Abstract
The recent increase of reported cyclosporiasis outbreaks associated with fresh produce has highlighted the need for understanding environmental transmission of Cyclospora cayetanensis in agricultural settings and facilities. Conducting such environmental investigations necessitates robust sample collection and analytical methods to detect C. cayetanensis in water samples. This study evaluated three sample collection methods for recovery of C. cayetanensis oocysts from water samples during seeded recovery experiments. Two filtration-based methods, dead-end ultrafiltration (DEUF) and USEPA Method 1623.1, were evaluated for oocyst recovery from irrigation water. A non-filter-based method, continuous flow centrifugation (CFC), was evaluated separately for recovery from creek water and spent produce wash water. Median C. cayetanensis recovery efficiencies were 17% for DEUF and 16–22% for Method 1623.1. The DEUF method proved to be more robust than Method 1623.1, as the recovery efficiencies were less variable and the DEUF ultrafilters were capable of filtering larger volumes of high-turbidity water without clogging. Median C. cayetanensis recovery efficiencies for CFC were 28% for wash water and 63% for creek water, making it a viable option for processing water with high turbidity or organic matter. The data from this study demonstrate the capability of DEUF and CFC as filter-based and non-filter-based options, respectively, for the recovery of C. cayetanensis oocysts from environmental and agricultural waters.
Keywords: Ultrafiltration, Cyclospora, Cryptosporidium, Irrigation water, Wash water, Food safety
Abbreviations: DEUF, Dead-end ultrafiltration; CFC, continuous flow centrifugation
Graphical abstract
Highlights
-
•
Large-volume sampling facilitates detection of Cyclospora in the environment.
-
•
Dead-end ultrafiltration is a robust method for agricultural water.
-
•
Continuous flow centrifugation is a robust method for produce wash water.
1. Introduction
Cyclospora cayetanensis is a coccidian parasite that infects the small intestine of humans and is transmitted through ingestion of fecally-contaminated food or water. Environmental surveillance studies conducted in the 1990s reported the detection of C. cayetanensis oocysts in wastewater in Peru and in rivers impacted by raw sewage outfalls in Guatemala, revealing the potential for C. cayetanensis transmission through water contaminated with human sewage or wastewater (Bern et al., 1999; Sturbaum et al., 1998). While water has been documented as an implicated transmission vehicle, sporadic cyclosporiasis cases and outbreaks are more often associated with the consumption of contaminated fresh produce (Brunkard et al., 2011; Casillas et al., 2019; Huang et al., 1995; Rabold et al., 1994). Between 2000 and 2017 a total of 39 foodborne outbreaks of cyclosporiasis were reported in the US (https://www.cdc.gov/parasites/cyclosporiasis/outbreaks/foodborneoutbreaks.html). Of these 39 outbreaks, the confirmed or suspected food vehicle was identified for 17 outbreaks, and all of these were a fresh produce item or combinations of produce items. However, the source of contamination of these food vehicles was not definitively determined. C. cayetanensis oocysts are shed in human stools and require 10 to 14 days outside the human host to sporulate and become infective. These oocysts can contaminate irrigation water, which may serve as a transmission vehicle for contamination of produce crops (Ortega and Sanchez, 2010).
There is a need for robust and user-friendly sample collection and processing methods for the detection of C. cayetanensis in environmental and agricultural waters as these can be used to understand the occurrence, survival, seasonality, and transmission of C. cayetanensis in water systems and in the environment. A method that enables large-volume sample collection is ideal, as pathogens are often present at low concentrations in the environment (as an obligate intracellular parasite, C. cayetanensis does not propagate outside the human host and, consequently, cannot proliferate in the environment). Additionally, many produce growing regions utilize surface waters with varying turbidity due to various influence such as surface runoff and animal inputs, so a robust method that can reliably handle high-tubidity water is needed. Methods used for previous environmental investigation studies have utilized varying sample collection and concentration methods, without the inclusion of performance data to gauge method recovery and reliability (Bern et al., 1999; Bilung et al., 2017; Galván et al., 2013; Giangaspero et al., 2015; Onstad et al., 2019; Sturbaum et al., 1998). Collection and concentration methods ranged from simple centrifugation, filtration and elution of yarn wound filter cartridges, overnight flocculation, and sucrose flotation and gradient centrifugation and versions of USEPA Method 1623.1, which is USEPA-approved for detection of the protozoan parasites Cryptosporidium and Giardia from drinking water sources. Procedures such as overnight flocculation and elution of woven filter cartridges are laborious and time consuming. A few previous studies utilized USEPA Method 1623.1, but these failed to include performance data for recovery of C. cayetanensis (Bilung et al., 2017; Galván et al., 2013).
