Abstract
Severe acute respiratory syndrome corona virus-2 (SARS-CoV-2) has first emerged from China in December 2019 and causes coronavirus induced disease 19 (COVID-19). Since then researchers worldwide have been struggling to detect the possible pathogenesis of this disease. COVID-19 showed a wide range of clinical behavior from asymptomatic to severe acute respiratory disease syndrome. However, the etiology of susceptibility to severe lung injury is not yet fully understood. Angiotensin-converting enzyme1 (ACE1) convert angiotensin I into Angiotensin II that was further metabolized by ACE 2 (ACE2). The binding ACE2 receptor to SARS-CoV-2 facilitate its enter into the host cell. The interaction and imbalance between ACE1 and ACE2 play a crucial role in the pathogenesis of lung injury. Thus, the aim of this study was to investigate the association of ACE1 I/D polymorphism with severity of Covid-19.
The study included RT-PCR confirmed 269 cases of Covid-19. All cases were genotyped for ACE1 I/D polymorphism using polymerase chain reaction and followed by statistical analysis (SPSS, version 15.0).
We found that ACE1 DD genotype, frequency of D allele, older age (≥46 years), unmarried status, and presence of diabetes and hypertension were significantly higher in severe COVID-19 patient. ACE1 ID genotype was significantly independently associated with high socio-economic COVID-19 patients (OR: 2.48, 95% CI: 1.331–4.609).
These data suggest that the ACE1 genotype may impact the incidence and clinical outcome of COVID-19 and serve as a predictive marker for COVID-19 risk and severity.
Keywords: Covid-19, SARS-COV2, ACE I/D polymorphism, ADRS, RT-PCR
1. Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was first identified in China that causes coronavirus induced disease 19 (COVID-19) (Lu et al., 2020). COVID19 has confronted a major threat to human health globally and posed a serious risk to the public healthcare systems. This pandemic has resulted in 45.9 million confirmed cases and 2 million deaths worldwide as of November 1, 2020 (WHO, 2020). The clinical spectrum of COVID-19 includes asymptomatic, mild symptom and severe acute respiratory distress syndrome (ARDS) with high mortality due to respiratory failure, stroke, thrombotic complications and multi organic failure (Connors and Levy, 2020; Hess et al., 2020; Zheng and Cao, 2020). Severity of COVID-19 patient increases with other comorbidities such as older age, diabetes, hypertension, and obesity (Sanyaolu et al., 2020). However, many cases without these comorbidities also have severe lung disease or ARDS (Richardson et al., 2020). Thus, underlying pathophysiological mechanism of COVID-19 is not yet fully understood.
The entry of SARS-CoV-2 into host cells is facilitated by the binding of viral spike protein (S-protein) to the extracellular peptidase domain of angiotensin converting enzyme 2 (ACE2), followed by the s-protein priming through a specific transmembrane serine protease 2 (TMPRSS2) (Lam et al., 2020). ACE2 converts angiotensin II to angiotensin 1–7 and prevent the effects of ACE1/angiotensin II axis. Angiotensin II can induce strong vasoconstriction, proinflammatory effects, and profibrotic effects, while angiotensin 1–7 exhibits antiproliferative, antiapoptotic, and mild vasodilating abilities and protect different cardiovascular effects such as anti–heart failure, anti-thrombosis, anti-myocardial hypertrophy, anti-fibrosis, anti-arrhythmia, anti-atherogenesis, and attenuating vascular dysfunction related to metabolic syndrome (Santos et al., 2018). Therefore, the coexistence in the ACE1 and ACE2 genes of inherited predispositions or common genetic polymorphisms of these genes that affect their levels of mutual expression can lead to increased capillary permeability, fibrosis, coagulation and apoptosis in the alveolar cells and accelerating lung damage. The role of ACE2 in susceptibility and pathogenesis of SARS-CoV infections has been deciphered by numerous reports in the mouse model, the entry rate of the virus is increased with overexpression of ACE-2 receptors, antibodies to ACE-2 receptors significantly block the entry of SARS-CoV and ACE2 knockout mice had lower pulmonary lesions compared to the wild type (Pati et al., 2020; Yang et al., 2007). All these studies showed the importance of ACE receptor in SARS-CoV pathogenesis. SARS-CoV shared approximately 76% amino acid sequence similarity with SARS-CoV-2 (Pati et al., 2020; Xu et al., 2020). The ACE gene consists of 26 exons which is located on chromosome 17q35. ACE insertion/deletion (I/D) genotypes of a 287-bp Alu repeat sequence of intron 16, can cause alternative splicing in ACE protein, with one active site for ACE I allele while two active sites for D allele. The I/D polymorphism has been reported to account for 47% of the variance in plasma ACE level, whereas the DD genotype is associated with the highest levels in most of the ethnic groups in reported series (Clarke and Turner, 2012; Gemmati et al., 2020). ACE2 expression improved significantly via administration of ACE inhibitors (Li et al., 2017; Pati et al., 2020). The association of the ‘D' allele of ACE with the occurrence of pneumonia in SARS patients and the death of subjects with acute respiratory distress syndrome (ARDS) was observed in different studies. Thus, the aim of this study was to determine the impact of ACE1 I/D polymorphism on the severity of COVID-19 patients in north Indian population.
