Abstract
A major event in embryonic development is the rearrangement of epigenetic information as the somatic genome is reprogrammed for a new round of organismal development. Epigenetic data are held in chemical modifications on DNA and histones, and there are dramatic and dynamic changes in these marks during embryogenesis. However, the mechanisms behind this intricate process and how it is regulating and responding to embryonic development remain unclear. As embryos develop from totipotency to pluripotency, they pass through several distinct stages that can be captured permanently or transiently in vitro. Pluripotent naïve cells resemble the early epiblast, primed cells resemble the late epiblast, and blastomere-like cells have been isolated, although fully totipotent cells remain elusive. Experiments using these in vitro model systems have led to insights into chromatin changes in embryonic development, which has informed exploration of pre-implantation embryos. Intriguingly, human and mouse cells rely on different signaling and epigenetic pathways, and it remains a mystery why this variation exists. In this review, we will summarize the chromatin rearrangements in early embryonic development, drawing from genomic data from in vitro cell lines, and human and mouse embryos.
Keywords: embryonic stem cells, naïve, primed, epigenetic, transposable elements, reprogramming
Introduction
Each cell contains the same DNA that is interpreted to provide specialized cell function, yet the interpretation of the DNA code is cell type-specific, and epigenetic barriers exist that impair and permit cell type conversions. Cell type conversions are surprisingly rare in the adult organism, outside some stem cells, which retain limited ability to derive various progeny (Avgustinova and Benitah, 2016). Despite the importance of understanding cell type control for regenerative therapies, exactly how this process is controlled remains surprisingly elusive (Hutchins et al., 2017). Epigenetic control mechanisms are a major contributor to this process; however, there are a wide array of overlapping and competing mechanisms, particularly histone and DNA chemical modifications (Ohbo and Tomizawa, 2015). Epigenetic modifications form barriers that can be permissive for some cell type transitions but intolerant for others. These epigenetic barriers can resist all cell type conversions but can also act as bidirectional valves, guiding cells toward a differentiated cell type but then blocking reversion to progenitor cells and locking cells into a fixed cell type (Arabaci et al., 2020). Epigenetic control is mediated by DNA-binding proteins, particularly transcription factors (TFs) (Fu et al., 2017), that recruit epigenetic enzymes to regulate chemical modifications on DNA and histones and reshape or remodel chromatin structure. This process both responds to cell type changes and controls them and is critical in normal developmental processes. During embryonic development of a new organism, epigenetic information is reset (Guo et al., 2014; Xia and Xie, 2020), and many epigenetic rearrangements occur during early pre-implantation development and accompany, and in some instances drive, developmental changes. Research to understand the epigenetic reconfigurations in embryonic development has been intense, as understanding epigenetic control could give fine-grained control of cell type states, which would be useful for a wide range of medical applications, from regeneration (Avgustinova and Benitah, 2016), and cell replacement therapy, to understanding how cell type control is disrupted in cancerous cells (Meacham and Morrison, 2013). In this review, we will discuss epigenetic control of early embryonic stages, using work from embryos, in vitro mimics of embryonic cells, and cell type conversion systems.
NaÏVe and Primed Cells in Mice
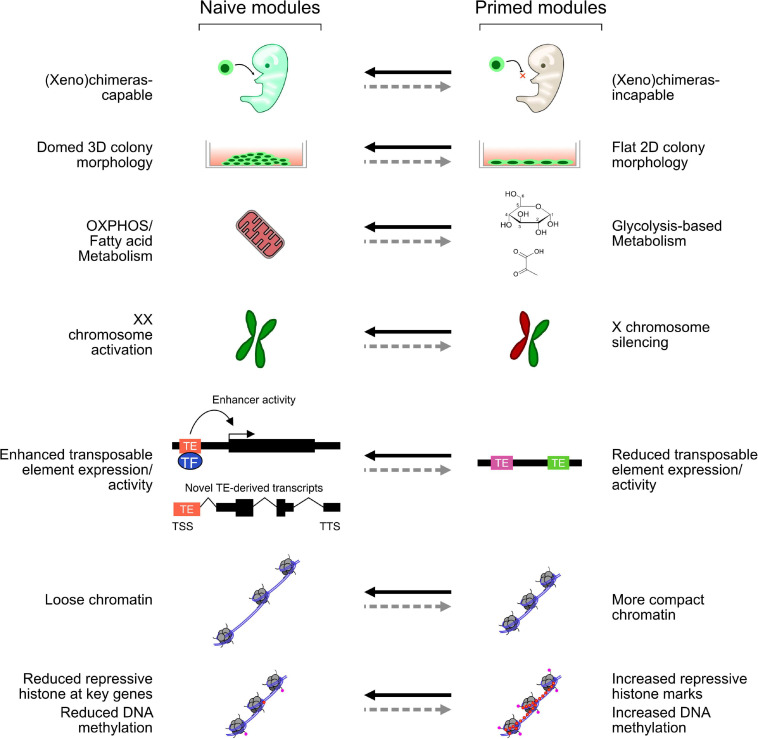
Amongst the first in vitro-derived embryonic cells were the human and mouse embryonal carcinoma (EC) cell lines, which can be maintained in vitro but are derived from teratocarcinomas and have multiple mutations and abnormal karyotypes (Martin, 1980). Several EC lines were derived, and while each line has a set of common embryonic features, they also have line-specific effects, such as restricted differentiation potential and different culture requirements (Andrews, 1988; Alonso et al., 1991). When ECs are injected into a mouse blastocyst, they can contribute to development but often lead to teratomas in the adult mice. Fully viable EC-derived chimeras have been reported (Hanaoka et al., 1987); however, considering what we now know about the rapid growth of normal, untransformed mouse embryonic stem cells (mESCs), it is not inconceivable that those EC cultures harbored small numbers of mESCs. Compared with ECs, mESCs are untransformed, contribute to chimeras at high frequency without generating teratomas in the adult, and can be grown in vitro in defined conditions indefinitely. In vitro, mESCs can be differentiated to all three germ lineages and have become a powerful model of the early stages of embryonic development (Rossant, 2011). mESCs are thought to most closely resemble the early epiblast (Boroviak et al., 2014). They express marker genes specific for the epiblast; and in female mESCs, both X chromosomes are active; and silencing is required to exit the pluripotent state (Schulz et al., 2014). mESCs were first derived in 1981 (Evans and Kaufman, 1981), but it was not until 1998 that human ESCs (hESCs) were reported (Thomson et al., 1998). However, there are several morphological and molecular differences between mESCs and hESCs: mESC colonies are domed, hESCs are flat, hESCs rely on glycolysis, and mESCs oxidate phosphorylation (Zhou et al., 2012; Sperber et al., 2015). There are also major differences in the ectopic signaling pathways that are required: mESCs rely on bone morphogenic protein 4 (BMP4) and leukemia inhibitory factor (LIF) signaling (Ying et al., 2003); however, hESCs rely on Activin A and fibroblast growth factors (FGFs) (Beattie et al., 2005); and not only is BMP signaling not required for hESCs, but inhibition of BMP signaling is even beneficial (Xu et al., 2005). Application of the hESC growth medium to mouse cells led to the isolation of epiblast stem cells (EpiSCs). These cells were derived from E6.5 pre-gastrulating embryos and are quite different in morphology, gene expression, and culture conditions than mESCs (Brons et al., 2007; Tesar et al., 2007). EpiSCs, instead, more closely resemble hESCs, and the pre-gastrulating epiblast, a later developmental stage than mESCs. To explain the properties of mESCs and EpiSCs, a two-phase model of embryonic development was proposed, consisting of a “naïve” (mESC) state, which is closer to the early epiblast, and a “primed” (EpiSC) state, which is closer to the late pre-gastrulating epiblast. Other cell types exist on a continuum between these two conditions and, sometimes, outside of this classification (Nichols and Smith, 2009). Several features distinguish the naïve and primed states (Figure 1). Primed and naïve cells have a distinctive morphology: primed cells are flat and more two-dimensional, while naïve cells have a domed shape and are more three-dimensional (3D). Primed cells are more glycolytic, while naïve cells rely more on oxidative phosphorylation (Zhou et al., 2012). In females, the X chromosomes are active in naïve but silent in primed cells. Finally, in the naïve state, the chromatin tends to be looser and, overall, less repressive, which enables more transposable element (TE) expression.
FIGURE 1.
Selected modules representing key features of naïve and primed cells. Each module can be switched on and off, perhaps independently, in naïve and primed mouse and human embryonic stem cells (hESCs). While mouse ESCs (mESCs) activate all these modules, different naïve-like hESCs may only activate some aspects.
The Naïve and Ground States in Mouse Embryonic Stem Cells
Typical mESCs are grown in serum supplemented with the cytokine LIF (Smith et al., 1988; Williams et al., 1988) and adopt a “naïve” state. mESCs can also be supported in medium referred to as ground state or “2iLIF” medium, which consists of PD0325901 (a MEK inhibitor), CHIR99021 (a GSK3 inhibitor), and LIF (Ying et al., 2008). Intriguingly, growth in 2iLIF improves the cells, making them easier to maintain, less heterogeneous, and less prone to spontaneous differentiation (Kolodziejczyk et al., 2015; Takashima et al., 2015). Cells grown in 2iLIF also have an altered chromatin state and reduced levels of repressive marks; for example, H3K27me3 levels are reduced at developmental genes (Marks et al., 2012) and lower levels of other repressive histone modifications (Sim et al., 2017). The change in DNA methylation is more drastic: serum + LIF-grown mESCs have slightly reduced DNA methylation levels, compared with somatic cells, but cells grown in 2iLIF are nearly completely demethylated (Habibi et al., 2013; Leitch et al., 2013). The mechanism appears to be mediated by one of the inhibitors in the 2i cocktail, the MEK inhibitor PD0325901, which indirectly reduces the levels of H3K9me2 and so blocks the recruitment of UHRF1 to chromatin. The loss of UHRF1 then leads to a failure to recruit the methyltransferase maintainer DNMT1, and so DNA is passively demethylated as cells divide (von Meyenn et al., 2016). A complementary mechanism posits a more active process involving MEK inhibitor indirectly stabilizing the histone H3K9me3 demethylase KDM4C (JMJD2C). This leads to demethylation of H3K9me3, the active conversion of 5mC to 5hmC by TET1 hydroxylase, and eventual demethylation (Sim et al., 2017). The global DNA demethylation in naïve cells is reminiscent of a similar DNA demethylation that occurs in early embryonic development between the two-cell (2C) and blastocyst stages (Guo et al., 2014; Smith et al., 2014). However, how close MEK inhibitor-driven DNA demethylation is to embryonic DNA hypomethylation is unclear. Another issue that is complex in 2iLIF-grown cells is that while H3K27me3 at gene promoters is reduced, 2iLIF cells have increased overall H3K27me3 levels and often have increased heterochromatic marks (van Mierlo et al., 2019). This suggests that 2iLIF cells have an overall more repressed chromatin state. One possibility is that the loss of DNA methylation leads to compensatory mechanisms that repress genes and generate heterochromatin by methylating histones. Indeed, H3K27me3 and H3K9me3 compensate for the loss of DNA methylation to repress TEs in mESCs (Walter et al., 2016).
The bromodomain-containing protein BRD4 is an epigenetic reader, which binds to acetylated histones. Brd4 knockout mice lack an inner cell mass, and when Brd4 was inhibited or knocked down in naïve cells, it led to differentiation, at least partly due to the loss of Nanog expression and other pluripotency genes (Di Micco et al., 2014; Liu et al., 2014; Horne et al., 2015). BRD4 also has a key role in maintaining enhancers, by recruiting CDK9 and the mediator complex (Di Micco et al., 2014). Beyond the direct regulation of pluripotent genes, BRD4 has a complex role in mediating the differences between the 2iLIF ground state and serum + LIF-grown naïve cells, as 2iLIF cells can tolerate the loss of Brd4, while serum + LIF cells cannot (Zhang et al., 2020b). The mechanism involves the inhibition of GSK3 in 2iLIF conditions, which leads to stabilization of beta-catenin, and the recruitment of BRD4 and a multimolecular protein complex to pluripotency genes to make 2iLIF-grown mESCs resistant to differentiation (Zhang et al., 2020b). This results in reduced pause-release of RNA polymerase II (polII) and more stable transcription, which helps explain previous observations that 2iLIF-grown cells have more homogenous gene expression than serum + LIF-grown cells (Kolodziejczyk et al., 2015). The mediator complex is a multiprotein complex that is important at integrating signals to activate nearby gene expression by recruiting polII. It has a key role in regulating super-enhancers, which are large regions of DNA with a potent enhancer activity that are important for regulating cell type-specific genes (Whyte et al., 2013). Mediator, in addition to being recruited by BRD4, also has a role in naïve and primed states, as chemical inhibition of two CDKs, CDK8/19, promotes the naïve state in both humans and mice (Lynch et al., 2020). This effect is driven by the hyperactivation of enhancers, as inhibition of CDK8/19 leads to derepression of mediator. BRD4 is not the only bromodomain-containing protein involved in naïve and primed state control. BRD9 was identified as a member of a non-canonical chromatin remodeling BAF complex, and when BRD9 was inhibited, the cells began to acquire aspects of EpiSCs, and, like BRD4, Brd9 is dispensable in 2iLIF conditions (Gatchalian et al., 2018).