In the present study, three sample collection and processing methods were evaluated for the recovery of C. cayetanensis in water: dead-end ultrafiltration (DEUF), USEPA Method 1623.1 (“Method 1623”), and continuous flow centrifugation (CFC). DEUF is a large-volume collection method that utilizes hollow-fiber dialysis filters with a 30 kDa pore size. The DEUF method is capable of co-capturing all classes of microbes, is field-deployable, and has demonstrated reliable and robust recovery from various water matrices (Mull and Hill, 2012; Smith and Hill, 2009). Method 1623 is a USEPA-approved method for detection of the protozoan parasites Cryptosporidium and Giardia from drinking water sources (USEPA, 2012). Method 1623 is also field-deployable, but it utilizes an Envirochek HV capsule filter (Pall Corporation, Port Washington, NY) with a 1 μm pore size membrane, and thus is rated for recovery of parasites only. Method 1623 was included in one of the FDA's standard methods for detection of C. cayetanensis and Cryptosporidium in the Bacteriological Analytical Manual (BAM), under chapter 19a (Orlandi et al., 2004). The CFC method evaluated in this study utilizes a portable continuous flow centrifuge in conjunction with single-use disposable centrifuge bowls with ports that allow water to be continuously fed into the spinning centrifuge through an influent port and out of the centrifuge through an effluent port (CFC Express, Scientific Methods Inc., Granger, IN). The parasites are retained in the bowl through centrifugal force as the water is pumped through, resulting in an increasingly concentrated sample in the bowl. The CFC Express can be deployed in the field, although the low feed rate of water through the unit puts a practical limitation on the volume of sample that can be processed. However, the potential benefit of the CFC method is that it may allow for processing water with high turbidity or organic matter that could clog a filter. For each method, C. cayetanensis was seeded into bulk water samples and percent recovery was calculated to evaluate method performance.
2. Materials and methods
2.1. Water samples
Four types of water samples were used in the present study: 1) “high turbidity” irrigation water, 2) “low turbidity” irrigation water, 3) spent produce wash water, and 4) creek water. Irrigation water was obtained from surface irrigation reservoirs in the Suwannee River watershed in Southeast Georgia. High turbidity water was collected from a surface water-fed reservoir and low turbidity water was collected from a groundwater-fed reservoir. Turbidity levels for the high turbidity water ranged from 3.0–8.3 NTU and for the low turbidity water the levels ranged from 0.38–1.2 NTU. Irrigation water was transported to the Centers for Disease Control (CDC) in Atlanta on the day of collection. Spent romaine wash water was collected from a domestic produce processing facility and shipped overnight for delivery to CDC the following morning. Wash water turbidity levels ranged from 14 to 18 NTU (17 NTU mean). Surface water used to evaluate the CFC method was collected from a local creek in Atlanta, GA and ranged in turbidity from 7.7–14 NTU. Water samples were held at 4 °C and brought to room temperature before use. Turbidity was measured immediately before an experiment using a Hach 2100P turbidimeter (Hach, Loveland, CO).
2.2. Oocyst preparation and seeding
C. cayetanensis oocysts were purified from anonymized human stool specimens using discontinuous sucrose and cesium chloride gradients as described previously (Qvarnstrom et al., 2018b). In addition to seeding of C. cayetanensis, Cryptosporidium parvum was included in recovery experiments as a benchmark for performance of Method 1623 for the water types used in this study. C. parvum oocysts were obtained from experimentally infected calves and prepared as previously described (Arrowood and Donaldson, 1996). Oocysts were stored in the dark at 4 °C in 2.5% aqueous potassium dichromate. C. cayetanensis and C. parvum oocysts were further purified by flow cytometry sorting using a BD Biosciences (San Jose, CA) FACSAria™ III as previously described (Arrowood et al., 1995; Qvarnstrom et al., 2018b). Flow-sorted oocysts were dispensed into low-retention microcentrifuge tubes (Phenix Research Products, Hayward, CA) containing 0.01 M phosphate buffered saline (PBS, pH 7.2) and 0.01% Tween 80 in aliquots of 15,000 oocysts each and stored in the dark at 4 °C. Before each recovery experiment, a tube was vortexed for 30 s and the contents seeded into the bulk water sample.