2. Methods and materials
2.1. Study subjects
All procedures in this study involving human participants were performed in accordance with the ethical standards of Era University, India. This study was approved by the Ethics Committee of the Era University, India. Informed consent for participation was obtained from all patients in an appropriate manner. A total of 269 COVID-19 patients admitted to the Eras Lucknow Medical College and Hospital (ELMC&H), Era University, Lucknow from August 2020 to September 2020 and diagnosed with COVID-19 were enrolled. All patients with inclusion criteria (COVID-19 patients confirmed by RT-PCR with age more than 20 years), and exclusion criteria (Pregnant patients, patients with known malignant disease) were selected.
2.2. Data collection and blood sampling
All clinical details of individuals were collected as per self-administered questionnaire which included demographic data, clinical history of diabetes and hypertension, family history and associated complications and other clinical data was collected from hospital records under supervision of expert clinician. The diagnosis of hypertension and diabetes mellitus was performed according to World Health Organization criteria.
After an informed consent from patient, 2 mL of blood sample all subjects were collected in tubes containing ethylene diamine tetra acetic acid (EDTA) and stored at −20 °C until further use. Further, samples were categorized in two groups (mild and severe), according to Indian Council of Medical Research (ICMR), New Delhi, India. Patients with respiratory rate less than 24 per minute and SpO2 > 94% on room air were considered as mild patient while patients with respiratory rate more than 30 per minute OR SpO2 < 90% on room air with pneumonia were categorized into severe patients.
2.3. Genotyping analysis
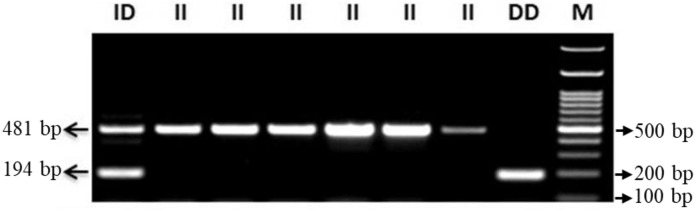
High molecular weight genomic DNA was extracted from peripheral blood leucocytes using commercially available kit (Nucleospin Blood) and quality/quantity was assessed by using spectrophotometer quantification and analyzed on gel electrophoresis. ACE1 gene polymorphisms (rs4646994) were done in COVID-19 patients via polymerase chain reaction- amplified fragment length polymorphism (PCR- AFLP) using specific primers (forward primer- 5′-CTGGAGACCACTCCCATCCTTTCT-3′ and reverse primer- 5′-GATGTGGCCATCACATTCGTCAGAT-3′). Reactions were performed in 15 μL volume containing 10 pmol of each primer, 1× PCR master mix (EmeraldAmp GT PCR master mix), 3 mM MgCl2, and 2 U Taq polymerase (G Biosciences). PCR sequence amplification was performed under the conditions: Initial denaturation at 95 °C for 5 min, followed by 35 cycles of denaturation at 95 °C for 45 s, annealing at 60 °C for 1.15 min, extension at 72 °C for 2.30 min and final extension at 72 °C for 10 min. PCR products were checked on 2.5% agarose gel and visualized by gel documentation system EZ, BIO-RAD (California), 481 bp product for allele I and 194 bp for allele D (Fig. 1 ).
Fig. 1.
Agarose gel showing PCR products different genotypes of ACEI/D (II: 481 bp; ID: 481, 194 bp; DD: 194 bp) M: DNA Ladder (100 bp).
2.4. Statistical analysis
The continuous variables of each group were summarized as mean ± SE and compared by Student's t-test. Allele and genotype frequencies in both groups were compared using 2 × 2 contingency table by Fisher's exact test by using the SPSS software (version 21). All ‘P' values were considered statistically significant for P < 0.05. Odds ratio (OR) at 95% confidence intervals (CI) were determined to describe the strength of association by logistic regression model.