Chromatin inside the cell is tightly packed into successive 3D layers that can be broadly divided into a hierarchy of three organizational features (Rowley and Corces, 2018). The first level is the A and B compartments, which, very roughly, correspond to euchromatin (A compartment) and heterochromatin (B compartment). At the second level, topologically associated domains (TADs) are megabase domains of chromatin that extensively interact within a TAD but weakly between TADs. Finally, at the third level, individual TFs and epigenetic factors form contacts between strands of DNA to form chromatin loops, which are often responsible for bringing distal enhancers together with promoters. mESCs have unique features at all three of these levels, which are suggestive of open and relaxed chromatin. As mESCs are differentiated to neurons, the A compartments decrease and the B compartments increase in interaction frequency, indicating the loss of active chromatin and the acquisition of repressed chromatin as mESCs differentiate (Bonev et al., 2017). In human cells, the situation is similar, and a high-resolution HiC dataset in hESCs and somatic cells showed many A to B compartment switches (Dixon et al., 2015). At the second level, TAD compartment structure strengthened as mESCs differentiated, and TADs containing actively expressed genes interacted weakly, while inactive TADs increased (Bonev et al., 2017). The chromatin state can also influence the 3D genome folding, as knockout of the H3K9me1/2 methyltransferase Ehmt2 led to reduced TAD boundary strength, although compartments were unaffected (Jiang et al., 2020).
Intriguingly, the 3D structure in developing embryos is initially undefined. From the zygote to the late 2C stage, the TADs and chromatin loops are nearly completely absent, and only compartments on the paternal genome are weakly present (Du et al., 2017; Ke et al., 2017). TADs and chromatin loops reestablish at the eight-cell to the morula stages (Du et al., 2017; Ke et al., 2017). TADs and compartments reform around the same time as zygotic genome activation (ZGA), and there is some evidence that the reestablishment of 3D structure can influence embryonic development. In somatic cell nuclear transfer (SCNT) experiments, the somatic nucleus inside the oocyte briefly retains TADs, which are relaxed at the 2C stage and match normal development. However, the brief window when TADs are erroneously present impairs minor ZGA and embryonic development at the 2C stage, which can be rescued by depleting cohesin to help disrupt TADs in the somatic nucleus (Zhang et al., 2020a). Based on these observations, the totipotent stages of embryonic development (zygote to late eight-cell stage) seem to require relaxed unstructured 3D chromatin. However, it is unclear if this is a necessary feature of totipotency or a consequence of epigenetic reprogramming in early embryogenesis or simply reflects rearrangements in chromatin that are independent of embryonic development. It would be useful to explore these issues in the more experimentally tractable mESCs, and several systems have been explored that lead to dissolution of the 3D genome. With the use of a degron system, the key cohesin complex member RAD21 was deleted in mESCs, leading to the near-complete loss of TADs but a slight strengthening of A/B compartments, and RING1B-mediated polycomb loops persisted (Rhodes et al., 2020). However, depletion of RAD21 in mESCs had surprisingly little effect on gene expression or cell phenotype (Rhodes et al., 2020). CTCF, a major 3D genome organizer, was similarly degraded in mESCs. A/B compartments were unaffected, but TADs and chromatin loops were disrupted; however, once again, the effect on the mESC phenotype was modest, although there was a proliferation defect if CTCF loss persisted (Nora et al., 2017). These results suggest that 3D structure is relatively uncoupled from cell type control, although the precise 3D structure of embryonic cells has not been fully recapitulated in mESCs and hence remains inconclusive.
A remarkable feature of mESCs is their tolerance for the loss of epigenetic regulatory enzymes with relatively few effects (He et al., 2019). For example, components of the polycomb repressor complex 1 and 2 (PRC1 and PRC2) are dispensable for mESCs (Chamberlain et al., 2008). Loss of Rybp, a member of an atypical PRC1 complex, also has little to no effect (Li et al., 2017b). Knockdown of Setdb1, a H3K9me3 methyltransferase, only predisposes cells to differentiation (Karimi et al., 2011). Co-activators also show the same pattern: Kmt2d (MLL2), a H3K4 methyltransferase, is dispensable (Lubitz et al., 2007). Epigenetic factor knockouts often do not substantially impact the pluripotent state, although they may make them more prone to spontaneous differentiation and alter the differentiation direction of the cells, or, as is often the case for epigenetic factor knockouts, lead to embryonic arrest at gastrulation, for example, in the Kdm1a (Macfarlan et al., 2011) or Setdb1 (Dodge et al., 2004) knockouts. A CRISPR/Cas9 screen identified around 40 epigenetic factors that, when knocked out, delayed the differentiation of mESCs, and just two epigenetic factor knockouts promoted mESC differentiation, Cbx7 and Sin3b (Li et al., 2018). As their screen was set up primarily to detect improved or impaired ability to differentiate, knockouts that did not affect differentiation would not be detected. This emphasizes the remarkable ability of mESCs to tolerate the widespread loss of epigenetic regulators. This tolerance may be related to the more open and active chromatin, which appears to be a feature of early embryonic cells (Boskovic et al., 2014; Schlesinger and Meshorer, 2019).
Transcriptional Control of Primed Mouse Epiblast Stem Cells
Primed EpiSCs are a distinct cell state, compared with mESCs. EpiSCs show both molecular and phenotypic differences, particularly colony morphology and the lack of ability to form chimeras (Figure 1). Despite these phenotypic differences, mESCs and EpiSCs have both shared and distinct transcriptional regulation. The core pluripotent network of OCT4, SOX2, and NANOG are active in both cell types (Weinberger et al., 2016), but they bind to different genomic loci (Buecker et al., 2014; Galonska et al., 2015; Matsuda et al., 2017). The core pluripotency network is coordinated by a different set of TFs in each cell type. For example, ESRRB, NR0B1, ZFP42, and TFCP2L1 are important in mESCs (Festuccia et al., 2012; Hutchins et al., 2013; Adachi et al., 2018; Atlasi et al., 2019), but ZIC2, ZIC3, POU3F1, and OTX2 are key in EpiSCs (Acampora et al., 2013; Matsuda et al., 2017; Yang et al., 2019c). Thus, there is a core gene expression module that is common to mESCs and EpiSCs, and divergent regulatory modules specific to each cell type. Both cell types are centered around SOX2-OCT4, but mESCs use TFs that are expressed in the early blastocyst (e.g., NANOG, ESRRB, and TFCP2L1), and EpiSC-specific TFs tend to be expressed in the gastrulating blastocyst (OTX2 and POU3F1). In addition to transcriptional differences, the chromatin and epigenetic states are also altered between EpiSCs and mESCs, and enhancer usage is dramatically altered between EpiSCs and mESCs. Even though only a few hundred genes change expression between mESCs and EpiSCs, several tens of thousands of enhancer marks are differentially regulated (Factor et al., 2014). Even genes that are expressed in both cell types can utilize different enhancers (Factor et al., 2014). This effect had been seen previously at the Pou5f1 locus, which has two enhancers, a distal enhancer that is active in preimplantation embryos and mESCs, and a proximal enhancer that is active in the epiblast and EpiSCs (Yeom et al., 1996). Yet the global profiling of chromatin highlighted how widespread this phenomenon is (Factor et al., 2014). This redistribution of enhancers is ultimately driven by changes in the TF activity, which decommission and activate panels of enhancers to control each cell state. A good example is OCT4 and SOX2, and both are expressed in mESCs and EpiSCs but are drastically altered in their binding patterns in the two-cell types (Matsuda et al., 2017). It is most likely that OCT4 and SOX2 binding is altered due to the activity of OTX2 and POU3F1, which becomes the dominant factors in EpiSCs (Matsuda et al., 2017).
Interconversion Between Mouse Naïve and Primed Cells
Mouse embryonic stem cells and EpiSCs can be interconverted in vitro, and while conversion of mESCs to EpiSCs is relatively efficient, the conversion of EpiSCs to ESCs remains inefficient without transgenes or epigenetic modulation (Zhou et al., 2010; Tosolini and Jouneau, 2016; Stuart et al., 2019). This indicates that EpiSCs are developmentally later, as, in general, cells can efficiently differentiate toward their progeny but are resistant to dedifferentiation to their precursor cell type. Epigenetic barriers, particularly unidirectional blocks, appear to permit the conversion of mESCs to EpiSCs but block the reverse. ZFP281 acts as just such a bidirectional valve, as it assists mESC to EpiSC conversion but blocks the reverse (Mayer et al., 2020). The mechanism involves ZFP281 co-binding with EHMT1 to methylate H3K9 and inhibit genes in the early stages of mESC to EpiSC conversion (Mayer et al., 2020), while in the reverse case ZFP281 binds the NuRD co-repressor complex to suppress Nanog expression and enable exit from pluripotency (Fidalgo et al., 2012). When mESCs are converted to EpiSCs, there is a global reconfiguring of chromatin (Factor et al., 2014). These properties have made the interconversion of mESCs to EpiSCs a powerful model to understand the epigenetic modulation of cell conversions.
Several factors have been identified that influence the conversion of mESCs to EpiSCs (Table 1). Most are TFs that promote the conversion of EpiSCs to mESCs. These TFs recruit other co-activators and co-repressors to influence the chromatin state, although their direct activity is not always clear, as TFs can often act as both activator and repressor in a context-specific manner. For example, NANOG is mainly an activator but can also work as a repressor (Heurtier et al., 2019). When NANOG binds to DNA with ZFP281, it recruits the NuRD histone deacetylase (HDAC) co-repressor complex (Fidalgo et al., 2012). Esrrb is a major requirement to convert EpiSCs to mESCs, as it participates in extensive chromatin remodeling (Adachi et al., 2018). Many naïve-specific enhancers are kept silent in EpiSCs by DNA methylation and inaccessible chromatin. ESRRB opens these naïve-specific enhancers by recruiting the p300 complex, displacing and phasing nucleosomes, and opening closed chromatin, making it accessible for other members of the pluripotency regulatory network, such as OCT4, SOX2, and NANOG, to bind (Adachi et al., 2018).
TABLE 1.
Selected factors that influence the interconversions of mESCs and EpiSCs.
| Molecule(s) | Function | Effect on mESCs, EpiSCs, and their interconversion | References |
| Brd9/BAF complex | Chromatin remodeler | BRD9, through a non-canonical BAF complex, blocks transition to a primed state | Gatchalian et al., 2018 |
| Epiblastin A | Inhibitor | Inhibits CK1 and induces EpiSC-to-mESC conversion | Illich et al., 2016 |
| Esrrb | Transcription factor | Overexpression sustains LIF-independent self-renewal in the absence of Nanog and reverts EpiSCs to mESCs | Festuccia et al., 2012; Adachi et al., 2018 |
| Hif1a | Transcription factor | HIF-1α activation switches mESC metabolism and pushes cells toward an EpiSC-like state | Zhou et al., 2012 |
| iPStat3 | Gp130Y118F receptor | Artificially induces STAT3 signaling and converts mESCs to EpiSCs | Stuart et al., 2019 |
| Kdm1a/Kmt2d (MLL4) | Histone methylation | Kmt2d (MLL4) is required to exit pluripotency; knockdown of Kdm1a restores the ability of mESCs to convert to EpiSCs | Cao et al., 2018 |
| Kdm6b (JMJD3) | Histone demethylase | Demethylates H3K27me2 or H3K27me3; Kdm6b facilitates a Klf4-driven EpiSC-to-ESC conversion | Huang et al., 2020 |
| Klf2, Klf4 | Transcription factor | Overexpression sustains LIF-independent self-renewal and can convert EpiSCs to mESCs | Guo et al., 2009 |
| Kmt2a (MLL1) | Epigenetic inhibitor | H3K4me1 methyltransferase, deletion of Kmt2a (MLL1) impairs mESC differentiation to EpiLCs; MLL1 Inhibition reprograms EpiSCs to mESCs | Zhang et al., 2016 |
| Nanog | Transcription factor | Transient transfection of Nanog mediates reprogramming of mESCs to EpiSCs | Silva et al., 2009 |
| Nr5a1, Nr5a2 | Transcription factor | Overexpression reprograms EpiSCs to mESCs | Guo and Smith, 2010 |
| Otx2 | Transcription factor | Required to stably establish EpiSCs | Acampora et al., 2013 |
| Prdm14 | Epigenetic factor | Overexpression drives EpiLCs to mESC-like cells | Okashita et al., 2016 |
| Sall1 | Transcription factor | Promotes reprogramming EpiSCs to mESCs | Yang et al., 2019a |
| Setdb1 | Histone methyltransferase | Methyltransferase for H3K9me3; Setdb1 loss enriches a transient 2C-like state in mESCs | Wu et al., 2020 |
| Smarcad1 | SWI/SNF helicase | Knockdown converts mESCs to EpiSC-like cells | Xiao et al., 2017 |
| Tcf3, Etv5, Rbpj | Transcription factors | mESCs lacking Tcf3, Etv5, and Rbpj are trapped in a naïve pluripotent condition and are difficult to differentiate | Kalkan et al., 2019 |
| Tfe3 | Transcription factor | Nuclear-localized TFE3 blocks mESCs from differentiating | Betschinger et al., 2013 |
| Wnt, Gsk3b | Transcription factor | Wnt signaling blocks the conversion of mESCs to EpiSCs and maintains mESCs in the naïve state | Ying et al., 2008; ten Berge et al., 2011 |
| Zbtb7a/b | Transcription factor | Knockdown converts EpiSCs to naïve mESCs | Yu et al., 2020b |
| Zfp281 | Transcription factor | Deletion of Zfp281 promotes EpiSCs reprogramming and acts downstream of Ehmt1 (G9a-like protein; a histone methyltransferase for H3K9me1/2) | Mayer et al., 2020 |
| Zfp706 | Transcription factor | Deletion of Zfp706 promotes mESC self-renewal and promotes EpiSC reprogramming | Leeb et al., 2014 |
mESC, mouse embryonic stem cell; EpiSC, epiblast stem cell; LIF, leukemia inhibitory factor; EpiLC, epiblast-like cell.
