Abstract
We hypothesized that epigenetics is a link between smoking/allergen exposures and the development of Asthma and chronic obstructive pulmonary disease (ACO). A total of 75 of 228 COPD patients were identified as ACO, which was independently associated with increased exacerbations. Microarray analysis identified 404 differentially methylated loci (DML) in ACO patients, and 6575 DML in those with rapid lung function decline in a discovery cohort. In the validation cohort, ACO patients had hypermethylated PDE9A (+ 30,088)/ZNF323 (− 296), and hypomethylated SEPT8 (− 47) genes as compared with either pure COPD patients or healthy non-smokers. Hypermethylated TIGIT (− 173) gene and hypomethylated CYSLTR1 (+ 348)/CCDC88C (+ 125,722)/ADORA2B (+ 1339) were associated with severe airflow limitation, while hypomethylated IFRD1 (− 515) gene with frequent exacerbation in all the COPD patients. Hypermethylated ZNF323 (− 296) / MPV17L (+ 194) and hypomethylated PTPRN2 (+ 10,000) genes were associated with rapid lung function decline. In vitro cigarette smoke extract and ovalbumin concurrent exposure resulted in specific DNA methylation changes of the MPV17L / ZNF323 genes, while 5-aza-2′-deoxycytidine treatment reversed promoter hypermethylation-mediated MPV17L under-expression accompanied with reduced apoptosis and decreased generation of reactive oxygen species. Aberrant DNA methylations may constitute a determinant for ACO, and provide a biomarker of airflow limitation, exacerbation, and lung function decline.
Subject terms: Genetics, Systems biology, Biomarkers, Diseases, Medical research, Molecular medicine, Pathogenesis
Introduction
A large proportion (15–30%) of patients with chronic airways disease has features of both asthma and chronic obstructive pulmonary disease (COPD) (Asthma and COPD Overlap, ACO). ACO patients experience more frequent exacerbations, have poorer quality of life, decline in lung function more rapidly, and consume a disproportionate amount of healthcare resources than asthma or COPD alone1,2. Up to date, no universal definition criteria exist3–7, and few studies have investigated the pathogenesis of this overlap syndrome8,9. Only 25% of life-long smokers develop COPD, and asthma exhibit a strong familial connection, suggesting genetic determinants in susceptibility to both COPD and asthma10,11. In the past three decades, extensive research to identify genetic determinants of COPD and asthma has shown that only a few single nucleotide polymorphisms (SNP) are independently and consistently associated with fixed airflow limitation12,13. Epigenetics, which refers to the process of influencing gene expression through other genetic mechanisms without affecting DNA sequences, may accounts for this discrepancy.
DNA methylation occurring at position 5 of the pyrimidine ring of cytosines in the context of the cytosine followed by guanine dinucleotide sequence (CpG) form the basis of epigenetic mechanisms through inhibiting the binding of transcription factors at the promoter regions or influencing transcriptional elongation and alternative splicing at the intragenetic regions. Gene promoter methylation often leads to transcriptional repression of the gene, whereas gene body methylation is frequently associated with high gene expression levels14,15. DNA methylation patterns are not only inheritable but also susceptible to change in response to environmental stimuli, such as smoking and allergens16. Additionally, SNPs in non-coding regions may simultaneously alter both the consensus sequence and its DNA methylation, if they alter or generate CpG dinucleotides17. Recent candidate gene and epigenome-wide association studies (EWAS) have identified several CpG site-specific aberrant DNA methylation changes associated with COPD and asthma individually18–21, but none has been performed in this overlap group22. We hypothesized that gene-specific CpG methylation profiles of peripheral blood mononuclear cells (PBMCs) may contribute to disease susceptibility, severity, and clinical phenotypes in ACO patients, with the goal of identifying novel epigenetic changes related to frequent exacerbation, rapid lung function decline, or severe airflow limitation.
Results
Clinical characteristics of the whole cohort
A total of 75 of the 228 COPD patients were identified as ACO, while the others classified as pure COPD (Table 1). Using the new GOLD 2019 staging system (A–D), ACO group (A: 20.8%, B: 38.9%, C: 13.9%, D: 26.4%) had a greater proportion of patients categorized as C and D (40.3% versus 24.8%, p = 0.018) compared with pure COPD group (A: 32.7%, B: 42.5%, C: 7.2%, D: 17.6%). The numbers of all (1.6 ± 2 versus 0.9 ± 1.3, p = 0.013) and moderate (1 ± 1.5 versus 0.5 ± 0.9, p = 0.009) exacerbations in the past one year were higher in ACO group versus pure COPD group. Among 156 patients who received follow-up 1 year later, the numbers of all (1.6 ± 2 versus 0.9 ± 1.3, p = 0.033) and moderate exacerbation (1 ± 1.39 versus 0.6 ± 0.9, p = 0.032) in the next one year were also higher in the ACO group (n = 55) versus the pure COPD group (n = 101). Stepwise forward multivariate linear regression analysis showed that the presence of ACO (co-efficient 0.519, 95% CI 0.06 to 0.518, p = 0.027) and a higher modified Medical Research Council (mMRC) value (co-efficient 0.355, 95% CI 0.169 to 0.541, p < 0.001) were independent factors associated with total number of exacerbations in the past one year, while the presence of ACO (co-efficient 1.212, 95% CI 0.507 to 1.918, p = 0.001), the use of inhaled corticosteroids (ICS) and long-acting β2 agonist (LABA) combination therapy (co-efficient 1.368, 95% CI 0.498 to 2.237, p = 0.003), and a lower post-bronchodilator (BD) forced expiratory volume in one second (FEV1) %predicted value at visit 2 (co-efficient − 0.026, 95% CI − 0.05 to − 0.003, p = 0.027) were independent factors associated with total number of exacerbations in the next one year.
Table 1.
Baseline characteristic of all study participants with asthma-COPD overlap (ACO) or pure COPD.
| ACO N = 75 | Pure COPD, N = 153 | P value | |
|---|---|---|---|
| Age, years | 69.2 (10.6) | 68.8 (10) | 0.794 |
| Smoking exposure, pack-years | 50.5 (31.9) | 55.7 (36.7) | 0.296 |
| Current smoker | 20 (26.7) | 76 (49.7) | 0.001 |
| Body mass index, Kg/m2 | 24.2 (5.3) | 24.2 (4.3) | 0.968 |
| Charlson co-morbidity index | 2.7 (1.6) | 2.5 (1.6) | 0.233 |
| Atopic disease, n (%) | 50 (66.7) | 54 (35.3) | < 0.001 |
| Asthma | 23 (30.7) | 17 (11.1) | < 0.001 |
| Allergic rhinitis | 40 (53.3) | 50 (32.7) | 0.003 |
| Atopic dermatitis | 4 (5.5) | 5 (3.3) | 0.433 |
| Lung function | |||
| Pre-BD FEV1/FVC, % | 54.2 (10.2) | 57.3 (13.8) | 0.06 |
| Pre-BD FEV1, %predicted | 55 (17.7) | 59.1 (19.8) | 0.124 |
| Pre-BD FEF25-75%, %predicted | 22.7 (11.2) | 27.4 (14.6) | 0.01 |
| Post-BD FEV1/FVC, % | 57.8 (11.1) | 58.8 (13.5) | 0.588 |
| Post-BD FEV1, %predicted | 61.5 (18.4) | 62.1 (19.4) | 0.821 |
| Post-BD FEF25-75%, %predicted | 27.9 (13.1) | 29.4 (15.5) | 0.473 |
| BD responsive | 75 (100) | 31 (22) | < 0.001 |
| Dyspnea score | |||
| mMRC at the first visit | 1.7 (1.1) | 1.6 (1.2) | 0.422 |
| CAT at the first visit | 11.4 (7.8) | 10.3 (7.3) | 0.322 |
| Blood and biochemistry test | |||
| Neutrophil, % | 58.1 (11.8) | 63.3 (13.3) | 0.005 |
| Eosinophil, % | 4.5 (4.3) | 2.7 (3.5) | 0.002 |
| Absolute neutrophil count, μL−1 | 4384 (2048) | 5054 (2654) | 0.063 |
| Absolute eosinophil count, μL−1 | 312 (273) | 213 (492) | 0.106 |
| Total cholesterol | 181.8 (37.4) | 182.5 (40.5) | 0.904 |
| Triglyceride | 115.8 (60.3) | 115.1 (71.1) | 0.947 |
| Uric acid | 7.6 (2.2) | 6.9 (1.7) | 0.018 |
| Glycohemoglobin | 5.9 (0.5) | 6.1 (1) | 0.014 |
| Controller medicines, n (%) | |||
| LAMA | 33 (44) | 68 (44.4) | 0.949 |
| LABA | 34 (45.3) | 65 (42.5) | 0.683 |
| ICS + LABA | 31 (41.3%) | 53 (34.6) | 0.325 |
| Theophylline | 46 (61.3) | 69 (45.1) | 0.021 |
| Exercise endurance test | |||
| Maximum inspiratory pressure, cmH2O | 70.1 (30.5) | 70.5 (27.1) | 0.922 |
| Maximum expiratory pressure, cmH2O | 101.9 (40.1) | 96.2 (35.9) | 0.324 |
| 6 min walking distance, m | 388.5 (103.1) | 368.6 (129.5) | 0.29 |
| 6 min walking distance, %predicted | 81.4 (21.3) | 76.9 (25.6) | 0.341 |
COPD chronic obstructive pulmonary disease, BD bronchodilator, FEV1 forced expiratory volume within first second, FVC forced expiratory vital capacity, FEF forced expiratory flow, LAMA = long acting muscarinic antagonist, LABA long acting β2 agonist, ICS inhaled corticosteroid, mMRC modified Medical Research Council, CAT COPD assessment test.