3. Results
3.1. General characteristics and genotype distribution of COVID19 patients
The comparison of demographic, clinical characteristics, genotype distribution, allele frequencies and carriage rate of COVID-19 with the estimated OR for each risk factor are summarized in Table 1 . Genotype patterns of ACE1 I/D polymorphism is represented in Fig. 1. In the univariate logistic regression model, the ACE1 DD genotype, frequency of D allele, older age (≥46 years), unmarried status, and presence of diabetes and hypertension were found to be high risk factors for causing disease severity among patients of COVID-19. The other variables such as sex, socio-economic status was not found significant between the two groups. The absence of the ‘I' allele of ACE1 I/D polymorphisms was significantly higher in severe COVID-19 patients (OR = 2.59; 95% CI: 1.348–4.970, P = 0.004). However, in multivariate logistic regression, while adjusting other factors including age, sex, marital status, income, diabetes, hypertension, the DD genotype was still significantly associated with COVID-19, conferring a 3-fold higher risk (OR = 3.69; 95% CI: 1.612–8.431). Older age, unmarried status and presence of diabetes were shown the similar pattern between the two groups of COVID-19 patients.
Table 1.
Univariate and multivariate analyses of COVID-19 risk factors.
| Parameters | Mild |
Severe |
Univariate |
p = value | Multivariate |
p = value |
|---|---|---|---|---|---|---|
| n = 149 | n = 120 | OR (95% CI) | OR (95% CI) | |||
| Age, (n %) | ||||||
| ≤45 | 75 (50.3) | 14 (11.7) | 1 (Ref.) | 1 (Ref.) | ||
| ≥46 | 74 (49.7) | 106 (88.3) | 7.67 (4.033–14.600) | <0.001 | 4.84 (2.391–9.816) | <0.001 |
| Gender, (n %) | ||||||
| Male | 97 (65.1) | 73 (60.8) | 1 (Ref.) | 1 (Ref.) | ||
| Female | 52 (34.9) | 47 (39.2) | 1.20 (0.730–1.976) | 0.471 | 1.22 (0.683–2.191) | 0.497 |
| Marital Status, (n %) | ||||||
| Married | 125 (83.9) | 119 (99.2) | 1 (Ref.) | 1 (Ref.) | ||
| Unmarried | 24 (16.1) | 1 (0.8) | 22.85 (3.043–171.560) | 0.002 | 6.61 (0.790–55.375) | 0.082 |
| Socio-economic status, (n %) | ||||||
| Low | 107 (71.8) | 91 (75.8) | 1 (Ref.) | 1 (Ref.) | ||
| High | 42 (28.2) | 29 (24.2) | 0.81 (0.469–1.407) | 0.457 | 0.65 (0.344–1.224) | 0.649 |
| Diabetes, (n %) | ||||||
| No | 136 (91.3) | 86 (71.7) | 1 (Ref.) | 1 (Ref.) | ||
| Yes | 13 (8.7) | 34 (28.3) | 4.14 (2.067–8.278) | <0.001 | 2.80 (1.290–6.052) | 0.009 |
| Hypertension, (n %) | ||||||
| No | 142 (95.3) | 100 (83.3) | 1 (Ref.) | 1 (Ref.) | ||
| Yes | 7 (4.7) | 20 (16.7) | 4.06 (1.653–9.959) | 0.002 | 1.83 (0.673–5.006) | 0.236 |
| ACE Genotype (n %) | ||||||
| II | 74 (49.7) | 42 (35.0) | 1 (Ref.) | 1 (Ref.) | ||
| ID | 58 (38.9) | 48 (40.0) | 1.46 (0.851–2.498) | 0.17 | 1.54 (0.835–2.850) | 0.166 |
| DD | 17 (11.4) | 30 (25.0) | 3.11 (1.536–6.294) | 0.002 | 3.69 (1.612–8.431) | 0.002 |
| Allele (n %) | ||||||
| I* | 206 (69.1) | 132 (55.0) | 1 (Ref.) | |||
| D* | 92 (30.9) | 108 (45.0) | 1.83 (1.286–2.609) | 0.001 | ||
| Carriage rate (n %) | ||||||
| I (+) | 132 (88.6) | 90 (75.0) | 1 (Ref.) | 1 (Ref.) | ||
| I (−) | 17 (11.4) | 30 (25)0.0 | 2.59 (1.348–4.970) | 0.004 | 3.00 (1.388–6.495) | 0.005 |
| D (+) | 75 (50.3) | 78 (65.0) | 1 (Ref.) | 1 (Ref.) | ||
| D (−) | 74 (49.7) | 42 (35.0) | 0.55 (0.333–0.894) | 0.016 | 0.50 (0.284–0.878) | 0.016 |
CI = Confidence interval; OR = Odds ratio; 1.0 (Reference), Alleles*, total number of chromosomes.