Epigenetic pathways have been directly implicated in the conversion of mESC to EpiSCs. Activatory epigenetic marks are also redistributed between mESCs and EpiSCs (Factor et al., 2014), and the enhancer mark H3K4me1 has been directly implicated in the conversion to mESCs. Inhibition of the histone methyltransferase that catalyzes H3K4me1, MLL1, drives EpiSCs back to a naïve state (Zhang et al., 2016). In addition to H3K4me1/MLL1 inhibition, an inhibitor of the histone H3K4/9 demethylase KDM1A (LSD1) was part of a cocktail of chemicals that could promote the conversion of EpiSCs to mESCs (Zhou et al., 2010), underlining the importance of epigenetic modulation in cell type conversions. Histone citrullination is the post-translational conversion of arginine to citrulline, and it can act as an epigenetic mark, although its functions are not well defined. In naïve mESCs, histone H1 is citrullinated and evicted from chromatin, decondensing chromatin and likely making it more accessible for TF binding (Christophorou et al., 2014). Histone H3 can also be citrullinated, and it can recruit the SWI/SNF chromatin remodeler SMARCAD1 to relax chromatin (Xiao et al., 2017). Knockdown of Smarcad1 led to H3K9me3 deposition and heterochromatin spreading, and the cells adopted features of EpiSCs (Xiao et al., 2017). This suggests that citrullination assists in the control of heterochromatin and the maintenance of the naïve state. Overall, these observations agree with the idea that naïve cells represent an “unprogrammed” ground state with lower levels of both repressor and enhancer marks and agree with the idea that less overall epigenetic regulation is a feature of naïve mESCs (Schlesinger and Meshorer, 2019).
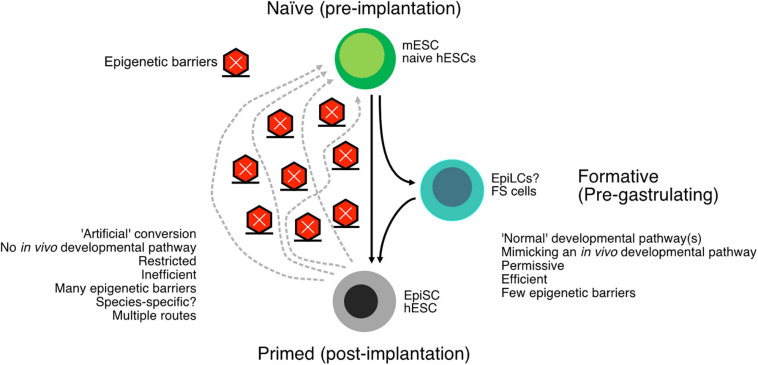
In summary, the naïve and primed states are relatively well described in mouse cells, and it is possible to interconvert the cell types. While conversion from mESCs to EpiSCs is relatively easy, the converse transition is difficult and often inefficient without transgenes. Indeed, there are multiple trajectories to convert EpiSCs back to mESCs, with some mechanisms passing cells through later developmental stages, such as mesoderm-like cells, and other conversion methods pass cells through earlier developmental stages (Stuart et al., 2019). It appears that there is a single main pathway in the differentiation of mESCs to EpiSCs but multiple pathways that EpiSCs must be forced along to revert to mESCs. These observations suggest the existence of epigenetic barriers, probably many, that act to impair the dedifferentiation of EpiSCs to mESCs (Figure 2). Ultimately, the interconversions of mESCs and EpiSCs are a crucial window into epigenetic controls that underlie cell type conversions.
FIGURE 2.
Schematic of the conversion of naïve embryonic stem cells (ESCs) to primed ESCs. The “downstream” pathway follows a normal developmental program and tends to be relatively easy and efficient. The reverse process, reverting primed cells to naïve cells, is arguably an artificial process and consequently more difficult and inefficient. Epigenetic barriers exist between the primed and naïve cells, and there may be more than one way to proceed through these epigenetic barriers. The many epigenetic barriers and the divergent pathways have likely contributed to the difficulty in generating human naïve ESCs. In addition to the naïve and primed states, there is also a formative state that exists intermediate to naïve and primed cells but is capable of primordial germ cell (PGC) formation.
An Intermediate, Formative Pluripotent State
Work on the naïve state in mice and humans has led to the emergence of a more subtle conception of naïve and primed states. Instead of existing as a binary state, either naïve or primed, there is instead a spectrum of states, some stable and others unstable, that exist between naïve and primed states. A recent proposal posits the existence of a critical intermediate state, the formative state, that exists between naïve and primed mESCs (Smith, 2017). The formative state represents the loss of naïve pluripotency but precedes lineage commitment. One of the first events in the in vitro differentiation of mESCs is the loss of the naïve-specific TFs, such as Tfcp2l1 and Esrrb. This matches a similar loss in the early post-implantation epiblast (Boroviak et al., 2014) and precedes lineage priming and acquisition of primed-specific genes. A formative state helps explain some curious phenomena that are not easily reconciled with a binary naïve and primed model. First, deriving primordial germ cells (PGCs) from both mESCs and EpiSCs is inefficient, despite both cell types being on the presumptive developmental path capable of generating PGCs (Hayashi et al., 2011). Second, a third type of in vitro epiblast cell, epiblast-like cells (EpiLCs), can give rise to PGCs at relatively high efficiency and can adopt a gene expression profile more reminiscent of E5.0–E6.0 pre-gastrulating blastocysts (Hayashi et al., 2011), which is similar to the timepoint for the specification of PGCs in the embryo (E5.5–E6.25) (Ohinata et al., 2009). This suggests that EpiLCs represent a transient window when PGCs are specified. An interesting aspect of the formative state is the extensive remodeling of chromatin that provides a blank slate for later lineage specification (Smith, 2017). Indeed, EpiLCs have lower levels of repressive histone marks, particularly H3K27me3 and H3K9me2, and have higher levels of bivalent promoters, marked by both H3K4me3 and H3K27me3 (Kurimoto et al., 2015; Yang et al., 2019b). This suggests the EpiLCs are poised for lineage commitment. A comprehensive multi-omic exploration of the conversion of mESCs to EpiLCs revealed a series of waves of gene expression changes that were preceded by widespread chromatin remodeling (Yang et al., 2019b). The conversion to a formative state relies on the expression and activity of Tcf3, Etv5, and Rbpj, as the deletion of all three impairs the differentiation of mESCs to EpiLCs (Kalkan et al., 2019). Of these three factors, their mechanism is unclear, but ETV5 binds to formative state-specific genes and promotes their expression, at least partly through histone acetylation (Kalkan et al., 2019), and all three combine to suppress the naïve pluripotency program.
A disadvantage of EpiLCs is their transient nature, and they cannot be captured in vitro like mESCs and EpiSCs. This makes a detailed exploration of the formative state challenging. Recently, two groups reported the isolation of cell lines that fulfill the properties of the formative state but can be maintained in culture (Kinoshita et al., 2020; Yu et al., 2020a). These cells [formative state (FS) and XPSCs] have a gene expression program distinct from mESCs and EpiSCs, yet they can partially contribute to mouse chimeras and can contribute to the germline (Kinoshita et al., 2020) and can generate PGCs in vitro (Yu et al., 2020a). Both FS and XPCs show increased bivalent genes, marked by H3K4me3 and H3K27me3 (Kinoshita et al., 2020; Yu et al., 2020a). Overall, in the naïve state, chromatin is open and lacking repressive marks, while in the FS, the cells begin to acquire bivalent chromatin marks that poise the cells for later lineage commitment.
NaÏVe and Primed States in Humans
Human ESCs were first derived in 1998 from blastocyst-stage embryos (Thomson et al., 1998). hESCs are quite different from mESCs and require Activin A and FGFs, rather than serum/2i and LIF that mESCs need. Such a large difference in growth requirements led to research into why hESCs and mESCs were so different. As described above, mouse EpiSCs could be derived that more closely resemble hESCs, based upon morphology and marker gene expression (Brons et al., 2007; Tesar et al., 2007; Rossant, 2011). However, this prompted a question: If EpiSCs resembled hESCs, is it possible to generate naïve hESCs? Subsequent research began from the basis that mESCs are easiest to derive and stable in ground-state 2iLIF media (Ying et al., 2008), and most naïve hESCs began with this cocktail. However, much as the conversion of EpiSCs to mESCs is challenging, likely due to multiple routes and potent epigenetic barriers (Figure 2), the transition of human primed to naïve pluripotency has also been challenging. The first reported naïve conditions for hESCs involved the transfection of ectopic OCT4, KLF2, and KLF4, along with 2iLIF (Hanna et al., 2010). Since then, there has been a veritable explosion of competing protocols for naïve hESCs, including many that do not require transgenes (Yilmaz and Benvenisty, 2019). However, the situation remains complex, and there is considerable argument about the nature of the putative naïve hESCs.
Transcriptional and Epigenetic Control of Human Naïve and Primed Cells
The naïve and primed states are well described in mice, but in humans, the situation remains complex (Davidson et al., 2015). Research using human cells has led to the development of a different model of naïve and primed states, which suggests instead of distinct naïve and primed cell states; there are instead modules that can be switched on and off relatively independently of one another (Figure 1). In this model, naïve ESCs switch on a set of modules, while primed ESCs switch on a different set. This view has emerged due to the difficulty in establishing a human version of the complete mouse naïve state and the existence of naïve-like cells that only partially fulfill the naïve criteria. Human naïve ESCs cannot be derived using only 2iLIF; instead, a large number of protocols have been developed that give rise to cells that mimic several aspects of mouse naïve cells (Chan et al., 2013; Gafni et al., 2013; Wang et al., 2014; Ware et al., 2014; Duggal et al., 2015; Carter et al., 2016; Theunissen et al., 2016). However, the competing protocols have distinct transcriptional profiles, cell surface markers, and epigenome states (Yang et al., 2019b), and no comprehensive model has emerged concerning the mechanisms controlling these states in humans. Indeed, just like the conversion of primed EpiSCs to naïve mESCs, there appear to be multiple routes from primed hESCs to naïve pluripotency (Duggal et al., 2015), and potent epigenetic barriers resist the transition. Both naïve and primed hESCs are regulated through several core TFs, for example, SOX2 and OCT4, which are common to both naïve and primed hESCs in mice and humans. However, the human cells have gene regulatory networks that utilize KLF5, KLF7, TFCP2L1, FOXR1, ZIC2, and TFAP2C, for the naïve state; and OTX2 and SALL2 for the primed state (Takashima et al., 2014; Weinberger et al., 2016; Pastor et al., 2018). Some of these are active in mouse naive mESCs (KLF5 and TFCP2l1); however, several seem unique to humans (e.g., TFAP2C), and some critical regulators in mice (e.g., Esrrb) are not typically upregulated in human naïve cells (Kisa et al., 2017; Rostovskaya et al., 2019). Consequently, there are substantial differences in transcriptional regulation in humans (Table 2), and the full naïve and primed regulatory networks remain to be elucidated.
TABLE 2.
Epigenetic factors implicated in naïve and primed hESC control.
| Gene name | Function | Effect on naïve or primed hESCs | Epigenetic pathway | References |
| EZH2 | Histone methyltransferase | Required to primed hESCs, but dispensable in naïve hESCs | H3K27me3 | Shan et al., 2017 |
| HDAC1/3 | Histone deacetylase | Histone deacetylase inhibition was required to establish naïve-like hESCs | Histone deacetylation/transcriptional regulation | Ware et al., 2014 |
| MCRS1, TET1, THAP11 | DNA demethylation (TET1) and epigenetic remodeling | Expressed in combination and can convert primed hESCs to naïve-like cells | 5mC to 5hmC conversion (TET1) and unknown | Durruthy-Durruthy et al., 2016 |
| NNMT | Nicotinamide N-methyltransferase | Knockout in primed hESCs leads to cells to acquire some naïve characteristics | Reduces histone methylation by removing the methyl-group donor SAM | Sperber et al., 2015 |
| SIRT2 | Histone/protein deacetylase | SIRT2 controls primed hESC state | Acetylation and regulation of glycolytic enzymes | Cha et al., 2017 |
| TNKS1/2 | Chromatin remodeling | Tankyrase 1/2 inhibition promotes naïve and extended pluripotency | Telomere elongation | Zimmerlin and Zambidis, 2020 |
| TET1/2 | DNA demethylation | Required for pluripotency in primed but not the naïve state | 5mC to 5hmC conversion | Finley et al., 2018 |
hESC, human embryonic stem cell; SAM, S-adenosylmethionine.