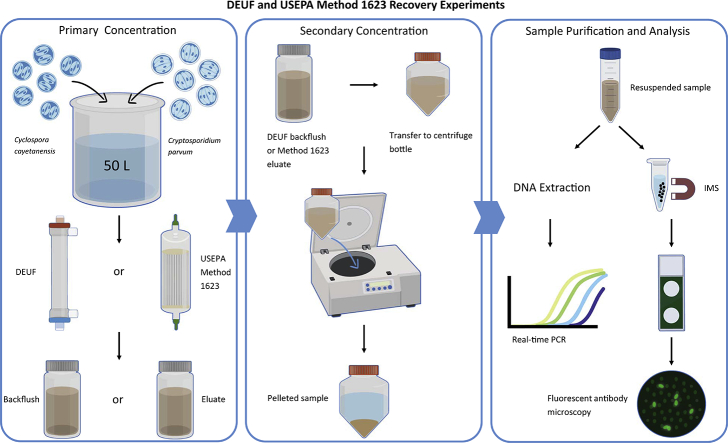
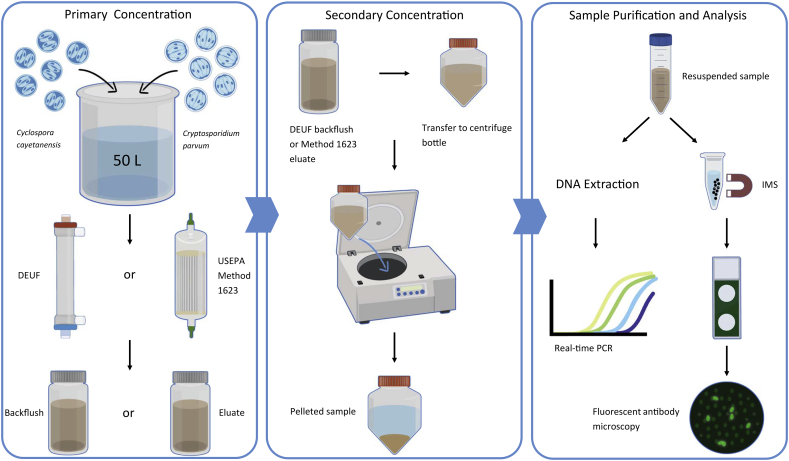
2.3. DEUF and Method 1623 recovery experiments
Method performance was assessed by determining percent recovery of seeded oocysts. Because DEUF and Method 1623 are filter-based methods, they were evaluated in parallel for their recovery performance for irrigation water (Fig. 1). Recovery efficiency experiments were conducted using 100-L irrigation water that was split into two 50-L portions for use in each method. Five replicate experiments each were conducted using high and low turbidity water. Immediately prior to each experiment the bulk water was seeded with 15,000 oocysts each of C. cayetanensis and C. parvum. For DEUF, the bulk water was concentrated as previously described and the resulting backflush was centrifuged at 4000xg for 30 min (Kahler and Hill, 2020). For Method 1623, the bulk water was concentrated using Envirochek HV capsules per the USEPA protocol (USEPA, 2012). The resulting eluate was centrifuged at 1500 xg for 15 min. After aspiration of the supernatant, the sample concentrates were resuspended in 0.01 M PBS containing 0.01% Tween 80 and 0.001% Antifoam Y-30 Emulsion.
Fig. 1.
Flow diagram showing procedures for DEUF and Method 1623 recovery experiments.
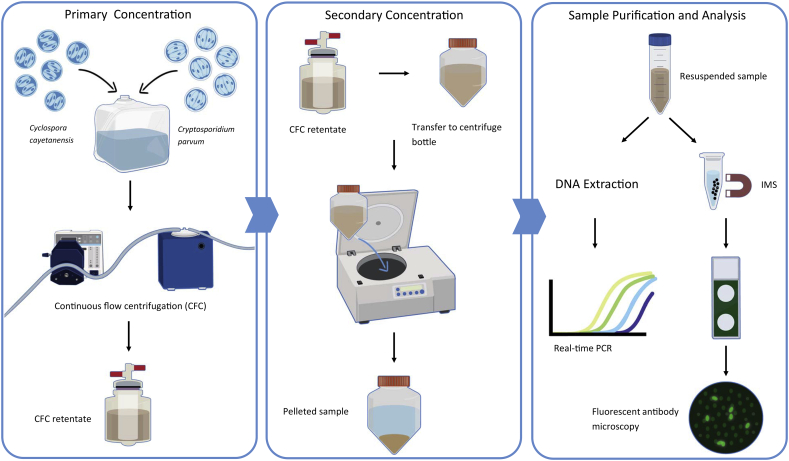
2.4. CFC recovery experiments
The CFC method was evaluated for recovery efficacy performance for creek water and spent produce wash water (Fig. 2). CFC experiments were conducted using 20-L spent romaine wash water and creek water, which served as a high turbidity environmental surface water. Five replicate experiments were conducted with each type of water. Immediately prior to each experiment the bulk water was seeded with 15,000 oocysts each of C. cayetanensis and C. parvum. Samples were concentrated by pumping the bulk water through a CFC Express unit at a feed rate of 750 mL/min using CFC 210 disposable bowls and CFC 220 disposable tubing. The resulting sample concentrate was decanted into a 250-mL centrifuge tube. The CFC bowl was eluted with 5 mL of DEUF backflush solution (500 mL of DI water containing 0.5% Tween 80, 0.01% sodium polyphosphate, and 0.001% Antifoam) and hand-mixed for 30 s. The eluate was decanted and combined with the sample concentrate and centrifuged at 4000 xg for 30 min. After aspiration of the supernatant, the concentrate was resuspended in 0.01 M PBS containing 0.01% Tween 80 and 0.001% Antifoam.
Fig. 2.
Flow diagram showing procedures for CFC recovery experiments.