3.2. Association of the ACE I/D polymorphism with the COVID-19 relevant risk factors
To further evaluate the etiologic effects of ACE I/D polymorphisms in COPVID-19, we evaluated the possible influence of ACE I/D polymorphisms on demographic and clinical characteristics in COVID-19 patients (Table 2, Table 3 ). The ACE ID genotypes were found to be statistically associated with high socio-economic COVID19 patients (OR: 2.39, 95% CI: 1.296–4.398). After adjusting for other risk factors, the ID genotype was still significantly independently associated with high socio-economic COVID-19 patients (OR: 2.48, 95% CI: 1.331–4.609). Whereas there was no statistical significance between the ACE I/D polymorphism and demographic & clinical characteristics (age, gender, marital status, diabetes, and hypertension).
Table 2.
Associations between ACE I/D polymorphisms with demographic and clinical characteristics in COVID-19 patients (univariate analyses).
| ACE genotypes |
II |
ID |
Univariate |
p = value | DD |
Univariate |
p = value |
|---|---|---|---|---|---|---|---|
| Parameters | (n = 116) | (n = 106) | OR (95% CI) | (n = 47) | OR (95% CI) | ||
| Age (n %) | |||||||
| ≤45 | 42 (36.2) | 35 (33.0) | 1 (Ref.) | 12 (25.5) | 1 (Ref.) | ||
| ≥46 | 74 (63.8) | 71 (67.0) | 1.15 (0.661–2.004) | 0.618 | 35 (74.5) | 1.66 (0.776–3.530) | 0.192 |
| Gender, (n %) | |||||||
| Male | 77 (66.4) | 63 (59.4) | 1 (Ref.) | 30 (63.8) | 1 (Ref.) | ||
| Female | 39 (33.6) | 43 (40.6) | 1.35 (0.780–2.328) | 0.285 | 17 (36.2) | 1.12 (0.551–2.273) | 0.756 |
| Marital Status (n %) | |||||||
| Married | 106 (91.4) | 96 (90.6) | 1 (Ref.) | 42 (89.4) | 1 (Ref.) | ||
| Unmarried | 10 (8.6) | 10 (9.4) | 0.91 (0.361–2.270) | 0.833 | 5 (10.6) | 0.79 (0.256–2.457) | 0.687 |
| Socio-economic status (n %) | |||||||
| Low | 94 (81.0) | 68 (64.2) | 1 (Ref.) | 36 (76.6) | 1 (Ref.) | ||
| High | 22 (19.0) | 38 (35.8) | 2.39 (1.296–4.398) | 0.005 | 11 (23.4) | 1.32 (0.575–2.962) | 0.524 |
| Diabetes (n %) | |||||||
| No | 97 (83.6) | 87 (82.1) | 1 (Ref.) | 38 (80.9) | 1 (Ref.) | ||
| Yes | 19 (16.4) | 19 (17.9) | 1.12 (0.554–2.242) | 0.76 | 9 (19.1) | 1.21 (0.503–2.907) | 0.671 |
| Hypertension (n %) | |||||||
| No | 107 (92.2) | 93 (87.7) | 1 (Ref.) | 42 (89.4) | 1 (Ref.) | ||
| Yes | 9 (7.8) | 13 (12.3) | 1.66 (0.680–4.064) | 0.266 | 5 (10.6) | 1.42 (0.448–4.470) | 0.554 |
CI = Confidence interval; OR = Odds ratio; 1.0 (Reference).
Table 3.
Associations between ACE I/D polymorphisms with demographic and clinical characteristics in COVID-19 patients (multivariate analyses).
