ABSTRACT
Mitophagy, the selective recycling of mitochondria through autophagy, is a crucial metabolic process induced by cellular stress, and defects are linked to aging, sarcopenia and neurodegenerative diseases. To therapeutically target mitophagy, the fundamental in vivo dynamics and molecular mechanisms must be fully understood. Here, we generated mitophagy biosensor zebrafish lines expressing mitochondrially targeted, pH-sensitive fluorescent probes, mito-Keima and mito-EGFP–mCherry, and used quantitative intravital imaging to illuminate mitophagy during physiological stresses, namely, embryonic development, fasting and hypoxia. In fasted muscle, volumetric mitolysosome size analyses documented organelle stress response dynamics, and time-lapse imaging revealed that mitochondrial filaments undergo piecemeal fragmentation and recycling rather than the wholesale turnover observed in cultured cells. Hypoxia-inducible factor (Hif) pathway activation through physiological hypoxia or chemical or genetic modulation also provoked mitophagy. Intriguingly, mutation of a single mitophagy receptor (bnip3) prevented this effect, whereas disruption of other putative hypoxia-associated mitophagy genes [bnip3la (nix), fundc1, pink1 or prkn (Parkin)] had no effect. This in vivo imaging study establishes fundamental dynamics of fasting-induced mitophagy and identifies bnip3 as the master regulator of Hif-induced mitophagy in vertebrate muscle.
KEY WORDS: Autophagy, Hypoxia, Fasting, Mitochondria, Lysosome
Highlighted Article: Live imaging of mitophagy reporter zebrafish shows mitophagy occurs via a piece-by-piece mechanism in vivo. Hypoxia induces mitophagy in muscle in a bnip3-dependent but pink1-independent manner.
INTRODUCTION
Mitophagy is the removal of damaged mitochondria via autophagy, the lysosomal pathway responsible for degrading and recycling cellular components. It prevents cellular damage and apoptosis (Youle, 2019), and defective mitophagy has been linked to both acute and chronic disease states, including muscular dysfunction (Sandri, 2013), neurodegenerative diseases (Pickrell and Youle, 2015) and drug-induced liver injury (Williams et al., 2015; Wrighton et al., 2019). Occurrence of baseline mitophagy has also been detected in a variety of cells, especially those with high metabolic rates (McWilliams et al., 2016, 2018; Sun et al., 2015). However, the full extent of basal mitophagy is incompletely studied, especially during embryonic development. This highlights the need for the development of accurate systems to monitor mitophagy in situ that can provide novel mechanistic insight into system dynamics and activation triggers and could enable effective targeting for interventional or therapeutic purposes.
Mitophagy occurs following mitochondrial damage when the autophagosome, a specialized double-membrane organelle, is recruited to the damaged mitochondrial fragment, and then fuses with a lysosome, initiating cargo degradation (Anding and Baehrecke, 2017). However, the organelle dynamics and interactions between mitochondria, autophagosomes, autolysosomes and lysosomes in response to physiological stresses are underexplored in vivo. Fasting is well documented to induce lysosome biogenesis and autophagy (Settembre et al., 2013; Raefsky and Mattson, 2017; Preidis et al., 2017; Sebastián and Zorzano, 2020). In contrast, dynamic changes in mitolysosome (lysosomes containing mitochondria for degradation) size or number in response to increasing stress in vivo are unknown. Furthermore, the temporal dynamics of mitochondrial recycling and trafficking to autolysosomes have not been verified via intravital imaging in any live vertebrate organism.
Multiple molecular pathways govern mitophagy depending on the cell type and nature of stress. PINK1 and PARKIN (also known as PRKN), genes with mutant alleles linked to Parkinson's disease, have been shown to facilitate mitophagy following mitochondrial membrane depolarization, and most molecular mechanisms governing mitophagy were initially described in cell lines overexpressing PARKIN in response to non-physiological stressors, such as exposure to mitochondrial uncouplers (Pickrell and Youle, 2015). In contrast, hypoxia is a physiological stress that induces mitophagy in cultured mouse embryonic fibroblasts (MEFs) (Zhang et al., 2008), as well as in murine liver (Sun et al., 2015), muscle (Zhang et al., 2008) and bone marrow (Zhang et al., 2016b). The mechanisms governing hypoxia-induced mitophagy remain to be fully interrogated, with conflicting evidence pointing towards activation of specific mitophagy receptors, such as BNIP3, FUNDC1 or BNIP3L (also known as NIX) (Palikaras et al., 2018; Zhang et al., 2016b), and being either dependent on (Kim et al., 2019; Zhang et al., 2016a) or independent (Lee et al., 2018) of the PINK1–PARKIN pathway. Thus, genetic epistasis analyses using in vivo models to dissect which mitophagy receptors or pathways are active in different tissues and under different stresses will further elucidate this process.
Here, we generate mitophagy biosensor zebrafish and define mitochondrial dynamics during vertebrate organ development and complex physiological stresses. Zebrafish develop completely externally and demonstrate a high degree of conservation with regards to mammalian tissue organization, organ development and molecular signaling (Cox and Goessling, 2015). This vertebrate model system offers the unique ability to illuminate and measure in vivo mitophagy trafficking and subcellular dynamics. In this study, we discover widespread basal mitophagy in many organs during development. Furthermore, using time-lapse imaging, we illuminate mitophagy dynamics in vivo, directly observing the trafficking of mitochondrial fragments to autolysosomes as it occurs in situ in fasted muscle. Using volumetric imaging, we then demonstrate that physiological hypoxia or chemical or genetic activation of the hypoxia inducible factor (Hif) pathway induces mitophagy in skeletal muscle. Importantly, using precise genetic loss-of-function analyses, we demonstrate that bnip3 is singularly necessary for Hif-induced mitophagy in muscle independently of other reported hypoxia-associated mitophagy receptors and independently of the pink1-prkn (Parkin) pathway. In sum, we established fundamental in vivo dynamics of fasting-induced mitophagy and elucidated the genetic and molecular mechanisms governing hypoxia-induced mitophagy.
RESULTS
Biosensor zebrafish lines report bona fide mitophagy during development
Recently, mitochondria-targeted, pH-sensitive fluorescent protein constructs – mito-mKeima (mito-Keima) (Katayama et al., 2011) and mito-EGFP–mCherry (mito-GR, for green and red) (Rojansky et al., 2016) – have been developed as sensitive readouts of mitophagy and incorporated into cell culture models and model organisms (Lee et al., 2018; McWilliams et al., 2016; Sun et al., 2015; Ordureau et al., 2020). Keima is a fluorescent, red light-emitting protein with a pH-sensitive excitation spectrum, enabling differentiation between Keima in a slightly basic pH environment, such as the healthy mitochondrial matrix (Porcelli et al., 2005), or an acidic environment, such as an autolysosome (Katayama et al., 2011). Whereas EGFP is sensitive to acid quenching and lysosomal degradation, mCherry and Keima are resistant to lysosomal degradation, allowing for signal accumulation, which is an indicator of the extent of mitophagic flux (Katayama et al., 2008).
Zebrafish lines Tg(ubi:mito-Keima) and Tg(ubi:mito-GR) were generated to express mito-Keima or mito-GR under the control of the ubiquitous ubiquitin promoter (Kwan et al., 2007; Mosimann et al., 2011). Importantly, these reporters are targeted to the mitochondrial matrix because reporters targeted to other mitochondrial locations can lead to false-positive signals (Katayama et al., 2020). Healthy mitochondria maintain a pH 7.8 matrix (Porcelli et al., 2005) and can be distinguished by either a high Keima440:Keima561 ratio (Keima signal when excited by a 440 nm laser divided by Keima signal when excited by a 561 nm laser) or presence of both EGFP and mCherry signal. In contrast, mitochondria undergoing degradation exist in acidic autolysosomes and are distinguished by a high Keima561:Keima440 ratio for mito-Keima or presence of mCherry signal alone for mito-GR.
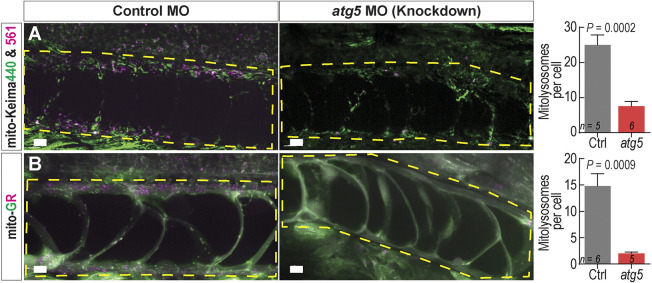
Ubi:mito-Keima and ubi:mito-GR transgenic zebrafish demonstrated no physiological abnormalities to suggest fluorophore toxicity and were generally analyzed as larvae from 1 to 8 days post fertilization (dpf). To test whether the ubi:mito-Keima construct faithfully reports mitophagy in vivo, we exposed ubi:mito-Keima larvae to MitoTracker Green dye, which labels all mitochondria regardless of polarization state, and performed confocal microscopy. Using 3D image analysis software, we generated Keima440high isosurface objects with high signal in the Keima440, and we generated Keima561high objects based on a ratiometric image produced by dividing the Keima561 image by the Keima440 image as described in the Materials and Methods. We performed object-based colocalization and found that Keima440high objects colocalized with MitoTracker Green signal in skin cells, whereas Keima561high objects did not. This indicates that Keima440high objects are mitochondria and that the Keima561high areas are representative of degraded mitochondria (Fig. S1A). In skeletal muscle cells, LysoTracker dye, which accumulates in lysosomes, colocalized with Keima561high puncta, but not with Keima440high puncta (Fig. S1B). Thus, areas with a high Keima561:Keima440 signal ratio represent mitolysosomes. Furthermore, we crossed ubi:mito-Keima fish with Tg(CMV:EGFP-map1lc3b)zf155 (EGFP–Lc3) fish, which have EGFP-labeled autophagosomes and autolysosomes due to Lc3 recruitment to these organelles during autophagy (He et al., 2009). In skeletal muscle, Keima561high fluorescence colocalized with the EGFP–Lc3 signal, indicating that, to induce degradation, mitochondrial fragments are shuttled to low-pH Lc3+ autolysosomes (Fig. S1C). Similarly, we incubated ubi:mito-GR embryos with LysoTracker deep red and observed colocalization with mCherry+;GFP− puncta (Fig. S1D). To ensure macroautophagy was necessary for the reporter signal, larvae were subjected to morpholino (MO)-mediated knockdown of the essential autophagy gene atg5. This induced cardiac and central nervous system defects in both ubi:mitoKeima and ubi:mito-GR embryos as previously described (Lee et al., 2016, 2014). However, notochords developed similarly at 48 hpf and displayed greatly reduced Keima561high (Fig. 1A) and mCherry+;EGFP− signals (Fig. 1B). Thus, both biosensor lines faithfully differentiate between healthy mitochondria and those undergoing mitophagy.
Fig. 1.
mito-Kiema and mito-GR zebrafish report bona fide mitophagy. (A) Maximum intensity projection (MIP) confocal images of 48 hpf ubi:mitoKeima and ubi:mito-GR larvae injected with control or atg5-targeting morpholinos. Yellow dashed boxes highlight the notochord. Control MO-injected larvae have prominent levels of mitophagy (indicated by magenta puncta), whereas atg5 MO-injected larvae have reduced mitophagy in the notochord. The graphs depict quantification of the number of Keima561high or mCherryhigh puncta per notochord cell. Data shown are mean±s.e.m. P-values were calculated with an unpaired, two-tailed Student's t-test. n, number of individual larvae. Scale bars: 10 µm.
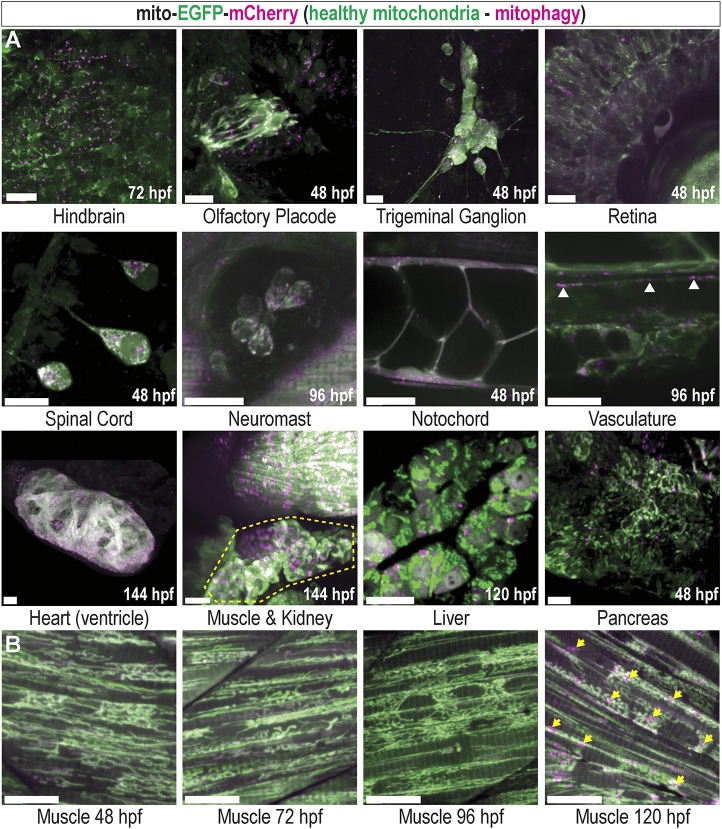
While basal mitophagy has been observed in many highly metabolic adult tissues (McWilliams et al., 2016), much less is known about mitophagy during vertebrate development. We utilized the mito-GR line to delineate basal levels of mitophagy during organogenesis and observed basal mitophagy in several larval tissues, indicated by dense mCherry+;EGFP− signal. Neural tissues, including the hindbrain, olfactory placode, trigeminal ganglion, retina, spinal cord and neuromasts, had high levels of basal mitophagy. The developing notochord, vasculature, heart (ventricle), kidney and liver all displayed high levels of basal mitophagy, and the pancreas displayed moderate levels (Fig. 2A). Prior to 4 dpf, larval skeletal muscle exhibited minimal mitophagy. However, after 5 dpf, larvae exhibited a dramatic increase in mitophagy (Fig. 2B, arrows). The timing of the onset of skeletal muscle mitophagy coincided with the stage during which zebrafish larvae must transition from subsisting on yolk sac-deposited nutrients to external food sources absorbed through their gut (Wilson, 2012) and suggests this observation may be caused by nutrient deprivation. These results reveal widespread basal mitophagy throughout development and identify zebrafish skeletal muscle as uniquely suited to investigate stress-induced mitophagy in vivo.
