Abstract
Precise delineation of the ischemic lesion from unimodal Magnetic Resonance Imaging (MRI) is a challenging task due to the subtle intensity difference between the lesion and normal tissues. Hence, multispectral MRI modalities are used for characterizing the properties of brain tissues. Traditional lesion detection methods rely on extracting significant hand-engineered features to differentiate normal and abnormal brain tissues. But the identification of those discriminating features is quite complex, as the degree of differentiation varies according to each modality. This can be addressed well by Convolutional Neural Networks (CNN) which supports automatic feature extraction. It is capable of learning the global features from images effectively for image classification. But it loses the context of local information among the pixels that need to be retained for segmentation. Also, it must provide more emphasis on the features of the lesion region for precise reconstruction. The major contribution of this work is the integration of attention mechanism with a Fully Convolutional Network (FCN) to segment ischemic lesion. This attention model is applied to learn and concentrate only on salient features of the lesion region by suppressing the details of other regions. Hence the proposed FCN with attention mechanism was able to segment ischemic lesion of varying size and shape. To study the effectiveness of attention mechanism, various experiments were carried out on ISLES 2015 dataset and a mean dice coefficient of 0.7535 was obtained. Experimental results indicate that there is an improvement of 5% compared to the existing works.
Keywords: Deep neural network, FCN, Attention, Ischemic lesion segmentation, MRI
Introduction
Ischemic stroke arises due to the accumulation of fatty deposits in the blood vessels of the brain. When the fatty particles accumulate at one spot, it affects the flow of blood and vital nutrients. This eventually leads to cell death in the occluded area. The primary treatment option in acute ischemic stroke concentrates on dealing with this occlusion quickly with the help of thrombolysis. This thrombolytic process will be safe and effective only if it is initiated within 3 to 4.5 h of symptom onset [1]. Neuroimaging techniques prove to be a gold standard in identifying the actual cause of infarction. The infarction can be either due to ischemia or hemorrhage inside the blood vessel. Application of machine learning techniques in neuro-radiology might assist the radiologist to rapidly analyze the changes like those brain tissues under infarction. Numerous approaches were presented in the past two decades for effective segmentation and characterization of stroke lesions [2–7].
Many supervised learning algorithms have been explored for the detection of ischemic stroke [8–10]. Oskar Maier et al. developed an approach for segmentation of ischemic lesion with simple image features and Extra Tree Forests [10]. Texture and morphological features were considered along with the ANFIS classifier for the segmentation of ischemic lesion in [9]. A combination of the Bayesian-Markov Random Field and Random Forest classifier has been suggested by Mitra et al. for segmentation [8]. All the above-discussed approaches for lesion detection rely on extracting handcrafted features that demand domain expertise in handling radiological aspects of brain anatomy.
Most of these methods initially select feature primitives that were relevant to the context and utilize them to generate the trained model. Convolutional Neural Network eliminates this requirement as there is no prerequisite of feature extraction. The network learns to accomplish automatic feature extraction and utilize them for classification. Hence, CNN is now quite popular in the computer vision community. The success rate of CNN is increasing in many application areas due to the effective use of graphical processing platforms, optimized activation functions like ReLU, and effective data augmentation techniques [11]. Due to the advantage of end to end training and automatic feature learning, CNN’s are now widely used for biomedical problems. Chin et al. presented an approach for ischemic stroke detection using CNN [12]. Principal Component Analysis was combined with CNN to detect brain tumors in [13]. Diniz et al. introduced an approach to detect white matter lesion by combining CNN with SLIC0 clustering [14]. CNN was optimized using Particle Swarm Optimization to detect stroke lesions in [15]. All the above methods were addressed towards the detection of the brain lesion. But further investigation is required to identify the precise location of the lesion in the input slices.
When CNN is used for image classification, a fully connected layer at the end is applied for learning global features. But it will not retain the local spatial arrangement of pixels in an image. Also, to fully reconstruct the desired region of interest for segmentation purposes, this spatial arrangement of pixels should be retained in addition to learning the local features of an input image. This was accomplished by including a set of convolutional layers in the place of a fully connected layer. Such networks are called Fully Convolutional Network (FCN). It learns representation from the input images and reconstructs the output based on local spatial input. The major highlight in FCN is that it learns end to end convolutional filters for segmentation. Even the decision-making layers at the end of the network are filters that try to reconstruct the desired output. Several approaches were proposed in recent years for semantic image segmentation using FCN [16–18]. Shaikh et al. proposed a segmentation approach for brain tumor by combining dense Conditional Random Field with FCN [19]. Shen et al. developed a boundary aware FCN by extracting multilevel contextual details from multimodal MRI [20]. Liang Cheng presented a FCN for lesion segmentation from Diffusion-Weighted images by combining two de-convolutional Neural Networks [21]. Another approach for ischemic lesion segmentation using three-dimensional FCN for multimodal MRI was presented in [22]. Another FCN based approach was introduced to segment ischemic lesion in [23]. Various hyper-parameters were analyzed to fine-tune the parameters of the model for precise segmentation.