| ACE genotypes |
II |
ID |
Multivariate |
P = value | DD |
Multivariate |
P = value |
|---|---|---|---|---|---|---|---|
| Parameters | n = 116 | n = 106 | OR (95% CI) | n = 47 | OR (95% CI) | ||
| Age (n %) | |||||||
| ≤45 | 42 (36.2) | 35 (33.0) | 1 (Ref.) | 12 (25.5) | 1 (Ref.) | ||
| ≥46 | 74 (63.8) | 71 (67.0) | 1.15 (0.605–2.185) | 0.669 | 35 (74.5) | 2.10 (0.821–5.369) | 0.122 |
| Gender, (n %) | |||||||
| Male | 77 (66.4) | 63 (59.4) | 1 (Ref.) | 30 (63.8) | 1 (Ref.) | ||
| Female | 39 (33.6) | 43 (40.6) | 1.31 (0.743–2.309) | 0.352 | 17 (36.2) | 1.15 (0.558–2.371) | 0.705 |
| Marital Status, (n %) | |||||||
| Married | 106 (91.4) | 96 (90.6) | 1 (Ref.) | 42 (89.4) | 1 (Ref.) | ||
| Unmarried | 10 (8.6) | 10 (9.4) | 1.50 (0.521–4.284) | 0.833 | 5 (10.6) | 0.41 (0.104–1.600) | 0.199 |
| Socio-economic status (n %) | |||||||
| Low | 94 (81.0) | 68 (64.2) | 1 (Ref.) | 36 (76.6) | 1 (Ref.) | ||
| High | 22 (19.0) | 38 (35.8) | 2.48 (1.331–4.609) | 0.004 | 11 (23.4) | 1.31 (0.562–3.045) | 0.533) |
| Diabetes, (n %) | |||||||
| No | 97 (83.6) | 87 (82.1) | 1 (Ref.) | 38 (80.9) | 1 (Ref.) | ||
| Yes | 19 (16.4) | 19 (17.9) | 1.03 (0.482–4.960) | 0.947 | 9 (19.1) | 0.99 (0.381–2.593) | 0.994 |
| Hypertension, (n %) | |||||||
| No | 107 (92.2) | 93 (87.7) | 1 (Ref.) | 42 (89.4) | 1 (Ref.) | ||
| Yes | 9 (7.8) | 13 (12.3) | 1.91 (0.738–4.960) | 0.182 | 5 (10.6) | 1.19 (0.343–4.097) | 0.787 |
CI = Confidence interval; OR = Odds ratio; 1.0 (Reference).
4. Discussion
In present study, 269 COVID-19 patients were selected with demographic and clinical characteristics. We observed that high frequency of diabetes patients (37%) as compared to hypertension (21.4%) (Table 1). Similarly, de Abajo et al., found higher prevalence of diabetes in 1339 COVID-19 cases from Madrid, Spain as compared with 13,390 matched controls (27.2% vs 20.3%; crude odds ratio, OR, 1.50) (de Abajo et al., 2020). In contrast, Singh et al., pooled different studies (n = 2209) and found higher percentage of hypertension (21%) as compared to diabetes (11%) and CVD (7%) (Singh et al., 2020). Similarly, a meta-analysis of COVID-19 Patients (n = 1576 patients) reported percentage of different comorbid conditions such as hypertension (17%) > diabetes (8%) > CVD (5%) (Yang et al., 2020).
COVID-19 is a highly contagious disease characterized by high mortality, especially for patients with severe comorbidities such as diabetes, hypertension, CVD and CKD (Zhou et al., 2020). The documented history of diabetes has been stated to be an independent indicator of morbidity and death in SARS patients (Li et al., 2020; Yang et al., 2006). Diabetes hyperglycemia is suspected to cause immune response dysfunction, which fails to regulate the spread of invasive pathogens. Therefore, diabetic individuals are considered to be more prone to infections and incidence of infectious diseases and it will increase associated comorbidities (Berbudi et al., 2020). A study was released by the Chinese Centre for Disease Control and Prevention, which showed elevated mortality rate in people with diabetes (2.3%, total and 7.3%, diabetes patients) study was performed in 72,314 cases of COVID-19 (Wu and McGoogan, 2020). Li et al. also found high mortality rate in diabetic individuals (14.5%) as compared to non-diabetic (5.7) COVID-19 cases (Li et al., 2020). Similarly, our study found that diabetic patients have 2.80-fold higher risk for having severity/mortality in COVID-19 (P < 0.001). When we classified the diabetes patients according to the severity of the COVID-19, we found 8.7% mild, and 28.3% severe cases (Table 1). Our study also suggested that hypertensive individual might have 4.24-fold higher risk of severity/mortality. When we classified our hypertensive COVID-19 cases, we found 4.7% cases in mild and 16.7% cases in severe. Similarly, a meta-analysis suggested that hypertension may be risk factor for severity in COVID-19 (Yang et al., 2020).