Whole-genome DNA methylation profiles and enrichment pathway analysis in the discovery cohort
Twelve ACO patients and 6 healthy non-smokers (HS) enrolled in the discovery cohort were matched in terms of age, BMI, and Charlson co-morbidity index (Supplementary Table S1). A total of 21 PBMC samples were grouped and analyzed in two different comparisons. The first comparison (I) was between 12 ACO patients and 6 HS, resulting in 125 hypermethylated differentially methylated loci (DMLs) and 279 hypomethylated DMLs (all p values < 0.0005, all q values < 0.3; Fig. 1A, Table 2). The second comparison (II) was before and after 1-year follow-up in 3 ACO patients with rapid lung function decline, resulting in 2432 hypermethylated DMLs and 4143 hypomethylated DMLs (all p values < 0.005, all q values < 0.3; Fig. 1B, Supplementary Table S2). For the 404 DMLs in comparison I, enrichment in previous EWAS signals was tested by using EWAS toolkit (https://bigd.big.ac.cn/ewas/toolkit)23. The results showed that there is high co-occurrence probability between the 404 probes and smoking, air pollution, aging, atopy, or autoimmune disease-related DNA methylation probes in previous EWAS signals. Furthermore, 19 DMLs in comparison I overlapped with asthma trait-related DNA methylation probes (Supplementary Table S3), but none overlapped with COPD trait-related probes. TNRC6B and MET were hypermethylated in both of our ACO patients and the asthma patients in previous EWAS, while DHX30, SFXN, C19orf28, and CLCN7 were hypomethylated.
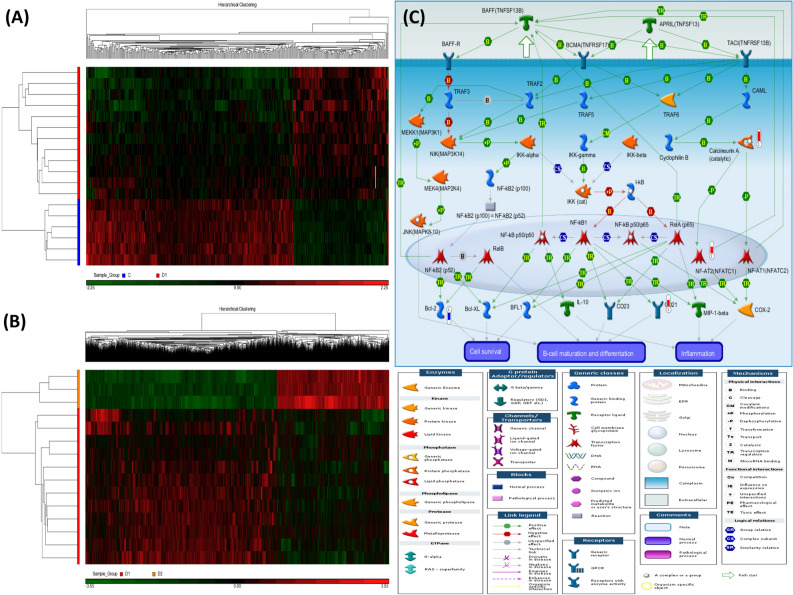
Figure 1.
Heatmaps and a representative enriched pathway of the differentially methylated loci (DML) for the two comparisons of the whole genome microarray experiment in the discovery cohort. Hierarchical clustering of DML in 21 samples classified into two comparisons: (A) ACO patients versus healthy subjects (comparison I). (B) ACO patients with rapid decline in lung function after 1-year follow-up versus at enrollment (comparison II). (C) Apoptosis and survival of APRIL and BAFF signaling pathway enriched in ACO patients (comparison I). The significantly hypermethylated genes were highlighted with a red-colored barometric bar, while hypomethylated genes in a blue-colored bar. The changes represent the differences between the mean β-values of normal and ACO patients. For example, the mean β-value for normal and ACO patients is 1.44 and 1.94, respectively, for calcineurin A (CACNA1C), indicating a higher methylation level (+ 0.5) in ACO. The image was created by the Metacore software.
Table 2.
Top differentially methylated loci in the comparison between patients with asthma and COPD overlap (ACO) and healthy non-smokers (comparison I) in the discovery cohort.
| Column ID | UCSC RefGene Name | UCSC RefGene Accession | UCSC RefGene Group | P value | q value | Mean difference 0f. β value |
|---|---|---|---|---|---|---|
| cg16093065 | TNRC6B | NM_001162501 | 3′UTR | 0.000430942 | 0.174101 | 0.179 |
| cg07674304 | WWOX | NM_016373 | Body | 0.000215471 | 0.0870503 | 0.173 |
| cg14150023 | TNRC6B | NM_001162501 | 3′UTR | 0.000215471 | 0.0870503 | 0.159 |
| cg13828808 | ROR2 | NM_004560 | Body | 0.000430942 | 0.174101 | 0.137 |
| cg19421218 | TIGIT | NM_173799 | TSS200 | 0.000107735 | 0.0435249 | 0.132 |
| cg26601310 | PRR5L | NM_001160167 | 5′UTR | 0.000215471 | 0.0870503 | 0.131 |
| cg14392772 | TRAF1 | NM_005658 | 3′UTR | 0.000430942 | 0.174101 | 0.127 |
| cg24450112 | PDE9A | NM_001001582 | Body | 0.000430942 | 0.174101 | 0.119 |
| cg22027471 | SLC5A4 | NM_014227 | TSS1500 | 0.000430942 | 0.174101 | 0.115 |
| cg05757530 | NLRC5 | NM_032206 | 5′UTR | 0.000430942 | 0.174101 | 0.113 |
| cg07563400 | ADORA2B | NM_000676 | Body | 0.000215471 | 0.0870503 | − 0.116 |
| cg17278447 | NPTX2 | NM_002523 | Body | 0.000430942 | 0.174101 | − 0.116 |
| cg22422264 | USP50 | NM_203494 | 3′UTR | 0.000430942 | 0.174101 | − 0.116 |
| cg13676763 | FAM125B | NM_033446 | Body | 0.000430942 | 0.174101 | − 0.118 |
| cg03707168 | PPP1R15A | NM_014330 | Body | 0.000430942 | 0.174101 | − 0.119 |
| cg13334727 | SEPT8 | NM_001098813 | TSS200 | 0.000215471 | 0.0870503 | − 0.119 |
| cg14459011 | NHEDC2 | NM_178833 | TSS1500 | 0.000107735 | 0.0435249 | − 0.121 |
| cg20861489 | HCRTR2 | NM_001526 | Body | 0.000430942 | 0.174101 | − 0.122 |
| cg01534527 | CTDSPL | NM_005808 | Body | 0.000215471 | 0.0870503 | − 0.123 |
| cg12655112 | EHD4 | NM_139265 | Body | 0.000430942 | 0.174101 | − 0.123 |
| cg07568296 | MAD1L1 | NM_003550 | Body | 0.000430942 | 0.174101 | − 0.123 |
| cg15243578 | PITPNM2 | NM_020845 | 3′UTR | 0.000430942 | 0.174101 | − 0.124 |