Epigenetic Influence of the Chemical Cocktails That Convert Primed to Naïve Cells
The human naïve cocktails begin with 2iLIF as a starting base, although LIF can be substituted for other molecules, and LIF is not strictly required in mouse naïve cells. Beyond 2i, many other inhibitors and signaling factors have been used, targeting JNK, MAPK, BRAF, SRC, and ROCK kinases. These inhibitors likely have widespread downstream effects on epigenetic regulation, although the pathways have not been fully explored. The two small molecules that have been directly implicated in epigenetic control are HDAC inhibitors, which are useful as a pretreatment of primed cells before conversion to naïve cells (Ware et al., 2014), and vitamin C. Vitamin C acts as a co-factor for TET and Jumonji (JMJ) domain-containing proteins. JMJ domain proteins are involved in histone demethylations, while TET domains convert 5mC to 5hmC, which is the initial step in the DNA demethylation pathway (Teslaa and Teitell, 2015). Overexpression of TET1, along with MCRS1 and THAP11, can drive cells toward a naïve-like state (Durruthy-Durruthy et al., 2016), indicating that TET1 is important in naïve cells. Nonetheless, how vitamin C modulates TETs and JMJs to promote the formation of naïve hESCs remains unclear. Other epigenetic pathways involved include the PRC2 component EZH2, which is required to maintain primed hESCs but is dispensable for naïve cells (Shan et al., 2017). As EZH2 is a key catalytic enzyme for the repressive histone mark H3K27me3, it suggests that human naïve cells may have reduced epigenetic repression, the same as mice, although, just as in mice, human naïve cells have higher overall levels of H3K27me3 (De Clerck et al., 2019). Possibly, the situation is similar to that of the mice, and H3K27me3 is lost at the promoters of critical genes, but overall H3K27me3 is elevated in response to reduced DNA methylation.
Ultimately, there remains argument over which of these naïve cocktails captures most of the naïve state. This led to an expansion of the model that the naïve and primed cells exist on a spectrum (Weinberger et al., 2016; Smith, 2017; Yang et al., 2019b) and led to ideas of multiple interchangeable naïve and primed modules that can be switched on or off under certain conditions (Figure 1; Theunissen et al., 2016; Cornacchia et al., 2019). This helps explain the differences in the naïve cocktails, as each cocktail can switch on certain modules but may fail to activate them all.
Extra/Expanded-Capability Cells With Totipotent-Like Properties
An extra complication for the naïve and primed model is the existence of “extra-capability cells” that describe ESC-like cells that are pluripotent and also have some aspects of totipotency (Yang et al., 2017a, b; Gao et al., 2019). These cells are grown under culture conditions similar to naïve cells, but they drop the MEK inhibitor from 2i and include WNT pathway inhibitors and then either SRC and tankyrase inhibitors (EPSCs) or ROCK inhibitor (EPSs). First described in mice, EPS/EPSCs (extra/expanded pluripotent stem cells) can contribute to the trophoblast, which is a property that both naïve and primed mESCs and hESCs lack. However, the gold standard test of totipotency, the derivation of a complete mouse using only these cells, has not yet been reported. This suggests that their totipotent properties remain incomplete or that they lack full totipotency. Indeed, there is argument that while mouse EPS/EPSCs can occasionally localize in the trophoblast, these cells lack trophoblast-specific markers, still express epiblast markers, and do not have totipotent properties (Posfai et al., 2021). Additionally, the DNA methylation state of EPS/EPSCs is somewhat contradictory. EPSCs have high levels of methylated DNA (Yang et al., 2017a), which does not match the DNA hypomethylation of totipotent embryonic cells and 2iLIF-grown naïve mESCs, although it should be noted that it is unclear if the hypomethylation in 2iLIF cells is a cell type effect, or is a side effect of PD0325901, a component of the 2iLIF cocktail. One advantage that the EPS/EPSCs do have is in the derivation of cells from species that have been previously resistant to the isolation of ESCs, for example, deriving porcine EPSCs (Gao et al., 2019). The EPS and EPSC cocktails have also been applied to human embryos to derive putative totipotent cells (Yang et al., 2017b; Gao et al., 2019). Overall, the identity of the EPS and EPSCs remains unclear, particularly how they correspond to in vivo development. EPS and EPSCs may hint at a further expansion to the module concept, where EPS and EPSCs are switching off some naïve modules and activating some totipotent modules. But like the naïve/primed split, these cell types are potentially activating only some of the totipotent modules, and only partial totipotency is achieved. A fascinating study of how biological phenotypes can act independently is the reprogramming of mESCs to oocyte-like cells (Hamazaki et al., 2020). In that study, oocyte-like cells could be derived without PGC specification, meiosis, or epigenetic reprogramming of DNA demethylation. This suggests that these aspects are independent and can be switched on and off in a module-specific fashion (Hamazaki et al., 2020).
Links Between Metabolism and Epigenetic Control of the Naïve and Primed States
Pluripotent stem cell fate transitions from naïve to primed are accompanied by a metabolic switch, from oxidative phosphorylation (oxphos) to mainly glycolysis, respectively (Figure 1; Teslaa and Teitell, 2015). This mirrors the developing embryo, which mainly uses pyruvate and oxphos from fertilization to the blastocyst stage, before transitioning to glucose-based glycolysis and anaerobic metabolism in the late blastocyst (Devreker and Englert, 2000; Chason et al., 2011). Although it should be noted that it is unclear if glycolysis is required to produce energy for embryos, instead, it may be needed for biosynthetic pathways (Smith and Sturmey, 2013). ESCs, conversely, are highly active cells that divide rapidly and need a lot of energy. Hence, the link between embryonic metabolism and ESC metabolism is not a complete match. There is nonetheless a close link between metabolism and epigenetic control that has not been thoroughly explored (Shyh-Chang and Ng, 2017). For example, SIRT1 is high in hESCs and acetylates and activates glycolytic enzymes (Cha et al., 2017), and in mice, HIF1A controls glycolytic/oxphos metabolism and influences cell state (Zhou et al., 2012). Many of the reactants required for epigenetic control are metabolic products. For example, acetyl-CoA is the main acetyl donor for histone acetylation, and intracellular levels of acetyl-CoA directly regulate global histone acetylation, and so function as a signal for overall cellular energy metabolism (Cai et al., 2011). An analysis of naïve and primed hESC states revealed that naïve cells express nicotinamide N-methyltransferase (NNMT) at high levels (Sperber et al., 2015), which is responsible for metabolizing S-adenosylmethionine (SAM), the major chemical donor for histone methyltransferases. As a consequence, naïve cells have low levels of SAM and correspondingly low levels of histone methylation, while primed cells have the inverse, high SAM and high histone methylation (Sperber et al., 2015). Importantly, these metabolic changes may be required for the naïve to primed transition, as NNMT knockout cells transition toward a primed state (Sperber et al., 2015). Similarly, 2iLIF-grown mESCs maintain high levels alpha-ketoglutarate that biases the cells toward DNA and histone demethylation by promoting the activity of JMJ-containing demethylases (Carey et al., 2015). In addition to these direct links between metabolism and chromatin, manipulation of metabolism by altering the growth medium also has strong effects on hESCs. Lipid deprivation of primed hESCs reverts them to an intermediate naïve-like state, and the reapplication of lipids pushes the cells toward a primed state (Cornacchia et al., 2019). The exact metabolic/epigenetic pathways behind this effect remain to be elucidated, but lipid deprivation may promote glucose utilization by glycolysis, making more acetyl-CoA available and leading to increased histone acetylation and gene activation. Similarly, mouse 2iLIF-grown cells can utilize fatty acid oxidation (FAO); and inhibition of FAO leads to a reversible quiescence (Khoa et al., 2020), reminiscent of diapause in mice where embryos can be paused if the developmental environment is unfavorable. This effect is driven by the activity of MOF, a histone acetyltransferase that acetylates histones at FAO-related genes and helps activate them (Khoa et al., 2020). Ultimately, there is an intimate interdependence between metabolism, and epigenetic control in embryonic cells and the embryo (Chason et al., 2011), which remains to be comprehensively explored (Betschinger, 2017).
Epigenetic Control of the X Chromosomes in Female Cells
The epigenetic status of the X chromosomes in female ESCs is a particularly important point when discussing human naïve cells and is considered something of a hallmark for the naïve state (Nichols and Smith, 2009; Theunissen et al., 2016). In mice, the situation is relatively straightforward; in naïve mESCs, both X chromosomes are active; Xist, a long non-coding RNA that silences one X chromosome is not expressed; and in primed EpiSCs, Xist is expressed and one X chromosome is epigenetically inactivated (Bao et al., 2009). This roughly matches the in vivo embryonic states: in the late inner cell mass, both X chromosomes are active, and during differentiation in the late epiblast, one random X chromosome is silenced (Okamoto et al., 2004, 2011). The situation in human cells is more complex. Female hESCs (i.e., primed state) have one active and one inactive X chromosomes, which is strong evidence that hESCs are developmentally later than the early blastocyst (Nichols and Smith, 2009). Human naïve cells, depending upon the protocol used, have varying states of X chromosome inactivation, including intermediate states of XIST expression and epigenetic silencing (Sahakyan et al., 2017). The discrepancy between mice and humans may, at least partly, be related to the mechanism of X chromosome inactivation in humans, which appears to be more complex than in mice (Patrat et al., 2020). Briefly, in humans, XIST is expressed in the inner cell mass, but its expression is not correlated with epigenetic suppression (Okamoto et al., 2011; Petropoulos et al., 2016). Instead, one X chromosome is “dampened” by an unclear mechanism, before later full X chromosome inactivation by chromatin silencing (Petropoulos et al., 2016), possibly as late as post-implantation. This added complexity in humans may explain the differences in X chromosome status in naïve and primed hESCs or may reflect species-specific epigenetic regulatory differences (Okamoto et al., 2011).
Ultimately, human naïve ESCs remain surprisingly elusive to pin down (Theunissen et al., 2016). A close comparison of gene expression profiles and epigenetic states indicates that naïve hESC protocols, to date, remain distinct from naïve mESCs (Yang et al., 2019b), suggesting that the current protocols only capture aspects of the naïve state. As the conversion of primed to naïve cells is an artificial conversion, not only are potent epigenetic barriers in place, but the route to true naïve cells is unclear (Figure 2). Another, perhaps uncomfortable, possibility is that the naïve mouse state has no clear mimic in humans, is species-specific, or is a transitory stage in humans that cannot be stably captured in vitro (Rossant and Tam, 2017; Yang et al., 2019b).
Epigenetic Control in Reprogramming Somatic Cells to Pluripotent Stem Cells
Interconversions between closely related embryonic states have helped inform our understanding of the epigenetic control of embryogenesis. Another, more drastic, cell conversion is the reprogramming of somatic cells to induced pluripotent stem cells (iPSCs) or by SCNT (Dean et al., 2003; Takahashi and Yamanaka, 2006). These two techniques have revolutionized the study of epigenetic regulation of the embryonic state, particularly in humans where early embryogenesis is hard to study. Both reprogramming techniques involve the global reconfiguration of gene expression patterns driven by epigenetic remodeling (Liu et al., 2020), and these methods have revealed potent epigenetic barriers that restrain cell type conversions (Xu et al., 2016). Chromatin is dramatically reorganized during reprogramming (Wang et al., 2017), as enhancer–promoter interactions and active and repressive sequences make new contacts and reorder the transcriptional program (Apostolou et al., 2013; Di Stefano et al., 2020; Lu et al., 2020b). During somatic cell reprogramming, the ectopic pluripotency transgenes Oct4, Sox2, Klf4, and Myc reconnect target enhancers to promoters to induce transcriptional change (Wei et al., 2013; Beagan et al., 2016; Stadhouders et al., 2018). More importantly, during this process, chromatin reorganization occurs prior to, or independent from, gene expression changes (Wei et al., 2013; Beagan et al., 2016). Indeed, changes in chromatin accessibility often precede changes in gene expression, often by several days (Li et al., 2017a).