2.5. Oocyst quantification and recovery calculations
Percent recovery of C. cayetanensis was calculated by dividing the gene copy quantity recovered by the input quantity and multiplying by 100. The C. cayetanensis input for each experiment was the average gene copyquantity of five flow-sorted oocyst stock tubes analyzed directly by qPCR. It was assumed the average gene copy quantity per tube was consistent, as all flow-sorted tubes were generated at one time with one oocyst stock preparation.
For C. cayetanensis quantification, nucleic acid was extracted from 750 μL of the sample concentrate for analysis by quantitative real-time PCR (qPCR) targeting the 18S rRNA gene as previously described (Hill et al., 2015; Murphy et al., 2017; Qvarnstrom et al., 2018a). Two and five microliters of template DNA were each analyzed in duplicate in 50-μL reaction volumes. The qPCR master mix consisted of 1× TaqMan™ Environmental Master Mix 2.0 (Applied Biosystems, Foster City, CA), 250 nM each primer, and 100 nM probe. Amplification was performed in a 7500 Real-Time PCR System with the following parameters: initial denaturation at 95 °C for 10 min; 45 cycles of denaturation at 95 °C for 15 s and annealing, extension, and fluorescence capture at 67 °C for 1 min. At least three no-template controls were included during each instrument run and the amplification threshold was set to 0.02 ΔRn units. Gene copies of the C. cayetanensis 18S rRNA gene (NCBI accession #: AF111183) were enumerated using qPCR standard curves using a synthetic gBlocks® gene fragment (HMgBlock135m, Integrated DNA Technologies, Coralville, CA) (Murphy et al., 2017). Ten-fold serial dilutions (105 to 100 copies per reaction) were analyzed in triplicate during each instrument run. Gene copy number per reaction was calculated by inputting the average quantification cycle (Cq) of the four sample replicates into the pooled standard curve equation. All no-template controls and standard curves performed as expected with a pooled efficiency of 98.7% and y-intercept of 39.35.
A portion of the final sample concentrate was subjected to immunomagnetic separation (IMS) to further concentrate and purify the sample for C. parvum quantification(USEPA, 2012). The volume subjected to IMS contained ~102 oocysts in order to enable direct quantification of the entire IMS concentrate. IMS was conducted using the Dynabeads anti-Cryptosporidium kit (Life Technologies) with two dissociation steps. The IMS concentrate was enumerated by fluorescent antibody microscopy (USEPA, 2012). Microscopy slides were stained using the EasyStain kit (BioPoint USA Inc., Pittsburgh, PA), and oocysts were enumerated at 200-400× magnification using a fluorescence microscope with an FITC filter. Percent recovery of C. parvum was calculated by dividing the oocyst quantity recovered by the input quantity and multiplying by 100. The input quantity for C. parvum oocysts was set at 15,000 oocysts, as determined by flow cytometry, as the microscopy procedure was not amenable to counting more than several hundred oocysts and enumeration of a fraction of the oocyst stock was not reliable.
2.6. Statistical analyses
The C. cayetanensis oocyst input level necessary to achieve a qPCR Cq between 30 and 35, even in instances of low recovery efficiency (~10%), was back-calculated to be approximately 15,000 oocysts. The seed level of C. parvum was matched at 15,000 oocysts to evaluate the recovery performance between the two parasites. Percent recoveries underwent an arcsine square root transformation to enable statistical analyses of bounded proportional data (McMinn et al., 2017). During two of the high-turbidity irrigation water experiments, the Method 1623 Envirochek HV filter clogged after processing approximately 30–40 lL of water. As such, recovery efficiency could only be determined for three of the five high-turbidity water experiments for Method 1623. Two-sample t-tests were conducted to evaluate differences in transformed mean percent recovery between DEUF and Method 1623 at each turbidity level. An F-test of variance equality was conducted for each condition to determine which type of t-test was appropriate. Paired t-tests were conducted to evaluate statistical differences between the performance of DEUF and Method 1623 for each water type and differences in percent recovery of C. cayetanensis and C. parvum. Statistical significance set at α = 0.05.
3. Results and discussion
The median recovery efficiencies for C. cayetanensis using DEUF were 17% for both high and low turbidity water, with the ranges for each water type shown in Table 1. The median recovery efficiencies for C. cayetanensis using Method 1623 were 16% for low turbidity water and 22% for high turbidity water. The median recovery efficiencies for C. parvum using DEUF were 55% for low turbidity water and 46% for high turbidity water, while the median recovery efficiencies for Method 1623 were 56% and 75% for low and high turbidity water, respectively. The C. parvum recovery efficiencies for Method 1623 met the performance criteria established by USEPA of 13% minimum recovery (USEPA, 2012). Additionally, both the DEUF and Method 1623 recovery efficiencies fell within the expected ranges compared to previously reported data from other surface water types (Francy et al., 2013; Kahler et al., 2015; Kimble et al., 2013; Kimble et al., 2012).
Table 1.