| cg08510456 | BSN | NM_003458 | TSS1500 | 0.000430942 | 0.174101 | − 0.125 |
| cg15545247 | GPR109B; | NM_006018 | 1stExon | 0.000430942 | 0.174101 | − 0.125 |
| cg02853948 | HCRTR2 | NM_001526 | Body | 0.000430942 | 0.174101 | − 0.125 |
| cg20705781 | SSH3 | NM_017857 | TSS1500 | 0.000430942 | 0.174101 | − 0.125 |
| cg18200150 | MYO1D | NM_015194 | Body | 0.000107735 | 0.0435249 | − 0.126 |
| cg04658021 | PER1 | NM_002616 | TSS1500 | 0.000430942 | 0.174101 | − 0.127 |
| cg01376079 | SSH3 | NM_017857 | TSS1500 | 0.000215471 | 0.0870503 | − 0.127 |
| cg07987148 | TP53RK | NM_033550 | TSS1500 | 0.000430942 | 0.174101 | − 0.127 |
| cg18688704 | PDGFC | NM_016205 | Body | 0.000430942 | 0.174101 | − 0.129 |
| cg07180646 | TMEM51 | NM_001136218 | 5′UTR | 0.000430942 | 0.174101 | − 0.13 |
| cg22331200 | MPO | NM_000250 | Body | 0.000430942 | 0.174101 | − 0.134 |
| cg06487194 | JARID2 | NM_004973 | Body | 0.000430942 | 0.174101 | − 0.135 |
| cg22499893 | SFRS13A | NM_054016 | TSS1500 | 0.000215471 | 0.0870503 | − 0.135 |
| cg01394781 | ABCC1 | NM_019862 | Body | 0.000430942 | 0.174101 | − 0.137 |
| cg12401918 | NOTCH4 | NM_022107 | TSS1500 | 0.000430942 | 0.174101 | − 0.137 |
| cg06815976 | NOTCH4 | NM_004557 | Body | 0.000430942 | 0.174101 | − 0.137 |
| cg15529344 | ANKRD58 | NM_001105576 | TSS1500 | 0.000430942 | 0.174101 | − 0.138 |
| cg26337070 | ATOH8 | NM_032827 | Body | 0.000430942 | 0.174101 | − 0.143 |
| cg24892069 | NRP1 | NM_001024628 | Body | 0.000215471 | 0.0870503 | − 0.146 |
| cg02607972 | ASXL2 | NM_018263 | 3′UTR | 0.000430942 | 0.174101 | − 0.147 |
| cg01836137 | INF2; | NM_022489 | 5′UTR | 0.000430942 | 0.174101 | − 0.152 |
| cg11615395 | MAML3 | NM_018717 | Body | 0.000107735 | 0.0435249 | − 0.152 |
| cg05655915 | NARF | NM_012336 | TSS1500; | 0.000430942 | 0.174101 | − 0.152 |
| cg00813999 | CYSLTR1 | NM_006639 | 1stExon | 0.000430942 | 0.174101 | − 0.153 |
| cg19351604 | ARHGEF10 | NM_014629 | Body | 0.000430942 | 0.174101 | − 0.154 |
| cg05413628 | CLCN7 | NM_001287 | Body | 0.000430942 | 0.174101 | − 0.157 |
| cg01870865 | TREX1 | NM_016381 | TSS200 | 0.000215471 | 0.0870503 | − 0.159 |
| cg25918947 | TMEM106A | NM_145041 | Body | 0.000430942 | 0.174101 | − 0.16 |
| cg07375836 | ACACA | NM_198839 | 5′UTR | 0.000430942 | 0.174101 | − 0.165 |
| cg26746309 | ERLIN1 | NM_001100626 | Body | 0.000215471 | 0.0870503 | − 0.165 |
| cg14481208 | RTKN | NM_001015055 | TSS1500 | 0.000430942 | 0.174101 | − 0.165 |
| cg09841842 | FRMD6 | NM_001042481 | 5′UTR | 0.000215471 | 0.0870503 | − 0.166 |
| cg08223235 | BCL2 | NM_000633 | Body | 0.000215471 | 0.0870503 | − 0.167 |
| cg20981848 | BTBD3 | NM_014962 | Body | 0.000215471 | 0.0870503 | − 0.167 |
| cg10718056 | TRIM27 | NM_006510 | Body | 0.000430942 | 0.174101 | − 0.167 |
| cg19628988 | CXXC5 | NM_016463 | 5′UTR | 0.000430942 | 0.174101 | − 0.169 |
| cg21141827 | ETF1 | NM_004730 | 3′UTR | 0.000215471 | 0.0870503 | − 0.179 |
| cg17514528 | MTHFR | NM_005957 | Body | 0.000215471 | 0.0870503 | − 0.184 |
| cg16672562 | HIF3A | NM_022462 | 1st Exon | 0.000430942 | 0.174101 | − 0.226 |
| cg10288111 | IFRD1 | NM_001007245 | TSS1500 | 0.000430942 | 0.174101 | − 0.243 |
| cg11307715 | DENND3 | NM_014957 | Body | 0.000430942 | 0.174101 | − 0.361 |
UTR un-translated region; TSS transcription start site.
The top-ranking pathways enriched in comparison I included apoptosis and survival of APRIL and BAFF signaling (Fig. 1C), immune response of NF-AT signaling in leukocyte interactions (Supplementary Fig. S1), and development role of HDAC and CaMK in control of skeletal myogenesis (Supplementary Table S4). The top-ranking pathways enriched in comparison II included NF-AT signaling, regulation of epithelial-to-mesenchymal transition, and TGF/WNT signaling for cytoskeletal remodeling (Supplementary Table S5).
Differential PDE9A, SEPT8, and ZNF323 gene methylations with respect to the presence of ACO in the validation cohort
The 22 ACO patients, 48 pure COPD patients, and 10 HS enrolled in the validation cohort were matched in terms of age, BMI, and Charlson co-morbidity index (Supplementary Table S6).
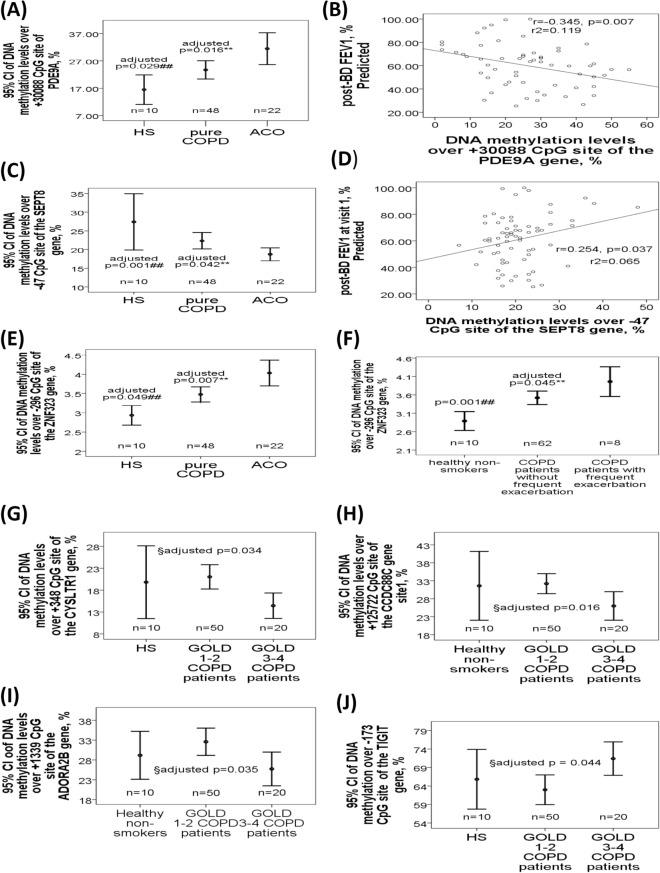
PDE9A gene (+ 30,088, Fig. 2A) was hypermethylated in ACO patients versus pure COPD patients or HS, and negatively correlated with post-BD FEV1%predicted (Fig. 2B). SEPT8 gene (− 47, Fig. 2C) was hypomethylated in ACO patients versus pure COPD patients or HS, and positively correlated with post-BD FEV1%predicted (Fig. 2D). ZNF323 gene (− 296, Fig. 2E) was hypermethylated in ACO patients versus pure COPD patients or HS, and increased in all COPD patients with frequent severe AE versus those without frequent severe AE or HS (Fig. 2F).