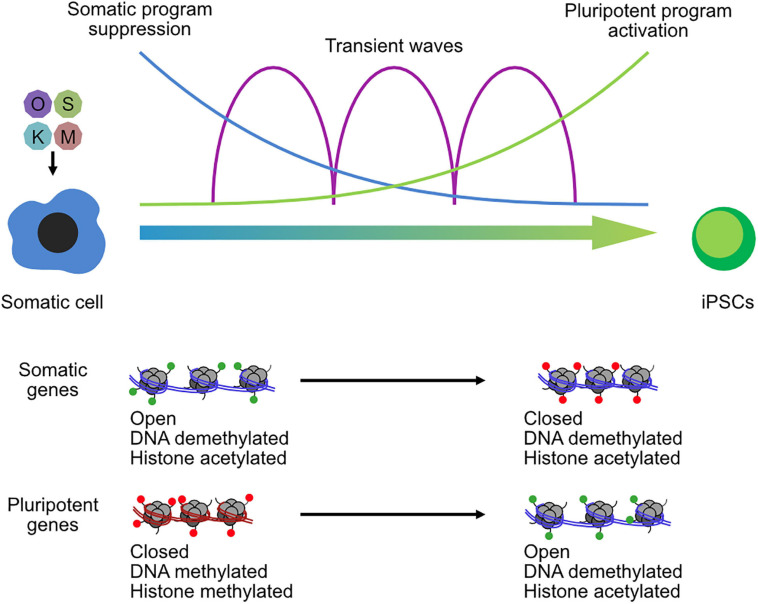
Broadly, somatic cells tend to have higher levels of repressive marks, which are reduced during the reprogramming to pluripotency. Vitamin C improves the reprogramming of somatic cells to pluripotency by modulating TET and JMJ domain-containing proteins (Wang et al., 2011; Chen et al., 2013a), leading to demethylation of DNA and H3K36me2/3. Other repressive epigenetic marks have been identified as major barriers for reprogramming (Arabaci et al., 2020), particularly the repressive histone modification H3K9me3 that is redistributed during iPSC reprogramming (Hawkins et al., 2010). Methyltransferases are downstream targets of BMPs and are a determinant for iPSC generation by regulating the methylation states at core pluripotency loci (Chen et al., 2013b). Similarly, the loss of the H3K9 methyltransferase Setdb1, or its co-factor Trim28, leads to improved reprogramming (Miles et al., 2017), although it may ultimately be deleterious as it causes spontaneous differentiation in the resulting iPSCs (Klimczak et al., 2017). Other H3K9me3 enzymes also impair the conversion of somatic cells to iPSCs, including Suv39h1/2 and Ehmt2 (G9a), along with the H3K79me3 methyltransferase Dot1l (Onder et al., 2012). However, the regulation of repressive histones is more subtle than just repressive mechanisms are bad for reprogramming. Reprogramming is a multi-phased program (Brambrink et al., 2008), and one of the earliest phases is the large-scale suppression of the somatic gene expression program (Chronis et al., 2017; Li et al., 2017a). Whether an epigenetic enzyme is beneficial or deleterious for reprogramming may ultimately depend upon the balance of its role in suppression or activation of the somatic and pluripotent programs (Figure 3). Outsize roles in suppression of the somatic or activation of the pluripotent program are likely to improve reprogramming, while the opposite is likely to shift the balance toward impairment. Consequently, epigenetic regulators have context-specific and temporal-specific effects during reprogramming. For example, knockdown of the co-repressors Ncor1/Ncor2 is deleterious for the early stages of reprogramming, due to reduced somatic gene suppression, but beneficial for the late stages due to reduced pluripotent gene repression (Zhuang et al., 2018). A similar pattern was observed for the histone H3K27 demethylase Kdm6b (JMJD3) (Huang et al., 2020) and the H3K27me3 methyltransferase Ezh2 (Rao et al., 2015). Other epigenetic regulators can be beneficial in both phases, although they may use different mechanisms to achieve this effect. For example, the H2AK119 ubiquitinase RYBP cooperates with PRC1 complex to suppress the somatic program via the histone demethylase KDM2B but cooperates with OCT4 to activate the pluripotent program (Li et al., 2017b). Epigenetic regulators can be something of a double-edged sword for somatic cell reprogramming (Onder et al., 2012; Li et al., 2017b; Zhuang et al., 2018).
FIGURE 3.
Chromatin reconfiguration in the reprogramming of somatic cells to induced pluripotent stem cells. OSKM transgenes are transfected into somatic cells and initiate a complex series of biological programs, including the suppression of the somatic program in the early phase of reprogramming and the activation of the pluripotent program in the late phase. Transient waves of gene expression programs occur between the two-cell type states. Chromatin is remodeled throughout the reprogramming process, and especially at somatic loci and pluripotent gene loci that open and close chromatin and lose or gain histone methylation.
A curious observation in studies of epigenetic reprogramming is that the loss of epigenetic regulators can have strong effects on the reprogramming process (Xu et al., 2016), yet the loss of the same factors in ESCs tends to have a much weaker or negligible effect. For example, knockdown of Ncor1/2 improves reprogramming but does not affect mESCs (Zhuang et al., 2018). Similarly, Sin3a/Sap30 loss impairs reprogramming (Li et al., 2017a; Saunders et al., 2017) but causes no change in mESCs. The short-term knockdown of a panel of 40 epigenetic regulators resulted in differentiation in only two knockdowns (He et al., 2019). These observations indicate that epigenetic pathways involved in the establishment of pluripotency may not always be involved in the maintenance of pluripotency or, if they are involved, can often be in contradictory ways. A good example is the knockdown of the PRC2 component Ezh2. Knockdown promotes reprogramming (Onder et al., 2012), but in mESCs, it affects self-renewal and makes the cells prone to differentiation. Overall, this suggests two patterns for epigenetic regulators in reprogramming and pluripotent maintenance: (1) once the reprogramming epigenetic barriers have been overcome, they are dispensable in ESCs, and (2) loss of epigenetic regulators makes ESCs unstable and more prone to differentiation. Epigenetic barriers are not always two-way and often act more like valves that can be easily traversed again if going in the opposite direction. This effect puts a limitation on screening technologies such as genome-wide knockdowns/outs and sgRNA screens. Epigenetic factors that impact the stable ESC state may not be relevant to the entirety of the reprogramming process. Consequently, challenging experiments must be performed during the reprogramming time course to understand the requirements for the temporal order of events. So far, these screens have focused on the early stages of reprogramming, which is more experimentally tractable (Borkent et al., 2016; Toh et al., 2016; Miles et al., 2017; Neganova et al., 2019). However, reprogramming is a curiously multistage-phased process, and epigenetic barriers may be transiently erected and disassembled, meaning that screening also needs to be timed to specific stages. Potentially, technologies such as Perturb-seq (Dixit et al., 2016), which provides candidate factors and phenotype readout simultaneously, may help in understanding the full range of epigenetic barriers blocking reprogramming.
Transposable Elements, the Early Embryo, and NaÏVe and Primed Embryonic States
Transposable elements are the single largest constituent of mammalian genomes (Hutchins and Pei, 2015), taking up around 40% of the total DNA sequence. They can be divided into four broad categories, DNA transposons, and three types of retrotransposon: long-interspersed elements (LINEs), short interspersed elements (SINEs), and the endogenous retroviruses/long-terminal repeats (ERVs/LTRs). TEs have been viewed as genetic parasites that are especially dangerous during embryogenesis when transposition duplications are capable of entering the germline. However, TEs can act as a source of evolutionary innovation, by duplicating TF binding sites, rewiring gene regulatory networks, altering splicing patterns, and many other effects on the genome and cell (Bourque et al., 2018), both beneficial and deleterious (Enriquez-Gasca et al., 2020). During early embryonic development, the genome is reprogrammed back to a naïve state. This process involves the global DNA demethylation of the genome, a process that is presumed to be a requirement for the correct execution of a new developmental program. However, DNA methylation is also one of the dominant mechanisms for the suppression of TEs in somatic tissues (Feng et al., 2010; Jonsson et al., 2019), and global DNA demethylation in the early embryo helps release waves of TE expression (Beraldi et al., 2006; Goke et al., 2015). Intriguingly, TE activity is dynamic in early embryonic cells (Wang et al., 2020) and is both stage and TE type-specific (Goke et al., 2015). In the early embryo and ESCs, instead of DNA methylation suppressing TE expression, other mechanisms take over, particularly the methylation of histone H3K9me3 by SETDB1, which is recruited to specific TEs by TRIM28 binding to KRAB-family zinc finger TFs (Ecco et al., 2016). However, there is also evidence that a wide range of epigenetic enzymes are involved in the suppression (or management) of the expression of TEs. Indeed, the early embryo can contain vast quantities of TE RNAs, a single MaLR LTR family TE can comprise up to 13% of the total oocyte RNA (Peaston et al., 2004), and SINE elements may make up a further 3% (Bachvarova, 1988). Functional roles for TEs in embryogenesis are less well explored, but LINE L1 expression is required for progression to the blastocyst stage (Percharde et al., 2018), while depletion of L1s in mESCs leads to the derepression of genes that are proximal to LINE L1s (Lu et al., 2020a). HERVKs are expressed and produce viroid-like particles in normal human embryos (Grow et al., 2015). Multiple lines of evidence indicate that TE expression and epigenetic activity, aside from retrotransposition, are involved in the embryonic process, although their full involvement, both beneficial and deleterious, is unclear.
Transposable Elements and Two-Cell-Like Cells
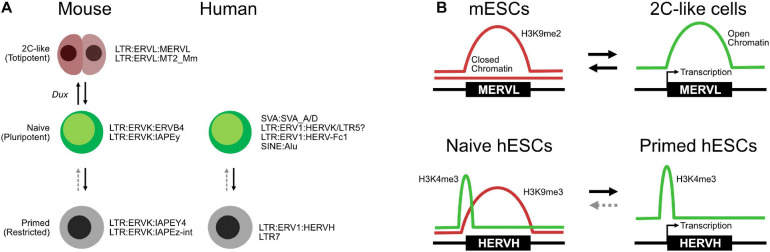
TE expression has found utility as both a marker of embryonic stages and also as a tool to isolate new cell types with enhanced features. In mESC cultures, there is a small subpopulation (about 1%) that expresses a mouse-specific MERVL ERV (Macfarlan et al., 2012). Intriguingly, MERVL is expressed in the 2C stage of the mouse embryo when the cells are still totipotent. Isolation of the MERVL expressing “2C-like” cells from an mESC culture resulted in a population of cells that cycle into and out of the mESC state and have some totipotent-like properties. For example, they can partially colonize the embryonic trophoblast, a capability that normal mESCs lack. The 2C-like cells are transient and cannot be maintained, but various cocktails and protocols have been developed that improve their derivation (Iturbide and Torres-Padilla, 2020). Mechanistically, 2C-like cells rely on a transcriptional network distinct from the OCT4–SOX2 pluripotency network. The details are still being worked out, but the 2C-like state centers around several families of TFs and microRNAs, including miR344, DPPA2/4, ZSCAN4-family, ZMYM2, NELFA, and GATA2 (De Iaco et al., 2019; Fu et al., 2019; Zhang et al., 2019; Hu et al., 2020; Yang et al., 2020). These pathways ultimately center on the expression of Dux (Figure 4A; Percharde et al., 2018), a key TF required for ZGA in the developing embryo (De Iaco et al., 2017). This 2C-like transcriptional network cooperates to remodel the epigenetic state, H3K9me3 and H2AK119ub1 (ubiquitination), and chromatin assembly by CAF1, which are all particularly critical (Ishiuchi et al., 2015). MERVLs themselves tend to lack H3K9me3 and are not bound by SETDB1 (Maksakova et al., 2013) but are marked by H3K9me2 and H3K56ac (Macfarlan et al., 2011; He et al., 2019). Knockdown of Ehmt2 (G9a) leads to the upregulation of MERVLs by a direct mechanism involving loss of H3K9me2 at MERVLs and the gain of open accessible chromatin (Figure 4B; Maksakova et al., 2013; Hendrickson et al., 2017). Similarly, Kdm1a, a histone demethylase, is important for suppressing the expression of MERVLs (Macfarlan et al., 2011), although the exact mechanism by which KDM1A suppresses MERVLs is not clear, as KDM1A can demethylate both H3K4me1 and H3K9me2. H3K4me1 marks enhancers and is generally associated with gene activation, while H3K9me2 is a repressive mark, often associated with heterochromatin and H3K9me3. H3K9me2 marks MERVLs, but H3K4me1 does not, suggesting that KDM1A may at least partially regulate MERVLs indirectly. Indeed, several direct and indirect chromatin modifiers regulate MERVLs. Both TRIM28 and RNF2 do not bind directly to MERVLs, and their corresponding marks, H3K9me3 and H2AK119ub, are not found either (Maksakova et al., 2013; He et al., 2019), but knockdown of Trim28 or Rnf2 leads to upregulation of MERVLs. Curiously, histone ubiquitination has two roles in both suppressing and activating MERVLs: knockdown of the histone H2A ubiquitinase Rnf2 leads to the deubiquitination and upregulation of MERVLs (Zhang et al., 2019), while conversely, knockdown of the H2B deubiquitinase Usp7 leads to H2B ubiquitination and upregulation of MERVLs (Chen et al., 2020). There is a similar pattern here to the conversion of primed cells to naïve cells: generally, the loss of repressive histone marks is beneficial for the conversion of cells to a 2C-like state. For example, knockdown of Setdb1 (Wu et al., 2020), Trim28 (KAP1) (Maksakova et al., 2013), Dnmt1 (Fu et al., 2019), and Rnf2 (RING1B) (He et al., 2019; Zhang et al., 2019) and inhibition of HDACs by trichostatin A (TSA) (Macfarlan et al., 2012) can all increase the number of 2C-like cells in an mESC culture.
FIGURE 4.
Transposable elements (TEs) in totipotent, naïve, and primed embryonic states. (A) Expression of specific TEs mark cell type states in both mice and humans, although the TEs involved are species-specific. (B) Selected chromatin transitions at specific MERVLs in mouse embryonic stem cells (mESCs) during the transition to two-cell (2C)-like cells and at HERVHs in naïve and primed human ESCs (hESCs).