Median recovery efficiencies (%) and ranges (%) for C. cayetanensis and C. parvum oocysts from 50-L irrigation water samples by DEUF and USEPA Method 1623.
| Water type |
C. cayetanensis (%) |
C. parvum (%) |
||||
|---|---|---|---|---|---|---|
| n | DEUF | Method 1623 | n | DEUF | Method 1623 | |
| Low-turbidity | 5 | 17 (12–25) | 16 (14–24) | 4a | 55 (19–87) | 56 (19–102)b |
| High-turbidity | 3b | 17 (9–23) | 22 (19–100) | 3c | 46 (39–60) | 75 (71–90) |
Could not calculate recovery efficiency for one experiment due to analyst error.
>100% recovery for one C. parvum experiment could be due to overestimation of recovered oocysts caused by non-homogenous distribution or clumping of oocysts in the sample concentrate, resulting in an overrepresentation of oocysts in the aliquot removed for IMS purification and microscopy.
Excluded data from the two experimental trials in which Method 1623 Envirochek filter clogged to keep the comparison between methods consistent. For example, although the DEUF ultrafilters did not clog during those two experiments, the recovery could have been significantly impacted by these water matrices.
Recovery efficiencies of both C. cayetanensis and C. parvum using DEUF were not significantly different between high and low turbidity water (p = 0.72C. cayetanensis, p = 0.75C. parvum). Using Method 1623, recovery efficiencies of C. cayetanensis and C. parvum were not significantly different between high and low turbidity water (p = 0.38C. cayetanensis, p = 0.57C. parvum). Pairwise comparisons for each water type revealed that the recovery efficiencies for both C. cayetanensis and C. parvum were not significantly different between DEUF and Method 1623 (p = 0.36C. cayetanensis, p = 0.053C. parvum). Taken together, these results indicate that both DEUF and Method 1623 effectively recovered oocysts from varying water types and that overall recovery efficiency was similar between methods. However, DEUF was more robust than Method 1623 for high-turbidity irrigation water samples, as it was capable of consistently filtering the entire 50 lL of all water samples without clogging and demonstrated more consistent recovery of oocysts, as demonstrated by the tighter recovery efficiency ranges. Surface waters can have highly variable turbidity levels which are impacted by factors such as rainfall runoff, composition of the sediment, and activities that can disturb benthic sediment. The inability of Envirochek HV filters to reliably filter 50 lL of the high-turbidity water represents a major pitfall for their use in performing Method 1623 for recovery of low-concentration parasites from environmental waters. This was highlighted in 2013 when FDA analysts had difficulty recovering Cyclospora oocysts from processing facility water samples when using the Method 1623 as recommended per FDA BAM Chapter 19a. Due to the presence of high levels of organic matter in the recycled wash tank water the Envirochek filters clogged rapidly (personal communication). CDC has previously utilized the DEUF method to successfully recover pathogens from irrigation water for environmental surveillance and outbreak response (FDA, 2018; Hill et al., 2017). These DEUF performance data, alongside the data obtained from this study, provided the evidence to justify the adaptation of this method for smaller sample volumes in FDA's BAM Chapter 19c (Durigan et al., 2020a; Durigan et al., 2020b).
The median recovery efficiencies for C. cayetanensis from creek water and spent produce wash water using CFC were 63% and 29%, respectively, with the ranges for each water type shown in Table 2. The median recovery efficiencies for C. parvum from creek water and spent produce wash water using CFC were 25% and 57%, respectively. Comparative data on the performance of CFC for recovering protozoan parasites is limited. However, two separate studies report recovery efficiencies for C. parvum from source water and C. cayetanensis from tap water with artificial turbidity, which are in line with the results reported here, considering methodological differences and differences in water type and volume (Borchardt et al., 2009; Zuckerman and Tzipori, 2006). The data reported here indicate that the CFC method can enable reliable detection for both C. cayetanensis and C. parvum from complex water matrices containing high organic content such as spent produce wash water. The limitation to the CFC method is the slow feed rate compared to the filtration rates for DEUF or Envirochek HV filters.
Table 2.
Median recovery efficiencies (%) and ranges (%) for C. cayetanensis and C. parvum oocysts from 20-L creek and produce wash water samples by CFC.
| Water type | n | C. cayetanensis (%) | C. parvum (%) |
|---|---|---|---|
| Creek water | 5 | 63 (34–123)a | 25 (22–35) |
| Wash water | 5 | 28 (17–43) | 57 (44–64) |
>100% recovery for one C. cayetanensis experiment could be due to overestimation of recovered oocysts caused by non-homogenous distribution or clumping of oocysts in the sample concentrate, resulting in an overrepresentation of oocysts in the aliquot removed for nucleic acid extraction.