Figure 2.
Differential DNA methylation patterns of the PDE9A, SEPT8, ZNF323, CYSLTR1, TIGIT, ADORA2B, and CCDC88C genes at cross sectional levels in the validation cohort. (A) DNA methylation levels of the PDE9A gene body (+ 30,088 CpG site) were increased in ACO patients versus either pure COPD patients or healthy subjects (HS), and (B) negatively correlated with post-BD FEV1%predicted. (C) DNA methylation levels of the SEPT8 gene promoter region (− 47 CpG site) were decreased in ACO patients versus either pure COPD patients or HS, and (D) positively correlated with post-BD FEV1%predicted. (E) DNA methylation levels of the ZNF323 gene promoter region (− 296 CpG site) were increased in ACO patients versus either pure COPD patients or HS, and (F) also increased in all the COPD patients with frequent exacerbation versus those without frequent exacerbation or HS. (G) DNA methylation levels of the CYSLTR1 gene promoter region (+ 348 CpG site) were decreased in GOLD III-IV COPD patients versus GOLD I-II COPD patients. (H) DNA methylation levels of the CCDC88C gene body (+ 125,722 CpG site) were decreased in GOLD III-IV COPD patients versus GOLD I-II COPD patients. (I) DNA methylation levels of the ADORA2B gene body (+ 1339 CpG site) were decreased in GOLD III-IV COPD patients versus GOLD I-II COPD patients. (J) DNA methylation levels of the TIGIT gene promoter region (− 172 CpG site) were increased in GOLD III-IV COPD patients versus GOLD I-II COPD patients. **Compared between ACO and pure COPD patients, and adjusted by multivariate linear regression. ##Compared between ACO and healthy non-smokers (HS), and adjusted by multivariate linear regression. §Compared between COPD patients with GOLD I-II and GOLD III-IV, and adjusted by multivariate linear regression.
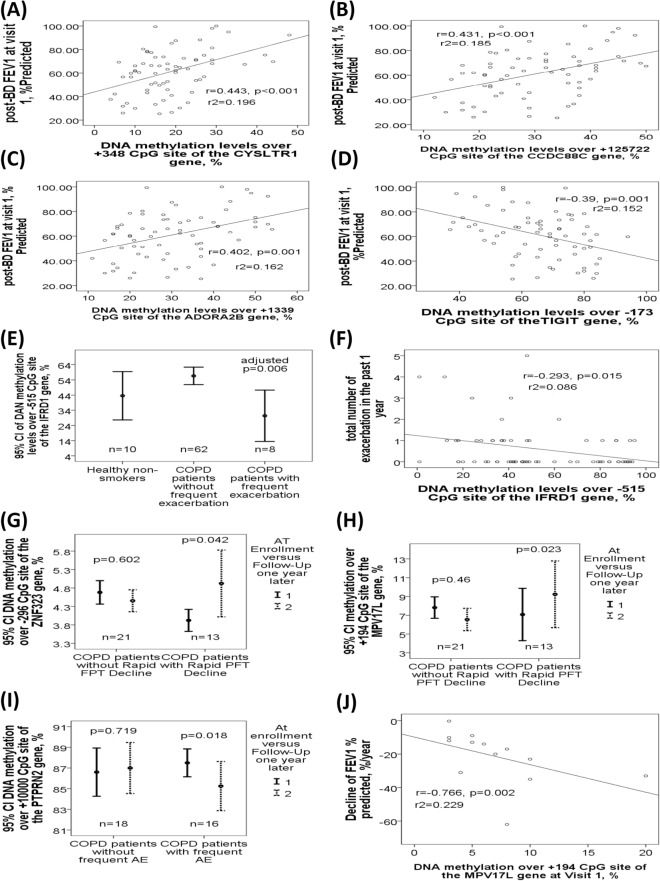
CYSLTR1 (+ 348, Fig. 2G), CCDC88C (+ 125,722, Fig. 2H), and ADORA2B (+ 1339, Fig. 2I) genes were all hypomethylated in COPD patients with severe to very severe airflow limitation (GOLD III-IV) versus those with mild to moderate airflow limitation (GOLD I-II), while TIGIT gene (− 173, Fig. 2J) was hypermethylated. CYSLTR1 (+ 348, Fig. 3A), CCDC88C (+ 125,722, Fig. 3B), and ADORA2B (+ 1339, Fig. 3C) gene methylations were all positively correlated with post-BD FEV1%predicted, while TIGIT gene methylation (− 173, Fig. 3D) was negatively correlated with post-BD FEV1%predicted. IFRD1 gene methylation (− 515, Fig. 3E) was decreased in all COPD patients with frequent severe AE versus those without frequent severe AE, and negatively correlated with exacerbation frequency (Fig. 3F).
Figure 3.
Differential DNA methylation patterns of the IRFD1, TIGIT, CysLTR1, ADORA2B, CCDC88C, ZNF323, and MPV17L, and PTPRN2 genes at cross sectional or longitudinal levels in the validation cohort. (A) DNA methylation levels of the CYSLTR1 gene promoter region (+ 348 CpG site) were positively correlated with post-BD FEV1%predicted value. (B) DNA methylation levels of the CCDC88C gene body (+ 125,722 CpG site) were positively correlated with post-BD FEV1%predicted value. (C) DNA methylation levels of the ADORA2B gene body (+ 1339 CpG site) were positively correlated with post-BD FEV1%predicted value. (D) DNA methylation levels of the TIGIT gene promoter region (− 172 CpG site) were negatively correlated with post-BD FEV1%predicted value. (E) DNA methylation levels of the IFRD1 gene promoter region (− 515 CpG site) were decreased in all the COPD patients with frequent exacerbation versus those without frequent exacerbation, and (F) negatively correlated with the number of exacerbations in the past one year. (G) DNA methylation levels of the ZNF323 gene promoter region (− 296 CpG site) were elevated after 1-year follow-up in COPD patients with rapid lung function decline versus that at enrollment. (H) DNA methylation levels of the MPV17L gene (+ 194 CpG site) were elevated after 1-year follow-up in COPD patients with rapid lung function decline versus that at enrollment. (I) DNA methylation levels of the MPV17L gene (+ 194 CpG site) at visit 1 were negatively correlated with the difference in FEV1% predicted values between visit 2 and visit 1. (J) DNA methylation levels over + 10,015 CpG site of the PTPRN2 gene were reduced after 1-year follow-up in COPD patients with frequent exacerbation versus that at enrollment.
Differential ZNF323, MPV17L, and PTPRN2 gene methylations with respect to rapid lung function decline in the validation cohort
DNA methylation levels over 7 CpG sites of 7 selected genes from comparison II were measured in 5 ACO patients and 8 pure COPD patients with rapid lung function decline after 1-year follow-up (FEV1%predicted 69.78 ± 12.15 versus 60.72 ± 12.18%, mean difference 9.06 ± 7.54%, p = 0.002), as well as in 12 ACO patients and 9 pure COPD patients without rapid lung function decline.
ZNF323 (− 296, Fig. 3G) and MPV17L gene methylations (+ 194, Fig. 3H) were both elevated after 1-year follow-up (visit 2) versus at enrollment (visit 1) in those with rapid lung function decline, but remained the same in those without rapid lung function decline, while MPV17L gene methylation at visit 1 and visit 2 (Fig. 3I) were both negatively correlated with the difference in FEV1% predicted values between visit 2 and visit 1. PTPRN2 gene methylation (+ 10,000, Fig. 3J) were reduced after 1-year follow-up in 16 patients with frequent moderate to severe AE, and remained the same in those without frequent moderate to severe exacerbation.