Fig. 2.
Basal mitophagy is widespread in developing zebrafish tissues. (A) Maximum intensity projection (MIP) confocal images of mito-GR zebrafish larvae at the indicated developmental time points, displaying various developing organs and tissues. Green indicates EGFPhigh mitochondria, which are healthy, and magenta indicates mCherryhigh puncta, representing mitolysosomes (mitophagy). White arrowheads show mitophagy occurring in the vasculature. Yellow outline highlights the developing kidney. (B) Developmental time series of mito-GR skeletal muscle in the tail. Mitophagy is low at 48, 72 and 96 hpf, but high at 120 hpf (arrows). All images are of live ubi:mito-GR larvae except the liver image, which is from a hepatocyte-specific fabp10a:mito-GR larva, and the heart image, which is a fixed whole-mount ubi:mito-GR larva. Scale bars: 15 µm.
Fasting induces quantifiable mitophagy and reveals mitolysosome dynamics
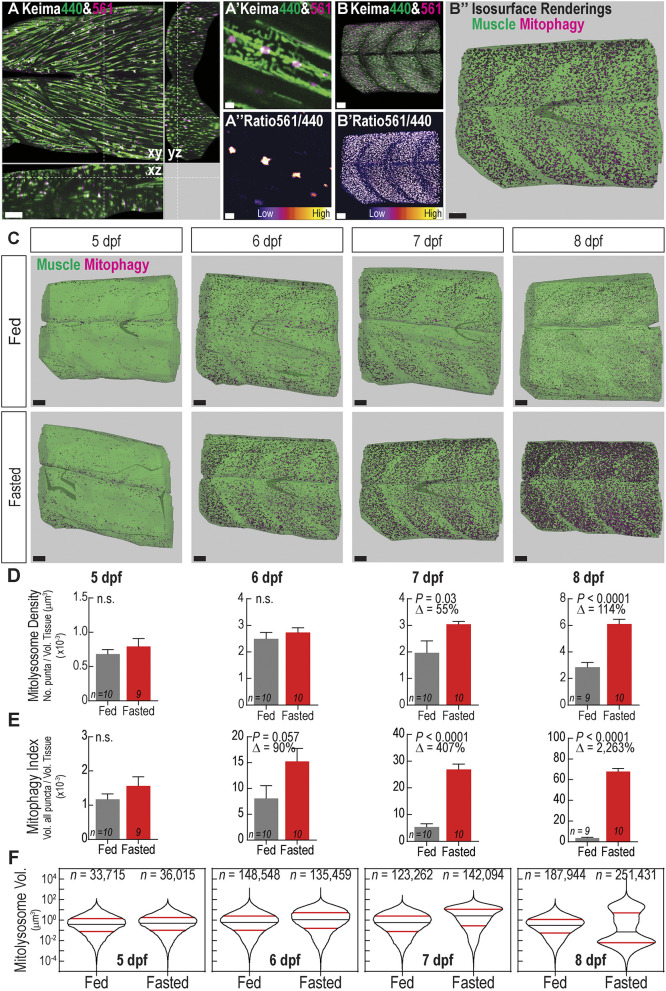
Skeletal muscle is a mitochondria-rich, highly metabolic tissue, which responds to nutrient deprivation by inducing autophagy (Mammucari et al., 2007); however, the regulation and dynamics of muscle mitophagy in vivo are not fully investigated. To precisely quantify mitophagy levels, we developed volumetric and ratiometric image acquisition and analysis methods to rapidly quantify the number and size of mitolysosomes within 3D tissue volumes. Spinning disc confocal fluorescence imaging enabled rapid generation of 3D images of zebrafish muscle. The images span more than six myotomes more than 150 µm deep (Fig. 3A) with resolution capable of distinguishing individual mitochondria, which are ∼0.35–1.5 µm in diameter (Jakobs, 2006) (Fig. 3A′; see methods for resolution calculations). The Keima561:(Keima440+background) signal ratio allows isolation of mitolysosomes (Fig. 3A″), the number and volume of which can be measured by applying an isosurface rendering algorithm. The addition of a small number approximately equal to background to the denominator allows for areas with low signal in both channels to be excluded. Normalizing either mitolysosome number or volume to the total tissue volume yields two mitophagy quantification metrics (Fig. 3B; Movie 1). This process is also applicable to mito-GR; however, at greater imaging depths the ubi:mito-Keima signal decayed more consistently and provided higher signal-to-noise. Thus, we applied this method to quantify fasting-induced mitophagy in ubi:mito-Keima zebrafish in two modes – mitolysosome density, defined as the number of mitolysosomes per tissue volume, and mitophagy index, defined as the proportion of total tissue volume occupied by mitolysosomes.
Fig. 3.
Fasting induces mitophagy in skeletal muscle and volumetric imaging reveals mitolysosome size dynamics. (A) Confocal microscopy cross-sectional image of 6 dpf ubi:mito-Keima zebrafish skeletal muscle demonstrates the typical imaging volume achieved for analyses. Scale bar: 100 µm. (A′) Magnified view of image in A demonstrating mitochondrial resolution achieved for all images and (A″) ratiometric images used to segment mitolysosomes. Scale bars: 3 µm. (B–B″) Representation of the image analysis process. (B) 3D reconstruction of the large volume image showing Keima440 and Keima561 signal throughout greater than six myotomes. (B′) Ratiometric image isolates Keima561high mitolysosomes. (B″) Isosurface renderings of muscle tissue volume (green) and the mitolysosomes (magenta) allow for volumetric analysis of mitophagy. Scale bars: 30 µm. (C) Isosurface renderings of muscle volume and mitolysosomes in 5–8 dpf zebrafish either fed or fasted. Green is the surface rendering of the muscle tissue, and magenta are surface renderings for individual mitolysosomes. Scale bars: 30 µm. (D) Quantification of mitolysosome density defined as the number of mitolysosomes divided by the total tissue volume. (E) Quantification of the mitophagy index defined as the sum of the volumes of all mitolysosomes divided by the total tissue volume. Data in D,E are shown are mean±s.e.m. P-values were calculated with an unpaired, two-tailed Student's t-test. n.s., not significant (P>0.05). Δ, percentage change of experimental cohort to control cohort. n indicates number of larvae in each cohort. (F) Violin plots demonstrating volume (µm3) distribution of individual mitolysosomes from all fed or fasted zebrafish in each cohort. Significant changes in the size distributions are evident at every time point (see Fig. S2). 50th percentile, black line; 25th and 75th percentiles, red lines. n, number of individual mitolysosomes combined from all larvae.
At 5 dpf, quantifiable mitophagy was observed in both fed and fasted zebrafish muscle, but there was not a significant difference in the extent as measured through mitolysosome density (Fig. 3D) or the mitophagy index (Fig. 3E). However, the mitophagy index was a more sensitive readout for mitophagy. Mitolysosome density was not significantly different between fed and fasted larvae until after 7 dpf, whereas the volumetric mitophagy index revealed increases by 6 dpf. Thus, mitophagy in skeletal muscle occurs after yolk depletion, but is abrogated by feeding.
To understand mitolysosome size dynamics during fasting, the volume distributions of individual mitolysosomes were assessed. In fasted skeletal muscle, the early response from 5–7 dpf was an increase in the volume distribution of mitolysosomes. Both fed and fasted fish had a monomodal mitolysosome volume distribution, with the fasted fish displaying an increase in 25th, 50th and 75th percentile mitolysosome volume (Fig. 3F; Fig. S2). However, by 8 dpf, when fasting entered a more advanced stage, mitolysosome size distribution became bimodal, with an increase in both large and small mitolysosomes (Fig. 3F). Surprisingly, this result is unique to mitolysosome dynamics. Although the size distribution of all LysoTracker+ lysosomes showed increased size and number in fasted larvae over time (Fig. S3A,B), all size distributions were monomodal and very small lysosomes were present at all stages (Fig. S3C). Together, these results reveal that the early fasting-induced mitophagy response primarily utilizes and expands existing autolysosomes within the cell as evident by an increase in mitophagy index and shift in the size distribution prior to significant changes in the number of mitolysosomes (mitolysosome density). During advanced fasting stages, however, the existing mitolysosomes become large, and new, small mitolysosomes appear. Taken together, these data validate the volumetric mitophagy quantification method and reveal novel mitolysosome size changes as a response to prolonged nutrient deprivation.
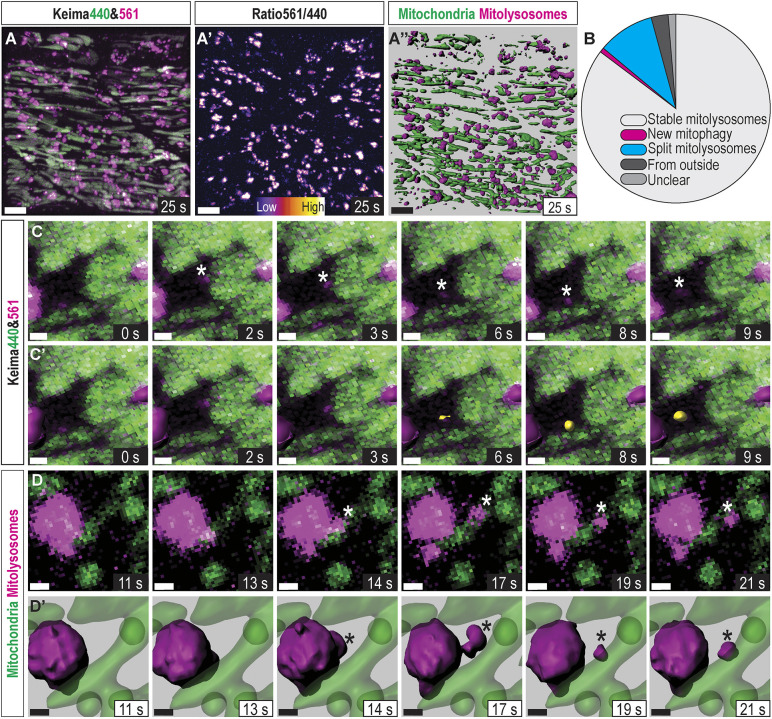
To determine the origin of the newly appearing small mitolysosomes found in advanced fasting stages, we performed rapid, continuous time-lapse analysis on a 7 dpf fasted mito-Keima larva (Movie 2). Application of an 3D-object tracking algorithm enabled identification of de novo mitophagy and mitolysosome fission events (Fig. 4A). Newly appearing Keima561high puncta were identified, and each event was binned into one of several categories. If the Keima561high punctum stemmed from a larger Keima440high mitochondrion, it was considered ‘new mitophagy’. If the punctum broke away from a different, larger Keima561high mitolysosome, it was considered a ‘split mitolysosome’. If the punctum migrated into the imaging volume near the edges, it was considered ‘from outside’ (Fig. 4B). Out of 381 Keima561high objects tracked for 28s, 325 were stable from the beginning to the end of the imaging period, 11 migrated from outside, only three were new mitophagy (Fig. 4C, asterisk and yellow surface object), and 37 puncta split from larger mitolysosomes (Fig. 4D, asterisk). Thus, de novo mitophagy rapidly occurs during this stage of fasting and accounts for some of the new, small mitolysosomes, but mitolysosome fission is the predominant source of the small mitolysosomes, which appear during advanced fasting.
Fig. 4.
In vivo mitophagy dynamics: mitophagy kinetics and mitolysosome fission. (A) Confocal 3D projection of the imaging volume in Movie 2. Images of skeletal muscle in a 7 dpf larva during advanced nutrient deprivation. Images were continuously acquired, finishing each volume approximately once per second and tracking skeletal muscle mitochondria in an approximately two-cell-deep volume for 28 s. (A′) Ratiometric image isolating mitolysosomes. (A″) Isosurface rendering of mitochondria (green) and mitolysosomes (magenta) used to track stable mitochondria, ongoing mitophagy, and mitolysosome fission. Scale bars: 10 µm. (B) Classification of each Keima561high object revealing that ∼85% were stable, 1% were newly formed and 10% split from existing, larger mitolysosomes within the 28 s timeframe. The remaining mitolysosomes were pre-existing or translocated from outside the imaging volume. (C) Magnified timeseries of a representative ‘new mitophagy’ event marked with (*). (C′) Same as C with isosurface objects shown for stable mitolysosomes (magenta) and the new Keima561high object (yellow). Scale bars: 1 µm. (D) Maximum intensity projection (MIP) magnified time series representative of a splitting mitolysosome (*). (D′) Same as D with isosurface renderings. Scale bars: 1 µm.
