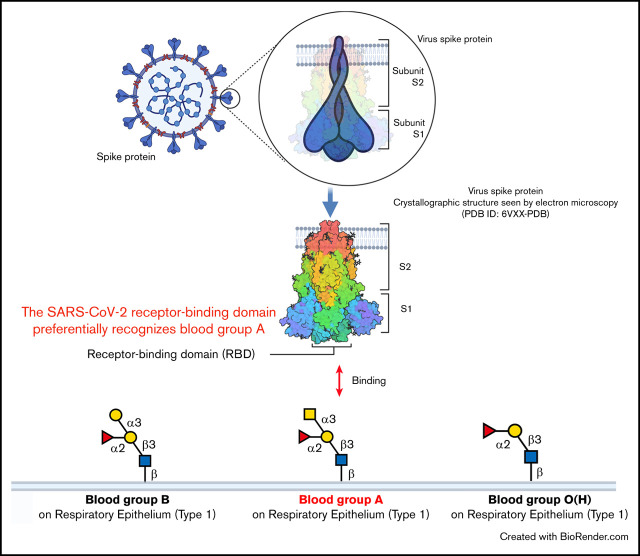
Key Points
The RBD of SARS-CoV-2 shares sequence similarity with an ancient lectin family known to bind blood group antigens.
SARS-CoV-2 RBD binds the blood group A expressed on respiratory epithelial cells, directly linking blood group A and SARS-CoV-2.
Visual Abstract
Introduction
Severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2), the cause of COVID-19, has resulted in a global pandemic, overwhelming modern health care systems and reshaping the world economy. Despite the devastating consequences of SARS-CoV-2, not all individuals seem to be equally susceptible to contracting the virus. Recent genome-wide association studies identified the locus responsible for ABO(H) blood group expression, the first polymorphism described in the human population well over a century ago, as one of the most significant genetic predictors of SARS-CoV-2 infection risk.1 Although previous and subsequent studies corroborate these results,2-6 additional data have failed to observe a similar association between ABO(H) blood group status and SARS-CoV-2 infection.7 Although differences in study population numbers and other variables may influence these outcomes, these collective studies in general warrant a direct examination of a possible association between ABO(H) blood group antigens and SARS-CoV-2.
ABO(H) blood groups are not only the first polymorphisms described in the human population, they are also the most well-recognized. Naturally occurring antibodies against the blood group ABO(H) antigens in individuals who do not express these same polymorphic structures can cause potentially fatal hemolytic transfusion reactions after transfusion and severe acute graft rejection after transplantation.8 It is possible that anti–blood group antibodies may also influence SARS-CoV-2 infection through engagement of putative ABO(H) blood group antigens on the surface of the virus.9 However, these antibodies can be found in individuals of multiple blood types (eg, anti–blood group B antibodies are present in both blood group A and blood group O individuals) and thus may not fully account for the propensity of blood group A individuals, in particular, to exhibit an increased risk for SARS-CoV-2 infection. Furthermore, although ABO(H) antigens may influence disease progression,10 early studies suggested that increased risk was primarily associated with the likelihood of initial infection.2-5,11 In this regard, the mechanism by which ABO(H) antigens, and particularly those of blood group A, influence the likelihood of infection is still unknown.
Methods
Each SARS-CoV receptor-binding domain (RBD), which is responsible for infection,12 was cloned and purified as previously outlined.13,14 The SARS-COV-2 RBD was incubated with HEK293 T cells, HEK293 T cells expressing angiotensin-converting enzyme 2(ACE2), or red blood cells (RBCs), followed by detection with anti-His antibody (Anti-His-Tag mAb-Alexa Fluor 647) and flow cytometric analysis. Anti-A antibody was similarly used to detect the A antigen on blood group A RBCs. For array analysis, the SARS-CoV-2 and SARS-CoV RBDs were incubated in phosphate-buffered saline containing 0.05% Tween 20 and 1% bovine serum albumin for 1 hour at room temperature on a glycan array populated with ABO(H) glycans synthesized and printed as outlined previously.15,16 RBD binding to glycan arrays was detected by anti-His antibody (Anti-His-Tag mAb-Alexa Fluor 647),15 followed by image generation by microarray scanner (ScanArray Express, PerkinElmer Life Sciences) and array analysis using ImaGene software (BioDiscovery). See supplemental Methods for additional details.