Though the above methods are capable of extracting lesion using FCN, the segmentation accuracy needs to be improved further. In FCN based methods, the features from each convolutional block need to be carefully filtered for precise reconstruction. Attention gates play a vital role in filtering such insignificant information. The concept of attention gates was proven to be successful in real-world problems [24–26]. The attention-based mechanism was explored for image classification by Jetley et al. [27] and Fei Wang et al. [28]. It was inferred that attention maps were able to learn and highlight the features of the region of interest effectively. These works clearly illustrate the significance of attention models in learning the context-aware features of natural images. Recently, the concept of attention mechanism was extended to medical images. Qinjgi et al. presented one such scheme using residual attention for classifying multi-label chest X-rays [29].
All the above attention-based methods give focus to learn and identify significant features for classification. But it can also be extended in FCN to filter the response from convolutional filters for effective segmentation. Schlemper et al. presented a novel attention gated mechanism for classification and segmentation of anatomical structures in CT images [30]. But it can be extended to segment the anatomical abnormalities present in brain images. This research places the first attempt to explore the performance of attention gated mechanism for the segmentation of ischemic lesion from brain MRI. A FCN with attention gates was employed to segment the region of the ischemic lesion from multimodal MRI.
The novel aspects of this research work are summarized as follows:
A fully convolutional network with an attention gated mechanism was proposed to delineate ischemic lesion from 2D multimodal MRI.
The architecture of FCN is enhanced by introducing attention gates at the decoder. This enables the network to filter prominent feature map activations that support precise reconstruction.
Materials and methods
Datasets
The proposed research utilized 28 samples from ISLES 2015 SISS Challenge [32]. Each dataset consists of four different MR imaging modality namely T1-weighted, T2-Weighted, Diffusion-Weighted imaging (DWI), and Fluid Attenuation Inverse Recovery (FLAIR). Ground truth labels were also provided for validation purposes. The proposed research is restricted to the axial plane due to computational requirements.
Pre-processing and data augmentation
Pre-processing of input images is an essential step as it could simplify the learning process in subsequent stages. The input images were already skull stripped, retaining the tissue portions alone for further processing.
Data augmentation process improves the sample size of the training data by using geometric deformation transforms to the input images. This step will apply these deformation functions to the input image and save the resultant output as a new augmented result. By doing so, the size of the input training vectors becomes considerably large with wide variation in the geometric properties. But, it introduces a huge computational overhead during the learning phase. Hence to have a trade-off between the data augmentation factor and the underlying memory requirements, the augmentation phase is slightly modified in this research. Instead of separately storing the augmented result as a new image, each epoch will apply these deformations with random scaling, shear and rotation factors and admit it for training. This step will not eventually increase the size of the input training vector, but it will adapt to learn for different possibilities as the elastic deformation parameters are changed in each epoch. By doing so, an efficient augmentation approach was involved in this research which will enable the network to learn for different possible combinations.
Proposed FCN architecture
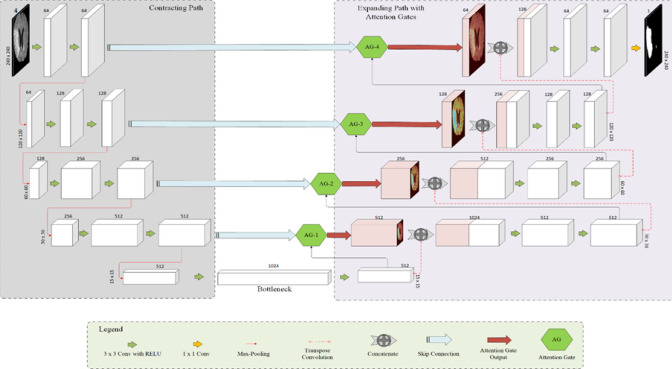
The architecture of the proposed FCN is presented in Fig. 1. This architecture was inspired by the one proposed for salient region segmentation from medical images [30]. But it was extended in this work to segment the ischemic lesion from brain MRI.