Fasting-induced mitophagy occurs piece-by-piece
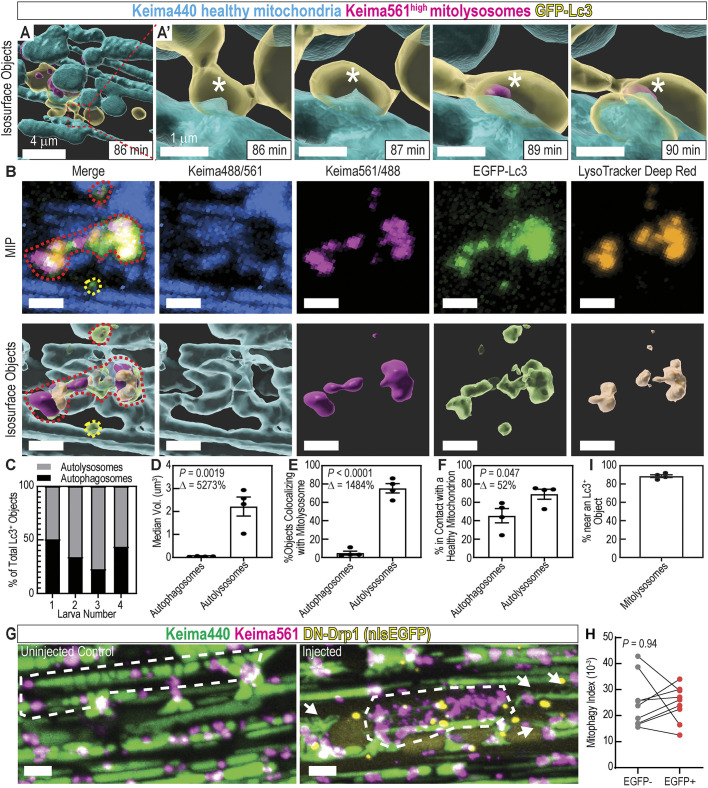
Because the mitophagy process has not been analyzed by time-lapse imaging in any live vertebrate, we next investigated how GFP–Lc3-labeled organelles interact with mitochondria in vivo. By performing time-lapse analysis in Tg(CMV:EGFP-map1lc3b)zf155;(ubi:mito-Keima) larvae, we could directly observe interactions between Lc3-labeled organelles (autophagosomes and autolysosomes) and mitochondrial fragments. Multiple mechanisms have been observed in cells cultured in vitro by which a mitochondrion is sequestered by the autophagy machinery. In the ‘wholesale’ mode, after a mitochondrial filament reaches a damage threshold, mitochondrial membrane depolarization and extensive fragmentation occurs, which is followed by recruitment of the autophagy machinery to degrade all of the fragments. In the ‘piecemeal’ mode, the damaged part of a mitochondrial filament is sequestered, and the autophagy machinery is localized to that point, which undergoes fission and degradation, leaving the filament largely intact (Youle, 2019; Yamashita et al., 2016; Burman et al., 2017). It is unknown whether in vivo mitophagy occurs via a wholesale or piecemeal mode. We performed time-lapse analysis in fasted muscle with ∼1 min/frame time resolution, and used isosurface renderings to identify and tracked the different organelle structures in three dimensions. Interestingly, we never observed massive mitochondrial fragmentation followed by mitophagy, indicating the wholesale mode of mitophagy is not active in this context. Mitophagy events (transition from Keima440high to Keima561high) occurred at discrete contact points between Lc3 and filamentous mitochondria (Fig. 5A,A′, asterisks; Movie 3), indicative of piecemeal mitophagy.
Fig. 5.
Mitophagy trafficking dynamics: mitophagy occurs piecemeal. (A) Isosurface object projections of confocal time-lapse imaging of fasted skeletal muscle in 5 dpf ubi:mito-Keima larva. Scale bar: 4 µm. (A′) Magnified view of region indicated in A; mitophagy occurs piecemeal. An EGFP–Lc3+ surface is interacting with a Keima440high filamentous mitochondrion (86 min; *). Then, a small portion of the longer filamentous mitochondrion becomes Keima561high, indicating pH change and mitophagy (89 min; *). Scale bars: 1 µm. (B) Maximum intensity projection (MIP) and isosurface object representations displaying healthy mitochondria (Keima488/561), mitolysosomes (Keima561/488), EGFP–Lc3+ objects, and LysoTracker Deep Red+ objects. Red dashed lines outline highlights an autolysosome containing degraded mitochondria. Yellow dashed lines outline highlights an autophagosome containing no mitochondria. Scale bars: 2 µm. (C) Classification of EGFP–Lc3+ objects as autophagosomes or autolysosomes. Those in contact with LysoTracker+ objects are autolysosomes, and the remaining are autophagosomes. (D) Volume quantification of autophagosomes and autolysosomes. (E) Colocalization analysis between mitolysosomes (Keima561high objects) and autophagosomes or autolysosomes. (F) Colocalization analysis between healthy mitochondria and autophagosomes or autolysosomes. In D–F and I data are shown as mean± s.e.m. P-values were calculated with an unpaired, two-tailed Student's t-test. Δ, percentage change between autophagosomes and autolysosomes. n=4 individual 6 dpf larvae. (G,H) MIP of 7 dpf ubi:mito-Keima larvae injected with an inducible DN-Drp1 construct. Compare the piecemeal mitophagy pattern in the control to the wholesale mitophagy pattern in the injected (white dotted outlines). Mitochondrial morphology is also altered in DN-Drp1-injected larvae (arrows). Graph (H) depicts mitophagy index calculations from EGFP+;DN-Drp1 cells and paired, neighboring EGFP−;WT cells. Each dot is an individual cell for a total of 9 pairs of cells from three different animals. P, paired, two-tailed Student's t-test. Scale bars: 4 µm. (I) Colocalization analysis between mitolysosomes and Lc3+ objects.
Intriguingly, the Keima561high mitolysosomes colocalized extensively with EGFP–Lc3 signal, indicating that EGFP–Lc3 labels both autophagosomes and autolysosomes in this system (Fig. S1C). To differentiate acidified autolysosomes from autophagosomes, Tg(CMV:EGFP-map1lc3b)zf155;(ubi:mito-Keima) larvae were stained with LysoTracker Deep Red, and object-based colocalization image analysis was performed. This analysis required activation of Keima with a 488 nm laser, causing additional overlap between the neutral/basic and acidic Keima signals. To accommodate for this, we defined healthy mitochondria by the Keima488/(Keima561+1) signal ratio, which isolated areas of high Keima488 signal and avoided errors caused by dividing by zero. Mitolysosomes were defined by the Keima561/(Keima440+background) signal ratio as in previous experiments, which isolated voxels of high Keima561 signal by rendering voxels with either Keima440high signal or low signal in both channels as low. We generated surface objects for healthy mitochondria, mitolysosomes, EGFP–Lc3+ objects and LysoTracker+ objects (Fig. 5B). First, object-based colocalization was employed to determine which EGFP–Lc3+ objects were autolysosomes – in contact with a LysoTracker+ object – or autophagosomes – not in contact with a LysoTracker+ object (Fig. 5C). Autolysosomes were substantially larger in volume than autophagosomes (Fig. 5D). Remarkably, nearly all autolysosomes contained (Keima561/488)high objects, classifying them as automitolysosomes, whereas nearly all autophagosomes did not (Fig. 5E). Approximately 45% of autophagosomes and ∼68% of autolysosomes were in direct contact with healthy mitochondria, supporting our observation that piecemeal mitophagy occurred at discrete contact points between EGFP–Lc3+ organelles and larger intact mitochondrial filaments.
Mitochondrial protein aggregates can be cleared via a piecemeal mitophagy mechanism in cultured cells, and knockout of Drp1 (also known as DNM1L) impairs mitochondrial fission, leading to wholesale mitophagy (Burman et al., 2017). We hypothesized that inhibition of Drp1 could similarly induce a wholesale mitophagy response in vivo. Thus, we introduced a heat shock-inducible, dominant-negative, human Drp1(K38A)-2A-nlsEGFP (DN-Drp1) construct (Smirnova et al., 1998) to ubi:mito-Keima larvae and induced expression during fasting in the resulting mosaic animals. At 7 dpf, cells expressing DN-Drp1 (identified by nuclear EGFP expression) had altered mitochondrial morphology with increased connections between filaments, indicating that expression of DN-Drp1 functionally reduced mitochondrial fission (Fig. 5G, arrows). Interestingly, mitophagy index levels were not altered in these cells compared to neighboring wild-type (WT) EGFP− cells, but in some EGFP+ cells, Keima561high signal occurred in a pattern indicative of wholesale mitophagy of large mitochondrial filaments (Fig. 5G,H). Together, these results indicate fasting-induced mitophagy in skeletal muscle occurs piecemeal, and that Drp1 inhibition does not prevent mitophagy but can induce wholesale recycling of mitochondrial filaments.
Mitochondrial substructures can also be removed piecemeal by an Lc3-independent mechanism utilizing mitochondria-derived vesicles (MDVs) (Sugiura et al., 2014; Cadete et al., 2016). However, 88.5% of mitolysosomes were measured to be within 0.5 µm of an Lc3+ object (Fig. 5I). Thus, in fasted muscle, the MDV pathway is not likely to play a major role in mitochondrial recycling. In sum, mitophagy in fasted skeletal muscle occurs primarily by a piecemeal mechanism utilizing Lc3+ organelles.
Hif activation induces mitophagy
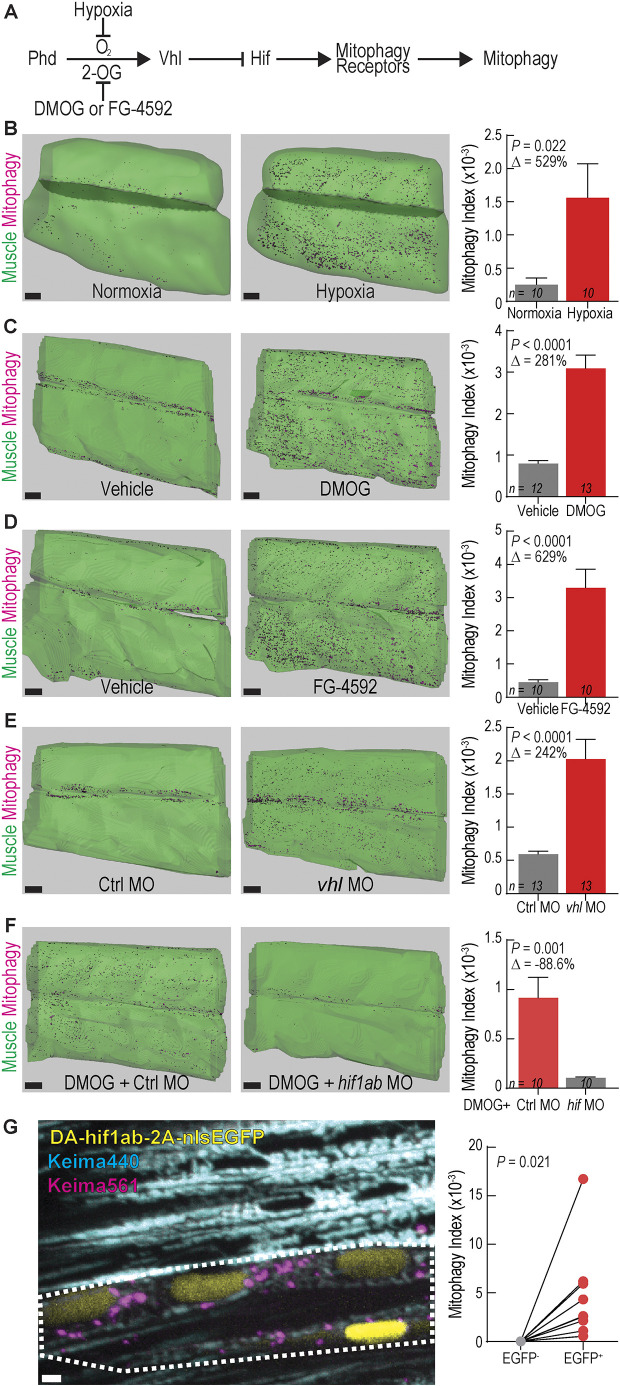
Tissue hypoxia (ischemia) occurs in muscle during exercise and other physiological and pathophysiological conditions (Leermakers and Gosker, 2016; Lee et al., 2019; Ferdinand and Roffe, 2016). This stress leads to production of excess reactive oxygen species, mitochondrial damage and mitophagy (Kaelin and McKnight, 2013). We used the zebrafish mitophagy reporter system to dissect the effects of hypoxia and activation of the Hif pathway on muscle mitophagy (Fig. 6A). To avoid fasting-induced mitophagy as a confounding variable, we examined larvae younger than 5 dpf. To induce physiological hypoxia, zebrafish larvae were raised in hypoxic fish water within a hypoxia chamber (4.7% O2) from 77–96 hpf, causing death in 11 of 35 larvae, indicative of the substantial hypoxic stress induced by this approach. Surviving larvae had a significant increase in mitophagy in skeletal muscle, although variation between animals was high (Fig. 6B). Next, we tested whether chemical activators of Hif signaling induced mitophagy in skeletal muscle. The prolyl hydrodylase (PHD) inhibitor dimethyloxalylglycine (DMOG) has been validated to induce Hif activation in similarly aged zebrafish larvae (Elks et al., 2013; Ivan et al., 2001; Iliopoulos et al., 1996). Fish exposed to DMOG from 3–4 dpf exhibited robust mitophagy compared to controls (Fig. 6C). This result was confirmed by exposing zebrafish to FG-4592 (Roxadustat), a chemically dissimilar PHD inhibitor evaluated for FDA approval, which caused a similar increase in mitophagy (Fig. 6D). Genetic knockdown of vhl using a previously validated MO (Harris et al., 2013) also induced mitophagy at 3 dpf (Fig. 6E). Knockdown of hif1ab using a previously validated MO (Harris et al., 2013; Lim et al., 2017) abrogated DMOG-induced mitophagy (Fig. 6F), indicating that DMOG treatment induces mitophagy via Hif signaling activation and not via off-target effects. To test whether Hif activation alone was sufficient to induce mitophagy in skeletal muscle in a cell autonomous manner, dominant-active Hif (DA-hif1ab) (Elks et al., 2011) and nuclear-localized EGFP were expressed under the control of the muscle-specific unc45b promoter (Berger and Currie, 2013). This resulted in mosaic expression of nuclear EGFP, marking the cells expressing DA-hif1ab to be compared to neighboring EGFP− WT cells within the same animal. EGFP+ cells had high levels of mitophagy, whereas surrounding EGFP− cells had little-to-no mitophagy (Fig. 6G). Together, these results confirm that hypoxia and Hif activation elicit mitophagy in a cell-autonomous fashion.