Results and discussion
Although the spike protein of SARS-CoV-2 can facilitate cell entry through well-known interactions between its RBD and ACE2,12 it is possible that the SARS-CoV-2 RBD may interact with other host molecules, including blood group antigens, which in turn may contribute to disease susceptibility. In particular, the RBD of SARS-CoV-2 contains general structural features similar to galectins, an ancient family of carbohydrate-binding proteins expressed in all metazoans. Accordingly, sequence alignment revealed sequence similarities between the SARS-CoV-2 RBD and human galectins (Figure 1A-B). Because galectins have been shown to exhibit high affinity for blood group antigens,15 we next examined whether the SARS-CoV-2 RBD may display similar blood group antigen recognition. To test this, we examined SARS-CoV-2 RBD binding with RBCs isolated from blood group A, B, or O individuals. Interestingly, the SARS-CoV-2 RBD exhibited only low-level binding to human RBCs of all types and failed to display any detectable preference for blood type A RBCs (Figure 1C; supplemental Figure 1). In contrast, the SARS-CoV-2 RBD readily bound ACE2-expressing HEK293 T cells, and anti-A antibody likewise bound blood group A RBCs (Figure 1D-E). Thus, significant binding of the SARS-CoV-2 RBD to the blood group A structures found on human blood group A RBCs does not seem to contribute to the increased likelihood of SARS-CoV-2 infection in blood group A individuals.
Figure 1.
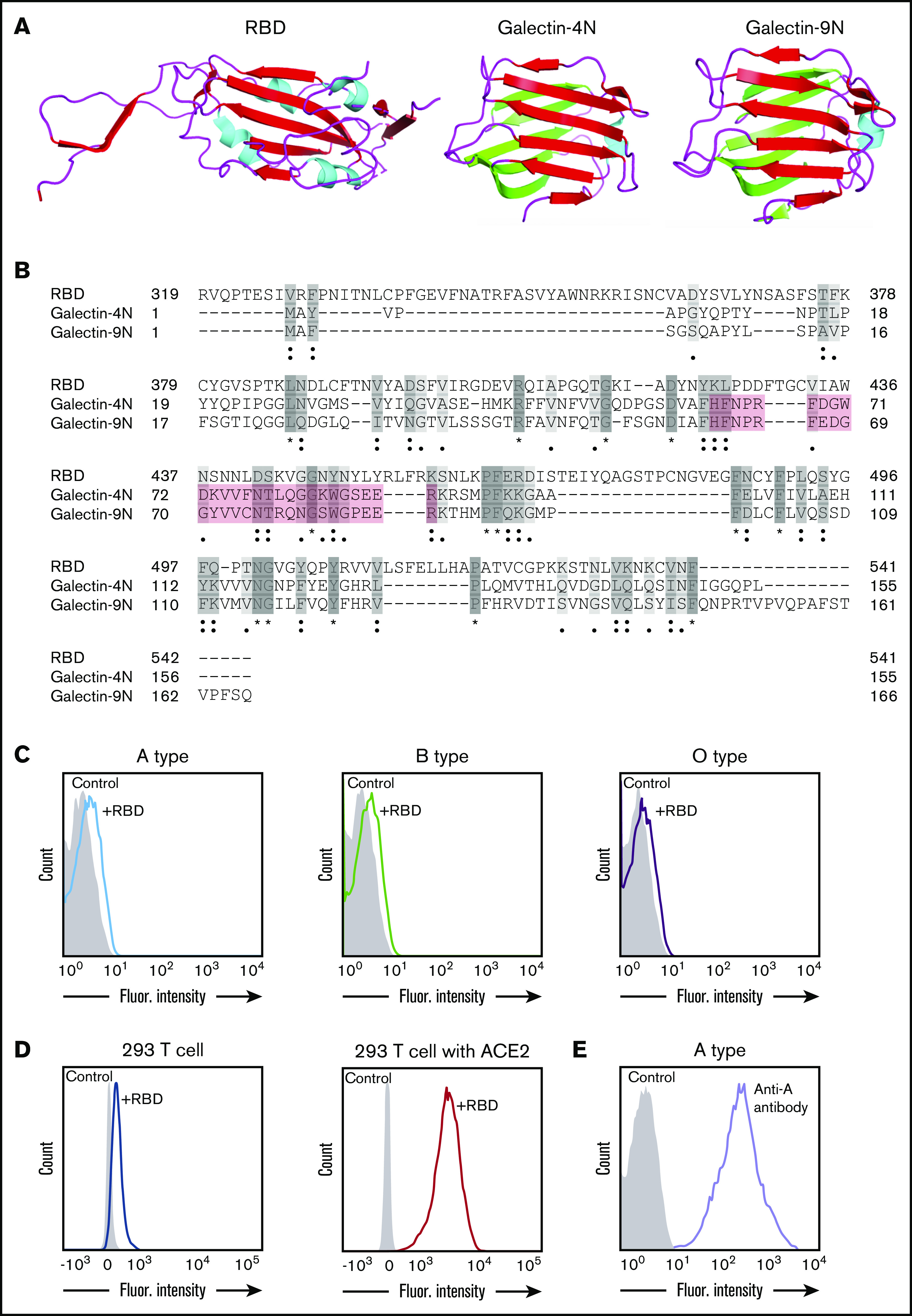
SARS-CoV-2 shares significant similarity to human galectins. (A) Structural representation of SARS-CoV-2 RBD (Protein Data Bank [PDB] ID: 6VXX), human galectin-4N-terminal domain (PDB: 5DUW), and human galectin-9N-terminal domain (PDB ID: 3LSD); β-sheet structures shown in red or green, α-helix shown in cyan. (B) Sequence alignment (Uniport align program) of SARS-CoV-2 RBD, human galectin-4N-terminal domain (galectin-4N) and human galectin-9 N-terminal domain (galectin-9N) indicating fully conserved residues (dark gray [*]), highly conserved residues (gray [:]), lowly conserved residues (light gray [.]); the galectin carbohydrate binding pocket is highlighted in light red. (C) Flow cytometric analysis of blood group A, B, or O type human RBCs after incubation with SARS-CoV-2 RBD for 1 hour followed by detection with anti-His antibody. (D) Flow cytometric analysis of HEK293 T cells alone or HEK293 T cells stably transduced to express ACE2 after a 1-hour incubation with the SARS-CoV-2 RBD and detection with anti-His antibody. (E) Flow cytometric examination of blood group A RBCs after incubation with anti-A antibody. Secondary antibody alone served as a control.