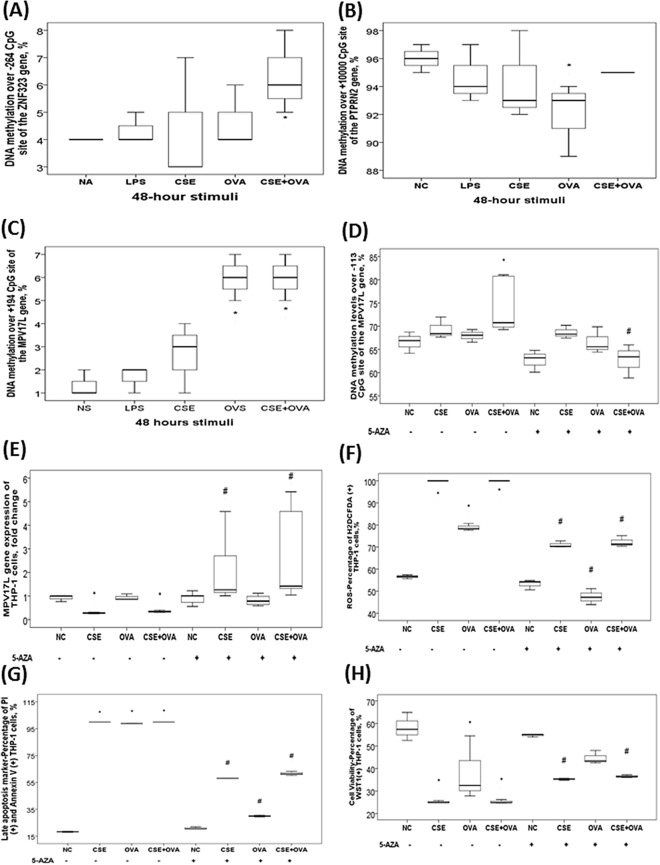
Effects of in vitro concurrent cigarette smoke extract (CSE) and ovalbumin (OVA) stimuli on DNA methylation levels or gene expressions of the 10 candidate genes
ZNF323 gene methylation (− 264) was increased in response to CSE plus OVA treatment (p < 0.05, Fig. 4A). PTPRN2 gene methylation (+ 10,000) was decreased with OVA stimuli (p < 0.05, Fig. 4B). MPV17L gene (+ 194) methylation was increased in response to CSE plus OVA treatment (p values < 0.05, Fig. 4C). Pre-treatment with de-methylation agent (5-aza-2ʹ-deoxycytidine; 5-aza) in the presence of CSE plus OVA stimuli resulted in decreased methylation over − 113 CpG site of the MPV17L gene, increased MPV17L gene expression, reduced reactive oxygen species production, reduced late apoptosis, and increased cell viability, as compared with CSE plus OVA treatment alone (all p values < 0.05, Fig. 4D–H), whereas DNA methylation levels of the other 8 candidate genes were not altered despite increased gene expressions (Supplementary Fig. S2).
Figure 4.
Aberrant DNA methylation and corresponding gene expression changes of the candidate genes in THP-1 cells in response to in vitro cigarette smoke extract (CSE) plus ovalbumin (OVA) allergen stimuli. (A) DNA methylation level of the ZNF323 gene (-264) was increased in response to CSE plus OVA treatment. (B) DNA methylation level of the PTPRN2 gene (+ 10,000) was decreased with OVA stimuli. (C) DNA methylation levels over + 194 CpG site of the MPV17L gene were increased in response to OVA alone or CSE plus OVA concurrent treatment. Pre-treatment with de-methylation agent (5-AZA) resulted in (D) decreased DNA methylation levels over -113 CpG site of the MPV17L gene, (E) increased MPV17L gene expression, (F) reduced reactive oxygen species (ROS) production (percentage of H2DCFDA positive cells), (G) reduced late apoptosis (percentage of Annexin V and PI double positive cells), and (H) increased cell viability (percentage of WST-1 positive cells), as compared with that of CSE, OVA, or CSE plus OVA treatment alone. *p < 0.05 compared between normal control (NC; culture medium) and specific stimuli by Kruskal Wallis H-test. #p < 0.05 compared between the comparative groups with and without 5-AZA supplement by Kruskal Wallis H-test.
Discussion
T helper type 2 immune gene signals associated with greater BD reversibility, eosinophilia, and better response to inhaled corticosteroids, have been identified in ACO patients8,24. Although one report is available on sputum DNA methylation changes of the PCDH20 and SULF2 genes in relation to ACO, and one genome-wide association study has identified SNPs in the CSMD1, SOX5 and GPR65 genes for ACO25,26, this is the first study to perform a whole genome DNA methylation analysis in ACO with replication of the principal findings, and identify a specific association of ACO and several clinical phenotypes with aberrant DNA methylation patterns.
Aberrant methylation patterns of the PDE9A, SEPT8, and ZNF323 genes showed the most significant associations with ACO. PDE9A is the most selective for cGMP degradation, and its inhibitor can enhance memory function through promoting synaptic plasticity and counter pathological remodeling of the heart27–30. Given that PDE-4 is responsible for metabolizing adenosine 3′,5′-cyclic monophosphate that reduces the activation of a wide range of inflammatory cells including eosinophils, targeting PDE9A signaling pathway may be a novel strategy for managing ACO31. SEPT8 contributes to kidney and liver fibrosis, and modulates the generation of toxic amyloid-beta peptides in Alzheimer's disease32,33. Given that SEPT8 functions in various biological processes of cell cytokinesis and migration, it may be another novel target for ACO34. Interestingly, using the BiosQTL database (https://genenetwork.nl/biosqtlbrowser/), there is evidence that the effect of the SEPT8 CpG site (cg13334727) methylation on whole blood may be genetically regulated by the cis-methylation quantitative trait loci (meQTL), rs39855. This SNP is associated with several respiratory phenotypes in the United Kingdom Biobank (UKBB) such as asthma, hay fever or allergic rhinitis, or it is in linkage disequilibrium with the lead variants associated with asthma and respiratory diseases based on the information from the OpenTarget Genetics database (https://genetics.opentargets.org/variant). Our in vitro experiments showed increased gene expressions but no significant changes in the methylation levels for 8 of the 11 selected genes, suggesting that the altered methylation patterns in patients may be inheriting epigenotypes rather than changes after cigarette smoke exposures. Several SNPs of the ZNF323 (ZSCAN31) gene are associated with lung function in asthmatic patients35,36, while its hypermethylation was noted both in ACO patients and in response to CSE plus OVA stimuli, suggesting a role of acquired ZNF323 hypermethylation in the pathogenesis of ACO.
Aberrant methylation patterns of the TIGIT, CYSLTR1, CCDC88C, and ADORA2B genes were associated with severe airflow limitation. TIGIT can enhance Th2 response37, and shows aberrant methylation in allergic asthma38. Interestingly, the probe (cg19421218) annotated to TIGIT is described as an expression quantitative trait methylation (eQTM) for the same gene in the BiosQTL database. CysLTR1 mediates bronchoconstriction and eosinophil migration in asthma39,40. CCDC88C genetic mutation is implicated in eosinophilia-associated myeloid/lymphoid neoplasms41. ADORA2B contributes to pulmonary fibrosis and pulmonary hypertension associated with COPD42–44. Based on the information from the BiosQTL and OpenTarget Genetics databases, DNA methylation of the ADORA2B CpG site (cg07563400) may be genetically regulated by two Cis-meQTL, rs3925260 and rs12452624, which have been linked to lung function parameters and asthma, respectively. Our results suggest that these epigenotypes related to allergic responses may be important determinates of lung function in early life and predispose individuals to COPD.
Aberrant methylation patterns of the MPV17L, ZNF323, and PTPRN2 genes associated with rapid lung function decline were identified, while altered methylations of the ZNF323 and IFRD1 with frequent exacerbation. MPV17L protects against mitochondrial oxidative stress and apoptosis by activation of Omi/HtrA2 protease, and its methylation may act as a biomarker for the prognosis of lung adenocarcinoma45–48. Our in vitro experiments showed that de-methylation agent could reverse promoter hypermethylation-mediated under-expression of the MPV17L gene and oxidative stress-mediated cell apoptosis in response to concurrent CSE and OVA stimuli, supporting the use of this epigenetic mark as potential therapeutic targets of ACO. Aberrant PTPRN2 gene methylation has been identified in smoking-related COPD patients in two previous EWAS49,50. IFRD1 regulates the pathogenesis of asthma and cystic fibrosis through mediating neutrophil function51,52.