Fig. 6.
Activating Hif signaling induces mitophagy in skeletal muscle. (A) Schematic of Hif signaling pathway. 2-OG, 2-oxoglutarate. All subsequent conditions in which Hif signaling is activated are coded red and those with Hif inactive are gray. (B–D) Isosurface renderings and mitophagy index quantification in skeletal muscle of 4 dpf ubi:mito-Keima zebrafish exposed to (B) 4.7% oxygen (hypoxia), (C) 60 µM DMOG or (D) 10 µM FG-4592 from 3-4 dpf. Data shown are mean±s.e.m. P-values were calculated with an unpaired, two-tailed Student's t-test. Δ, percentage change of experimental cohort to control cohort. n indicates number of larvae in each cohort. Scale bars: 30 µm. (E) Isosurface renderings and quantification of mitophagy in skeletal muscle of 3 dpf zebrafish injected with control MO (Ctrl) or vhl antisense MO. Scale bars: 30 µm. Data shown are mean±s.e.m. P-values were calculated with an unpaired, two-tailed Student's t-test. Δ, percentage change of experimental cohort to control cohort. n indicates number of larvae in each cohort. (F) Isosurface renderings and quantification of the mitophagy index in skeletal muscle of 3 dpf zebrafish injected with control MO (Ctrl) or hif1ab MO and treated with 100 µm DMOG from 2–3 dpf. Scale bars: 30 µm. Data shown are mean±s.e.m. P-values were calculated with an unpaired, two-tailed Student's t-test. Δ, percentage change of control cohort to hif1ab MO cohort. n indicates number of larvae in each cohort. (G) Confocal microscopy images of 4 dpf ubi:mito-Keima expressing dominant active (DA) hif1ab in a mosaic fashion. Keima440 is cyan. Keima561 is magenta. Nuclear EGFP is yellow and indicates the cells that express DA-hif1ab. White dotted line outlines two adjacent myotubes expressing DA-hif1ab and undergoing mitophagy. Graph on right shows values for individual cells for a total of 10 pairs of cells from three different animals. P-value was calculated with an paired, two-tailed Student's t-test. Scale bar: 2 µm.
Disruption of bnip3 alone diminishes Hif-induced mitophagy
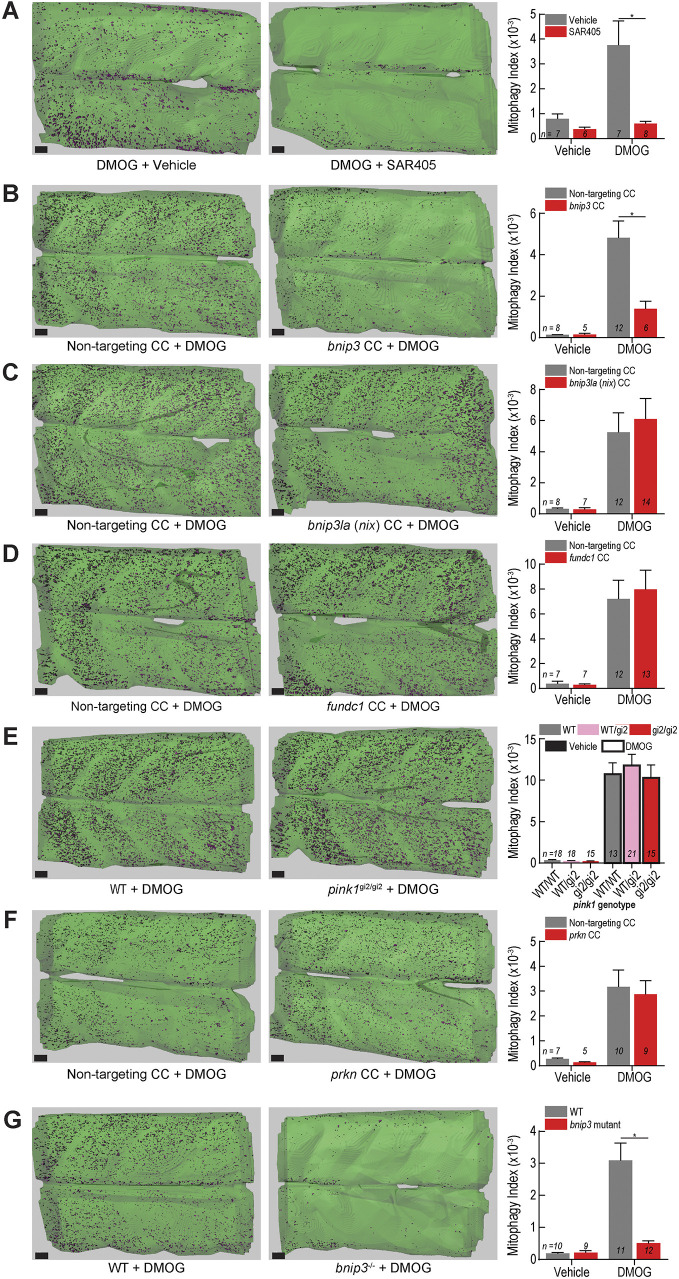
Next, we used the volumetric in vivo mitophagy quantification method and high-efficiency genome editing to dissect the molecular mechanisms required for Hif-induced mitophagy in vertebrate skeletal muscle. As a positive control, we tested whether the mitophagy signal resulting from DMOG exposure was dependent on macroautophagy by assessing dependence on Vps34 function, which is necessary for autophagosome formation and vital for macroautophagy (Ronan et al., 2014). A cohort of DMOG-exposed larvae (both ubi:mitoKeima and ubi:mito-GR in independent experiments) was treated with the Vps34 inhibitor SAR405 (Ronan et al., 2014). Exposure to SAR405 abrogated DMOG-induced mitophagy (Fig. 7A; Fig. S4), indicating that macroautophagy is vital for Hif-induced mitophagy.
Fig. 7.
bnip3 regulates Hif-induced mitophagy in muscle independently of bnip3la, fundc1, pink1 and prkn. (A) Mitophagy index quantification of larvae treated with 100 µM DMOG or vehicle (DMSO) and 20 µM SAR405 or vehicle (DMSO). Representative isosurface rendering images show the effect of SAR405 treatment on DMOG-induced mitophagy. (B–D,F) Mitophagy index quantification and representative isosurface rendering images of DMOG-induced mitophagy in larvae injected with CRISPR cutters (CC) targeting (B) bnip3, (C) bnip3la (nix), (D) fundc1, or (F) prkn. (E) Mitophagy index quantification and representative isosurface rendering images of DMOG-induced mitophagy in larvae carrying a null pink1 allele (gi2) and siblings. (G) Mitophagy index quantification and representative isosurface rendering images of DMOG-induced mitophagy in larvae carrying a 5-bp deletion mutation in the bnip3 gene. Data shown are mean±s.e.m. *P<0.01 (two-way ANOVA with Holm–Sidak's multiple comparisons test). Each comparison between vehicle- and DMOG-treated samples was also significant (P<0.05) except in (A) vehicle controls to DMOG+SAR405, (B) vehicle controls to DMOG+bnip3 CC, and (G) vehicle controls to bnip3−/−+DMOG. Scale bars: 20 µm. See Figs S4, S6 and S7 for replicates and full statistical analyses. n indicates number of larvae in each cohort.
HIF-induced mitophagy is thought to rely on the function of specific mitophagy receptors BNIP3, BNIP3L and FUNDC1, which contain LC3-interacting regions (LIRs) (Birgisdottir et al., 2013; Yoo and Jung, 2018; Hanna et al., 2012). Furthermore, the PINK1–PARKIN pathway governs mitochondrial depolarization-induced mitophagy (Pickles et al., 2018; Harper et al., 2018), but its role in hypoxia-induced mitophagy remains controversial with evidence in other model organisms both for (Kim et al., 2019; Zhang et al., 2016a) and against (Lee et al., 2018) a role. To test the requirement of these factors for Hif-induced mitophagy, we performed a targeted genetic epistasis screen. bnip3, bnip3la (nix) and fundc1 were disrupted in F0 larvae using high efficiency CRISPR methodology, an established and rapid method to assess loss of gene function (Fig. S5) (Burger et al., 2016; DiNapoli et al., 2020). Surprisingly, larvae injected with bnip3-targeted CRISPR cutter (CC) reagents reliably exhibited dramatic reduction in DMOG-induced mitophagy, indicating that singular disruption of the bnip3 locus diminishes Hif-induced mitophagy in vertebrate skeletal muscle (Fig. 7B; Fig. S6). Disruption of other receptors implicated in hypoxia-induced mitophagy, specifically bnip3la (Fig. 7C) and fundc1 (Fig. 7D), had no effect on DMOG-induced mitophagy. These results indicate that bnip3, a single mitophagy receptor, is necessary for the Hif-induced mitophagy response in vertebrate skeletal muscle.
BNIP3 has been shown to interact directly with PINK1, promoting mitophagy via the PINK1–PARKIN pathway (Zhang et al., 2016a), and a study in flies showed that Pink1 was necessary for hypoxia-induced mitophagy (Kim et al., 2019). However, there were no significant differences in Hif-induced mitophagy in skeletal muscle between fish homozygous for an established pink1 null allele (gi2) (Zhang et al., 2017) and their WT or heterozygous siblings (Fig. 7E; Fig. S7A). Larvae injected with a validated pink1 MO (Priyadarshini et al., 2013) also showed no difference in DMOG-induced mitophagy compared to controls (Fig. S7B). Furthermore, disruption of prkn had no effect (Fig. 7F). We sought to confirm the F0 CRISPR results by generating a stable bnip3 mutant line, containing a 5-bp frame-shift deletion in exon 2 and an early stop codon (Fig. S7C,D). Larvae homozygous for the mutant bnip3 allele had drastically reduced mitophagy capacity compared to WT when challenged with DMOG (Fig. 7G; Fig. S7E). Interestingly, the bnip3-dependent mitophagy response was specific to Hif-induced mitophagy. Fasted fish homozygous for the bnip3 or pink1 mutant alleles displayed no difference in mitophagy index compared to WT animals (Fig. S7F,G). In sum, bnip3, a single Hif-responsive mitophagy receptor, mediates Hif-induced mitophagy in skeletal muscle, independently of the pink1-prkn pathway.
DISCUSSION
In this study, we reveal fundamental characteristics of in vivo mitophagy during normal organ development and complex physiological stresses using mitophagy biosensor zebrafish combined with time-lapse and quantitative imaging. Basal mitophagy occurred in many developing organs. In fasted skeletal muscle, mitolysosomes initially increased in size, followed by a period of concurrent mitophagy and mitolysosome fission. Time-lapse imaging revealed fasting-induced mitophagy to occur in vivo via a piecemeal mechanism. Hypoxia and Hif-activation induced mitophagy in skeletal muscle. Hif-induced mitophagy requires bnip3 function, but pink1, prkn, bnip3la and fundc1 are dispensable. In sum, these results reveal in vivo subcellular dynamics and molecular mechanisms regulating physiological-stress induced mitophagy.
Many organs exhibit mitophagy during development
Transgenic animals expressing mitophagy reporter constructs have begun to reveal basal mitophagy levels in vivo. Murine studies have demonstrated that adult tissues with high metabolic demands – brain, liver, kidney, spleen and skeletal muscle – have high basal mitophagy levels (McWilliams et al., 2016; Sun et al., 2015). Similarly, studies in flies show basal mitophagy in the central nervous system, epidermis and muscle (Lee et al., 2018). The developing mouse heart and kidney display high mitophagy (McWilliams et al., 2016), but more extensive developmental analyses have not been performed. Recently, nipsnap1 mutant zebrafish were found to have decreased basal mitophagy in skin and defects in dopamine neuron development (Abudu et al., 2019). Our study detects widespread mitophagy in embryonic and larval zebrafish during the development of many organs, including several that were previously unrecognized, such as the spinal cord, notochord, vasculature, pancreas and liver.