Fig. 1.
The architecture of the proposed system (The dimension indicated above each block presents the number of feature maps generated)
In Fully Convolutional Networks, both encoder and decoder will have ‘n’ convolutional filters, each with depth ‘D’. Let the input image stack be represented as I = {I1, I2, … ID} and the convolutional filter banks be denoted as F = {F1, F2, …., Fn}.
Encoder (contracting path)
The layers present in the encoding side will apply a series of convolution operations with an input stack ‘I’ and the convolutional filter banks ‘FE’. This is followed by a non-linear activation function. This sequence of steps will result in an output stack ‘O’ as represented in Eq. 1.
| 1 |
where, ‘2 k + 1’ and ‘2 k-1’ are the parameters of filter side length, ‘a’ is the non-linear activation function, and ‘bm’ is the bias of mth feature map.
The feature maps generated as per the above relation will act as the encoded version of the input ‘I’. This can serve as the parameters used to generate the feature map ‘Om’. This step is followed by a max-pooling operation to reduce the dimension of the feature map by a factor of ‘2’.
The above-discussed steps are repeated in the encoder side of the network until it reaches the bottleneck layer or latent space representation ‘Zm’. This layer consists of a stack of significant feature maps used to represent the input image ‘I’.
Decoder (expanding path)
The ‘n’ feature maps ‘Zm=1,2,…n’ will be used by the decoder to reconstruct the desired segmented region ‘R’. It is subjected to a convolution operation to select and learn a prominent stack of filters. Then, it is up-sampled and concatenated with the feature response obtained through skip connection ‘Si’ for precise localization of pixels. But the information from skip connection might also pass some irrelevant feature response to the decoder. To filter such irrelevant feature response and allow only vital feature response to the decoder, attention gates are employed in this research. It filters the irrelevant feature response derived from the background details of the image (non-lesion) and admits only the significant activations of the foreground (lesion) to the decoder for further reconstruction.
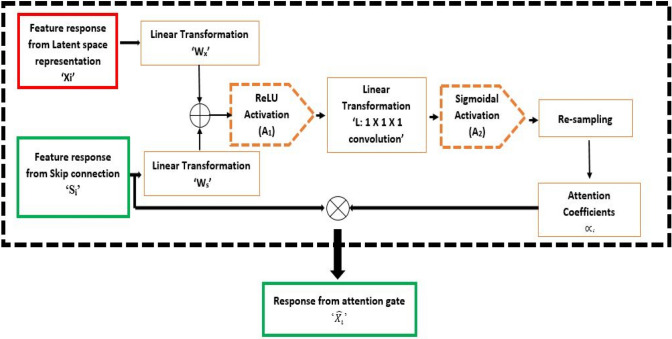
The feature response ‘Si’ is filtered with the help of attention gates as follows:
A convolutional operation is applied to ‘Zm’ to select and retain a significant stack of filters. These details are up-sampled to match the dimension of ‘Si’ to generate ‘Xi’. Both ‘Si’ and ‘Xi’ are subjected to linear transformations with ‘Ws’ and ‘Wx’ respectively. The transformed vectors of ‘Si’ and ‘Xi’ are added and ReLU activation is applied to the result for retaining the significant activation. Then, another linear transformation ‘’ with 1 × 1x1 convolution is applied to generate ‘Ci’ as presented in Eq. 2
| 2 |
where A1(x) = max (0,x) is the ReLU activation, ‘Ws’ & ‘Wx’ are the linear transformations that apply channel-wise convolution with filter dimensions of ‘Si’ and ‘Xi’ respectively, and ‘’ is the bias.
The intermediate output in the attention gate ‘Ci’ is then subjected to sigmoidal activation ‘A’2and re-sampled to generate the spatial attention coefficients ‘’ as per the relation presented in Eq. 3
| 3 |
where, is the sigmoidal activation generating values in , and is the function that re-samples the input to match the dimension of ‘Xi’.
Finally, the output from the attention gate ‘’ is generated as per the relation presented in Eq. 4.
| 4 |
Hence, now contains relevant feature response based on ‘Xi’ and the spatial attention coefficients ‘’. These coefficients suppress the irrelevant features for the background and help in learning the foreground object details alone for further reconstruction. The sequence of steps employed to generate attention coefficients is presented in Fig. 2.
Fig. 2.