TE activity, both as transcribed RNAs and as enhancers, is linked with the 2C-like state. In oocytes, a key factor is DPPA3 (Dppa3/Stella), a maternally inherited protein that is essential for the transition from the maternal to the zygotic gene expression program (Huang et al., 2017). When Dppa3/DPPA3 was removed from the maternal pool, MERVLs failed to be upregulated, and the embryos arrested at the 2C stage (Huang et al., 2017). Intriguingly, the authors found that microinjection of small interfering RNAs (siRNAs) targeting MERVLs led to a reduction in MERVL-derived Gag proteins and developmental impairment (Huang et al., 2017). These results suggest that MERVL expression is not simply a marker for the 2C stage but is also functionally relevant. MERVL sequences are spliced into other transcripts as TE–gene chimeras (Huang et al., 2017; Chen et al., 2020), and MERVL expression may also be driving transcript expression. MERVL sequences can act as an enhancer to recruit TFs to promote transcription (Huang et al., 2017; Zhang et al., 2019), and the MERVL sequence can act as a promoter (Jiang et al., 2020). In addition to MERVLs, LINE L1 RNAs silence Dux expression by recruiting TRIM28 to induce heterochromatin via H3K9me3 (Percharde et al., 2018). Knockdown of LINE L1 RNAs leads to reactivation of the 2C-like gene expression program and particularly reactivation of Dux (Percharde et al., 2018). This points to a surprisingly complex relationship between TE expression, transcriptional regulation, and epigenetic control of heterochromatin. LINE L1 RNAs can reactivate Dux, which then appears to lead to deregulation of H3K9me3, which activates MERVLs, which are spliced into key 2C-like transcripts and may also act as enhancers for genes required for the 2C-like state. Ultimately, the causal relationship between TE activation, 2C-like gene expression programs, and transcriptional and epigenetic control still needs to be unpicked, but it is a fascinating model system for the establishment of totipotency.
Transposable Elements and Human Naïve Cells
The expression of TEs has also found utility as markers of the embryonic state in hESCs (Theunissen et al., 2016). hESCs express the primate-specific HERVH ERV, and their accompanying LTR (LTR7) can act as pluripotent-specific transcription start sites (Fort et al., 2014). This pattern of TE expression has been proposed as one of several criteria that define the naïve and primed hESC states (Theunissen et al., 2016). Briefly, primed hESCs express HERVH/LTR7 RNAs, while naïve cells express a more mixed set of TEs, but particularly SVAs, LTR5, and HERVK. HERVH are marked by H3K4me3 in both naïve and primed cells but are typically marked by the repressive H3K9me3 mark in primed cells (Figure 4B; Theunissen et al., 2016), although another study suggests that high levels of HERVH specifically mark naïve hESCs (Figure 4A; Wang et al., 2014). An interesting observation of that study was the splicing of HERVH directly into hESC chimeric transcripts. This is similar to the splicing of MERVLs seen in 2C-like cells, suggesting that a robust understanding of TE splicing patterns may lead to insights into embryonic cell states.
Transposable elements can be expressed as fragmentary RNA, and an area that remains poorly explored is the splicing of TEs into other transcripts. TEs can be expressed as individual units within the cell, but they can be spliced into longer transcripts, often as part of long non-coding transcripts but also into normal coding transcripts to generate novel chimeric transcripts (Bourque et al., 2018). To date, exploring the contribution of TEs to the normal transcriptome of a cell has been hampered by the use of short-reads to assemble transcripts (Babarinde et al., 2019). Nonetheless, the chimeric splicing of TEs into transcripts is a feature of pre-implantation embryonic cell types. The mouse-specific MERVLs that are transcribed in 2C-like cells are spliced into other coding and non-coding transcripts (Macfarlan et al., 2012; Huang et al., 2017; He et al., 2019). Similarly, the HERVH human-specific ERVs that are a feature of pluripotent stem cells are also spliced into other transcripts (Fort et al., 2014; Wang et al., 2014). Intriguingly, TEs are spliced into pluripotent transcripts in cancerous cells (Jang et al., 2019), the implication being that these TEs are activating pluripotent genes and converting them to oncogenes. However, HERVH activation appears not to be a general feature of cancer (Zapatka et al., 2020).
When TEs are still retrotranspositionally active, it poses a danger to the cell; however, once the coding sequences are mutated, and they are no longer functional, and epigenetic suppression mechanisms should decline due to a lack of evolutionary pressure to suppress TEs. Yet TEs maintain complex epigenetic regulatory patterns that are TE-type specific and are present long after they have stopped being capable of retrotransposition and are several million years old (Bourque et al., 2008; He et al., 2019). This suggests regulatory function and co-option for legitimate biological function. A good example is H3K9me3, a critical epigenetic mark responsible for silencing TEs in mESCs (Rowe et al., 2013; Yang et al., 2015), which is intimately involved in 2C-like cells, naïve cells, and reprogramming (Chen et al., 2013b; Bao et al., 2015; Xiao et al., 2017; Wang et al., 2018; Wu et al., 2020). H3K9me3 is remodeled during embryonic development, particularly at LTRs and ERVs (Wang et al., 2018). Knockdown of several H3K9me3-related factors, Setdb1 and Trim28, impaired mouse embryonic development to the blastocyst, but Chaf1a (a modulator of H3K9me3 and part of the CAF1 complex) knockdown nearly completely blocked embryos from progressing past the morula stage (Wang et al., 2018). Consequently, H3K9me3 seems to be performing double duty as a major repressive mark for LTRs and ERVs, as well as erecting epigenetic barriers between cell type conversions. Ultimately, there is a tight integration between epigenetic control of TE activity and cell fate, and they should be considered as a unified mechanism with overlapping activities.
Conclusion
Epigenetic reconfiguration during early embryonic development is a critical process that resets the cells and makes them capable of a new round of development. The epigenetic rearrangements on chromatin are widespread and encompass changes in histone modifications, nucleosome positioning, 3D structure, and DNA modifications. A complex system of epigenetic regulators is involved in this process, and there are many distinct stages that cells transition through during normal development. Some of these states can be captured in vitro and have informed our understanding of the mechanisms behind embryonic development, and how autonomous and exogenous signaling, transcriptional control, and epigenetics combine to regulate development. Many mysteries remain, particularly in the role of epigenetic control in maintaining and blocking cell type conversions. Understanding this process in detail will lead to an enhanced understanding of cell type transitions that will inform potential medical treatments, particularly cell replacement therapy.
Author Contributions
LS, XF, and GM compiled the tables and prepared the figures. All authors were involved in manuscript writing. AH wrote and approved the final text, and funded the study.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by the Science and Technology Planning Project of Guangdong Province (2019A050510004), the National Natural Science Foundation of China (31970589 and 31850410463), the Shenzhen Science, Technology and Innovation Commission (JCYJ20200109141018712), the Shenzhen Peacock plan (201701090668B), and the Center for Computational Science and Engineering of the Southern University of Science and Technology.
References
- Acampora D., Di Giovannantonio L. G., Simeone A. (2013). Otx2 is an intrinsic determinant of the embryonic stem cell state and is required for transition to a stable epiblast stem cell condition. Development 140 43–55. 10.1242/dev.085290 [DOI] [PubMed] [Google Scholar]
- Adachi K., Kopp W., Wu G., Heising S., Greber B., Stehling M., et al. (2018). Esrrb Unlocks Silenced Enhancers for Reprogramming to Naive Pluripotency. Cell Stem Cell 23 266–275e266. 10.1016/j.stem.2018.05.020 [DOI] [PubMed] [Google Scholar]
- Alonso A., Breuer B., Steuer B., Fischer J. (1991). The F9-EC cell line as a model for the analysis of differentiation. Int. J. Dev. Biol. 35 389–397. [PubMed] [Google Scholar]
- Andrews P. W. (1988). Human teratocarcinomas. Biochim. Biophys. Acta 948 17–36. 10.1016/0304-419x(88)90003-0 [DOI] [PubMed] [Google Scholar]
- Apostolou E., Ferrari F., Walsh R. M., Bar-Nur O., Stadtfeld M., Cheloufi S., et al. (2013). Genome-wide chromatin interactions of the Nanog locus in pluripotency, differentiation, and reprogramming. Cell Stem Cell 12 699–712. 10.1016/j.stem.2013.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabaci D. H., Terzioglu G., Bayirbasi B., Onder T. T. (2020). Going up the Hill: Chromatin-based Barriers to Epigenetic Reprogramming. FEBS J. 2020:15628. 10.1111/febs.15628 [DOI] [PubMed] [Google Scholar]
- Atlasi Y., Megchelenbrink W., Peng T., Habibi E., Joshi O., Wang S. Y., et al. (2019). Epigenetic modulation of a hardwired 3D chromatin landscape in two naive states of pluripotency. Nat. Cell Biol. 21 568–578. 10.1038/s41556-019-0310-9 [DOI] [PubMed] [Google Scholar]
- Avgustinova A., Benitah S. A. (2016). Epigenetic control of adult stem cell function. Nat. Rev. Mol. Cell Biol. 17 643–658. 10.1038/nrm.2016.76 [DOI] [PubMed] [Google Scholar]
- Babarinde I. A., Li Y., Hutchins A. P. (2019). Computational Methods for Mapping, Assembly and Quantification for Coding and Non-coding Transcripts. Comput. Struct. Biotechnol. J. 17 628–637. 10.1016/j.csbj.2019.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachvarova R. (1988). Small B2 RNAs in mouse oocytes, embryos, and somatic tissues. Dev. Biol. 130 513–523. 10.1016/0012-1606(88)90346-6 [DOI] [PubMed] [Google Scholar]
- Bao S., Tang F., Li X., Hayashi K., Gillich A., Lao K., et al. (2009). Epigenetic reversion of post-implantation epiblast to pluripotent embryonic stem cells. Nature 461 1292–1295. 10.1038/nature08534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X., Wu H., Zhu X., Guo X., Hutchins A. P., Luo Z., et al. (2015). The p53-induced lincRNA-p21 derails somatic cell reprogramming by sustaining H3K9me3 and CpG methylation at pluripotency gene promoters. Cell Res. 25 80–92. 10.1038/cr.2014.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beagan J. A., Gilgenast T. G., Kim J., Plona Z., Norton H. K., Hu G., et al. (2016). Local Genome Topology Can Exhibit an Incompletely Rewired 3D-Folding State during Somatic Cell Reprogramming. Cell Stem Cell 18 611–624. 10.1016/j.stem.2016.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie G. M., Lopez A. D., Bucay N., Hinton A., Firpo M. T., King C. C., et al. (2005). Activin A maintains pluripotency of human embryonic stem cells in the absence of feeder layers. Stem Cells 23 489–495. 10.1634/stemcells.2004-0279 [DOI] [PubMed] [Google Scholar]
- Beraldi R., Pittoggi C., Sciamanna I., Mattei E., Spadafora C. (2006). Expression of LINE-1 retroposons is essential for murine preimplantation development. Mol. Reprod. Dev. 73 279–287. 10.1002/mrd.20423 [DOI] [PubMed] [Google Scholar]
- Betschinger J. (2017). Charting Developmental Dissolution of Pluripotency. J. Mol. Biol. 429 1441–1458. 10.1016/j.jmb.2016.12.017 [DOI] [PubMed] [Google Scholar]
- Betschinger J., Nichols J., Dietmann S., Corrin P. D., Paddison P. J., Smith A. (2013). Exit from pluripotency is gated by intracellular redistribution of the bHLH transcription factor Tfe3. Cell 153 335–347. 10.1016/j.cell.2013.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonev B., Mendelson Cohen N., Szabo Q., Fritsch L., Papadopoulos G. L., Lubling Y., et al. (2017). Multiscale 3D Genome Rewiring during Mouse Neural Development. Cell 171 557–572e524. 10.1016/j.cell.2017.09.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkent M., Bennett B. D., Lackford B., Bar-Nur O., Brumbaugh J., Wang L., et al. (2016). A Serial shRNA Screen for Roadblocks to Reprogramming Identifies the Protein Modifier SUMO2. Stem Cell Rep. 6 704–716. 10.1016/j.stemcr.2016.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boroviak T., Loos R., Bertone P., Smith A., Nichols J. (2014). The ability of inner-cell-mass cells to self-renew as embryonic stem cells is acquired following epiblast specification. Nat. Cell Biol. 16 516–528. 10.1038/ncb2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boskovic A., Eid A., Pontabry J., Ishiuchi T., Spiegelhalter C., Raghu Ram E. V., et al. (2014). Higher chromatin mobility supports totipotency and precedes pluripotency in vivo. Genes Dev. 28 1042–1047. 10.1101/gad.238881.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque G., Burns K. H., Gehring M., Gorbunova V., Seluanov A., Hammell M., et al. (2018). Ten things you should know about transposable elements. Genome Biol. 19:199. 10.1186/s13059-018-1577-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque G., Leong B., Vega V. B., Chen X., Lee Y. L., Srinivasan K. G., et al. (2008). Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 18 1752–1762. 10.1101/gr.080663.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambrink T., Foreman R., Welstead G. G., Lengner C. J., Wernig M., Suh H., et al. (2008). Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell 2 151–159. 10.1016/j.stem.2008.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brons I. G. M., Smithers L. E., Trotter M. W. B., Rugg-Gunn P., Sun B. W., Lopes S. M. C. D., et al. (2007). Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448 U191–U197. 10.1038/nature05950 [DOI] [PubMed] [Google Scholar]