Mitophagy occurs via a piecemeal process in vivo
The in vivo dynamics of mitophagy have not been previously studied via live imaging. The canonical mitophagy sequence involves mitochondrial fission occurring prior to engulfment by preformed autophagosomes (Anding and Baehrecke, 2017). Conversely, in HeLa cells, mitophagy receptors localized to the point of damage on mitochondria can recruit the ULK1 complex, initiating autophagosome biogenesis locally (Vargas et al., 2019) and concurrently with mitochondrial division (Yamashita et al., 2016). In live zebrafish skeletal muscle, we found that many autophagosomes and most autolysosomes were in direct contact with intact mitochondrial filaments, indicating that much of autophagosome biogenesis may occur locally, and that much of the fasting-induced autophagy occurring is mitophagy. Time-lapse analysis indicated that mitophagy, as defined by a transition from Keima440high to Keima561high, occurred near these points in a piecemeal fashion. This also reveals that the process of autophagosome acidification in skeletal muscle is rapid during this advanced stage of fasting.
The piecemeal nature of mitophagy in skeletal muscle may be an adaptation necessary to allow for muscle mitochondria to exist as large, fused networks to more efficiently generate ATP and other metabolites (Romanello and Sandri, 2013; Friedman and Nunnari, 2014). Interestingly, there are studies linking mitochondrial fission to both increased and decreased mitophagy. In MEFs, downregulation of Drp1 (also known as Dnm1l), a GTPase vital for mitochondrial fission, occurs after nutrient starvation leading to elongated mitochondria, and mitochondrial fusion-deficient Opa1-knockout cells are primed for mitophagy (Rambold et al., 2011). Increased mitochondrial fission and mitophagy caused by Drp1 activation during mid-life increases lifespan in Drosophila (Rana et al., 2017). In C. elegans and mice, PUM2 is induced upon aging and inhibits mitochondrial fission factor (Mff), which, in turn, inhibits mitochondrial fission and mitophagy (D'Amico et al., 2019). However, in separate study in mice, skeletal muscle-specific deletion of Opa1, a protein necessary for inner mitochondrial membrane fusion, leads to fragmented mitochondria and mitophagy defects (Rodríguez-Nuevo et al., 2018). Indeed, another study demonstrated that loss of Drp1 caused wholesale turnover of mitochondria, leading to an increase in the rate of mitophagy and a decrease in the specificity of clearing the damaged mitochondrial compartments (Burman et al., 2017). In zebrafish muscle, we found that expression of DN-Drp1 led to a more tubular mitochondrial morphology. We found no immediate effect on mitophagy levels in fasted zebrafish muscle but did observe instances of wholesale mitochondrial turnover. It will be important to investigate under which circumstances Drp1-facilitated piecemeal mitophagy and Drp1-independent wholesale mitophagy occur, and how they are linked to physiological or pathophysiological outcomes. It will also be important to determine why mitophagy occurs at specific points along the mitochondrial filament in vivo. The topology and shape of mitochondria have been implicated as driving forces for starvation-induced mitophagy in osteosarcoma cells (Zhou et al., 2020). Differential segregation and phosphorylation of mitochondrial matrix proteins was also shown to determine mitophagy rates of different mitochondrial proteins (Abeliovich et al., 2013; Kolitsida et al., 2019). It is exciting to speculate that different mitochondrial proteins could segregate to specific subdomains and dictate the location of mitochondrial fission and mitophagy.
Different types of piecemeal selective autophagy have been discovered. A LIR-containing protein TEX264 was recently demonstrated to be necessary for piecemeal selective autophagy of the ER in HEK293 cells (Abeliovich et al., 2013), proving that receptor-mediated selective autophagy can also occur piecemeal. In HeLa cells, a homeostatic piecemeal mitophagy mechanism relies on p62 (SQSTM1) and LC3C (MAP1LC3C) but not PINK1–PARKIN to regulate oxidative phosphorylation (Le Guerroué et al., 2017). A recent study explored mitophagy kinetics using mild oxidative stress to induce more physiological levels of mitophagy in cultured rat neurons, suggesting that mild mitochondrial stresses may better model the in vivo process (Evans and Holzbaur, 2020). It is possible the wholesale mechanism only occurs in dying cells or those with defects in mitochondrial fission, or that it is primarily an in vitro event caused by unphysiologically high mitochondrial stress levels. It will be important for future work to analyze the mitophagy trafficking dynamics and mechanisms in vivo in other cell types and under different physiological stresses to uncover endogenous mitophagy kinetics and to determine whether every instance of mitophagy occurs via a piecemeal mechanism.
bnip3 is required for hypoxia-induced mitophagy
Many genes have been implicated in both ubiquitin-dependent and ubiquitin-independent mitophagy (Rodger et al., 2018; Anding and Baehrecke, 2017; Harper et al., 2018). These studies, however, were primarily performed in cell culture with limited confirmation of conserved mechanisms in vivo. BNIP3, BNIP3L and FUNDC1 have all been linked to hypoxia-induced mitophagy and HIF signaling (Yoo and Jung, 2018; Hanna et al., 2012). They contain domains to localize them to the mitochondrial outer membrane, and LIRs, which bind to LC3 and recruit autophagosomal membranes (Birgisdottir et al., 2013). BNIP3 mediates hypoxia-induced mitophagy in MEFs and murine lung tissue (Zhang et al., 2008). In murine skeletal muscle, FoxO signaling causes fasting-induced autophagy via BNIP3, although neither mitophagy nor hypoxia were analyzed (Mammucari et al., 2007). BNIP3 also facilitates mitophagy in murine cortical neurons in a stroke model (Shi et al., 2014) and in murine hearts and HL1 cardiac muscle cells after myocardial ischemia/reperfusion injury (Hamacher-Brady et al., 2007). BNIP3L is linked to hypoxia-induced mitophagy in ischemic brain injury and heart ischemia/reperfusion injury in mice (Yuan et al., 2017). FUNDC1 is thought to facilitate hypoxia-mediated mitophagy in HeLa cells (Liu et al., 2012; Chen et al., 2014), several murine tissues including platelets, liver, heart, and muscle (Zhang et al., 2016b), and also in cardiac ischemia/reperfusion injury (Zhou et al., 2017). In light of these studies, it might be expected that multiple, redundant mitophagy receptors facilitate the hypoxia response in skeletal muscle. In contrast, we found that mutation of the singular receptor bnip3 prevented Hif-induced mitophagy in skeletal muscle, whereas disruption of bnip3la or fundc1 had no effect. Our results add mechanistic insight into the tissue- and stress-dependent contexts for vertebrate mitophagy mechanisms.
There is conflicting evidence for how BNIP3 induces mitophagy. In MEFs, BNIP3 functions by facilitating the release of Beclin-1 from Bcl-2, allowing Beclin-1 to initiate autophagy independently of the PINK1–PARKIN pathway (Zhang et al., 2008). However, a separate study in MEFs and HEK293 cells showed BNIP3 interacted with and stabilized PINK1, which recruited PARKIN to the mitochondrial outer membrane and initiated PINK1–PARKIN-dependent mitophagy (Zhang et al., 2016a). Separately, Pink1 was shown to be necessary for hypoxia-induced mitophagy in flies (Kim et al., 2019). In zebrafish, however, we tested an established mutant pink1 allele, pink1 morphants and prkn disruption via CRISPR, and found that none of these interventions influenced or prevented Hif-induced mitophagy in zebrafish muscle. This supports the conclusion that in vertebrates, bnip3-induced mitophagy is independent of the Pink1–Parkin ubiquitination system. In flies, Pink1 mutation causes muscle degeneration (Clark et al., 2006; Yang et al., 2006; Park et al., 2006), and BNIP3 overexpression can rescue the phenotype (Zhang et al., 2016a). In humans (Gasser, 2009), monkeys (Yang et al., 2019), mice (McWilliams et al., 2018) and zebrafish (Zhang et al., 2017), there is no evidence of direct muscle degeneration due to PINK1 mutation, indicating that there are important differences between vertebrate and invertebrate mitophagy mechanisms. Because there are few yeast homologs to vertebrate mitophagy receptors (Mao et al., 2011), and due to the observed differences in mitophagy mechanisms between vertebrate and invertebrate models, analysis of mitophagy reporter zebrafish provides a unique avenue toward deeper understanding of mitochondrial quality control dynamics and mechanisms. In summary, our study revealed widespread mitophagy during development, fundamental piecemeal mitophagy dynamics during fasting and that hypoxia-induced mitophagy in muscle is dependent on bnip3.
MATERIALS AND METHODS
Animal studies
Zebrafish (Danio rerio; WT strains, Tübigen/TU and Tüpfel long fin/TL) were maintained according to institutional animal care and use committee (IACUC-BIDMC #506-2015) protocols. Adult fish and larva were raised at 28.5°C. All studies were performed in larval zebrafish before sex can be determined, and therefore sex was not taken into consideration. All embryos and larvae were raised and experimentally treated in E3 salt solution. For experiments involving larval feeding, embryos were provided live paramecia and larval AP-100 fish food (Zeigler) ad libitum.
Molecular cloning and transgenesis
Transgenic lines Tg(ubi:mito-Keima,cryaa:Cerulean), Tg(ubi:mito-GR) and Tg(fabp10a:mito-GR) were generated using tol2 transgenesis (Mosimann and Zon, 2011; Mosimann et al., 2011; Her et al., 2003). Plasmids generated or used are listed in Table S1. Middle entry vector pENTR:mito-Keima was generated via PCR using pHAGE mt-Keima IRES Puro as template and pENTR/D-TOPO kit (Invitrogen) following the manufacturer's instructions. A Gateway reaction using LR Clonase II Plus (Invitrogen) was used to generate the pTol2-ubi:mito-Keima,cryaa:Cerulean vector following manufacturer's instructions. Middle entry vector pME-mito-GR was generated using BP Gateway cloning using pCLBW cox8 EGFP mCherry (Rojansky et al., 2016) and pDONR221 (Invitrogen) using Gateway BP Clonase II Enzyme (Invitrogen) mix following manufacturer's instructions. LR Gateway cloning was used to generate pTol2-ubi:mito-GR and pTol2-2.8fabp10a:mito-GR. Middle entry vector pENTR-DA-hif1ab(noStop) was generated using PCR to amplify DA-hif1ab minus the Stop codon from a plasmid containing DA-hif1ab (Elks et al., 2011), which was a gift from Phil Elks (Department of Infection, Immunity and Cardiovascular Disease, University of Sheffield, UK). Middle entry vector pENTR-DN-Drp1(noStop) was generated similarly using a plasmid containing Drp1(K38A) (Smirnova et al., 1998). LR Gateway reactions generated the pTol2-503unc45b:DA-hif1ab-2A-nlsEGFP,cryaa:mCherry, pTol2-hsp70l:DN-Drp1-2A-nlsEGFP,cryaa:mCherry, and pTol2-hsp70l:mito-Keima,cryaa:mCherry vectors following manufacturer's instructions. Tol2 mRNA was synthesized using the mMessage mMachine SP6 in vitro transcription kit (Invitrogen) following the manufacturer's instructions. To generate stable transgenic lines, one-cell stage TU embryos were injected with the transgenesis vector and tol2 transposase mRNA. Positive embryos were screened either for cyan fluorescent lenses (ubi:mito-Keima) or for EGFP fluorescence (ubi:mito-GR and fabp10a:mito-GR) and raised to adulthood. F0 adults were outcrossed to TL mates, the F1 generation clutches were screened for bright fluorescence, and potential founders were raised. Potential F1 founders were outcrossed again to TU or TL mates and screened for bright fluorescence and 1:1 ratio of WT to transgenic offspring indicative of single copy insertion (Mosimann and Zon, 2011).
Fasting and feeding protocols
Fasted fish were kept in E3 medium cleaned of any dead larvae and shed chorions. Fed controls were provided live paramecia and AP100 dry larval diet (Zeigler) ad libitum. For heat-shock induction of mito-Keima and DN-Drp1, injected F0 mosaic larvae were placed into a 50 ml conical tube with 35 ml of E3 then incubated for 45 min at 38°C before being returned to room temperature E3.
Hypoxia and chemical treatments
For MitoTracker analysis, larvae were exposed to 1 µM MitoTracker Green FM (Cell Signaling Technology) overnight in E3, washed in E3 twice and prepared for imaging. For LysoTracker analysis, larvae were exposed to 1 µM LysoTracker Green DND-26 (Cell Signaling Technology) or LysoTracker Deep Red (Thermo Fisher Scientific) overnight in E3, washed in E3 twice and prepared for imaging. For all drug treatment experiments, single clutches were used and split between experimental groups. If additional embryos were necessary, two clutches from sibling breeding pairs were combined and mixed to split randomly. For hypoxia experiments, E3 was reduced to ∼1.7 mg/l O2 by bubbling in a mixture of ∼96% N2 and 4% O2 for ∼10 min. The larvae E3 in a 10 cm dish was replaced with about 40 ml of the hypoxic water, and the dish was moved to a hypoxia chamber, which was then flushed with the same mixture of ∼96% N2 and 4% O2 for about 10 min until an O2 sensor (DO210, Extech Instruments) read 4.7%. The chamber was sealed and moved into the 28.5°C incubator. For chemical hypoxia experiments, larvae were exposed to 60 µM or 100 µM DMOG (Tocris) or 10 µM FG-4592 (Cayman Chemical) in E3 from ∼70 hpf to ∼98 hpf. The exception is the pink1 MO experiment, for which the treatment window was from 48–72 hpf because MO knockdown is most effective during the first 3 dpf before additional cell divisions dilute the MO (Bill et al., 2009). For SAR405 experiments, larvae were split into cohorts receiving vehicle (DMSO; Sigma Aldrich) or 100 µM DMOG and then those cohorts were split into cohorts receiving additional vehicle (DMSO) or 10 µM SAR405 (EMD Millipore). The SAR405 treatment experiment and each high-efficiency CRIPSR experiment was replicated.