In addition to the ABO(H) blood group antigens expressed on RBCs, called type II ABO(H) blood group antigens, ABO(H) antigens are also expressed on various other tissues throughout the body including the respiratory epithelium.17,18 However, the types of ABO(H) antigens expressed along respiratory epithelium, referred to as type I ABO(H) antigens, are subtly, yet fundamentally different than their type II ABO(H) counterparts because of a distinct linkage configuration between the penultimate galactose residue and N-acetylglucosamine (type I, β1-3; type II, β1-4) (Figure 2A).19 Given its aerosolized route of infection, SARS-CoV-2 could have evolved a specific preference for the types of ABO(H) antigens expressed along respiratory epithelial cells. However, to specifically detect this possible preference, a completely distinct approach must be used. Because carbohydrates by definition are posttranslational modifications, they cannot easily be cloned and therefore rapidly expressed to explore these types of interactions. Instead, their synthesis often requires a complex approach of chemoenzymatic synthesis to generate distinct glycan libraries. To overcome this challenge, glycan microarrays populated with distinct glycan determinants have been generated to elucidate the fine specificity of carbohydrate-binding proteins for glycan determinants; such an approach has accurately predicted interactions with numerous cell types.20 As a result, we next turned to a glycan microarray format to define the possible specificity of SARS-CoV-2 for the distinct type I ABO(H) antigens expressed along the respiratory epithelium. In accordance with the lack of preferential blood group binding of the SARS-CoV-2 RBD to RBCs isolated from blood group A, B, or O individuals, no significant binding was observed toward the type II structures of A, B, or O(H) individuals (Figure 2B). In contrast, the SARS-CoV-2 RBD exhibited high preference for the same type of blood group A (type I) expressed on respiratory epithelial cells (Figure 2B). Earlier studies demonstrated that the association of ABO(H) blood group expression and infection is likewise shared by SARS-CoV.21 However, the underlying mechanism responsible for the increased propensity of blood group A individuals to become infected with SARS-CoV, similar to that in SARS-CoV-2, remained incompletely understood. Given the binding preference of SARS-CoV-2 RBD for type I blood group A, we next sought to determine whether a similar binding specificity existed for the SARS-CoV RBD. Despite sharing only 73% of its identity with SARS-CoV-2, SARS-CoV exhibited the same ABO(H) glycan-binding preference for the A antigen expressed in the respiratory tract (Figure 2C).
