanmin Wu, S.M. Research Inc., Richmond Hill, Canada - Jiaao Yu, University of Alberta, Edmonton, Canada.
Acknowledgments
This work was supported by multiple funding sources. The work performed by the uOttawa-CHEO group was supported by a CHEO (Children's Hospital of Eastern Ontario) CHAMO (Children's Hospital Academic Medical Organization) grant, awarded to Dr. Alex E. MacKenzie. The work performed by University of Saskatchewan was supported by the “Next generation solutions to ensure healthy water resources for future generations” funded by the Global Water Futures program, Canada First Research Excellence Fund (#419205). Dr. Giesy was supported by the Canada Research Chairs Program of the Natural Sciences and Engineering Research Council of Canada (NSERC). The work performed at École Polytechnique was supported by funding from NSERC Discovery and Strategic Grant Programs. Dr. Hrudey was supported by funding from an NSERC Discovery Grant. The University of Windsor group was supported by the NSERC Alliance COVID-19 Grant and by Mitacs through the Mitacs Accelerate program. The group led by Dr. Pang was supported by Canadian Institutes of Health Research (CIHR), Alberta Innovates, Alberta Health-Water for Life Strategy. Dr. Prystajecky's group was supported by the BC center for Disease Control, BC center for Disease Control Foundation for Public Health and Metro Vancouver. The University of Waterloo group was supported by the “Next generation solutions to ensure healthy water resources for future generations” funded by the Global Water Futures program, Canada First Research Excellence Fund (#419205). Dr. Servos was supported by the Canada Research Chairs Program of NSERC.
We would also like to express our thanks to the members of the Canadian COVID-19 Wastewater Coalition's research advisory group and public health advisory group, who have generously shared their time and expertise. The City of Winnipeg is also thanked for their coordination and provision of wastewater samples. The authors would also like to thank Liana Kreamer who proofread an earlier version of this manuscript.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jes.2021.01.029.
Appendix A. Supplementary materials
References
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O'Brien J.W. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bivins A., Bibby K., Farkas K., Gathercole A. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bivins A., Bertsch P.M., Bibby K., Choi P.M., Farkas K. Surveillance of SARS-CoV-2 RNA in wastewater: methods optimization and quality control are crucial for generating reliable public health information. Curr. Opin. Environ. Sci. Health. 2020 doi: 10.1016/j.coesh.2020.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpaslan Kocamemi B., Kurt H., Sait A., Sarac F., Saatci A.M., Pakdemirli B. SARS-CoV-2 detection in Istanbul wastewater treatment plant sludges. MedRxiv. 2020 doi: 10.1101/2020.05.12.20099358. [DOI] [Google Scholar]
- Balboa S., Mauricio-Iglesias M., Rodríguez S., Martínez-Lamas L., Vasallo F.J., Regueiro B. The fate of SARS-CoV-2 in wastewater treatment plants points out the sludge line as a suitable spot for incidence monitoring. MedRxiv. 2020 doi: 10.1101/2020.05.25.20112706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt W.W., Schmitz B.W., Innes G.K., Brown K.M.P., Prasek S.M., Stark E.R. MedRxiv. 2020. Wastewater-based epidemiology for averting COVID-19 outbreaks on the university of Arizona campus. [DOI] [Google Scholar]
- Bibby K., Viau E., Peccia J. Viral metagenome analysis to guide human pathogen monitoring in environmental samples. Lett. Appl. Microbiol. 2011;52(4):386–392. doi: 10.1111/j.1472-765X.2011.03014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A., Greaves J., Fischer R., Yinda K.C., Ahmed W., Kitajima M. Persistence of SARS-CoV-2 in water and wastewater. Environ. Sci. Technol. Lett. 2020;7(12):937–942. doi: 10.1021/acs.estlett.0c00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Canchola J.A., Tang S., Hemyari P., Paxinos E., Marins E. Correct use of percent coefficient of variation (%CV) formula for log-transformed data. MOJ Proteom. Bioinform. 2017;6(4) doi: 10.15406/mojpb.2017.06.00200. [DOI] [Google Scholar]