- Buecker C., Srinivasan R., Wu Z., Calo E., Acampora D., Faial T., et al. (2014). Reorganization of enhancer patterns in transition from naive to primed pluripotency. Cell Stem Cell 14 838–853. 10.1016/j.stem.2014.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L., Sutter B. M., Li B., Tu B. P. (2011). Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol. Cell 42 426–437. 10.1016/j.molcel.2011.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao K. X., Collings C. K., Morgan M. A., Marshall S. A., Rendleman E. J., Ozark P. A., et al. (2018). An Mll4/COMPASS-Lsd1 epigenetic axis governs enhancer function and pluripotency transition in embryonic stem cells. Sci. Adv. 4:eaa8747. 10.1126/sciadv.aap8747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey B. W., Finley L. W., Cross J. R., Allis C. D., Thompson C. B. (2015). Intracellular alpha-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature 518 413–416. 10.1038/nature13981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter M. G., Smagghe B. J., Stewart A. K., Rapley J. A., Lynch E., Bernier K. J., et al. (2016). A Primitive Growth Factor, NME7AB, Is Sufficient to Induce Stable Naive State Human Pluripotency; Reprogramming in This Novel Growth Factor Confers Superior Differentiation. Stem Cells 34 847–859. 10.1002/stem.2261 [DOI] [PubMed] [Google Scholar]
- Cha Y., Han M. J., Cha H. J., Zoldan J., Burkart A., Jung J. H., et al. (2017). Metabolic control of primed human pluripotent stem cell fate and function by the miR-200c-SIRT2 axis. Nat. Cell Biol. 19 445–456. 10.1038/ncb3517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain S. J., Yee D., Magnuson T. (2008). Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells 26 1496–1505. 10.1634/stemcells.2008-0102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y. S., Goke J., Ng J. H., Lu X., Gonzales K. A., Tan C. P., et al. (2013). Induction of a human pluripotent state with distinct regulatory circuitry that resembles preimplantation epiblast. Cell Stem Cell 13 663–675. 10.1016/j.stem.2013.11.015 [DOI] [PubMed] [Google Scholar]
- Chason R. J., Csokmay J., Segars J. H., DeCherney A. H., Armant D. R. (2011). Environmental and epigenetic effects upon preimplantation embryo metabolism and development. Trends Endocrinol. Metab. 22 412–420. 10.1016/j.tem.2011.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Zhang W., Xie D., Gao T., Dong Z., Lu X. (2020). Histone chaperone FACT represses retrotransposon MERVL and MERVL-derived cryptic promoters. Nucleic Acids Res. 48 10211–10225. 10.1093/nar/gkaa732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Guo L., Zhang L., Wu H., Yang J., Liu H., et al. (2013a). Vitamin C modulates TET1 function during somatic cell reprogramming. Nat. Genet. 45 1504–1509. 10.1038/ng.2807 [DOI] [PubMed] [Google Scholar]
- Chen J., Liu H., Liu J., Qi J., Wei B., Yang J., et al. (2013b). H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat. Genet. 45 34–42. 10.1038/ng.2491 [DOI] [PubMed] [Google Scholar]
- Christophorou M. A., Castelo-Branco G., Halley-Stott R. P., Oliveira C. S., Loos R., Radzisheuskaya A., et al. (2014). Citrullination regulates pluripotency and histone H1 binding to chromatin. Nature 507 104–108. 10.1038/nature12942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chronis C., Fiziev P., Papp B., Butz S., Bonora G., Sabri S., et al. (2017). Cooperative Binding of Transcription Factors Orchestrates Reprogramming. Cell 168 442–459e420. 10.1016/j.cell.2016.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornacchia D., Zhang C., Zimmer B., Chung S. Y., Fan Y., Soliman M. A., et al. (2019). Lipid Deprivation Induces a Stable, Naive-to-Primed Intermediate State of Pluripotency in Human PSCs. Cell Stem Cell 25 120–136e110. 10.1016/j.stem.2019.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson K. C., Mason E. A., Pera M. F. (2015). The pluripotent state in mouse and human. Development 142 3090–3099. 10.1242/dev.116061 [DOI] [PubMed] [Google Scholar]
- De Clerck L., Taelman J., Popovic M., Willems S., Van der Jeught M., Heindryckx B., et al. (2019). Untargeted histone profiling during naive conversion uncovers conserved modification markers between mouse and human. Sci. Rep. 9:17240. 10.1038/s41598-019-53681-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Iaco A., Coudray A., Duc J., Trono D. (2019). DPPA2 and DPPA4 are necessary to establish a 2C-like state in mouse embryonic stem cells. EMBO Rep. 20:201847382. 10.15252/embr.201847382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Iaco A., Planet E., Coluccio A., Verp S., Duc J., Trono D. (2017). DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat. Genet. 49 941–945. 10.1038/ng.3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean W., Santos F., Reik W. (2003). Epigenetic reprogramming in early mammalian development and following somatic nuclear transfer. Semin. Cell Dev. Biol. 14 93–100. 10.1016/s1084-9521(02)00141-6 [DOI] [PubMed] [Google Scholar]
- Devreker F., Englert Y. (2000). In vitro development and metabolism of the human embryo up to the blastocyst stage. Eur. J. Obstet. Gynecol. Reprod. Biol. 92 51–56. 10.1016/s0301-2115(00)00425-5 [DOI] [PubMed] [Google Scholar]
- Di Micco R., Fontanals-Cirera B., Low V., Ntziachristos P., Yuen S. K., Lovell C. D., et al. (2014). Control of embryonic stem cell identity by BRD4-dependent transcriptional elongation of super-enhancer-associated pluripotency genes. Cell Rep. 9 234–247. 10.1016/j.celrep.2014.08.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Stefano M., Stadhouders R., Farabella I., Castillo D., Serra F., Graf T., et al. (2020). Transcriptional activation during cell reprogramming correlates with the formation of 3D open chromatin hubs. Nat. Commun. 11:2564. 10.1038/s41467-020-16396-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit A., Parnas O., Li B., Chen J., Fulco C. P., Jerby-Arnon L., et al. (2016). Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell 167 1853–1866e1817. 10.1016/j.cell.2016.11.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon J. R., Jung I., Selvaraj S., Shen Y., Antosiewicz-Bourget J. E., Lee A. Y., et al. (2015). Chromatin architecture reorganization during stem cell differentiation. Nature 518 331–336. 10.1038/nature14222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodge J. E., Kang Y. K., Beppu H., Lei H., Li E. (2004). Histone H3-K9 methyltransferase ESET is essential for early development. Mol. Cell Biol. 24 2478–2486. 10.1128/mcb.24.6.2478-2486.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z., Zheng H., Huang B., Ma R., Wu J., Zhang X., et al. (2017). Allelic reprogramming of 3D chromatin architecture during early mammalian development. Nature 547 232–235. 10.1038/nature23263 [DOI] [PubMed] [Google Scholar]
- Duggal G., Warrier S., Ghimire S., Broekaert D., Van der Jeught M., Lierman S., et al. (2015). Alternative Routes to Induce Naive Pluripotency in Human Embryonic Stem Cells. Stem Cells 33 2686–2698. 10.1002/stem.2071 [DOI] [PubMed] [Google Scholar]
- Durruthy-Durruthy J., Wossidlo M., Pai S., Takahashi Y., Kang G., Omberg L., et al. (2016). Spatiotemporal Reconstruction of the Human Blastocyst by Single-Cell Gene-Expression Analysis Informs Induction of Naive Pluripotency. Dev. Cell 38 100–115. 10.1016/j.devcel.2016.06.014 [DOI] [PubMed] [Google Scholar]
- Ecco G., Cassano M., Kauzlaric A., Duc J., Coluccio A., Offner S., et al. (2016). Transposable Elements and Their KRAB-ZFP Controllers Regulate Gene Expression in Adult Tissues. Dev. Cell 36 611–623. 10.1016/j.devcel.2016.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enriquez-Gasca R., Gould P. A., Rowe H. M. (2020). Host Gene Regulation by Transposable Elements: The New, the Old and the Ugly. Viruses 12:12101089. 10.3390/v12101089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M. J., Kaufman M. H. (1981). Establishment in culture of pluripotential cells from mouse embryos. Nature 292 154–156. 10.1038/292154a0 [DOI] [PubMed] [Google Scholar]
- Factor D. C., Corradin O., Zentner G. E., Saiakhova A., Song L., Chenoweth J. G., et al. (2014). Epigenomic comparison reveals activation of “seed” enhancers during transition from naive to primed pluripotency. Cell Stem Cell 14 854–863. 10.1016/j.stem.2014.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Jacobsen S. E., Reik W. (2010). Epigenetic reprogramming in plant and animal development. Science 330 622–627. 10.1126/science.1190614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festuccia N., Osorno R., Halbritter F., Karwacki-Neisius V., Navarro P., Colby D., et al. (2012). Esrrb is a direct Nanog target gene that can substitute for Nanog function in pluripotent cells. Cell Stem Cell 11 477–490. 10.1016/j.stem.2012.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidalgo M., Faiola F., Pereira C. F., Ding J., Saunders A., Gingold J., et al. (2012). Zfp281 mediates Nanog autorepression through recruitment of the NuRD complex and inhibits somatic cell reprogramming. Proc. Natl. Acad. Sci. U S A. 109 16202–16207. 10.1073/pnas.1208533109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley L. W. S., Vardhana S. A., Carey B. W., Alonso-Curbelo D., Koche R., Chen Y., et al. (2018). Pluripotency transcription factors and Tet1/2 maintain Brd4-independent stem cell identity. Nat. Cell Biol. 20 565–574. 10.1038/s41556-018-0086-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fort A., Hashimoto K., Yamada D., Salimullah M., Keya C. A., Saxena A., et al. (2014). Deep transcriptome profiling of mammalian stem cells supports a regulatory role for retrotransposons in pluripotency maintenance. Nat. Genet. 46 558–566. 10.1038/ng.2965 [DOI] [PubMed] [Google Scholar]
- Fu X., He F., Li Y., Shahveranov A., Hutchins A. P. (2017). Genomic and molecular control of cell type and cell type conversions. Cell Regen. 6 1–7. 10.1016/j.cr.2017.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X., Wu X., Djekidel M. N., Zhang Y. (2019). Myc and Dnmt1 impede the pluripotent to totipotent state transition in embryonic stem cells. Nat. Cell Biol. 21 835–844. 10.1038/s41556-019-0343-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafni O., Weinberger L., Mansour A. A., Manor Y. S., Chomsky E., Ben-Yosef D., et al. (2013). Derivation of novel human ground state naive pluripotent stem cells. Nature 504 282–286. 10.1038/nature12745 [DOI] [PubMed] [Google Scholar]
- Galonska C., Ziller M. J., Karnik R., Meissner A. (2015). Ground State Conditions Induce Rapid Reorganization of Core Pluripotency Factor Binding before Global Epigenetic Reprogramming. Cell Stem Cell 17 462–470. 10.1016/j.stem.2015.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Nowak-Imialek M., Chen X., Chen D., Herrmann D., Ruan D., et al. (2019). Establishment of porcine and human expanded potential stem cells. Nat. Cell Biol. 21 687–699. 10.1038/s41556-019-0333-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatchalian J., Malik S., Ho J., Lee D. S., Kelso T. W. R., Shokhirev M. N., et al. (2018). A non-canonical BRD9-containing BAF chromatin remodeling complex regulates naive pluripotency in mouse embryonic stem cells. Nat. Commun. 9:5139. 10.1038/s41467-018-07528-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goke J., Lu X., Chan Y. S., Ng H. H., Ly L. H., Sachs F., et al. (2015). Dynamic transcription of distinct classes of endogenous retroviral elements marks specific populations of early human embryonic cells. Cell Stem Cell 16 135–141. 10.1016/j.stem.2015.01.005 [DOI] [PubMed] [Google Scholar]
- Grow E. J., Flynn R. A., Chavez S. L., Bayless N. L., Wossidlo M., Wesche D. J., et al. (2015). Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature 522 221–225. 10.1038/nature14308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G., Smith A. (2010). A genome-wide screen in EpiSCs identifies Nr5a nuclear receptors as potent inducers of ground state pluripotency. Development 137 3185–3192. 10.1242/dev.052753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G., Yang J., Nichols J., Hall J. S., Eyres I., Mansfield W., et al. (2009). Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development 136 1063–1069. 10.1242/dev.030957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Zhu P., Yan L., Li R., Hu B., Lian Y., et al. (2014). The DNA methylation landscape of human early embryos. Nature 511 606–610. 10.1038/nature13544 [DOI] [PubMed] [Google Scholar]