Figure 2.
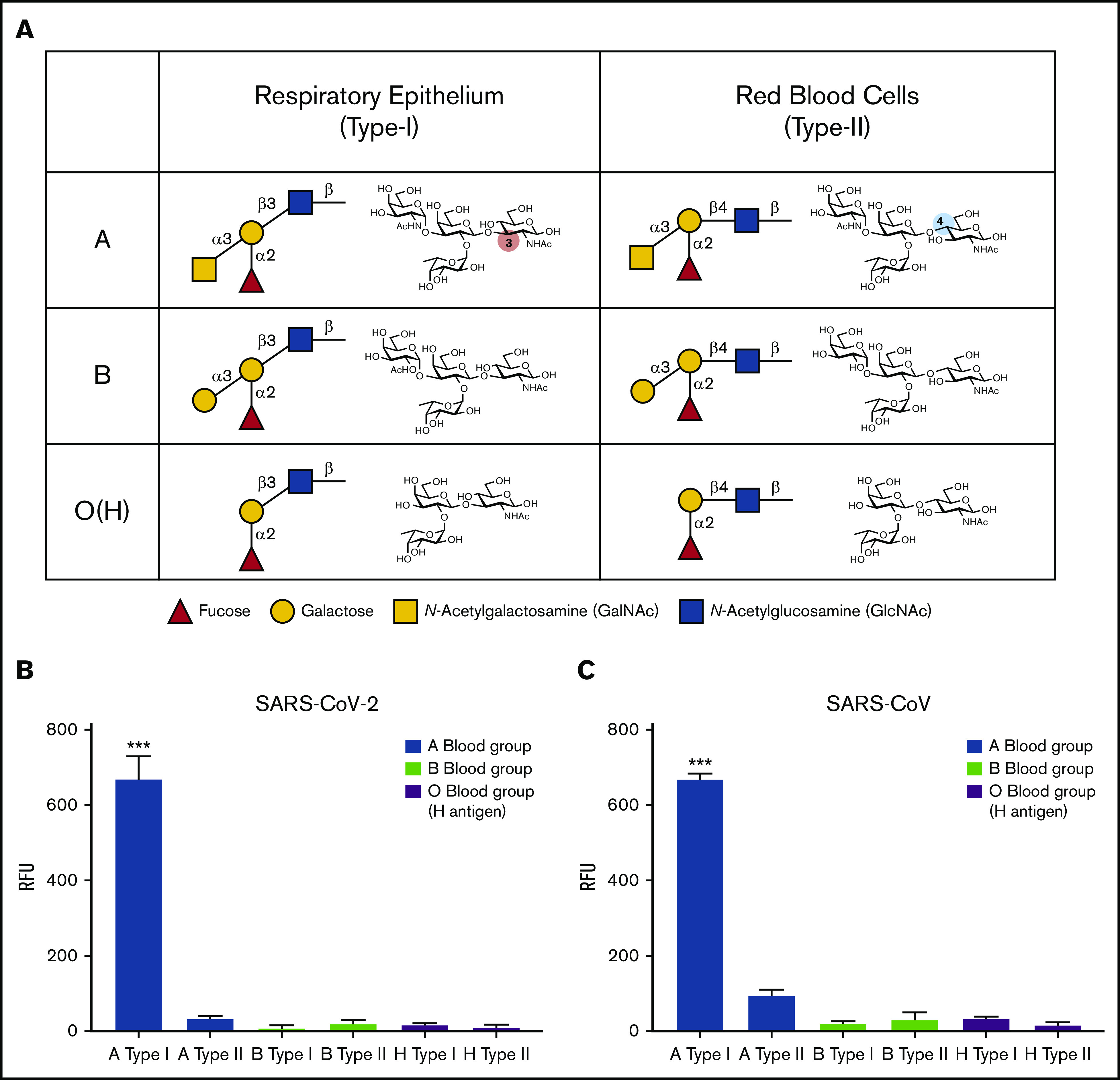
SARS-CoV-2 RBD preferentially binds type I blood group A antigen. (A) Representation of human blood group antigens present on respiratory epithelium (type I) and RBCs (type II). Type I vs type II structures differ based on the linkage between the galactose and N-acetylglucosamine (type I: galactose β1-3 N-acetylglucosamine; type II: galactose β1-4 N-acetylglucosamine). (B-C) ABO(H) glycan microarray data obtained after incubation of SARS-CoV-2 RBD (B) or SARS-CoV RBD (C) with the corresponding glycans shown. Data are representative of 2 independent experiments. Error bars represent mean ± standard deviation. Statistics were generated by one-way analysis of variance with a post hoc Tukey’s multiple comparison. RFU, relative fluorescence unit. ***P < .001 for comparison between RBD binding to A type I vs all other glycans.
The capacity of the RBDs of SARS-CoV and SARS-CoV-2 that are responsible for initial host-cell contact to preferentially recognize the type of blood group A antigen uniquely expressed on respiratory epithelial cells may provide some insight into the apparent preference of SARS-CoV-2 and perhaps other severe corona viruses (SARS-CoV) for blood group A individuals. However, because these results do not definitively demonstrate that blood group A directly contributes to SARS-CoV-2 infection, future studies will certainly be needed to expand upon these initial findings, including an examination of the overall affinity and residues within the RBD responsible for blood group A interactions. Additional studies will likewise be needed to determine whether ABO(H) expression influences viral adhesion to different