Median recovery efficiencies of C. cayetanensis from irrigation water were lower than for C. parvum, but a statistically significant difference was only seen for high turbidity water using DEUF, in which the median recovery for C. cayetanensis was 38% lower than for C. parvum (p = 0.13 and p = 0.46 for Method 1623 low and high, p = 0.084 and p = 0.024 for DEUF low and high). Recovery efficiencies of C. cayetanensis from creek water using CFC were similar to C. parvum (p = 0.074), but were lower than C. parvum for wash water (p = 0.029). It is not uncommon for recovery efficacy to vary between microbes of the same taxonomic group (Hill et al., 2007; Hill et al., 2009; Kahler et al., 2015). Based on our experimental design, it was not possible to ascertain where losses occurred during each step of the recovery experiments. However, differences in recovery efficiencies may be due in part to microbial properties, such as surface characteristics, and the way those properties interact with the constituents of the water matrix or other surfaces. These phenomena may have caused differential amounts of oocyst clumping, resulting in non-homogenous distribution, or differential adherence of oocysts to the surfaces of tubes or bottles, both of which may have resulted in an underrepresentation of oocysts in the aliquot removed for subsequent processing or purification steps. Additionally, the different purification procedures for C. cayetanensis (nucleic acid extraction) and C. parvum (IMS) may have resulted in unequal oocyst loses, which could have contributed to differences in the overall recovery efficiencies for the two microbes.
As with any method performance evaluation, there are several limitations to these data. First, the reported recovery efficiencies incorporated each procedural step within the method and therefore included oocyst losses during seeding, transfer and, when applicable, nucleic acid extraction. However, any uncertainty or inaccuracy in the quantification of oocysts or gene copies were reduced or nullified by converting the quantification estimates to a ratio of output to input as express by percent recovery. Additionally, quantification of the oocyst input may have varied due to the nucleic acid extraction inefficiencies and adherence of oocysts to the tube walls after flow sorting, but this was addressed by averaging the input quantity for C. cayetanensis and setting C. parvum input at 15,000 oocysts. Finally, the CFC method was not evaluated using the same water samples as DEUF and Method 1623 due to logistical constraints. To directly compare performance of the CFC method against a filter-based method, further evaluation is needed using equivalent water samples and volumes.
For recovery of low-concentration pathogens such as C. cayetanensis from irrigation water, the DEUF method was more robust (i.e., not clogging) compared to Method 1623 using Envirochek HV filters. Additionally, medical-grade dialysis filters are substantially less expensive than Envirochek HV filters, resulting in a more cost-effective method. While the performance of the DEUF and CFC methods were not directly compared, each method has unique advantages. DEUF dialysis filters are robust enough to filter significant volumes of environmental water types, such as irrigation water. For surface water, 50 lL is sufficient to facilitate detection of low-concentration pathogens (≤ 101 organisms per L), although filtration of volumes larger than 50 lL is possible for low-turbidity water. The DEUF method is readily field-deployable, allowing for shipment of filters to the processing laboratory instead of bulky containers of water. The DEUF method is also capable of co-concentrating all classes of microbes, including bacteria and viruses, if detection of multiple pathogens is desired. The CFC method is also field-portable, but would require amendments to be fully operational in all field settings. The CFC method demonstrated reliable recovery of oocysts from waters with high turbidity and organic matter and can provide a useful alternative to a filter-based method which may clog during processing of these low-quality sample types. Further evaluation is needed to determine the detection limits for these methods for various water matrices.
Declaration of Competing Interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Use of trade names and commercial sources is for identification only and does not imply endorsement by the Centers for Disease Control and Prevention (CDC), the U.S. Food and Drug Administration (FDA) or the U.S. Department of Health and Human Services (HHS). A portion of the funding for this study was provided through Interagency Agreement 224-15-2010S with the Center for Food Safety and Applied Nutrition (CFSAN) at the US FDA (IAA Program Official: Alex DaSilva). This study was part of the intramural project number IF01253 as listed in the FDA's Component Automated Research Tracking System (CARTS). We also gratefully acknowledge the technical support and guidance of CDC and FDA scientists, Dr. Michael Arrowood, Delynn Moss and Dr. Mauricio Durigan. The findings and conclusions in this report are those of the authors and do not necessarily represent those of CDC or FDA.