There are several limitations in the present study. First, the sample size of the discovery cohort is relatively small and the analyses of both the DMLs and enriched pathways would have not met a more stringent threshold for significance of false discovery rate < 0.5 or 0.1. Three fifths of the results in the comparison I would not remain significant after applying a more stringent threshold of q-value of 0.1 to control the rate of false positives, but all the results in the comparison II remain significant. There would be 162 DMLs with 118 hypomethylated and 44 hypermethylated in the comparison I, if q value is less than 0.1. However, we used a 2-tiered approach to verify some of the findings. Moreover, testing for differentially methylated regions where the cumulative effect of probes included in the same genomic region will be conducted in the future. Second, the cause and effect relationship could not be straightforward determined in this association study. CSE and OVA co-exposure resulted in under-expression of the MPV17L gene and hypermethylation of its promoter region, which were in accordance with the findings in the clinical samples and fit the scientific consensus of an anti-correlation between promoter methylation and gene expression. Third, peripheral blood mononuclear cells are comprised of a mixture of cell types, which may contribute to different methylation changes. Single-cell multi-omic strategies will enhance the specificity and sensitivity of the analysis of DNA methylation patterns over both CpG and non-CpG sites, and site-specific methylome editing will serve as a key technique for the study of 5-methylcytosine function, although both were still un-mature 5 years ago when the current study started. The percentage of monocytes, T cells and B cells in the PBMC samples was not determined, so we could not make corrections for cell-type composition in the best way53. However, the results open the possibility of using de-methylation in the treatment of ACO.
Conclusions
We reported a novel association of ACO in adults of Asian origin with aberrant DNA methylation in the promoter or body regions of the PDE9A, SEPT8, and ZNF323 genes. The findings extend reports linking hypomethylated CYSLTR1/ADORA2B/CCDC88C and hypermethylated TIGIT with more severe fixed airflow limitation in COPD patients, identifying hypomethylated IFRD1/hypermethylated ZNF323 as biomarkers of frequent exacerbation, and providing direct evidence that perturbation of MPV17L signaling through epigenetic programming may play a role in the mediation of both inflammatory and allergic responses in ACO. Our findings provide a new direction for this disease and might establish novel biological insights into the development and effective treatment of ACO.
Materials and methods
This study was approved by the Institutional Review Board of Chang Gung Memorial Hospital, Taiwan (certificate number: 103-3366B). The study participants were recruited from the pulmonary clinics of Kaohsiung Chang Gung Memorial Hospital from October 2014 to July 2017. All the participants were Taiwanese Han people in ancestry. Written informed consent was obtained from each subject participating in the study. The enrollment and exclusion criteria for COPD, and the definition of acute exacerbation (AE) were in accordance with GOLD guideline (Supplementary-Appendix 1 Text). ACO was defined by the presence of three elements: (I) COPD diagnosis, (II) positive bronchodilator (BD) test, and (III) blood eosinophil > 3%, or history of atopic diseases, including asthma, allergic rhinitis, or atopic dermatitis. A total of 364 subjects were screened, and 228 COPD patients were enrolled for final analysis. The discovery cohort used for the EWAS microarray experiment included 12 ACO patients and 6 healthy non-smokers (HS) with normal lung function. The non-overlapping validation cohort included 22 ACO patients, 48 patients with pure COPD, and 10 HS. The experiment, enrolment and exclusion criteria for COPD, and the definition of acute exacerbation (AE) were in accordance with GOLD guideline.
DNA methylation measurement and analysis
Genome-wide DNA methylation profiles were measured by Infinium HumanMethylation 450 BeadChip v1.2 microarray method (San Diego, CA, USA). We filter out 485 CpG sites that have bead count smaller than 3 in 5% of total samples and filter out 901 CpG sites that have detection p-value greater than 0.01 in 5% of the total samples with R package wateRmelon54. We then transfer the methylation β value into M values55 which has better statistical properties for latter non-parametric statistical analysis. For those differentially methylated CpG sites, their corresponding gene symbols were used for pathway analysis and interaction networks contruction by MetaCore software (Thomson Reuters Incorporation, Philadelphia, USA). The significance threshold was a p < 0.0005 and a false discovery rate (q) < 0.3. All methylation datasets have been deposited in the NCBI Gene Expression Omnibus with the accession number GSE118468. Significantly differentially methylated CpG sites with at least a 10% difference in their β value (large effect size) and known biological or functional relevance were selected for further verification and validation by pyrosequencing method using PyroMark Q24 1.010 (Qiagen)56 (Supplementary Appendix 1 Text).
In vitro human monocytic THP-1 cell culture under the stimuli with cigarette CSE and OVA allergen
THP-1 immortalized monocyte-like cell lines are derived from the peripheral blood of a childhood case of acute monocytic leukemia (M5 subtype) and represent valuable tools for investigating circulatory monocytes, which are one of the main sources of inflammatory cytokines in response to allergens and smoking exposures57. DNA Methyl-Transferase (DNMT) 3A and 3B mediate de novo deposition of C5-methylcytosine to establish methylation marks in CpG sites, while 5-aza is a chemical nucleoside analog of cytidine, which can incorporate into DNA, and trap DNMTs through an irreversible covalent interaction58. Thus, THP-1 and 5-aza were adopted in the in vitro experiments. THP-1 were treated with normal medium, 100 ng/ml lipopolysaccharide (LPS), 2.5% CSE, 25 ug OVA, or CSE (2.5%) plus OVA (25 ug) mix for 48 h, and also treated with 1uM 5-aza (Sigma-Aldrich Corp) in advance for 24 h. Gene expressions were measured by quantitative real-time reverse transcription-PCR method with Taqman probe and specific primers (Supplementary Table S7). Relative expression levels were calculated using the ∆∆Ct method. (Supplementary Appendix 1 Text).
Statistical analysis
Data were expressed as the mean ± standard deviation. One-way analysis of variance were be used for comparing mean values of more than two experimental groups. Categorical variables were analyzed using Chi-square test. Stepwise multivariate linear regression analysis was used to adjust for confounding factors and obtain adjusted p values. The differences of continuous variables between study enrollment and follow-up after one year were analyzed by paired t-test. Pearson’s correlation was used to determine the relationship between selected variables. A p-value of less than 0.05 is considered statistically significant.
Ethics approval and consent to participate
This study was approved by the Institutional Review Board of Chang Gung Memorial Hospital, Taiwan (certificate number: 103-3366B). Written informed consent was obtained from each subject participating in the study.
Supplementary Information
Acknowledgements
The authors acknowledge the technical support provided by the Genomic and Proteomic Core Laboratory, and the Internal Medicine Core Facility of the Kaohsiung Chang Gung Memorial Hospital. We also acknowledge the support of bioinformatics analysis from Professor Petrus Tang, PhD (Molecular Medicine Research Center, and Bioinformatics Center of Chang Gung University, Taiwan), and appreciate the Biostatistics Center, Kaohsiung Chang Gung Memorial Hospital, for statistics work.