regions of the nasopharyngeal and respiratory tracts, actual infection of these cells in an ACE2-dependent or independent manner, or the overall stability of the virus along the mucosal surface. Furthermore, it should be noted that in addition to potentially influencing SARS-CoV-2 interactions with host cells, the impact of ABO(H) antigen expression on von Willebrand factor levels may likewise influence thromboembolic complications and therefore COVID-19 disease progression.22 Whatever the possible contribution of ABO(H) antigens to infection and possible disease progression, the ability of the SARS-CoV-2 to directly interact with the blood group A antigen uniquely expressed on respiratory epithelial cells provides clear evidence of a direct association between SARS-CoV-2 and the ABO(H) genetic locus.
Supplementary Material
The full-text version of this article contains a data supplement.
Acknowledgment
The authors thank the Emory Cloning Center and Oskar Laur for assistance with cloning.
Footnotes
To request data, please contact Sean R. Stowell by e-mail at srstowell@bwh.harvard.edu.
Authorship
Contribution: S.-C.W. and S.R.S. designed the study; S.-C.W. and J.W. performed the experiments and analyzed the results with assistance from C.M.A., H.V., C.D.J., D.K., and J.D.R.; R.D.C. provided critical support; S.-C.W., C.M.A., and S.R.S. wrote the manuscript; and all remaining authors helped edit and commented on the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Sean R. Stowell, Joint Program in Transfusion Medicine, Brigham and Women’s Hospital, Harvard Medical School, 630E New Research Building, 77 Avenue Louis Pasteur, Boston, MA 02115; e-mail: srstowell@bwh.harvard.edu.