Blood pressure homeostasis maintained by renin–angiotensin system (RAS) (Boehm and Nabel, 2002; Skeggs et al., 1980). RAS system modulated by ACE1 and ACE2. Angiotensin I is converted into angiotensin II (ATII) by ACE1 and degraded bioactive bradykinin (Baudin, 2002). Insertion/deletion (I/D) polymorphism has been correlated with levels of circulating and tissue ACE1 and influences almost half of the variability of serum ACE levels in the general population. ‘D' allele of ACEI/D is associated with higher ACE activity (Tiret et al., 1992). Which means individuals with DD genotype showed approximately twice ACE activity levels as compared to II genotype individuals (Rigat et al., 1990). ‘D' allele of ACE1 gene is significantly associated with hypoxemic group as compared to non-hypoxemic group (Itoyama et al., 2004). However, later on, Delanghe et al. have recently found that the prevalence of COVID-19 in 33 countries has been substantially associated with ACE1 I/D polymorphism (Delanghe et al., 2020b). To date, few studies have been published that investigate the relationship between ACE1 gene polymorphism and COVID-19 severity, but we are still lacking definite results (Delanghe et al., 2020a; Devic Pavlic et al., 2020; Gemmati et al., 2020; Hatami et al., 2020). In present study, we observed that individual with ‘DD' genotype showed significantly 3.69-fold higher risk of COVID-19 severity (P = 0.002, Table 1). Similarly, other studies found that ACE1 D/D-genotype showed association with COVID-19 related mortality (Annunziata et al., 2020; Yamamoto et al., 2020). Gomez et al. found D allele was significantly associated with hypoxemic as compared to non-hypoxemic patients; however, ‘DD' genotype individuals did not show any association with COVID-19 infection (Gomez et al., 2020).
5. Conclusion
Based on our results, we conclude that the severity of the COVID-19 patients may depend on age, diabetes, hypertension, and ACE gene polymorphism. Our results suggest a prospective paradigm of ‘DD' genotype that has the potential to help explain the susceptibility of the host response to SARS-CoV-2 infection and involve in numerous pathological. Thus, ACE I/D polymorphism may be a useful tool to predict the development of disease and may have an influence on the treatment outcomes against the COVID-19 to establish a population-based therapeutic development. Further studies involving a larger cohort and a control group should be carried out to better understand the association between COVID-19 severity and different genotypes of ACE with treatment response.
Funding
The work was supported by intramural research grants (ELMC&H/R_Cell/EC/2020/272 dated 31/12/2020) from Era University, Lucknow, India.
Author contributions
Conceived and designed the experiments: SVa1, MA, ZS, FM. performed the experiments: SVa1, MA, IA, SVa. Analyzed the data: MA, STR, SVa1.Wrote the paper: SVa1, MA, FHK.
Declaration of Competing Interest
No potential conflict of interest was reported by the authors.
Acknowledgements
Authors are thankful to Era University for giving the lab facility and financial support.
References
- Annunziata S., Bauckneht M., Albano D., Argiroffi G., Calabro D., Abenavoli E., Linguanti F., Laudicella R., Young Committee of the Italian Association of Nuclear, M Impact of the COVID-19 pandemic in nuclear medicine departments: preliminary report of the first international survey. Eur. J. Nucl. Med. Mol. Imaging. 2020;47:2090–2099. doi: 10.1007/s00259-020-04874-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin B. New aspects on angiotensin-converting enzyme: from gene to disease. Clin. Chem. Lab. Med. 2002;40:256–265. doi: 10.1515/CCLM.2002.042. [DOI] [PubMed] [Google Scholar]
- Berbudi A., Rahmadika N., Tjahjadi A.I., Ruslami R. Type 2 diabetes and its impact on the immune system. Curr. Diabetes Rev. 2020;16:442–449. doi: 10.2174/1573399815666191024085838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm M., Nabel E.G. Angiotensin-converting enzyme 2--a new cardiac regulator. N. Engl. J. Med. 2002;347:1795–1797. doi: 10.1056/NEJMcibr022472. [DOI] [PubMed] [Google Scholar]