Working of Attention mechanism
The output response from attention gate ‘’ is concatenated with ‘Xi’ and subjected to a series of de-convolutional filters to generate ‘Ri’. The reconstructed output in each layer ‘Ri’ will again be the result of convolution between the volume of activation maps ‘Zm’ with de-convolutional filters ‘FD’ as presented in Eq. 5.
| 5 |
The reconstructed output from each layer ‘Ri’ is up-sampled and concatenated with the filtered information from the attention gates. This step of determining ‘R’ and the subsequent up-sampling process will be repeated for ‘p’ layers until it reaches the dimension of the input image ‘I’. As a final step, the loss between the ground truth ‘G’ and the segmented output ‘R’ is calculated as per the relation presented in Eq. 6.
| 6 |
Based on the loss obtained, the filter parameters ‘F’ will be tuned to refine the segmented output.
Model training
To assess the performance of attention gates in improving the segmentation performance, it is included in each convolutional block of the decoder. These gates filter the response from the skip connection and admit only the significant features for further reconstruction. This attention gated FCN architecture was trained for 150 epochs with a learning rate of 0.0001. It took around eight hours to train the model. Four-fold cross-validation was performed with this approach to assess its performance under multiple runs. A dice coefficient of 0.928 was obtained for training datasets in terms of the dice coefficient.
Results and discussion
The proposed experiments were implemented on a workstation with 7th Gen Intel QuadCore 3.6 GHz Processor, Nvidia Quadro P4000 GPU 8 GB, 32 GB RAM, and Ubuntu 16.04 LTS platform. TensorFlow numerical computation Library was used for the implementation of deep learning function calls.
Effectiveness of attention gating mechanism
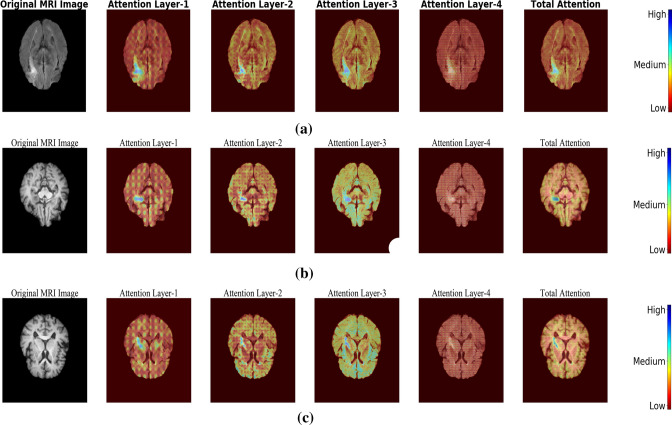
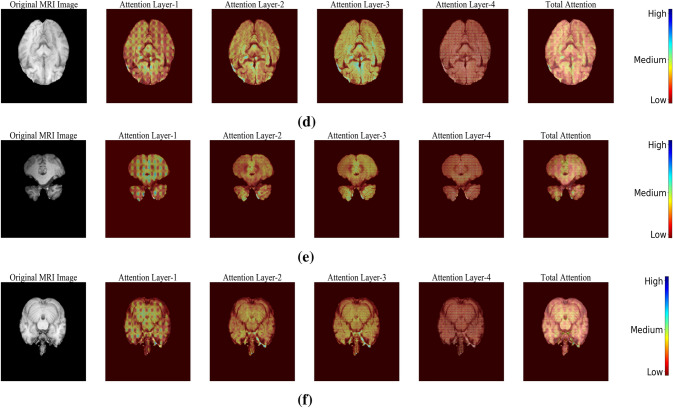
To clearly illustrate the effectiveness of attention gating mechanism, the response from each gate before applying sigmoidal activation is captured and the resultant feature maps are highlighted in Fig. 3. From samples (a) to (c), it could be observed that the attention gates were very effective in capturing the lesion related details. Sample (d) to (f) presents the inference for a normal sample, where the contribution made by these attention gates was less when compared to abnormal samples with the lesion.
Fig. 3.
Visualization of attention weights generated for a–c Samples with lesion and d–f Normal samples without lesion
From Fig. 3, it was evident that attention gates were highly efficient in focusing the lesion pixels of the image. Also, it helps in reconstructing the lesion which has different morphological properties like, size, shape, etc.
Performance analysis
The performance of the proposed FCN architecture for ischemic lesion segmentation was analyzed in this section. Figure 4 illustrates the change in values of loss parameter obtained during each fold of the training phase.
Fig. 4.