References
- 1.Ellinghaus D, Degenhardt F, et al. ; Severe Covid-19 GWAS Group . Genomewide association study of severe Covid-19 with respiratory failure. N Engl J Med. 2020;383(16):1522-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zietz M, Zucker JE, Tatonetti NP. Testing the association between blood type and COVID-19 infection, intubation, and death [published online ahead of print 10 September 2020]. medRxiv. doi.org/ 10.1101/2020.04.08.20058073. [DOI]
- 3.Zhao J, Yang Y, Huang H, et al. Relationship between the ABO blood group and the COVID-19 susceptibility. Clin Infect Dis. 2020;ciaa1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fan Q, Zhang W, Li B, Li DJ, Zhang J, Zhao F. Association between ABO blood group system and COVID-19 susceptibility in Wuhan. Front Cell Infect Microbiol. 2020;10:404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu BB, Gu DZ, Yu JN, Yang J, Shen WQ. Association between ABO blood groups and COVID-19 infection, severity and demise: A systematic review and meta-analysis. Infect Genet Evol. 2020;84:104485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnkob MB, Pottegård A, Støvring H, et al. Reduced prevalence of SARS-CoV-2 infection in ABO blood group O. Blood Adv. 2020;4(20):4990-4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Latz CA, DeCarlo C, Boitano L, et al. Blood type and outcomes in patients with COVID-19. Ann Hematol. 2020;99(9):2113-2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooling L. Blood groups in infection and host susceptibility. Clin Microbiol Rev. 2015;28(3):801-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breiman A, Ruvën-Clouet N, Le Pendu J. Harnessing the natural anti-glycan immune response to limit the transmission of enveloped viruses such as SARS-CoV-2. PLoS Pathog. 2020;16(5):e1008556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li J, Wang X, Chen J, Cai Y, Deng A, Yang M. Association between ABO blood groups and risk of SARS-CoV-2 pneumonia. Br J Haematol. 2020;190(1):24-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu Y, Feng Z, Li P, Yu Q. Relationship between ABO blood group distribution and clinical characteristics in patients with COVID-19. Clin Chim Acta. 2020;509:220-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amanat F, Stadlbauer D, Strohmeier S, et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med. 2020;26(7):1033-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stadlbauer D, Amanat F, Chromikova V, et al. SARS-CoV-2 seroconversion in humans: A detailed protocol for a serological assay, antigen production, and test setup. Curr Protoc Microbiol. 2020;57(1):e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stowell SR, Arthur CM, Dias-Baruffi M, et al. Innate immune lectins kill bacteria expressing blood group antigen. Nat Med. 2010;16(3):295-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stowell SR, Arthur CM, McBride R, et al. Microbial glycan microarrays define key features of host-microbial interactions. Nat Chem Biol. 2014;10(6):470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oriol R, Le Pendu J, Mollicone R. Genetics of ABO, H, Lewis, X and related antigens. Vox Sang. 1986;51(3):161-171. [DOI] [PubMed] [Google Scholar]
- 18.Fujitani N, Liu Y, Okamura T, Kimura H. Distribution of H type 1-4 chains of the ABO(H) system in different cell types of human respiratory epithelium. J Histochem Cytochem. 2000;48(12):1649-1656. [DOI] [PubMed] [Google Scholar]
- 19.Stowell CP, Stowell SR. Biologic roles of the ABH and Lewis histo-blood group antigens Part I: infection and immunity. Vox Sang. 2019;114(5):426-442. [DOI] [PubMed] [Google Scholar]
- 20.Arthur CM, Cummings RD, Stowell SR. Using glycan microarrays to understand immunity. Curr Opin Chem Biol. 2014;18:55-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng Y, Cheng G, Chui CH, et al. ABO blood group and susceptibility to severe acute respiratory syndrome. JAMA. 2005;293(12):1450-1451. [DOI] [PubMed] [Google Scholar]
- 22.O’Sullivan JM, Ward S, Fogarty H, O’Donnell JS. More on “Association between ABO blood groups and risk of SARS-CoV-2 pneumonia”. Br J Haematol. 2020;190(1):27-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.