- Clarke N.E., Turner A.J. Angiotensin-converting enzyme 2: the first decade. Int. J. Hypertens. 2012;2012:307315. doi: 10.1155/2012/307315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connors J.M., Levy J.H. COVID-19 and its implications for thrombosis and anticoagulation. Blood. 2020;135:2033–2040. doi: 10.1182/blood.2020006000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Abajo F.J., Rodriguez-Martin S., Lerma V., Mejia-Abril G., Aguilar M., Garcia-Luque A., Laredo L., Laosa O., Centeno-Soto G.A., Angeles Galvez M., Puerro M., Gonzalez-Rojano E., Pedraza L., de Pablo I., Abad-Santos F., Rodriguez-Manas L., Gil M., Tobias A., Rodriguez-Miguel A., Rodriguez-Puyol D., Group, M.-A.C.s Use of renin-angiotensin-aldosterone system inhibitors and risk of COVID-19 requiring admission to hospital: a case-population study. Lancet. 2020;395:1705–1714. doi: 10.1016/S0140-6736(20)31030-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delanghe J.R., Speeckaert M.M., De Buyzere M.L. ACE polymorphism and COVID-19 outcome. Endocrine. 2020;70:13–14. doi: 10.1007/s12020-020-02454-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delanghe J.R., Speeckaert M.M., De Buyzere M.L. COVID-19 infections are also affected by human ACE1 D/I polymorphism. Clin. Chem. Lab. Med. 2020;58:1125–1126. doi: 10.1515/cclm-2020-0425. [DOI] [PubMed] [Google Scholar]
- Devic Pavlic S., Nadalin S., Starcevic Cizmarevic N., Buretic-Tomljanovic A., Radojcic Badovinac A., Ristic S. Could angiotensin-converting enzyme 1 I/D polymorphism be a modificator of COVID-19 response in different populations, diseases, and/or conditions? J. Renin-Angiotensin-Aldosterone Syst. 2020;21 doi: 10.1177/1470320320957157. (1470320320957157) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemmati D., Bramanti B., Serino M.L., Secchiero P., Zauli G., Tisato V. COVID-19 and individual genetic susceptibility/receptivity: role of ACE1/ACE2 genes, immunity, inflammation and coagulation. Might the double X-chromosome in females be protective against SARS-CoV-2 compared to the single X-chromosome in males? Int J Mol Sci. 2020;21 doi: 10.3390/ijms21103474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez J., Albaiceta G.M., Garcia-Clemente M., Lopez-Larrea C., Amado-Rodriguez L., Lopez-Alonso I., Hermida T., Enriquez A.I., Herrero P., Melon S., Alvarez-Arguelles M.E., Boga J.A., Rojo-Alba S., Cuesta-Llavona E., Alvarez V., Lorca R., Coto E. Angiotensin-converting enzymes (ACE, ACE2) gene variants and COVID-19 outcome. Gene. 2020;762:145102. doi: 10.1016/j.gene.2020.145102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatami N., Ahi S., Sadeghinikoo A., Foroughian M., Javdani F., Kalani N., Fereydoni M., Keshavarz P., Hosseini A. Worldwide ACE (I/D) polymorphism may affect COVID-19 recovery rate: an ecological meta-regression. Endocrine. 2020;68:479–484. doi: 10.1007/s12020-020-02381-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess D.C., Eldahshan W., Rutkowski E. COVID-19-related stroke. Transl. Stroke Res. 2020;11:322–325. doi: 10.1007/s12975-020-00818-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoyama S., Keicho N., Quy T., Phi N.C., Long H.T., Ha L.D., Ban V.V., Ohashi J., Hijikata M., Matsushita I., Kawana A., Yanai H., Kirikae T., Kuratsuji T., Sasazuki T. ACE1 polymorphism and progression of SARS. Biochem. Biophys. Res. Commun. 2004;323:1124–1129. doi: 10.1016/j.bbrc.2004.08.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam S.D., Bordin N., Waman V.P., Scholes H.M., Ashford P., Sen N., van Dorp L., Rauer C., Dawson N.L., Pang C.S.M., Abbasian M., Sillitoe I., Edwards S.J.L., Fraternali F., Lees J.G., Santini J.M., Orengo C.A. SARS-CoV-2 spike protein predicted to form complexes with host receptor protein orthologues from a broad range of mammals. Sci. Rep. 2020;10:16471. doi: 10.1038/s41598-020-71936-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.C., Zhang J., Zhuo J.L. The vasoprotective axes of the renin-angiotensin system: physiological relevance and therapeutic implications in cardiovascular, hypertensive and kidney diseases. Pharmacol. Res. 2017;125:21–38. doi: 10.1016/j.phrs.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Deng Q., Feng J., Li F., Xiong N., He Q. Clinical characteristics of diabetic patients with COVID-19. J. Diabetes Res. 2020;2020:1652403. doi: 10.1155/2020/1652403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H., Stratton C.W., Tang Y.W. Outbreak of pneumonia of unknown etiology in Wuhan, China: the mystery and the miracle. J. Med. Virol. 