Confocal microscopy
Microscopy was performed using an inverted Ti2 (Nikon) microscope equipped with a Yokogawa CSUW1 spinning disc confocal unit, a CFI Apo LWD Lambda S 40XC WI (1.15 NA) objective lens (Nikon), and a Zyla 4.2+ sCMOS camera (ANDOR). Keima images were captured by exciting with 440 nm laser light and 561 laser light with ET Cy3/TRITC emission filter (Chroma) in triggered acquisition mode. Mito-GR images were captured using 488 nm laser light and 561 nm laser light with ET FITC/TRITC dual emission filter (Chroma) in triggered acquisition mode. For MitoTracker Green, LysoTracker Green, EGFP–Lc3, and nlsEGFP imaging, 488 nm excitation was utilized with a 525/36 FITC emission filter (Chroma). For Lysotraker Deep Red, a similar microscope equipped with a 640 nm laser and ET705/72m emission filter (Chroma) was utilized. For images involving both Keima and EGFP signal, the EGFP images were uniformly frame-shifted (Nikon Elements) based on control images to account for the shift caused by using different dichroic mirrors. For the in vivo mitophagy dynamics time-lapse imaging (Fig. 4; Movie 2), an iXon Life 888 EMCCD (ANDOR) was used for its increased light sensitivity and frame rate capacity, allowing greater time resolution. Resolution was calculated by 1.22×(wavelength)/(2×NA). For the 440 nm laser, this yields 233.4 nm. For the 561 nm laser, this yields 297.6 nm. Axial resolution is determined by 2×(wavelength)×(refractive index of the medium)/(NA2). For the 440 nm laser, this yields 886.3 nm. For the 561 nm laser, this yields 1130 nm. Imaris software (Bitplane) deconvolution algorithms were also applied to further improve axial resolution for colocalization analyses. Larvae were anesthetized with 0.16 mg/ml Tricaine-S (Western Chemical, Inc.) and mounted on glass-bottom dishes (MatTek) in 0.8% low-melt point agarose in E3-containing Tricane. After the agar solidified, additional E3 plus tricane was added to keep the gel hydrated during imaging. For skeletal muscle imaging, the collection volume was centered on the ninth and tenth myotomes. For all quantitative experiments, images of all samples were collected with identical laser power, camera gain and exposure time.
Image analysis
All mito-Keima image analysis was conducting using Imaris (Bitplane) software with either FIJI (ImageJ) or MATLAB (MathWorks) plugins to enable image mathematics. Raw images from Nikon Elements (.nd2) were converted to Imaris (.ims) file format using Imaris Image Converter (Bitplane) and imported to Imaris. A surface mask was drawn manually using the Surfaces function to segment muscle tissue away from skin. A ratiometric image (FIJI or MATLAB) was generated by changing the data type to 32 Bit Float and applying the following equation: (Keima561 channel)/(Keima440 channel+150). The 150 addition was chosen as a constant that is higher than background signal. This ensured that areas of low signal from both Keima561 and Keima440 were excluded. The ratiometric image normalizes for the decrease in signal intensity as imaging depth into the tissue increases and for small intensity differences between samples. Then, an isosurfaces thresholding algorithm was applied to the ratio channel, with 0.15 µm surface grain (smoothing) to eliminate noise, using a threshold value determined for each experiment from images of control fish. A size cutoff removed objects smaller than 2 voxels to eliminate additional noise. Obviously spurious surfaces were removed manually. Rarely, individual slices were removed from z-stack images to account for fish movement, which caused edge artifacts in some ratio images. Mitolysosome density was determined by dividing the number of ratio surface objects by the total tissue surface volume. The mitophagy index was determined by dividing the sum of all ratio surface object volumes by the total tissue surface volume. For mito-GR images, a deconvolution algorithm (Imaris) to adjust for the behavior of light of differing emission wavelengths (green and red) was used reduce edge artifacts. Object tracking in time-lapse microscopy (Fig. 4; Movie 2) was performed using Imaris software (Bitplane). Mitophagy isosurface masks were generated as described above. Mitochondria surfaces were generated using a 0.325 µm surface grain (smoothing), background subtraction (diameter largest sphere=1.219 µm), a manually determined florescence intensity threshold, and a size cutoff eliminating objects smaller than 15 voxels. Object tracking was performed on ratio surface objects, using autoregressive motion, a maximum distance of 1.003 µm, a maximum gap size of 3 (without gap filling), and a track duration minimum of 5.534 s. We manually assessed each newly formed mitophagy isosurface object, defined as a Ratiohigh tracked object that was not present at time zero; other mitophagy objects were considered stable. Using a combination of microscopy views, new objects were manually classified as new mitophagy, split mitolysosomes or from outside. For lysosome size analysis, LysoTracker Green z-stack images were acquired covering 8.8 µm depth within the skeletal muscle of the fish and analyzed using Imaris (Bitplane). First, total tissue volume was determined by manually drawing a surface around the evenly illuminated muscle tissue and excluding signal from other tissues. Then, the surface was used to mask the image, and an additional isosurface generation algorithm was applied to measure the number and volume of LysoTracker Green objects. Smoothing of 0.125 µm and background subtraction allowing for objects with an estimated diameter of 0.4 µm was included in the processing along with a filter for objects >2 voxels to remove additional background objects. Lysosome surface number, lysosome surface volume and total tissue volume were exported and used for analyses. For object-based colocalization experiments (Fig. 5B–F,I), Keima was excited using 488 nm and 561 nm lasers using a microscope equipped with a 640 nm laser needed to excite LysoTracker Far Red (this microscope does not have a 440 nm laser); 488 nm exposure excites both acidic and neutral/basic mitochondria, whereas 561 nm excitation heavily favors excitation of acidic mitochondria. Thus, healthy mitochondria were segmented using an image generated using the following equation: (Keima488 channel)/(Keima561 channel+1). Because the background signal was higher in the Keima561 channel than the Keima488 channel, a small non-zero number could be used. One myotome was manually segmented, and all objects were counted within. Mitolysosomes were segmented using an image generated using the following equation (Keima561 channel)/(Keima488 channel+150). EGFP–Lc3+ and LysoTracker Deep Red+ objects were segmented as described above. Object-based colocalization was performed using distance transformations from each object category, and objects near the edges of the somite or image border were excluded. Lc3+ objects were considered autolysosomes if there was a LysoTracker Deep Red object within 1 nm of it (filtered by Signal Intensity >1 in the LysoTracker Deep Red objects distance transformation channel). If not, they were considered autophagosomes. An autophagosome or autolysosome was considered ‘in contact’ with a healthy mitochondrion if there was a Keima488/561 object within 0 nm of it (filtered by Signal Intensity >0 in the Keima488/561 objects distance transformation channel). A mitolysosome was considered near an Lc3+ object if it was within 0.5 µm (filtered by Signal Intensity >500 in the Lc3+ objects distance transformation channel). In Fig. S1A, isosurface renderings for MitoTracker+ objects, Keima440high objects, and Ratio objects [Keima561/(Keima440+150)] were generated. The percentage of MitoTracker+ surfaces colocalizing with either Keima surface was determined by combining the Keima440 objects and Ratio objects, generating a distance transformation channel. A MitoTracker+ object was considered colocalized if it was located less than 1 nm from a Keima+ objects (Minimum Signal intensity in the distance transformation channel <1). Similar calculations were performed for Keima440high or Ratiohigh objects colocalizing with MitoTracker surfaces, and for LysoTracker+ objects colocalizing with EGFP or Ratiohigh objects in ubi:mito-GR embryos (Fig. S1D).
Morpholino injections
Morpholinos (GeneTools, LLC) were reconstituted to 1 mM stock solutions in water. The injection mixes were prepared at 0.9 mM (atg5), 0.4 mM (vhl), 0.2 mM (hif1ab), or 0.2 mM (pink1), and ∼2 nl was injected into the yolk of one-cell stage embryos. The sequences are listed in the Table S1.
CRISPR
All CRISPR genome editing was performed using a variation of the high-efficiency protocol described previously (Burger et al., 2016). Guide RNA sequences were designed using CHOPCHOP (Labun et al., 2019) and chosen to target a 5′ exonic region with the potential to introduce STOP codons in the event of frameshift. Alt-R crRNA (IDT) constructs were reconstituted and annealed with Alt-R tracrRNA (IDT) to form gRNA duplexes according to the manufacturer's instructions. Injection mixes were prepared by mixing 1 µl the gRNA duplex, 1 µl Duplex Buffer (IDT) and 1 µl ENGEN Spy Cas9 NLS (NEB), and allowing the mix to incubate at room temperature for at least 10 min. Approximately 2 nl of injection mix was injected into both the cell and yolk of one-cell stage embryos. Efficient editing was confirmed by PCR and Sanger sequencing. For experiments using F0 CRISPR editing, the Sanger sequencing trace of each imaged larva was used to determine the extent of editing by a researcher who was blind to the experimental conditions. Editing was considered sufficient if trace files showed a mixture of multiple traces emerging near the PAM site. Images from larvae found to have low or no editing were removed from analysis (Fig. S5). To isolate the bnip3 mutants, F0 parents were outcrossed and PCR and Sanger Sequencing was performed on gDNA isolated from pooled larvae and TOPO cloned into pCR4-TOPO TA vector (Invitrogen). Sequences were analyzed for the presence of alleles leading to premature stop codons, and clutches containing such alleles were raised to adulthood and outcrossed. Adult F2 fish were tail clipped and subjected to similar TOPO sequencing to identify heterozygous founders.
Genotyping
DNA from individual larvae was extracted using proteinase K (Invitrogen) and amplified by PCR with the primer pairs (IDT) listed in the Table S1. The 5′ primer of the pink1gi2 PCR genotyping primer pair was synthesized with a 5′ 6-FAM (Fluorescein) fluorescent dye attachment necessary for DNA fragment analysis, in which small, fluorescently labeled DNA fragments are electrophoretically separated such that precise changes in size (down to 1 bp) can be detected. The pink1gi2 (Zhang et al., 2017) and bnip3 mutant fish were identified by the presence of a small deletion, which was detected using DNA Fragment Analysis (MGH CCIB DNA Core). Efficient editing in F0 high-efficiency CRISPR experiments was confirmed by Sanger sequencing, as described in the CRISPR section.
Statistical analyses
All graphs and statistical analyses were generated using Prism (GraphPad). All bar graphs display the mean±s.e.m. unless otherwise stated in the figure legend. For comparisons between only two conditions, standard, unpaired, two-tailed Student's t-tests were employed. For comparisons within groups with four conditions, two-way ANOVA with Holm–Sidak's multiple comparisons test were employed. P values or an asterisk indicating significant differences as defined in the figure legends were included in each figure containing statistical analyses. Sample size was determined based on similar experiments in the zebrafish field and practical considerations of the maximum number of animals that could be injected or imaged within time constraints. No animals were excluded from analyses. Each experiment was performed on randomly chosen larvae from a single clutch from a single mating pair or from randomly combined clutches from sibling mating pairs. Figs S4, S6 and S7 were included for specific quantitative experiments to demonstrate reproducibility and provide full statistical analyses.
Acknowledgements
We thank Tom Schwarz for helpful discussions. We thank Nadine Budrow for maintaining our zebrafish colonies. We thank Phil Elks for sharing DA-hif plasmids, Donghun Shin for sharing the p5E-2.8fabp10a plasmid, and Daniel Hesselson for sharing the pink1gi2 line. We thank the Harvard Medical School MicRoN team for access to microscopes and imaging expertise.
Footnotes
Competing interests
J.W.H. is a founder and consultant to Caraway Therapeutics and is a founding scientific advisory board member for Interline Therapeutics Inc. W.G. is a consultant to Camp4 Therapeutics and Amagma Therapeutics and receives royalties from FATE Therapeutics.
Author contributions
Conceptualization: P.J.W., J.-M.H., J.W.H., W.G.; Methodology: P.J.W., A.S., J.-M.H.; Investigation: P.J.W., J.-M.H., E.D.Q., K.A.L.; Writing - original draft: P.J.W., W.G.; Writing - review & editing: P.J.W., A.S., J.-M.H., E.D.Q., J.W.H., W.G.; Visualization: P.J.W., A.S., W.G.; Supervision: J.W.H.; Project administration: W.G.; Funding acquisition: P.J.W., J.W.H., W.G.