Abbreviations
- ACO
Asthma COPD overlap
- ADORA2B
Adenosine A2b receptor
- AE
Acute exacerbation
- 5-aza
5-Aza-2′-deoxycytidine
- BD
Bronchodilator
- BMI
Body mass index
- CCDC88C
Coiled-coil domain containing 88C
- COPD
Chronic obstructive pulmonary disease
- CpG
Cytosine guanine dinucleotides
- CSE
Cigarette smoke extract
- CYSLTR1
Cysteinyl leukotriene receptor 1
- DML
Differentially methylated locus
- EWAS
Epigenome-wide association study
- FEV1
Forced expiratory volume in the first second
- FVC
Forced expiratory vital capacity
- HS
Healthy non-smoker
- IFRD1
Interferon related developmental regulator 1
- MPV17L
MPV17 mitochondrial inner membrane protein like
- OVA
Ovalbumin
- PCR
Polymerase chain reaction
- PDE9A
Phosphodiesterase 9A
- PTPRN2
Protein tyrosine phosphatase receptor type N2
- SEPT8
Septin 8
- SNP
Single nucleotide polymorphism
- TIGIT
T cell immunoreceptor with Ig and ITIM domains
- ZNF323
Zinc finger and SCAN domain containing 31
Author contributions
T.W.C. performed the analysis of the microarray data. S.F.L., C.C.W., W.F.F., and T.Y.C. analyzed and interpreted the patient data regarding the COPD. H.C.C., C.C.T., Y.P.C., Y.F.W., and C.C.Wang. assisted with data collection. C.P.L. and P.Y.H. performed the pyrosequencing and quantitative RT-PCR measurements of the blood and cell line samples. Y.C.C., M.C.L. and Y.H.T. contributed to the conceptualization and supervision of this study. Y.C.C. was a major contributor in writing the manuscript and the guarantor of the paper, taking responsibility for the integrity of the work as a whole. All authors read and approved the final manuscript.
Funding
This work was supported by grants from the Ministry of Science and Technology, Taiwan (101-2325-B-002-064/102-2325-B-002-087/103-2325-B-002-027/104-2325-B-002-035/105-2325-B-002-030/105-2314-B-182A-092-MY3 to M.C. Lin) and from Chang Gung Memorial Hospital, Taiwan (CMRPG8D1572/CMRPG8F1641/CMRPG8I0151/CMRPG8F1321/CMRPG8I0152 to Y.C. Chen). The funding body has no role in the design of the study and collection, analysis, and interpretation of data, or in writing the manuscript.
Data availability
All methylation datasets have been deposited in the NCBI Gene Expression Omnibus with the accession number GSE118468.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
A comprehensive list of consortium members appears at the end of the paper.
Contributor Information
Yung-Che Chen, Email: yungchechen@yahoo.com.tw.
Meng-Chih Lin, Email: linmengchih@hotmail.com.
Taiwan Clinical Trial Consortium of Respiratory Disease (TCORE) group:
Chong-Jen Yu, Hao-Chien Wang, Chi-Huei Chiang, Diahn-Warng Perng, Shih-Lung Cheng, Jeng-Yuan Hsu, Wu-Huei Hsu, Tzuen-Ren Hsiue, Hen-I. Lin, Cheng-Yi Wang, Yeun-Chung Chang, Chung-Ming Chen, Cing-Syong Lin, Likwang Chen, and Inn-Wen Chong
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-83185-1.
References
- 1.Alshabanat A, Zafari Z, Albanyan O, Dairi M, FitzGerald JM. Asthma and COPD overlap syndrome (ACOS): A systematic review and meta analysis. PLoS ONE. 2015;10:e0136065. doi: 10.1371/journal.pone.0136065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Marco R, et al. Asthma, COPD and overlap syndrome: A longitudinal study in young European adults. Eur. Respir. J. 2015;46:671–679. doi: 10.1183/09031936.00008615. [DOI] [PubMed] [Google Scholar]
- 3.Bateman ED, Reddel HK, van Zyl-Smit RN, Agusti A. The asthma-COPD overlap syndrome: Towards a revised taxonomy of chronic airways diseases? The Lancet. 2015;3:719–728. doi: 10.1016/S2213-2600(15)00254-4. [DOI] [PubMed] [Google Scholar]
- 4.Cosio BG, et al. Defining the asthma-COPD overlap syndrome in a COPD cohort. Chest. 2016;149:45–52. doi: 10.1378/chest.15-1055. [DOI] [PubMed] [Google Scholar]
- 5.Bonten TN, et al. Defining asthma-COPD overlap syndrome: a population-based study. Eur. Respir. J. 2017 doi: 10.1183/13993003.02008-2016. [DOI] [PubMed] [Google Scholar]
- 6.Montes de Oca M, et al. Asthma-COPD overlap syndrome (ACOS) in primary care of four Latin America countries: The PUMA study. BMC Pulmon. Med. 2017;17:69. doi: 10.1186/s12890-017-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plaza V, et al. Consensus on the asthma-COPD overlap syndrome (ACOS) between the Spanish COPD Guidelines (GesEPOC) and the Spanish Guidelines on the Management of Asthma (GEMA) Arch. Bronconeumol. 2017 doi: 10.1016/j.arbres.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 8.Cosio BG, et al. Th-2 signature in chronic airway diseases: towards the extinction of asthma-COPD overlap syndrome? Eur. Respir. J. 2017 doi: 10.1183/13993003.02397-2016. [DOI] [PubMed] [Google Scholar]
- 9.Vaz Fragoso CA, Murphy TE, Agogo GO, Allore HG, McAvay GJ. Asthma-COPD overlap syndrome in the US: A prospective population-based analysis of patient-reported outcomes and health care utilization. Int. J. Chron. Obstruct. Pulmon. Dis. 2017;12:517–527. doi: 10.2147/COPD.S121223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lokke A, Lange P, Scharling H, Fabricius P, Vestbo J. Developing COPD: A 25 year follow up study of the general population. Thorax. 2006;61:935–939. doi: 10.1136/thx.2006.062802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mathias RA. Introduction to genetics and genomics in asthma: genetics of asthma. Adv. Exp. Med. Biol. 2014;795:125–155. doi: 10.1007/978-1-4614-8603-9_9. [DOI] [PubMed] [Google Scholar]
- 12.Resendiz-Hernandez JM, Falfan-Valencia R. Genetic polymorphisms and their involvement in the regulation of the inflammatory response in asthma and COPD. Adv. Clin. Exp. Med. 2018;27:125–133. doi: 10.17219/acem/65691. [DOI] [PubMed] [Google Scholar]
- 13.Hall R, Hall IP, Sayers I. Genetic risk factors for the development of pulmonary disease identified by genome-wide association. Respirology. 2019;24:204–214. doi: 10.1111/resp.13436. [DOI] [PubMed] [Google Scholar]
- 14.Lev Maor G, Yearim A, Ast G. The alternative role of DNA methylation in splicing regulation. Trends Genet. 2015;31:274–280. doi: 10.1016/j.tig.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 15.Maunakea AK, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dick KJ, et al. DNA methylation and body-mass index: A genome-wide analysis. Lancet. 2014;383:1990–1998. doi: 10.1016/S0140-6736(13)62674-4. [DOI] [PubMed] [Google Scholar]
- 17.Wang H, Lou D, Wang Z. Crosstalk of genetic variants, allele-specific DNA methylation, and environmental factors for complex disease risk. Front. Genet. 2018;9:695. doi: 10.3389/fgene.2018.00695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edris A, den Dekker HT, Melen E, Lahousse L. Epigenome-wide association studies in asthma: A systematic review. Clin. Exp. Allergy. 2019;49:953–968. doi: 10.1111/cea.13403. [DOI] [PubMed] [Google Scholar]
- 19.Wan ES, et al. Smoking-associated site-specific differential methylation in buccal mucosa in the COPDGene study. Am. J. Respir. Cell Mol. Biol. 2015;53:246–254. doi: 10.1165/rcmb.2014-0103OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vucic EA, et al. DNA methylation is globally disrupted and associated with expression changes in chronic obstructive pulmonary disease small airways. Am. J. Respir. Cell Mol. Biol. 2014;50:912–922. doi: 10.1165/rcmb.2013-0304OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qiu W, et al. Variable DNA methylation is associated with chronic obstructive pulmonary disease and lung function. Am. J. Respir. Crit. Care Med. 2012;185:373–381. doi: 10.1164/rccm.201108-1382OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao X, Jia M, Zhang Y, Breitling LP, Brenner H. DNA methylation changes of whole blood cells in response to active smoking exposure in adults: A systematic review of DNA methylation studies. Clin. Epigenet. 2015;7:113. doi: 10.1186/s13148-015-0148-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li M, et al. EWAS Atlas: A curated knowledgebase of epigenome-wide association studies. Nucleic Acids Res. 2019;47:D983–D988. doi: 10.1093/nar/gky1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Christenson SA, et al. Asthma-COPD overlap. Clinical relevance of genomic signatures of type 2 inflammation in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2015;191:758–766. doi: 10.1164/rccm.201408-1458OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sood A, et al. Methylated genes in sputum among older smokers with asthma. Chest. 2012;142:425–431. doi: 10.1378/chest.11-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hardin M, et al. The clinical and genetic features of COPD-asthma overlap syndrome. Eur. Respir. J. 2014;44:341–350. doi: 10.1183/09031936.00216013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dunkerly-Eyring B, Kass DA. Myocardial phosphodiesterases and their role in cGMP regulation. J. Cardiovasc. Pharmacol. 2020;75:483–493. doi: 10.1097/FJC.0000000000000773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bray N. Cardiovascular disease: PDE9A inhibition mends broken hearts. Nat. Rev. Drug Discov. 2015;14:310. doi: 10.1038/nrd4618. [DOI] [PubMed] [Google Scholar]