References
- Arrowood M.J., Donaldson K. Improved purification methods for calf-derived Cryptosporidium parvum oocysts using discontinuous sucrose and cesium chloride gradients. J. Eukaryot. Microbiol. 1996;43:89. doi: 10.1111/j.1550-7408.1996.tb05015.x. [DOI] [PubMed] [Google Scholar]
- Arrowood M.J., Hurd M.R., Mead J.R. A new method for evaluating experimental cryptosporidial parasite loads using immunofluorescent flow cytometry. J. Parasitol. 1995;81:404–409. [PubMed] [Google Scholar]
- Bern C., Hernandez B., Lopez M.B., Arrowood M.J., Alvarez De Mejia M., De Merida A.M., Hightower A.W., Venczel L., Herwaldt B.L., Klein R.E. Epidemiologic studies of Cyclospora cayetanensis in Guatemala. Emerg. Infect. Dis. 1999;5:766–774. doi: 10.3201/eid0506.990604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilung L.M., Tahar A.S., Yunos N.E., Apun K., Lim Y.A., Nillian E., Hashim H.F. Detection of Cryptosporidium and Cyclospora oocysts from environmental water for drinking and recreational activities in Sarawak, Malaysia. Biomed. Res. Int. 2017:4636420. doi: 10.1155/2017/4636420. 2017/12/14 ed, p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchardt M.A., Spencer S.K., Bertz P.D., Ware M.W., Dubey J.P., Alan Lindquist H.D. Concentrating Toxoplasma gondii and Cyclospora cayetanensis from surface water and drinking water by continuous separation channel centrifugation. J. Appl. Microbiol. 2009;107:1089–1097. doi: 10.1111/j.1365-2672.2009.04316.x. [DOI] [PubMed] [Google Scholar]
- Brunkard J.M., Ailes E., Roberts V.A., Hill V., Hilborn E.D., Craun G.F., Rajasingham A., Kahler A., Garrison L., Hicks L., Carpenter J., Wade T.J., Beach M.J., Yoder J.S. Surveillance for waterborne disease outbreaks associated with drinking water - United States, 2007-2008. MMWR Morbid. Mortal. Wkly. Rep. 2011;60:38–68. [PubMed] [Google Scholar]
- Casillas S.M., Hall R.L., Herwaldt B.L. Cyclosporiasis surveillance - United States, 2011-2015. MMWR Surveill. Summ. 2019;68:1–16. doi: 10.15585/mmwr.ss6803a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durigan M., Murphy H.R., Da Silva A. Detection of Cyclospora cayetanensis in agrigultural water using dead-end ultrafiltration (DEUF) and DNA-based methods. Appl. Environ. Microbiol. 2020 doi: 10.1128/AEM.01595-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durigan M., Murphy H.R., Deng K., Kmet M., Lindemann S., Newkirk R., Patel V.Y., Ulaszek J., Warren J., Ewing L., Reddy R., Da Silva A. US Food and Drug Administration; 2020. BAM Chapter 19c: Dead-End Ultrafiltration for the Detection of Cyclospora cayetanensis from Agricultural Water. [Google Scholar]
- FDA . U.S. Food and Drug Administration; 2018. Environmental assessment of factors potentially contributing to the contamination of romaine lettuce implicated in a multi-state outbreak of E. coli O157:H7. [Google Scholar]
- Francy D.S., Stelzer E.A., Brady A.M.G., Huitger C., Bushon R.N., Ip H.S., Ware M.W., Villegas E.N., Gallardo V., Lindquist H.D.A. Comparison of filters for concentrating microbial indicators and pathogens in lake water samples. Appl. Environ. Microbiol. 2013;79:1342–1352. doi: 10.1128/AEM.03117-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galván A.L., Magnet A., Izquierdo F., Fenoy S., Rueda C., adillo C.F.V., Henriques-Gil N., del Aguila C. Molecular characterization of human-pathogenic microsporidia and Cyclospora cayetanensis isolated from various water sources in Spain: a year-long longitudinal study. Appl. Environ. Microbiol. 2013;79:449–459. doi: 10.1128/AEM.02737-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giangaspero A., Marangi M., Koehler A.V., Papini R., Normanno G., Lacasella V., Lonigro A., Gasser R.B. Molecular detection of Cyclospora in water, soil, vegetables and humans in southern Italy signals a need for improved monitoring by health authorities. Int. J. Food Microbiol. 2015;211:95–100. doi: 10.1016/j.ijfoodmicro.2015.07.002. [DOI] [PubMed] [Google Scholar]
- Hill V.R., Kahler A.M., Jothikumar N., Johnson T.B., Hahn D., Cromeans T.L. Multistate evaluation of an ultrafiltration-based procedure for simultaneous recovery of enteric microbes in 100-liter tap water samples. Appl. Environ. Microbiol. 2007;73:4218–4225. doi: 10.1128/AEM.02713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill V.R., Polaczyk A.L., Kahler A.M., Cromeans T.L., Hahn D., Amburgey J.E. Comparison of hollow-fiber ultrafiltration to the USEPA VIRADEL technique and USEPA method 1623. J. Environ. Qual. 2009;38:822–825. doi: 10.2134/jeq2008.0152. [DOI] [PubMed] [Google Scholar]
- Hill V.R., Narayanan J., Gallen R.R., Ferdinand K.L., Cromeans T., Vinjé J. Development of a nucleic acid extraction procedure for simultaneous recovery of DNA and RNA from diverse microbes in water. Pathogens. 2015;4:335–354. doi: 10.3390/pathogens4020335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill V.R., Vellidis G., Levy K. 2017. Improved Sampling and Analytical Methods for Testing Agricultural Water for Pathogens, Surrogates, and Source Tracking Indicators. [Google Scholar]