Funding
This work was supported by the National Institutes of Health (F32AA025271 to P.J.W., R37 NS083524 to J.W.H, R01DK090311, R01DK105198 and R24OD017870 to W.G.), by the Michael J. Fox Foundation for Parkinson's Research [#15866.01 to J.W.H.], by the American Liver Foundation (Postdoctoral Fellowship to P.J.W.), and the Pew Charitable Trusts (Pew Scholar to W.G.). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at https://jcs.biologists.org/lookup/doi/10.1242/jcs.256255.supplemental
References
- Abeliovich, H., Zarei, M., Rigbolt, K. T. G., Youle, R. J. and Dengjel, J. (2013). Involvement of mitochondrial dynamics in the segregation of mitochondrial matrix proteins during stationary phase mitophagy. Nat. Commun. 4, 2789. 10.1038/ncomms3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abudu, Y. P., Pankiv, S., Mathai, B. J., Håkon Lystad, A., Bindesbøll, C., Brenne, H. B., Yoke Wui Ng, M., Thiede, B., Yamamoto, A., Mutugi Nthiga, T.et al. (2019). NIPSNAP1 and NIPSNAP2 act as “Eat Me” signals for mitophagy. Dev. Cell 49, 509-525.e12. 10.1016/j.devcel.2019.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anding, A. L. and Baehrecke, E. H. (2017). Cleaning house: selective autophagy of organelles. Dev. Cell 41, 10-22. 10.1016/j.devcel.2017.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, J. and Currie, P. D. (2013). 503Unc, a small and muscle-specific zebrafish promoter. Genesis 51, 443-447. 10.1002/dvg.22385 [DOI] [PubMed] [Google Scholar]
- Bill, B. R., Petzold, A. M., Clark, K. J., Schimmenti, L. A. and Ekker, S. C. (2009). A primer for morpholino use in zebrafish. Zebrafish 6, 69-77. 10.1089/zeb.2008.0555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birgisdottir, A., Lamark, T. and Johansen, T. (2013). The LIR motif – crucial for selective autophagy. J. Cell Sci. 126, 3237-3247. [DOI] [PubMed] [Google Scholar]
- Burger, A., Lindsay, H., Felker, A., Hess, C., Anders, C., Chiavacci, E., Zaugg, J., Weber, L. M., Catena, R., Jinek, M.et al. (2016). Maximizing mutagenesis with solubilized CRISPR-Cas9 ribonucleoprotein complexes. Development 143, 2025-2037. 10.1242/dev.134809 [DOI] [PubMed] [Google Scholar]
- Burman, J. L., Pickles, S., Wang, C., Sekine, S., Vargas, J. N. S., Zhang, Z., Youle, A. M., Nezich, C. L., Wu, X., Hammer, J. A.et al. (2017). Mitochondrial fission facilitates the selective mitophagy of protein aggregates. J. Cell Biol. 216, 3231-3247. 10.1083/jcb.201612106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadete, V. J. J., Deschênes, S., Cuillerier, A., Brisebois, F., Sugiura, A., Vincent, A., Turnbull, D., Picard, M., McBride, H. M. and Burelle, Y. (2016). Formation of mitochondrial-derived vesicles is an active and physiologically relevant mitochondrial quality control process in the cardiac system. J. Physiol. 594, 5343-5362. 10.1113/JP272703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, G., Han, Z., Feng, D., Chen, Y., Chen, L., Wu, H., Huang, L., Zhou, C., Cai, X., Fu, C.et al. (2014). A regulatory signaling loop comprising the PGAM5 phosphatase and CK2 controls receptor-mediated mitophagy. Mol. Cell 54, 362-377. 10.1016/j.molcel.2014.02.034 [DOI] [PubMed] [Google Scholar]
- Clark, I. E., Dodson, M. W., Jiang, C., Cao, J. H., Huh, J. R., Seol, J. H., Yoo, S. J., Hay, B. A. and Guo, M. (2006). Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature 441, 1162-1166. 10.1038/nature04779 [DOI] [PubMed] [Google Scholar]
- Cox, A. G. and Goessling, W. (2015). The lure of zebrafish in liver research: regulation of hepatic growth in development and regeneration. Curr. Opin. Genet. Dev. 32, 153-161. 10.1016/j.gde.2015.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amico, D., Mottis, A., Potenza, F., Sorrentino, V., Li, H., Romani, M., Lemos, V., Schoonjans, K., Zamboni, N., Knott, G.et al. (2019). The RNA-binding protein pum2 impairs mitochondrial dynamics and mitophagy during aging. Mol. Cell 73, 775-787.e10. 10.1016/j.molcel.2018.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNapoli, S. E., Martinez-McFaline, R., Gribbin, C. K., Wrighton, P. J., Balgobin, C. A., Nelson, I., Leonard, A., Maskin, C. R., Shwartz, A., Quenzer, E. D.et al. (2020). Synthetic CRISPR/Cas9 reagents facilitate genome editing and homology directed repair. Nucleic Acids Res. 48, e38. 10.1093/nar/gkaa085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elks, P. M., Van Eeden, F. J., Dixon, G., Wang, X., Reyes-Aldasoro, C. C., Ingham, P. W., Whyte, M. K. B., Walmsley, S. R. and Renshaw, S. A. (2011). Activation of hypoxia-inducible factor-1α (hif-1α) delays inflammation resolution by reducing neutrophil apoptosis and reverse migration in a zebrafish inflammation model. Blood 118, 712-722. 10.1182/blood-2010-12-324186 [DOI] [PubMed] [Google Scholar]
- Elks, P. M., Brizee, S., van der Vaart, M., Walmsley, S. R., van Eeden, F. J., Renshaw, S. A. and Meijer, A. H. (2013). Hypoxia inducible factor signaling modulates susceptibility to mycobacterial infection via a nitric oxide dependent mechanism. PLoS Pathog. 9, e1003789. 10.1371/journal.ppat.1003789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, C. S. and Holzbaur, E. L. F. (2020). Degradation of engulfed mitochondria is rate-limiting in Optineurin-mediated mitophagy in neurons. eLife 9, e50260. 10.7554/eLife.50260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdinand, P. and Roffe, C. (2016). Hypoxia after stroke: a review of experimental and clinical evidence. Exp. Transl. Stroke Med. 8, 9. 10.1186/s13231-016-0023-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman, J. R. and Nunnari, J. (2014). Mitochondrial form and function. Nature 505, 335-343. 10.1038/nature12985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser, T. (2009). Mendelian forms of Parkinson's disease. Biochim. Biophys. Acta Mol. Basis Dis. 1792, 587-596. 10.1016/j.bbadis.2008.12.007 [DOI] [PubMed] [Google Scholar]
- Hamacher-Brady, A., Brady, N. R., Logue, S. E., Sayen, M. R., Jinno, M., Kirshenbaum, L. A., Gottlieb, R. A. and Gustafsson, Å. B. (2007). Response to myocardial ischemia/reperfusion injury involves Bnip3 and autophagy. Cell Death Differ. 14, 146-157. 10.1038/sj.cdd.4401936 [DOI] [PubMed] [Google Scholar]
- Hanna, R. A., Quinsay, M. N., Orogo, A. M., Giang, K., Rikka, S. and Gustafsson, Å. B. (2012). Microtubule-associated protein 1 light chain 3 (LC3) interacts with Bnip3 protein to selectively remove endoplasmic reticulum and mitochondria via autophagy. J. Biol. Chem. 287, 19094-19104. 10.1074/jbc.M111.322933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper, J. W., Ordureau, A. and Heo, J.-M. (2018). Building and decoding ubiquitin chains for mitophagy. Nat. Rev. Mol. Cell Biol. 19, 93-108. 10.1038/nrm.2017.129 [DOI] [PubMed] [Google Scholar]
- Harris, J. M., Esain, V., Frechette, G. M., Harris, L. J., Cox, A. G., Cortes, M., Garnaas, M. K., Carroll, K. J., Cutting, C. C., Khan, T.et al. (2013). Glucose metabolism impacts the spatiotemporal onset and magnitude of HSC induction in vivo. Blood 121, 2483-2493. 10.1182/blood-2012-12-471201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, C., Bartholomew, C. R., Zhou, W. and Klionsky, D. J. (2009). Assaying autophagic activity in transgenic GFP-Lc3 and GFP-Gabarap zebrafish embryos. Autophagy 5, 520-526. 10.4161/auto.5.4.7768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Her, G. M., Chiang, C.-C., Chen, W.-Y. and Wu, J.-L. (2003). In vivo studies of liver-type fatty acid binding protein (L-FABP) gene expression in liver of transgenic zebrafish (Danio rerio). FEBS Lett. 538, 125-133. 10.1016/S0014-5793(03)00157-1 [DOI] [PubMed] [Google Scholar]
- Iliopoulos, O., Levy, A. P., Jiang, C., Kaelin, W. G. and Goldberg, M. A. (1996). Negative regulation of hypoxia-inducible genes by the von Hippel-Lindau protein. Proc. Natl. Acad. Sci. USA 93, 10595-10599. 10.1073/pnas.93.20.10595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivan, M., Kondo, K., Yang, H., Kim, W., Valiando, J., Ohh, M., Salic, A., Asara, J. M., Lane, W. S. and Kaelin, W. G., Jr. (2001). HIFα targeted for VHL-mediated destruction by proline hydroxylation: Implications for O2 sensing. Science 292, 464-468. 10.1126/science.1059817 [DOI] [PubMed] [Google Scholar]
- Jakobs, S. (2006). High resolution imaging of live mitochondria. Biochim. Biophys. Acta 1763, 561-575. 10.1016/j.bbamcr.2006.04.004 [DOI] [PubMed] [Google Scholar]
- Kaelin, W. G. and McKnight, S. L. (2013). Influence of metabolism on epigenetics and disease. Cell 153, 56-69. 10.1016/j.cell.2013.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama, H., Yamamoto, A., Mizushima, N., Yoshimori, T. and Miyawaki, A. (2008). GFP-like proteins stably accumulate in lysosomes. Cell Struct. Funct. 33, 1-12. 10.1247/csf.07011 [DOI] [PubMed] [Google Scholar]
- Katayama, H., Kogure, T., Mizushima, N., Yoshimori, T. and Miyawaki, A. (2011). A sensitive and quantitative technique for detecting autophagic events based on lysosomal delivery. Chem. Biol. 18, 1042-1052. 10.1016/j.chembiol.2011.05.013 [DOI] [PubMed] [Google Scholar]
- Katayama, H., Hama, H., Nagasawa, K., Kurokawa, H., Sugiyama, M., Ando, R., Funata, M., Yoshida, N., Homma, M., Nishimura, T.et al. (2020). Visualizing and modulating mitophagy for therapeutic studies of neurodegeneration. Cell 181, 1176-1187.e16. 10.1016/j.cell.2020.04.025 [DOI] [PubMed] [Google Scholar]
- Kim, Y. Y., Um, J.-H., Yoon, J.-H., Kim, H., Lee, D.-Y., Lee, Y. J., Jee, H. J., Kim, Y. M., Jang, J. S., Jang, Y.-G., et al. (2019). Assessment of mitophagy in mt-Keima Drosophila revealed an essential role of the PINK1-Parkin pathway in mitophagy induction in vivo. FASEB J. 33, 9742-9751. 10.1096/fj.201900073R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolitsida, P., Zhou, J., Rackiewicz, M., Nolic, V., Dengjel, J. and Abeliovich, H. (2019). Phosphorylation of mitochondrial matrix proteins regulates their selective mitophagic degradation. Proc. Natl. Acad. Sci. USA 116, 20517-20527. 10.1073/pnas.1901759116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan, K. M., Fujimoto, E., Grabher, C., Mangum, B. D., Hardy, M. E., Campbell, D. S., Parant, J. M., Yost, H. J., Kanki, J. P. and Chien, C.-B. (2007). The Tol2kit: a multisite gateway-based construction Kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 236, 3088-3099. 10.1002/dvdy.21343 [DOI] [PubMed] [Google Scholar]
- Labun, K., Montague, T. G., Krause, M., Torres Cleuren, Y. N., Tjeldnes, H. and Valen, E. (2019). CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 47, W171-W174. 10.1093/nar/gkz365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guerroué, F., Eck, F., Jung, J., Starzetz, T., Mittelbronn, M., Kaulich, M. and Behrends, C. (2017). Autophagosomal content profiling reveals an LC3C-dependent piecemeal mitophagy pathway. Mol. Cell 68, 786-796.e6. 10.1016/j.molcel.2017.10.029 [DOI] [PubMed] [Google Scholar]
- Lee, E., Koo, Y., Ng, A., Wei, Y., Luby-Phelps, K., Juraszek, A., Xavier, R. J., Cleaver, O., Levine, B. and Amatruda, J. F. (2014). Autophagy is essential for cardiac morphogenesis during vertebrate development. Autophagy 10, 572-587. 10.4161/auto.27649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, E., Wei, Y., Zou, Z., Tucker, K., Rakheja, D., Levine, B. and Amatruda, J. F. (2016). Genetic inhibition of autophagy promotes p53 loss-of-heterozygosity and tumorigenesis. Oncotarget 7, 67919-67933. 10.18632/oncotarget.12084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. J., Sanchez-Martinez, A., Zarate, A. M., Benincá, C., Mayor, U., Clague, M. J. and Whitworth, A. J. (2018). Basal mitophagy is widespread in Drosophila but minimally affected by loss of Pink1 or parkin. J. Cell Biol. 217, 1613-1622. 10.1083/jcb.201801044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. W., Ko, J., Ju, C. and Eltzschig, H. K. (2019). Hypoxia signaling in human diseases and therapeutic targets. Exp. Mol. Med. 51, 1-13. 10.1038/s12276-019-0299-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leermakers, P. A. and Gosker, H. R. (2016). Skeletal muscle mitophagy in chronic disease: implications for muscle oxidative capacity. Curr. Opin Clin. Nutr. Metab. Care 19, 427-433. 10.1097/MCO.0000000000000319 [DOI] [PubMed] [Google Scholar]