- Chen J., Kadlubar F.F., Chen J.Z. DNA supercoiling suppresses real-time PCR: a new approach to the quantification of mitochondrial DNA damage and repair. Nucl. Acids Res. 2007;35(4):1377–1388. doi: 10.1093/nar/gkm010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Aoust P.M., Graber T.E., Mercier E., Montpetit D., Alexandrov I., Neault N. Catching a resurgence: Increase in SARS-CoV-2 viral RNA identified in wastewater 48 h before COVID-19 clinical tests and 96 h before hospitalizations. Sci. Total Environ. 2021;770 doi: 10.1016/j.scitotenv.2021.145319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Aoust P.M., Mercier E., Montpetit D., Jia J.-.J., Alexandrov I., Neault N. Quantitative analysis of SARS-CoV-2 RNA from wastewater solids in communities with low COVID-19 incidence and prevalence. MedRxiv. 2020 doi: 10.1101/2020.08.11.20173062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughton C.G. Wastewater surveillance for population-wide Covid-19: the present and future. Sci. Total Environ. 2020;736 doi: 10.1016/j.scitotenv.2020.139631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhart L., Bach J., Ban J., Tschachler E. Melanin binds reversibly to thermostable DNA polymerase and inhibits its activity. Biochem. Biophys. Res. Commun. 2000;271(3):726–730. doi: 10.1006/bbrc.2000.2716. [DOI] [PubMed] [Google Scholar]
- Emelko M.B., Schmidt P.J., Roberson J.A. Quantification of uncertainty in microbial data—reporting and regulatory implications. J. (Am. Water Works Assoc.) 2008:94–104. [Google Scholar]
- Esper F., Ou Z., Huang Y.T. Human coronaviruses are uncommon in patients with gastrointestinal illness. J. Clin. Virol. 2010;48(2):131–133. doi: 10.1016/j.jcv.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foladori P., Cutrupi F., Segata N., Manara S., Pinto F., Malpei F. SARS-CoV-2 from faeces to wastewater treatment: what do we know? A review. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham K., Loeb S., Wolfe M., Catoe D., Sinnott-Armstrong N., Kim S. SARS-CoV-2 in wastewater settled solids is associated with COVID-19 cases in a large urban sewershed. MedRxiv. 2020 doi: 10.1101/2020.09.14.20194472. [DOI] [PubMed] [Google Scholar]
- Green H., Wilder M., Middleton F.A., Collins M., Fenty A., Gentile K. Quantification of SARS-CoV-2 and cross-assembly phage (crAssphage) from wastewater to monitor coronavirus transmission within communities. MedRxiv. 2020 doi: 10.1101/2020.05.21.20109181. [DOI] [Google Scholar]
- Hietala S.K., Crossley B.M. Armored RNA as virus surrogate in a real-time reverse transcriptase PCR assay proficiency panel. J. Clin. Microbiol. 2006;44(1):67–70. doi: 10.1128/JCM.44.1.67-70.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y., Zhang H., Miranda L., Lin S. Serious overestimation in quantitative PCR by circular (supercoiled) plasmid standard: microalgal pcna as the model gene. PLoS One. 2010;5(3):e9545. doi: 10.1371/journal.pone.0009545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrudey S.E., Ashbolt N.J., Isaac-Renton J.L., McKay R.M., Servos M.R. Vol. 23. Royal Society of Canada; 2020. https://rsc-src.ca/en/voices/epidemiology-for-sars-cov-2 (Wastewater-based Epidemiology For SARS-CoV-2). [Google Scholar]
- Huggett J., Dheda K., Bustin S., Zumla A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005;6(4):279–284. doi: 10.1038/sj.gene.6364190. [DOI] [PubMed] [Google Scholar]
- Jafferali M.H., Khatami K., Atasoy M., Birgersson M., Williams C., Cetecioglu Z. Benchmarking virus concentration methods for quantification of SARS-CoV-2 in raw wastewater. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.142939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings W.C., Gálvez-Arango E., Prieto A.L., Boehm A.B. CrAssphage for fecal source tracking in Chile: covariation with norovirus, HF183, and bacterial indicators. Water Res. X. 2020;9 doi: 10.1016/j.wroa.2020.100071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan E.H., Wang D., Wang M., Malik A.A., Zulli A., Peccia J.H. Aligning SARS-CoV-2 Indicators via an epidemic model: application to hospital admissions and RNA detection in sewage sludge. MedRxiv. 2020 doi: 10.1101/2020.06.27.20141739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M., Ahmed W., Bibby K., Carducci A., Gerba C.P., Hamilton K.A. SARS-CoV-2 in wastewater: state of the knowledge and research needs. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Patel A.K., Shah A.V., Raval J., Rajpara N., Joshi M. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total Environ. 2020;746 doi: 10.1016/j.scitotenv.2020.141326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Bonadonna L, Lucentini L, Kenmoe S., Suffredini E. Coronavirus in water environments: occurrence, persistence and concentration methods - a scoping review. Water Res. 2020;179 doi: 10.1016/j.watres.2020.115899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Liu S.-.M., Yu X.-.H., Tang S.-.L., Tang C.-.K. Coronavirus disease 2019 (COVID-19): current status and future perspectives. Int. J. Antimicrob. Agents. 2020;55(5) doi: 10.1016/j.ijantimicag.2020.105951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W., Zhang X., Tan Y., Li P., Ren Y. Can water quality indicators and biomarkers be used to estimate real-time population? Sci. Total Environ. 2019;660:603–610. doi: 10.1016/j.scitotenv.2018.12.390. [DOI] [PubMed] [Google Scholar]