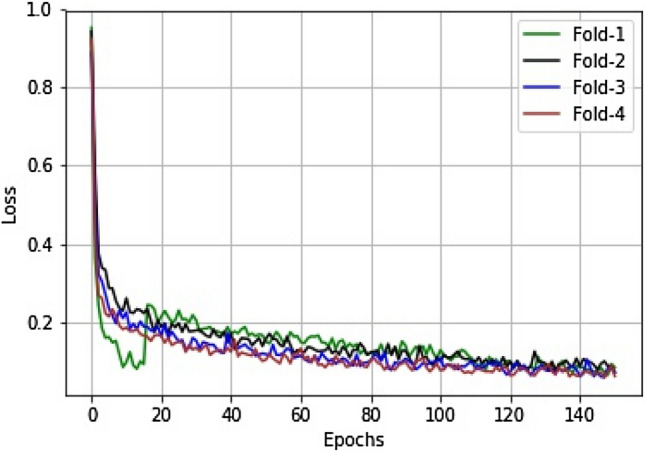
The decay of loss values during the training phase for the FCN with an attention gating mechanism
The observations obtained during the cross-validation phase were presented in Table 1.
Table 1.
Cross-validation performance of FCN with and without attention mechanism
| S.No | FCN without attention mechanism | FCN with attention mechanism | ||||||
|---|---|---|---|---|---|---|---|---|
| Time taken for training | Time taken for testing (in Sec) | Mean dice coefficient | Time taken for training | Time taken for testing (in Sec) | Mean dice coefficient | |||
| Training | Validation | Training | Validation | |||||
| 1 | 7 h, 55 min | 0.953 | 0.8723 | 0.8032 | 8 h, 31 min | 0.993 | 0.9175 | 0.8432 |
| 2 | 7 h, 53 min | 0.952 | 0.8829 | 0.8173 | 8 h, 33 min | 0.992 | 0.9292 | 0.8573 |
| 3 | 7 h, 51 min | 0.953 | 0.8709 | 0.7823 | 8 h, 29 min | 0.993 | 0.9309 | 0.7930 |
| 4 | 7 h, 52 min | 0.951 | 0.8807 | 0.7634 | 8 h, 31 min | 0.993 | 0.9397 | 0.7731 |
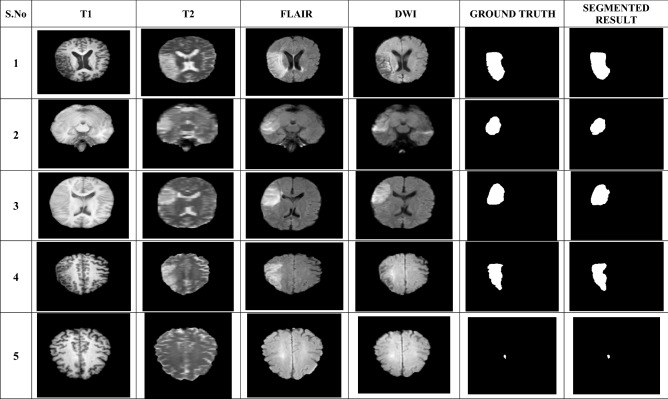
A mean Dice coefficient of 0.7535 was obtained for testing datasets when attention mechanism was integrated with FCN architecture. The FCN with attention mechanism was applied to different datasets and the segmented regions are presented in Table 2. It could be observed that the attention gated mechanism has produced appreciable results in extracting the lesion segments with different structural properties.
Table 2.
Segmented results of the attention-based FCN
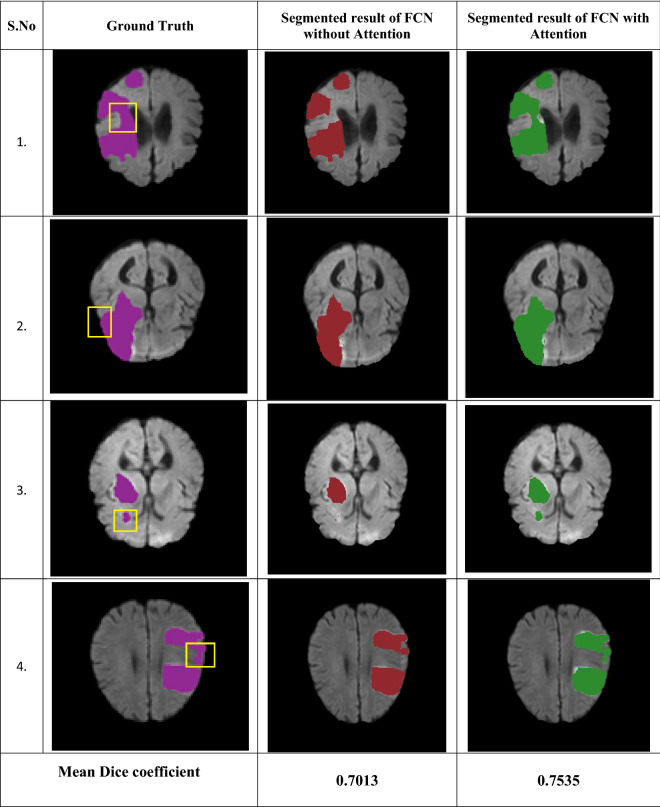
To highlight the effectiveness of attention mechanism in lesion segmentation, the segmented regions obtained using FCN with and without attention gates are presented in Table 3. It could be observed from these images that, the segmented region by the proposed approach correlates well with the ground truth reference. A bounding box was employed to visually indicate what portion of the region has been missed by FCN without attention mechanism and how it is captured by including attention mechanism with FCN.