2020;92:401–402. doi: 10.1002/jmv.25678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pati A., Mahto H., Padhi S., Panda A.K. ACE deletion allele is associated with susceptibility to SARS-CoV-2 infection and mortality rate: an epidemiological study in the Asian population. Clin. Chim. Acta. 2020;510:455–458. doi: 10.1016/j.cca.2020.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson S., Hirsch J.S., Narasimhan M., Crawford J.M., McGinn T., Davidson K.W., the Northwell, C.-R.C, Barnaby D.P., Becker L.B., Chelico J.D., Cohen S.L., Cookingham J., Coppa K., Diefenbach M.A., Dominello A.J., Duer-Hefele J., Falzon L., Gitlin J., Hajizadeh N., Harvin T.G., Hirschwerk D.A., Kim E.J., Kozel Z.M., Marrast L.M., Mogavero J.N., Osorio G.A., Qiu M., Zanos T.P. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York City Area. JAMA. 2020;323:2052–2059. doi: 10.1001/jama.2020.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigat B., Hubert C., Alhenc-Gelas F., Cambien F., Corvol P., Soubrier F. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J. Clin. Invest. 1990;86:1343–1346. doi: 10.1172/JCI114844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos R.A.S., Sampaio W.O., Alzamora A.C., Motta-Santos D., Alenina N., Bader M., Campagnole-Santos M.J. The ACE2/angiotensin-(1-7)/MAS Axis of the renin-angiotensin system: focus on angiotensin-(1-7) Physiol. Rev. 2018;98:505–553. doi: 10.1152/physrev.00023.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyaolu A., Okorie C., Marinkovic A., Patidar R., Younis K., Desai P., Hosein Z., Padda I., Mangat J., Altaf M. Comorbidity and its impact on patients with COVID-19. SN Compr Clin Med. 2020:1–8. doi: 10.1007/s42399-020-00363-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A.K., Gupta R., Misra A. Comorbidities in COVID-19: outcomes in hypertensive cohort and controversies with renin angiotensin system blockers. Diabetes Metab Syndr. 2020;14:283–287. doi: 10.1016/j.dsx.2020.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skeggs L.T., Dorer F.E., Levine M., Lentz K.E., Kahn J.R. The biochemistry of the renin-angiotensin system. Adv. Exp. Med. Biol. 1980;130:1–27. doi: 10.1007/978-1-4615-9173-3_1. [DOI] [PubMed] [Google Scholar]
- Tiret L., Rigat B., Visvikis S., Breda C., Corvol P., Cambien F., Soubrier F. Evidence, from combined segregation and linkage analysis, that a variant of the angiotensin I-converting enzyme (ACE) gene controls plasma ACE levels. Am. J. Hum. Genet. 1992;51:197–205. [PMC free article] [PubMed] [Google Scholar]
- WHO . World Health Organization; 2020. Coronavirus Disease (COVID-19) Dashboard. [Google Scholar]
- Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- Xu X., Chen P., Wang J., Feng J., Zhou H., Li X., Zhong W., Hao P. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 2020;63:457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto N., Ariumi Y., Nishida N., Yamamoto R., Bauer G., Gojobori T., Shimotohno K., Mizokami M. SARS-CoV-2 infections and COVID-19 mortalities strongly correlate with ACE1 I/D genotype. Gene. 2020;758:144944. doi: 10.1016/j.gene.2020.144944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.K., Feng Y., Yuan M.Y., Yuan S.Y., Fu H.J., Wu B.Y., Sun G.Z., Yang G.R., Zhang X.L., Wang L., Xu X., Xu X.P., Chan J.C. Plasma glucose levels and diabetes are independent predictors for mortality and morbidity in patients with SARS. Diabet. Med. 2006;23:623–628. doi: 10.1111/j.1464-5491.2006.01861.x. [DOI] [PubMed] [Google Scholar]
- Yang X.H., Deng W., Tong Z., Liu Y.X., Zhang L.F., Zhu H., Gao H., Huang L., Liu Y.L., Ma C.M., Xu Y.F., Ding M.X., Deng H.K., Qin C. Mice transgenic for human angiotensin-converting enzyme 2 provide a model for SARS coronavirus infection. Comp Med. 2007;57:450–459. [PubMed] [Google Scholar]
- Yang J., Zheng Y., Gou X., Pu K., Chen Z., Guo Q., Ji R., Wang H., Wang Y., Zhou Y. Prevalence of comorbidities and its effects in patients infected with SARS-CoV-2: a systematic review and meta-analysis. Int. J. Infect. Dis. 2020;94:91–95. doi: 10.1016/j.ijid.2020.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H., Cao J.J. Angiotensin-converting enzyme gene polymorphism and severe lung injury in patients with coronavirus disease 2019. Am. J. Pathol. 2020;190:2013–2017. doi: 10.1016/j.ajpath.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Ye S., Wang W., Li S., Hu Q. Clinical features of COVID-19 patients with diabetes and secondary hyperglycemia. J. Diabetes Res. 2020;2020:3918723. doi: 10.1155/2020/3918723. [DOI] [PMC free article] [PubMed] [Google Scholar]