- Lorenzo M., Picó Y. Wastewater-based epidemiology: current status and future prospects. Curr. Opin. Environ. Sci. Health. 2019;9:77–84. doi: 10.1016/j.coesh.2019.05.007. [DOI] [Google Scholar]
- Lu D., Huang Z., Luo J., Zhang X., Sha S. Primary concentration–The critical step in implementing the wastewater based epidemiology for the COVID-19 pandemic: a mini-review. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.141245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Been F., Heijnen L., Petterson S. Implementation of environmental surveillance for SARS-CoV-2 virus to support public health decisions: opportunities and challenges. Curr. Opin. Environ. Sci. Health. 2020 doi: 10.1016/j.coesh.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-coronavirus-2 in sewage. MedRxiv, 2020 doi: 10.1101/2020.03.29.20045880. [DOI] [PubMed] [Google Scholar]
- Monteiro L., Bonnemaison D., Vekris A., Petry K.G., Bonnet J., Vidal R. Complex polysaccharides as PCR inhibitors in feces: helicobacter pylori model. J. Clin. Microbiol. 1997;35(4):995–998. doi: 10.1128/jcm.35.4.995-998.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A. SARS-CoV-2 RNA concentrations in primary municipal sewage sludge as a leading indicator of COVID-19 outbreak dynamics. MedRxiv. 2020 doi: 10.1101/2020.05.19.20105999. [DOI] [Google Scholar]
- Pecson B.M., Darby E., Haas C.N., Amha Y., Bartolo M., Danielson R. Reproducibility and sensitivity of 36 methods to quantify the SARS-CoV-2 genetic signal in raw wastewater: findings from an interlaboratory methods evaluation in the US. MedRxiv. 2020 doi: 10.1039/d0ew00946f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pocock G., Coetzee L., Mans J., Taylor M., Genthe B. Proof of Concept Study: application of wastewater-based surveillance to monitor SARS-CoV-2 prevalence in South African communities. Water Res. Comm. 2020 [Google Scholar]
- Randazzo W., Cuevas-Ferrando E., Sanjuán R., Domingo-Calap P., Sánchez G. Metropolitan wastewater analysis for COVID-19 epidemiological surveillance (SSRN Scholarly Paper ID 3586696) Soc. Sci. Res. Netw. 2020 doi: 10.2139/ssrn.3586696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimoldi S.G., Stefani F., Gigantiello A., Polesello S., Comandatore F., Mileto D. Presence and infectivity of SARS-CoV-2 virus in wastewaters and rivers. Sci. Total Environ. 2020;744 doi: 10.1016/j.scitotenv.2020.140911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusiñol M., Martínez-Puchol S., Forés E., Itarte M., Girones R., Bofill-Mas S. Concentration methods for the quantification of coronavirus and other potentially pandemic enveloped virus from wastewater. Curr. Opin. Environ. Sci. Health. 2020;17:21–28. doi: 10.1016/j.coesh.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader C., Schielke A., Ellerbroek L., Johne R. PCR inhibitors – occurrence, properties and removal. J. Appl. Microbiol. 2012;113(5):1014–1026. doi: 10.1111/j.1365-2672.2012.05384.x. [DOI] [PubMed] [Google Scholar]
- Scipioni A., Bourgot I., Mauroy A., Ziant D., Saegerman C., Daube G. Detection and quantification of human and bovine noroviruses by a TaqMan RT-PCR assay with a control for inhibition. Mol. Cell. Probes. 2008;22(4):215–222. doi: 10.1016/j.mcp.2008.02.003. [DOI] [PubMed] [Google Scholar]
- Scipioni A., Mauroy A., Ziant D., Saegerman C., Thiry E. A SYBR Green RT-PCR assay in single tube to detect human and bovine noroviruses and control for inhibition. Virol. J. 2008;5(1):94. doi: 10.1186/1743-422X-5-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachler E., Kelty C., Sivaganesan M., Li X., Bibby K., Shanks O.C. Quantitative CrAssphage PCR assays for human fecal pollution measurement. Environ. Sci. Technol. 2017;51(16):9146–9154. doi: 10.1021/acs.est.7b02703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street R., Malema S., Mahlangeni N., Mathee A. Wastewater surveillance for COVID-19: an African perspective. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widjojoatmodjo M.N., Fluit A.C., Torensma R., Verdonk G.P., Verhoef J. The magnetic immuno polymerase chain reaction assay for direct detection of salmonellae in fecal samples. J. Clin. Microbiol. 1992;30(12):3195–3199. doi: 10.1128/jcm.30.12.3195-3199.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Xiao A., Zhang J., Gu X., Lee W.L., Kauffman K. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. MedRxiv. 2020 doi: 10.1101/2020.04.05.20051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.