Table 3.
Comparative analysis of segmentation results with and without attention gates
The performance of the proposed method is compared against various existing works for ischemic lesion detection and the results are highlighted in Table 4.
Table 4.
Comparative analysis of proposed work with existing works
| S.No | Source | Modality | Methodology | Mean Dice coefficient (DC) |
|---|---|---|---|---|
| 1. | Mitra et al. [8] | Multi-modal MRI | Bayesian-Markov Random Field (MRF) and Random Forests | 0.60 |
| 2. | Maier et al. [10] | FLAIR | Extra Tree forests | 0.65 |
| 3. | Kamnitsas et al. [22] | Multi-modal MRI | CNN | 0.66 |
| 4. | Chen et al. [21] | DWI | CNN | 0.67 |
| 5. | O. Maier et al. [31] | Multi-modal MRI | Decision trees, CNN | 0.67 |
| 6. | Karthik et al. [23] | Multi-modal MRI | FCN | 0.70 |
| 7. | Proposed approach | Multi-modal MRI | FCN with attention mechanism | 0.75 |
Supervised learning approaches like Random forests, Extra tree forests have been applied for ischemic lesion detection in [8, 10]. These methods primarily extract hand-engineered features to differentiate the properties of brain tissues. Hence, the dice coefficient was in the range of 0.60–0.65. To address this research gap, deep neural architectures using CCN and FCN were employed as it supports automatic feature extraction [21] 22, 23, and 31]. The DC of these approaches were in the range of 0.64–0.70. To further improve the performance of lesion segmentation, a deep supervised FCN based architecture with attention gating mechanism was presented in this research. The original FCN architecture was improved by including attention gates at the decoder. The information obtained from the individual skip connections might also pass some irrelevant feature responses to the decoder for reconstruction. To filter such irrelevant feature response and allow only significant feature response to the decoder, attention gates were employed in the network. This enables the network to filter prominent feature map activations that support precise reconstruction. Accuracy of 0.7535 was obtained by combining the advantages of automatic feature learning in CNN with the filtering mechanism of attention gates.
To further enhance the performance of segmentation, this attention mechanism can be directly tried with 3D CNN. This can support effective 3D volumetric visualization of the lesion structures. Also, it can be tried with other datasets like ISLES 2018, to analyze the effectiveness of the proposed architecture for other advanced imaging modalities like Perfusion CT.
Conclusion
In this research, an FCN was applied to segment ischemic lesion from multimodal MRI. The attention gating mechanism was included in FCN to efficiently delineate the ischemic lesion. The major highlight of this research is the application of attention gates which filters the irrelevant activations generated from multiple skip connections. Hence, it admits only the salient features to further levels for reconstruction. An extensive analysis was performed to analyze the performance of the proposed approach with the traditional network without attention models. The proposed network was able to segment ischemic lesion with different morphological properties like size, shape, etc. Also, the proposed method was successful in delineating scattered lesions with heterogeneous intensity profile.
It could be observed that the attention gated network was able to segment lesion with a dice coefficient of 0.75. An overall improvement of 17% has been achieved when compared to the existing methods. The proposed attention gated network was quite effective in delineating hyper-intense abnormal structures from brain MRI. Hence, this architecture can be extended to other neurological diseases.
Compliance with ethical standards
Conflict of interest
The authors do not have any conflict of interest.