- 29.Wang PX, et al. C33(S), a novel PDE9A inhibitor, protects against rat cardiac hypertrophy through upregulating cGMP signaling. Acta Pharmacol. Sin. 2017;38:1257–1268. doi: 10.1038/aps.2017.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosenbrock H, et al. The novel phosphodiesterase 9A inhibitor BI 409306 increases cyclic guanosine monophosphate levels in the brain, promotes synaptic plasticity, and enhances memory function in rodents. J. Pharmacol. Exp. Ther. 2019;371:633–641. doi: 10.1124/jpet.119.260059. [DOI] [PubMed] [Google Scholar]
- 31.Kwak HJ, et al. Discovery of a novel orally active PDE-4 inhibitor effective in an ovalbumin-induced asthma murine model. Eur J Pharmacol. 2012;685:141–148. doi: 10.1016/j.ejphar.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 32.Neubauer K, Neubauer B, Seidl M, Zieger B. Characterization of septin expression in normal and fibrotic kidneys. Cytoskeleton (Hoboken) 2019;76:143–153. doi: 10.1002/cm.21473. [DOI] [PubMed] [Google Scholar]
- 33.Kurkinen KM, et al. SEPT8 modulates beta-amyloidogenic processing of APP by affecting the sorting and accumulation of BACE1. J. Cell. Sci. 2016;129:2224–2238. doi: 10.1242/jcs.185215. [DOI] [PubMed] [Google Scholar]
- 34.Akhmetova KA, Chesnokov IN, Fedorova SA. Functional characterization of septin complexes. Mol. Biol. (Mosk) 2018;52:155–171. doi: 10.7868/S0026898418020015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li X, et al. Genome-wide association study identifies TH1 pathway genes associated with lung function in asthmatic patients. J. Allergy Clin. Immunol. 2013;132:313–320. doi: 10.1016/j.jaci.2013.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo XJ, et al. Systematic integration of brain eQTL and GWAS identifies ZNF323 as a novel schizophrenia risk gene and suggests recent positive selection based on compensatory advantage on pulmonary function. Schizophr. Bull. 2015;41:1294–1308. doi: 10.1093/schbul/sbv017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kourepini E, et al. TIGIT enhances antigen-specific Th2 recall responses and allergic disease. J. Immunol. 2016;196:3570–3580. doi: 10.4049/jimmunol.1501591. [DOI] [PubMed] [Google Scholar]
- 38.Yang IV, et al. DNA methylation and childhood asthma in the inner city. J. Allergy Clin. Immunol. 2015;136:69–80. doi: 10.1016/j.jaci.2015.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pniewska E, et al. Exacerbating factors induce different gene expression profiles in peripheral blood mononuclear cells from asthmatics, patients with chronic obstructive pulmonary disease and healthy subjects. Int. Arch. Allergy Immunol. 2014;165:229–243. doi: 10.1159/000370067. [DOI] [PubMed] [Google Scholar]
- 40.Kim SH, et al. Differential contribution of the CysLTR1 gene in patients with aspirin hypersensitivity. J. Clin. Immunol. 2007;27:613–619. doi: 10.1007/s10875-007-9115-x. [DOI] [PubMed] [Google Scholar]
- 41.Gosenca D, et al. Identification and functional characterization of imatinib-sensitive DTD1-PDGFRB and CCDC88C-PDGFRB fusion genes in eosinophilia-associated myeloid/lymphoid neoplasms. Genes Chromosom. Cancer. 2014;53:411–421. doi: 10.1002/gcc.22153. [DOI] [PubMed] [Google Scholar]
- 42.Dammen R, et al. The stimulatory adenosine receptor ADORA2B regulates serotonin (5-HT) synthesis and release in oxygen-depleted EC cells in inflammatory bowel disease. PLoS ONE. 2013;8:e62607. doi: 10.1371/journal.pone.0062607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Philip K, et al. HIF1A up-regulates the ADORA2B receptor on alternatively activated macrophages and contributes to pulmonary fibrosis. FASEB J. 2017;31:4745–4758. doi: 10.1096/fj.201700219R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karmouty-Quintana H, et al. Adenosine A2B receptor and hyaluronan modulate pulmonary hypertension associated with chronic obstructive pulmonary disease. Am. J. Respir. Cell Mol. Biol. 2013;49:1038–1047. doi: 10.1165/rcmb.2013-0089OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li R, et al. Methylation and transcriptome analysis reveal lung adenocarcinoma-specific diagnostic biomarkers. J. Transl. Med. 2019;17:324. doi: 10.1186/s12967-019-2068-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iida R, Ueki M, Yasuda T. Identification of interacting partners of Human Mpv17-like protein with a mitigating effect of mitochondrial dysfunction through mtDNA damage. Free Radic. Biol. Med. 2015;87:336–345. doi: 10.1016/j.freeradbiomed.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 47.Krick S, et al. Mpv17l protects against mitochondrial oxidative stress and apoptosis by activation of Omi/HtrA2 protease. Proc. Natl. Acad. Sci. U.S.A. 2008;105:14106–14111. doi: 10.1073/pnas.0801146105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iida R, Ueki M, Yasuda T. Knockout of Mpv17-Like Protein (M-LPH) gene in human hepatoma cells results in impairment of mtDNA integrity through reduction of TFAM, OGG1, and LIG3 at the protein levels. Oxid. Med. Cell Longev. 2018;2018:6956414. doi: 10.1155/2018/6956414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wielscher M, et al. Diagnostic performance of plasma DNA methylation profiles in lung cancer, pulmonary fibrosis and COPD. EBioMedicine. 2015;2:929–936. doi: 10.1016/j.ebiom.2015.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alkhaled Y, et al. Impact of cigarette-smoking on sperm DNA methylation and its effect on sperm parameters. Andrologia. 2018 doi: 10.1111/and.12950. [DOI] [PubMed] [Google Scholar]
- 51.Ehrnhoefer DE. IFRD1 modulates disease severity in cystic fibrosis through the regulation of neutrophil effector function. Clin. Genet. 2009;76:148–149. doi: 10.1111/j.1399-0004.2009.01247_2.x. [DOI] [PubMed] [Google Scholar]
- 52.Guan Y, et al. Uncovering potential key genes associated with the pathogenesis of asthma: A microarray analysis of asthma-relevant tissues. Allergol. Immunopathol. 2017;45:152–159. doi: 10.1016/j.aller.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 53.Teschendorff AE, Zheng SC. Cell-type deconvolution in epigenome-wide association studies: A review and recommendations. Epigenomics. 2017;9:757–768. doi: 10.2217/epi-2016-0153. [DOI] [PubMed] [Google Scholar]
- 54.Pidsley R, et al. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genom. 2013;14:293. doi: 10.1186/1471-2164-14-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Du P, et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinform. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Michels KB, et al. Recommendations for the design and analysis of epigenome-wide association studies. Nat. Methods. 2013;10:949–955. doi: 10.1038/nmeth.2632. [DOI] [PubMed] [Google Scholar]
- 57.Bosshart H, Heinzelmann M. THP-1 cells as a model for human monocytes. Ann. Transl. Med. 2016;4:438. doi: 10.21037/atm.2016.08.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mehdipour P, Murphy T, De Carvalho DD. The role of DNA-demethylating agents in cancer therapy. Pharmacol. Ther. 2020;205:107416. doi: 10.1016/j.pharmthera.2019.107416. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All methylation datasets have been deposited in the NCBI Gene Expression Omnibus with the accession number GSE118468.