- Lim, S.-E., Esain, V., Kwan, W., Theodore, L. N., Cortes, M., Frost, I. M., Liu, S. Y. and North, T. E. (2017). HIF1α-induced PDGFRβ signaling promotes developmental HSC production via IL-6 activation. Exp. Hematol. 46, 83-95.e6. 10.1016/j.exphem.2016.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, L., Feng, D., Chen, G., Chen, M., Zheng, Q., Song, P., Ma, Q., Zhu, C., Wang, R., Qi, W.et al. (2012). Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat. Cell Biol. 14, 177-185. 10.1038/ncb2422 [DOI] [PubMed] [Google Scholar]
- Mammucari, C., Milan, G., Romanello, V., Masiero, E., Rudolf, R., Del Piccolo, P., Burden, S. J., Di Lisi, R., Sandri, C., Zhao, J.et al. (2007). FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 6, 458-471. 10.1016/j.cmet.2007.11.001 [DOI] [PubMed] [Google Scholar]
- Mao, K., Wang, K., Zhao, M., Xu, T. and Klionsky, D. J. (2011). Two MAPK-signaling pathways are required for mitophagy in Saccharomyces cerevisiae. J. Cell Biol. 193, 755-767. 10.1083/jcb.201102092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliams, T. G., Prescott, A. R., Allen, G. F. G., Tamjar, J., Munson, M. J., Thomson, C., Muqit, M. M. K. and Ganley, I. G. (2016). Mito-QC illuminates mitophagy and mitochondrial architecture in vivo. J. Cell Biol. 214, 333-345. 10.1083/jcb.201603039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliams, T. G., Prescott, A. R., Montava-Garriga, L., Ball, G., Singh, F., Barini, E., Muqit, M. M. K., Brooks, S. P. and Ganley, I. G. (2018). Basal mitophagy occurs independently of PINK1 in mouse tissues of high metabolic demand. Cell Metab. 27, 439-449.e5. 10.1016/j.cmet.2017.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosimann, C. and Zon, L. I. (2011). Advanced zebrafish transgenesis with Tol2 and application for Cre/lox recombination experiments. Methods Cell Biol. 104, 173-194. 10.1016/B978-0-12-374814-0.00010-0 [DOI] [PubMed] [Google Scholar]
- Mosimann, C., Kaufman, C. K., Li, P., Pugach, E. K., Tamplin, O. J. and Zon, L. I. (2011). Ubiquitous transgene expression and Cre-based recombination driven by the ubiquitin promoter in zebrafish. Development 138, 169-177. 10.1242/dev.059345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordureau, A., Paulo, J. A., Zhang, J., An, H., Swatek, K. N., Cannon, J. R., Wan, Q., Komander, D. and Harper, J. W. (2020). Global landscape and dynamics of parkin and USP30-dependent ubiquitylomes in ineurons during mitophagic signaling. Mol. Cell 77, 1124-1142.e10. 10.1016/j.molcel.2019.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palikaras, K., Lionaki, E. and Tavernarakis, N. (2018). Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat. Cell Biol. 20, 1013-1022. 10.1038/s41556-018-0176-2 [DOI] [PubMed] [Google Scholar]
- Park, J., Lee, S. B., Lee, S., Kim, Y., Song, S., Kim, S., Bae, E., Kim, J., Shong, M., Kim, J.-M.et al. (2006). Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature 441, 1157-1161. 10.1038/nature04788 [DOI] [PubMed] [Google Scholar]
- Pickles, S., Vigié, P. and Youle, R. J. (2018). Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr. Biol. 28, R170-R185. 10.1016/j.cub.2018.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell, A. M. and Youle, R. J. (2015). The roles of PINK1, Parkin, and mitochondrial fidelity in parkinson's disease. Neuron 85, 257-273. 10.1016/j.neuron.2014.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcelli, A. M., Ghelli, A., Zanna, C., Pinton, P., Rizzuto, R. and Rugolo, M. (2005). pH difference across the outer mitochondrial membrane measured with a green fluorescent protein mutant. Biochem. Biophys. Res. Commun. 326, 799-804. 10.1016/j.bbrc.2004.11.105 [DOI] [PubMed] [Google Scholar]
- Preidis, G. A., Kim, K. H. and Moore, D. D. (2017). Nutrient-sensing nuclear receptors PPARα and FXR control liver energy balance. J. Clin. Invest. 127, 1193-1201. 10.1172/JCI88893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyadarshini, M., Tuimala, J., Chen, Y. C. and Panula, P. (2013). A zebrafish model of PINK1 deficiency reveals key pathway dysfunction including HIF signaling. Neurobiol. Dis. 54, 127-138. 10.1016/j.nbd.2013.02.002 [DOI] [PubMed] [Google Scholar]
- Raefsky, S. M. and Mattson, M. P. (2017). Adaptive responses of neuronal mitochondria to bioenergetic challenges: Roles in neuroplasticity and disease resistance. Free Radic. Biol. Med. 102, 203-216. 10.1016/j.freeradbiomed.2016.11.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambold, A. S., Kostelecky, B., Elia, N. and Lippincott-Schwartz, J. (2011). Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc. Natl. Acad. Sci. USA 108, 10190-10195. 10.1073/pnas.1107402108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana, A., Oliveira, M. P., Khamoui, A. V., Aparicio, R., Rera, M., Rossiter, H. B. and Walker, D. W. (2017). Promoting Drp1-mediated mitochondrial fission in midlife prolongs healthy lifespan of Drosophila melanogaster. Nat. Commun. 8, 448. 10.1038/s41467-017-00525-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodger, C. E., Mcwilliams, T. G., Ganley, I. G. and Ganley, I. G. (2018). Mammalian mitophagy – from in vitro molecules to in vivo models. FEBS J. 285, 1185-1202. 10.1111/febs.14336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Nuevo, A., Díaz-Ramos, A., Noguera, E., Díaz-Sáez, F., Duran, X., Muñoz, J. P., Romero, M., Plana, N., Sebastián, D., Tezze, C.et al. (2018). Mitochondrial DNA and TLR9 drive muscle inflammation upon Opa1 deficiency. EMBO J. 37, 1-18. 10.15252/embj.201796553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojansky, R., Cha, M.-Y. and Chan, D. C. (2016). Elimination of paternal mitochondria in mouse embryos occurs through autophagic degradation dependent on PARKIN and MUL1. eLife 5, e17896. 10.7554/eLife.17896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanello, V. and Sandri, M. (2013). Mitochondrial biogenesis and fragmentation as regulators of protein degradation in striated muscles. J. Mol. Cell. Cardiol. 55, 64-72. 10.1016/j.yjmcc.2012.08.001 [DOI] [PubMed] [Google Scholar]
- Ronan, B., Flamand, O., Vescovi, L., Dureuil, C., Durand, L., Fassy, F., Bachelot, M.-F., Lamberton, A., Mathieu, M., Bertrand, T.et al. (2014). A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy. Nat. Chem. Biol. 10, 1013-1019. 10.1038/nchembio.1681 [DOI] [PubMed] [Google Scholar]
- Sandri, M. (2013). Protein breakdown in muscle wasting: Role of autophagy-lysosome and ubiquitin-proteasome. Int. J. Biochem. Cell Biol. 45, 2121-2129. 10.1016/j.biocel.2013.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastián, D. and Zorzano, A. (2020). Self-eating for muscle fitness: autophagy in the control of energy metabolism. Dev. Cell 54, 268-281. 10.1016/j.devcel.2020.06.030 [DOI] [PubMed] [Google Scholar]
- Settembre, C., Fraldi, A., Medina, D. L. and Ballabio, A. (2013). Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat. Rev. Mol. Cell Biol. 14, 283-296. 10.1038/nrm3565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, R.-Y., Zhu, S.-H., Li, V., Gibson, S. B., Xu, X.-S. and Kong, J.-M. (2014). BNIP3 interacting with LC3 triggers excessive mitophagy in delayed neuronal death in stroke. CNS Neurosci. Ther. 20, 1045-1055. 10.1111/cns.12325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova, E., Shurland, D.-L., Ryazantsev, S. N. and Van Der Bliek, A. M. (1998). A human dynamin-related protein controls the distribution of mitochondria. J. Cell Biol. 143, 351-358. 10.1083/jcb.143.2.351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura, A., McLelland, G. L., Fon, E. A. and McBride, H. M. (2014). A new pathway for mitochondrial quality control: mitochondrial–derived vesicles. EMBO J. 33, 2142-2156. 10.15252/embj.201488104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, N., Yun, J., Liu, J., Malide, D., Liu, C., Rovira, I. I., Holmström, K. M., Fergusson, M. M., Yoo, Y. H., Combs, C. A.et al. (2015). Measuring in vivo mitophagy. Mol. Cell 60, 685-696. 10.1016/j.molcel.2015.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas, J. N. S., Wang, C., Bunker, E., Hao, L., Maric, D., Schiavo, G., Randow, F. and Youle, R. J. (2019). Spatiotemporal control of ULK1 activation by NDP52 and TBK1 during selective autophagy. Mol. Cell 74, 347-362.e6. 10.1016/j.molcel.2019.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, J. A., Ni, H.-M., Haynes, A., Manley, S., Li, Y., Jaeschke, H. and Ding, W.-X. (2015). Chronic deletion and acute knockdown of Parkin have differential responses to acetaminophen-induced mitophagy and liver injury in mice. J. Biol. Chem. 290, 10934-10946. 10.1074/jbc.M114.602284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, C. (2012). Aspects of larval rearing. ILAR J. 53, 169-178. 10.1093/ilar.53.2.169 [DOI] [PubMed] [Google Scholar]
- Wrighton, P. J., Oderberg, I. M. and Goessling, W. (2019). There is something fishy about liver cancer: zebrafish models of hepatocellular carcinoma. Cell. Mol. Gastroenterol. Hepatol. 8, 347-363. 10.1016/j.jcmgh.2019.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita, S.-I., Jin, X., Furukawa, K., Hamasaki, M., Nezu, A., Otera, H., Saigusa, T., Yoshimori, T., Sakai, Y., Mihara, K.et al. (2016). Mitochondrial division occurs concurrently with autophagosome formation but independently of Drp1 during mitophagy. J. Cell Biol. 215, 649-665. 10.1083/jcb.201605093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y., Gehrke, S., Imai, Y., Huang, Z., Ouyang, Y., Wang, J.-W., Yang, L., Beal, M. F., Vogel, H. and Lu, B. (2006). Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc. Natl. Acad. Sci. USA 103, 10793-10798. 10.1073/pnas.0602493103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, W., Liu, Y., Tu, Z., Xiao, C., Yan, S., Ma, X., Guo, X., Chen, X., Yin, P., Yang, Z.et al. (2019). CRISPR/Cas9-mediated PINK1 deletion leads to neurodegeneration in rhesus monkeys. Cell Res. 29, 334-336. 10.1038/s41422-019-0142-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, S.-M. and Jung, Y.-K. (2018). A molecular approach to mitophagy and mitochondrial dynamics. Mol. Cells 41, 18-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle, R. J. (2019). Mitochondria—Striking a balance between host and endosymbiont. Science 365, eaaw9855. 10.1126/science.aaw9855 [DOI] [PubMed] [Google Scholar]
- Yuan, Y., Zheng, Y., Zhang, X., Chen, Y., Wu, X., Wu, J., Shen, Z., Jiang, L., Wang, L., Yang, W.et al. (2017). BNIP3L/NIX-mediated mitophagy protects against ischemic brain injury independent of PARK2. Autophagy 13, 1754-1766. 10.1080/15548627.2017.1357792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., Bosch-Marce, M., Shimoda, L. A., Tan, Y. S., Baek, J. H., Wesley, J. B., Gonzalez, F. J. and Semenza, G. L. (2008). Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J. Biol. Chem. 283, 10892-10903. 10.1074/jbc.M800102200 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhang, T., Xue, L., Li, L., Tang, C., Wan, Z., Wang, R., Tan, J., Tan, Y., Han, H., Tian, R.et al. (2016a). BNIP3 protein suppresses PINK1 kinase proteolytic cleavage to promote mitophagy. J. Biol. Chem. 291, 21616-21629. 10.1074/jbc.M116.733410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W., Ren, H., Xu, C., Zhu, C., Wu, H., Liu, D., Wang, J., Liu, L., Li, W., Ma, Q.et al. (2016b). Hypoxic mitophagy regulates mitochondrial quality and platelet activation and determines severity of I/R heart injury. eLife 5, e21407. 10.7554/eLife.21407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Nguyen, D. T., Olzomer, E. M., Poon, G. P., Cole, N. J., Puvanendran, A., Phillips, B. R. and Hesselson, D. (2017). Rescue of Pink1 deficiency by stress-dependent activation of autophagy. Cell Chem. Biol. 24, 471-480.e4. 10.1016/j.chembiol.2017.03.005 [DOI] [PubMed] [Google Scholar]
- Zhou, H., Zhu, P., Guo, J., Hu, N., Wang, S., Li, D., Hu, S., Ren, J., Cao, F. and Chen, Y. (2017). Ripk3 induces mitochondrial apoptosis via inhibition of FUNDC1 mitophagy in cardiac IR injury. Redox. Biol. 13, 498-507. 10.1016/j.redox.2017.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y., Long, Q., Wu, H., Li, W., Qi, J., Wu, Y., Xiang, G., Tang, H., Yang, L., Chen, K.et al. (2020). Topology-dependent, bifurcated mitochondrial quality control under starvation. Autophagy, 16, 562-574. 10.1080/15548627.2019.1634944 [DOI] [PMC free article] [PubMed] [Google Scholar]