Ethical approval
This article does not contain any studies with animals performed by any of the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
R. Karthik, Email: r.karthik@vit.ac.in
Menaka Radhakrishnan, Email: menaka.r@vit.ac.in.
R. Rajalakshmi, Email: rajalakshmi.r@vit.ac.in
Joel Raymann, Email: joelraymann@gmail.com.
References
- 1.Cheng NT, Kim AS. Intravenous thrombolysis for acute ischemic stroke within 3 hours versus between 3 and 4.5 hours of symptom onset. Neurohospitalist. 2015;5(3):101–109. doi: 10.1177/1941874415583116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karthik R, Menaka R. Computer-aided detection and characterization of stroke lesion—a short review on the current state-of-the art methods. Imaging Sci J. 2018;66(1):1–22. doi: 10.1080/13682199.2017.1370879. [DOI] [Google Scholar]
- 3.Artzi M, Aizenstein O, Jonas-Kimchi T, Myers V, Hallevi H, Ben Bashat D. FLAIR lesion segmentation: application in patients with brain tumors and acute ischemic stroke. Eur J Radiol. 2013;82(9):1512–1518. doi: 10.1016/j.ejrad.2013.05.029. [DOI] [PubMed] [Google Scholar]
- 4.Karthik R, Menaka R. A multi-scale approach for detection of ischemic stroke from brain MR images using discrete curvelet transformation. Measurement. 2017;100:223–232. doi: 10.1016/j.measurement.2017.01.001. [DOI] [Google Scholar]
- 5.Karthik R, Menaka R. Statistical characterization of ischemic stroke lesions from MRI using discrete wavelet transformation. Trans Electr Eng Electron Commun. 2016;14(2):57–64. [Google Scholar]
- 6.Karthik R, Menaka R. A novel brain MRI analysis system for detection of stroke lesions using discrete wavelets. J Telecommun Electron Comput Eng. 2016;8(5):49–53. [Google Scholar]
- 7.Karthik R, Menaka R. A critical appraisal on wavelet based features from brain MR images for efficient characterization of ischemic stroke injuries. Electron Lett Comput Vis Image Anal. 2016;15(3):1–16. [Google Scholar]
- 8.Mitra J, Bourgeat P, Fripp J, Ghose S, Rose S, Salvado O, Connelly A, Campbell B, Palmer S, Sharma G, Christensen S, Carey L. Lesion segmentation from multimodal MRI using random forest following ischemic stroke. NeuroImage. 2014;98:324–335. doi: 10.1016/j.neuroimage.2014.04.056. [DOI] [PubMed] [Google Scholar]
- 9.Sivakumar P, Ganeshkumar P. An efficient automated methodology for detecting and segmenting the ischemic stroke in brain MRI images. Int J Imaging Syst Technol. 2017;27(3):265–272. doi: 10.1002/ima.22231. [DOI] [Google Scholar]
- 10.Maier O, Wilms M, von der Gablentz J, Krämer U, Münte T, Handels H. Extra Tree forests for sub-acute ischemic stroke lesion segmentation in MR sequences. J Neurosci Methods. 2015;240:89–100. doi: 10.1016/j.jneumeth.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 11.Kajbakhsh N, Shin JY, Gurudu S, Hurst RT, Kendall CB, Gotway MB, Liang J. Convolutional neural networks for medical image analysis: full training or fine tuning? IEEE Trans Med Imaging. 2016;35(5):1299–1312. doi: 10.1109/TMI.2016.2535302. [DOI] [PubMed] [Google Scholar]
- 12.Chin C et al. An automated early ischemic stroke detection system using CNN deep learning algorithm. In: 2017 IEEE 8th International conference on awareness science and technology (iCAST), Taichung, 2017. pp. 368–72.
- 13.Mohsen H, El-Dahshan E, El-Horbaty E, Salem A. Classification using deep learning neural networks for brain tumors. Future Comput Inform J. 2018;3(1):68–71. doi: 10.1016/j.fcij.2017.12.001. [DOI] [Google Scholar]
- 14.Diniz P, Valente T, Diniz J, Silva A, Gattass M, Ventura N, Muniz B, Gasparetto E. Detection of white matter lesion regions in MRI using SLIC0 and convolutional neural network. Comput Methods Programs Biomed. 2018;167:49–63. doi: 10.1016/j.cmpb.2018.04.011. [DOI] [PubMed] [Google Scholar]
- 15.Pereira DR, Filho PPR, de Rosa GH, Papa JP, de Albuquerque VHC. Stroke lesion detection using convolutional neural networks. In: 2018 International joint conference on neural networks (IJCNN), Rio de Janeiro, 2018, pp. 1–6.