- Huang P., Weber J.T., Sosin D.M., Griffin P.M., Long E.G., Murphy J.J., Kocka F., Peters C., Kallick C. The first reported outbreak of diarrheal illness associated with Cyclospora in the United States. Ann. Intern. Med. 1995;123:409–414. doi: 10.7326/0003-4819-123-6-199509150-00002. [DOI] [PubMed] [Google Scholar]
- Kahler A.M., Hill V.R. Detection of Cryptosporidium recovered from large-volume water samples using dead-end ultrafiltration. Methods Mol. Biol. 2020:23–41. doi: 10.1007/978-1-4939-9748-0_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahler A.M., Johnson T.B., Hahn D., Narayanan J., Derado G., Hill V.R. Evaluation of an ultrafiltration-based procedure for simultaneous recovery of diverse microbes in source waters. Water. 2015;7:1202–1216. doi: 10.3390/w7031202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble G.H., Amburgey J.E., Hill V.R. Comparison of hollow-fiber ultrafilters with pleated capsule filters for surface and tap water samples using U.S. EPA method 1623. J. Environ. Eng. 2012;138:899–901. doi: 10.1061/(ASCE)EE.1943-7870.0000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble G., Amburgey J., Hilger H. Improvements in Cryptosporidium recovery and variability through modifications to United States Environmental Protection Agency Method 1623. Water Environ. J. 2013;27:269–274. [Google Scholar]
- McMinn B.R., Huff E.M., Rhodes E.R., Korajkic A. Concentration and quantification of somatic and F+ coliphages from recreational waters. J. Virol. Methods. 2017;249:58–65. doi: 10.1016/j.jviromet.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mull B., Hill V.R. Recovery of diverse microbes in high turbidity surface water samples using dead-end ultrafiltration. J. Microbiol. Methods. 2012;91:429–433. doi: 10.1016/j.mimet.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy H.R., Lee S., Da Silva A.J. Evaluation of an improved U.S. food and drug administration method for the detection of Cyclospora cayetanensis in produce using real-time PCR. J. Food Prot. 2017;80:1133–1144. doi: 10.4315/0362-028X.JFP-16-492. [DOI] [PubMed] [Google Scholar]
- Onstad N.H., Beever J.E., Miller M.R., Green M.L., Witola W.H., Davidson P.C. Cyclospora cayetanensis presence in the environment—a case study in the Chicago metropolitan area. Environments. 2019;6 [Google Scholar]
- Orlandi P.A., Frazar C., Carter L., Chu D.T. U.S. Food and Drug Administration; 2004. BAM Chapter 19a: Detection of Cyclospora and Cryptosporidium from Fresh Produce: Isolation and Identificatoin by Polymerase Chain Reaction (PCR) and Microscopic Analysis. [Google Scholar]
- Ortega Y.R., Sanchez R. Update on Cyclospora cayetanensis, a food-borne and waterborne parasite. Clin. Microbiol. Rev. 2010;23:218–234. doi: 10.1128/CMR.00026-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qvarnstrom Y., Benedict T., Marcet P.L., Wiegand R.E., Herwaldt B.L., da Silva A.J. Molecular detection of Cyclospora cayetanensis in human stool specimens using UNEX-based DNA extraction and real-time PCR. Parasitology. 2018;145:865–870. doi: 10.1017/S0031182017001925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qvarnstrom Y., Wei-Pridgeon Y., Van Roey E., Park S., Srinivasamoorthy G., Nascimento F.S., Moss D.M., Talundzic E., Arrowood M.J. Purification of Cyclospora cayetanensis oocysts obtained from human stool specimens for whole genome sequencing. Gut Pathog. 2018:10. doi: 10.1186/s13099-018-0272-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabold J.G., Hoge C.W., Shlim D.R., Kefford C., Rajah R., Echeverria P. Cyclospora outbreak associated with chlorinated drinking water. Lancet. 1994;344:1360–1361. doi: 10.1016/s0140-6736(94)90716-1. [DOI] [PubMed] [Google Scholar]
- Smith C.M., Hill V.R. Dead-end hollow-fiber ultrafiltration for recovery of diverse microbes from water. Appl. Environ. Microbiol. 2009;75:5284–5289. doi: 10.1128/AEM.00456-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturbaum G.D., Ortega Y.R., Gilman R.H., Sterling C.R., Cabrera L., Klein D.A. Detection of Cyclospora cayetanensis in wastewater. Appl. Environ. Microbiol. 1998;64:2284–2286. doi: 10.1128/aem.64.6.2284-2286.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USEPA . Office of Water; Washington, D.C: 2012. Method 1623.1: Cryptosporidium and Giardia in water by filtration/IMS/FA. pp. EPA 816-R-812-001. [Google Scholar]
- Zuckerman U., Tzipori S. Portable continuous flow centrifugation and Method 1623 for monitoring of waterborne protozoa from large volumes of various water matrices. J. Appl. Microbiol. 2006;100:1220–1227. doi: 10.1111/j.1365-2672.2006.02874.x. [DOI] [PubMed] [Google Scholar]
Web references
- https://www.cdc.gov/parasites/cyclosporiasis/outbreaks/foodborneoutbreaks.html Accessed on April 30, 2020.