- 16.Cui S, Mao L, Xiong S. Brain tumor automatic segmentation using fully convolutional networks. J Med Imaging Health Inform. 2017;7(7):1641–1647. doi: 10.1166/jmihi.2017.2179. [DOI] [Google Scholar]
- 17.WangY, Sun Z, Liu V, Peng V, Zhang J. MRI image segmentation by fully convolutional networks. In: 2016 IEEE international conference on mechatronics and automation, Harbin, 2016. pp. 1697–702.
- 18.Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells W, Frangi A, editors. Medical image computing and computer-assisted intervention—MICCAI 2015. MICCAI 2015. Cham: Springer; 2015. [Google Scholar]
- 19.Shaikh M, Anand G, Acharya G, Amrutkar A, Alex V, Krishnamurthi G. Brain tumor segmentation using dense fully convolutional neural network. In: Crimi A, Bakas S, Kuijf H, Menze B, Reyes M, editors. Brainlesion: Glioma, multiple sclerosis, stroke and traumatic brain injuries. BrainLes 2017. Cham: Springer; 2018. [Google Scholar]
- 20.Shen H, Wang R, Zhang J, McKenna SJ. Boundary-aware fully convolutional network for brain tumor segmentation. In: Descoteaux M, Maier-Hein L, Franz A, Jannin P, Collins D, Duchesne S, editors. Medical image computing and computer-assisted intervention—MICCAI 2017. Cham: Springer; 2017. [Google Scholar]
- 21.Chen L, Bentley P, Rueckert D. Fully automatic acute ischemic lesion segmentation in DWI using convolutional neural networks. Neuro image Clin. 2017;15:633–643. doi: 10.1016/j.nicl.2017.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamnitsas K, Chen L, Ledig C, Rueckert D, Glocker B. Multi-scale 3D convolutional neural networks for lesion segmentation in brain MRI. In: Proc of ISLES-MICCAI, 2015.
- 23.Karthik R, Gupta U, Jha A, Rajalakshmi R, Menaka R. A deep supervised approach for ischemic lesion segmentation from multimodal MRI using fully convolutional network. Appl Soft Comput. 2019;84:105685. doi: 10.1016/j.asoc.2019.105685. [DOI] [Google Scholar]
- 24.Liu S, Shen F, Komandur Elayavilli R, Wang Y, Rastegar-Mojarad M, Chaudhary, V, Liu H. Extracting chemical–protein relations using attention-based neural networks. Database, 2018. [DOI] [PMC free article] [PubMed]
- 25.Chen Y, Zhao D, Lv L, Li C. A visual attention based convolutional neural network for image classification. In: 2016 12th World congress on intelligent control and automation (WCICA), Guilin, 2016. pp. 764–69.
- 26.Yu D, Fu , Mei T, Rui Y. Multi-level attention networks for visual question answering. In: 2017 IEEE conference on computer vision and pattern recognition (CVPR), Honolulu, HI, 2017. pp. 4187–95.
- 27.Jetley S, Lord NA, Lee N, Torr P. Learn to pay attention. In: International conference on learning representations, 2018.
- 28.Wang F et al. Residual attention network for image classification. In: 2017 IEEE conference on computer vision and pattern recognition (CVPR), Honolulu, HI, 2017. pp. 6450–58.
- 29.Guan Q, Huang Y. Multi-label chest x-ray image classification via category-wise residual attention learning. Pattern Recognit Lett. 2020;130:259–266. doi: 10.1016/j.patrec.2018.10.027. [DOI] [Google Scholar]
- 30.Schlemper J, Oktay O, Schaap M, Heinrich M, Kainz B, Glocker B, Rueckert D. Attention gated networks: learning to leverage salient regions in medical images. Med Image Anal. 2019;53:197–207. doi: 10.1016/j.media.2019.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maier O, Schröder C, Forkert ND, Martinetz T, Handels H. Classifiers for ischemic stroke lesion segmentation: a comparison study. PLoS ONE. 2015;10(12):e0145118. doi: 10.1371/journal.pone.0145118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maier O, Menze BH, von der Gablentz J, H¨ani L, Heinrich MP, Liebrand M, Winzeck S, Basit A, Chen L, et al. Isles 2015-A public evaluation benchmark for ischemic stroke lesion segmentation from multispectral MRI. Med Image Anal. 2017;35:250–269. doi: 10.1016/j.media.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]