- Habibi E., Brinkman A. B., Arand J., Kroeze L. I., Kerstens H. H., Matarese F., et al. (2013). Whole-genome bisulfite sequencing of two distinct interconvertible DNA methylomes of mouse embryonic stem cells. Cell Stem Cell 13 360–369. 10.1016/j.stem.2013.06.002 [DOI] [PubMed] [Google Scholar]
- Hamazaki N., Kyogoku H., Araki H., Miura F., Horikawa C., Hamada N., et al. (2020). Reconstitution of the oocyte transcriptional network with transcription factors. Nature 589 264–269. 10.1038/s41586-020-3027-9 [DOI] [PubMed] [Google Scholar]
- Hanaoka K., Hayasaka M., Noguchi T., Kato Y. (1987). Viable Chimeras between Embryonal Carcinoma Cells and Mouse Embryos: Comparison of Aggregation and Injection Methods. Dev. Growth Different. 29 263–270. 10.1111/j.1440-169X.1987.00263.x [DOI] [PubMed] [Google Scholar]
- Hanna J., Cheng A. W., Saha K., Kim J., Lengner C. J., Soldner F., et al. (2010). Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc. Natl. Acad. Sci. U S A. 107 9222–9227. 10.1073/pnas.1004584107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins R. D., Hon G. C., Lee L. K., Ngo Q., Lister R., Pelizzola M., et al. (2010). Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell 6 479–491. 10.1016/j.stem.2010.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K., Ohta H., Kurimoto K., Aramaki S., Saitou M. (2011). Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell 146 519–532. 10.1016/j.cell.2011.06.052 [DOI] [PubMed] [Google Scholar]
- He J., Fu X., Zhang M., He F., Li W., Abdul M. M., et al. (2019). Transposable elements are regulated by context-specific patterns of chromatin marks in mouse embryonic stem cells. Nat. Commun. 10:34. 10.1038/s41467-018-08006-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson P. G., Dorais J. A., Grow E. J., Whiddon J. L., Lim J. W., Wike C. L., et al. (2017). Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat. Genet. 49 925–934. 10.1038/ng.3844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurtier V., Owens N., Gonzalez I., Mueller F., Proux C., Mornico D., et al. (2019). The molecular logic of Nanog-induced self-renewal in mouse embryonic stem cells. Nat. Commun. 10:1109. 10.1038/s41467-019-09041-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horne G. A., Stewart H. J., Dickson J., Knapp S., Ramsahoye B., Chevassut T. (2015). Nanog requires BRD4 to maintain murine embryonic stem cell pluripotency and is suppressed by bromodomain inhibitor JQ1 together with Lefty1. Stem Cells Dev. 24 879–891. 10.1089/scd.2014.0302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Tan D. E. K., Chia G., Tan H., Leong H. F., Chen B. J., et al. (2020). Maternal factor NELFA drives a 2C-like state in mouse embryonic stem cells. Nat. Cell Biol. 22 175–186. 10.1038/s41556-019-0453-8 [DOI] [PubMed] [Google Scholar]
- Huang Y., Kim J. K., Do D. V., Lee C., Penfold C. A., Zylicz J. J., et al. (2017). Stella modulates transcriptional and endogenous retrovirus programs during maternal-to-zygotic transition. Elife 6:22345. 10.7554/eLife.22345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Zhang H., Wang L., Tang C., Qin X., Wu X., et al. (2020). JMJD3 acts in tandem with KLF4 to facilitate reprogramming to pluripotency. Nat. Commun. 11:5061. 10.1038/s41467-020-18900-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins A. P., Pei D. (2015). Transposable elements at the center of the crossroads between embryogenesis, embryonic stem cells, reprogramming, and long non-coding RNAs. Sci. Bull. 60 1722–1733. 10.1007/s11434-015-0905-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins A. P., Choo S. H., Mistri T. K., Rahmani M., Woon C. T., Ng C. K., et al. (2013). Co-motif discovery identifies an Esrrb-Sox2-DNA ternary complex as a mediator of transcriptional differences between mouse embryonic and epiblast stem cells. Stem Cells 31 269–281. 10.1002/stem.1279 [DOI] [PubMed] [Google Scholar]
- Hutchins A. P., Yang Z., Li Y., He F., Fu X., Wang X., et al. (2017). Models of global gene expression define major domains of cell type and tissue identity. Nucleic Acids Res. 45 2354–2367. 10.1093/nar/gkx054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illich D. J., Zhang M., Ursu A., Osorno R., Kim K. P., Yoon J., et al. (2016). Distinct Signaling Requirements for the Establishment of ESC Pluripotency in Late-Stage EpiSCs. Cell Rep. 15 787–800. 10.1016/j.celrep.2016.03.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiuchi T., Enriquez-Gasca R., Mizutani E., Boskovic A., Ziegler-Birling C., Rodriguez-Terrones D., et al. (2015). Early embryonic-like cells are induced by downregulating replication-dependent chromatin assembly. Nat. Struct. Mol. Biol. 22 662–671. 10.1038/nsmb.3066 [DOI] [PubMed] [Google Scholar]
- Iturbide A., Torres-Padilla M. E. (2020). A cell in hand is worth two in the embryo: recent advances in 2-cell like cell reprogramming. Curr. Opin. Genet. Dev. 64 26–30. 10.1016/j.gde.2020.05.038 [DOI] [PubMed] [Google Scholar]
- Jang H. S., Shah N. M., Du A. Y., Dailey Z. Z., Pehrsson E. C., Godoy P. M., et al. (2019). Transposable elements drive widespread expression of oncogenes in human cancers. Nat. Genet. 51 611–617. 10.1038/s41588-019-0373-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q., Ang J. Y. J., Lee A. Y., Cao Q., Li K. Y., Yip K. Y., et al. (2020). G9a Plays Distinct Roles in Maintaining DNA Methylation, Retrotransposon Silencing, and Chromatin Looping. Cell Rep. 33:108315. 10.1016/j.celrep.2020.108315 [DOI] [PubMed] [Google Scholar]
- Jonsson M. E., Ludvik Brattas P., Gustafsson C., Petri R., Yudovich D., Pircs K., et al. (2019). Activation of neuronal genes via LINE-1 elements upon global DNA demethylation in human neural progenitors. Nat. Commun. 10:3182. 10.1038/s41467-019-11150-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalkan T., Bornelöv S., Mulas C., Diamanti E., Lohoff T., Ralser M., et al. (2019). Complementary Activity of ETV5, RBPJ, and TCF3 Drives Formative Transition from Naive Pluripotency. Cell Stem Cell 24 785.e–801.e. 10.1016/j.stem.2019.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M. M., Goyal P., Maksakova I. A., Bilenky M., Leung D., Tang J. X., et al. (2011). DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell 8 676–687. 10.1016/j.stem.2011.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Y., Xu Y., Chen X., Feng S., Liu Z., Sun Y., et al. (2017). 3D Chromatin Structures of Mature Gametes and Structural Reprogramming during Mammalian Embryogenesis. Cell 170 367–381e320. 10.1016/j.cell.2017.06.029 [DOI] [PubMed] [Google Scholar]
- Khoa L. T. P., Tsan Y. C., Mao F., Kremer D. M., Sajjakulnukit P., Zhang L., et al. (2020). Histone Acetyltransferase MOF Blocks Acquisition of Quiescence in Ground-State ESCs through Activating Fatty Acid Oxidation. Cell Stem Cell 27 441–458e410. 10.1016/j.stem.2020.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita M., Barber M., Mansfield W., Cui Y., Spindlow D., Stirparo G. G., et al. (2020). Capture of Mouse and Human Stem Cells with Features of Formative Pluripotency. Cell Stem Cell 2020:5. 10.1016/j.stem.2020.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisa F., Shiozawa S., Oda K., Yoshimatsu S., Nakamura M., Koya I., et al. (2017). Naive-like ESRRB(+) iPSCs with the Capacity for Rapid Neural Differentiation. Stem Cell Rep. 9 1825–1838. 10.1016/j.stemcr.2017.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimczak M., Czerwinska P., Mazurek S., Sozanska B., Biecek P., Mackiewicz A., et al. (2017). TRIM28 epigenetic corepressor is indispensable for stable induced pluripotent stem cell formation. Stem Cell Res. 23 163–172. 10.1016/j.scr.2017.07.012 [DOI] [PubMed] [Google Scholar]
- Kolodziejczyk A. A., Kim J. K., Tsang J. C., Ilicic T., Henriksson J., Natarajan K. N., et al. (2015). Single Cell RNA-Sequencing of Pluripotent States Unlocks Modular Transcriptional Variation. Cell Stem Cell 17 471–485. 10.1016/j.stem.2015.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimoto K., Yabuta Y., Hayashi K., Ohta H., Kiyonari H., Mitani T., et al. (2015). Quantitative Dynamics of Chromatin Remodeling during Germ Cell Specification from Mouse Embryonic Stem Cells. Cell Stem Cell 16 517–532. 10.1016/j.stem.2015.03.002 [DOI] [PubMed] [Google Scholar]
- Leeb M., Dietmann S., Paramor M., Niwa H., Smith A. (2014). Genetic exploration of the exit from self-renewal using haploid embryonic stem cells. Cell Stem Cell 14 385–393. 10.1016/j.stem.2013.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch H. G., McEwen K. R., Turp A., Encheva V., Carroll T., Grabole N., et al. (2013). Naive pluripotency is associated with global DNA hypomethylation. Nat. Struct. Mol. Biol. 20 311–316. 10.1038/nsmb.2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Liu J., Yang X., Zhou C., Guo J., Wu C., et al. (2017a). Chromatin Accessibility Dynamics during iPSC Reprogramming. Cell Stem Cell 21 819–833e816. 10.1016/j.stem.2017.10.012 [DOI] [PubMed] [Google Scholar]
- Li H., Lai P., Jia J., Song Y., Xia Q., Huang K., et al. (2017b). RNA Helicase DDX5 Inhibits Reprogramming to Pluripotency by miRNA-Based Repression of RYBP and its PRC1-Dependent and -Independent Functions. Cell Stem Cell 20:571. 10.1016/j.stem.2017.03.014 [DOI] [PubMed] [Google Scholar]
- Li M., Yu J. S. L., Tilgner K., Ong S. H., Koike-Yusa H., Yusa K. (2018). Genome-wide CRISPR-KO Screen Uncovers mTORC1-Mediated Gsk3 Regulation in Naive Pluripotency Maintenance and Dissolution. Cell Rep. 24 489–502. 10.1016/j.celrep.2018.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Stein P., Cheng X., Yang W., Shao N. Y., Morrisey E. E., et al. (2014). BRD4 regulates Nanog expression in mouse embryonic stem cells and preimplantation embryos. Cell Death Differ. 21 1950–1960. 10.1038/cdd.2014.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Ouyang J. F., Rossello F. J., Tan J. P., Davidson K. C., Valdes D. S., et al. (2020). Reprogramming roadmap reveals route to human induced trophoblast stem cells. Nature 586 101–107. 10.1038/s41586-020-2734-6 [DOI] [PubMed] [Google Scholar]
- Lu J. Y., Shao W., Chang L., Yin Y., Li T., Zhang H., et al. (2020a). Genomic Repeats Categorize Genes with Distinct Functions for Orchestrated Regulation. Cell Rep. 30 3296–3311e3295. 10.1016/j.celrep.2020.02.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Liu X., Huang W. K., Giusti-Rodriguez P., Cui J., Zhang S., et al. (2020b). Robust Hi-C Maps of Enhancer-Promoter Interactions Reveal the Function of Non-coding Genome in Neural Development and Diseases. Mol. Cell 79 521–534e515. 10.1016/j.molcel.2020.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubitz S., Glaser S., Schaft J., Stewart A. F., Anastassiadis K. (2007). Increased apoptosis and skewed differentiation in mouse embryonic stem cells lacking the histone methyltransferase Mll2. Mol. Biol. Cell 18 2356–2366. 10.1091/mbc.e06-11-1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch C. J., Bernad R., Martinez-Val A., Shahbazi M. N., Nobrega-Pereira S., Calvo I., et al. (2020). Global hyperactivation of enhancers stabilizes human and mouse naive pluripotency through inhibition of CDK8/19 Mediator kinases. Nat. Cell Biol. 22 1223–1238. 10.1038/s41556-020-0573-1 [DOI] [PubMed] [Google Scholar]
- Macfarlan T. S., Gifford W. D., Agarwal S., Driscoll S., Lettieri K., Wang J., et al. (2011). Endogenous retroviruses and neighboring genes are coordinately repressed by LSD1/KDM1A. Genes Dev. 25 594–607. 10.1101/gad.2008511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan T. S., Gifford W. D., Driscoll S., Lettieri K., Rowe H. M., Bonanomi D., et al. (2012). Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487 57–63. 10.1038/nature11244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksakova I. A., Thompson P. J., Goyal P., Jones S. J., Singh P. B., Karimi M. M., et al. (2013). Distinct roles of KAP1, HP1 and G9a/GLP in silencing of the two-cell-specific retrotransposon MERVL in mouse ES cells. Epigenet.Chromat. 6:15. 10.1186/1756-8935-6-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H., Kalkan T., Menafra R., Denissov S., Jones K., Hofemeister H., et al. (2012). The transcriptional and epigenomic foundations of ground state pluripotency. Cell 149 590–604. 10.1016/j.cell.2012.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G. R. (1980). Teratocarcinomas and mammalian embryogenesis. Science 209 768–776. 10.1126/science.6250214 [DOI] [PubMed] [Google Scholar]
- Matsuda K., Mikami T., Oki S., Iida H., Andrabi M., Boss J. M., et al. (2017). ChIP-seq analysis of genomic binding regions of five major transcription factors highlights a central role for ZIC2 in the mouse epiblast stem cell gene regulatory network. Development 144 1948–1958. 10.1242/dev.143479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer D., Stadler M. B., Rittirsch M., Hess D., Lukonin I., Winzi M., et al. (2020). Zfp281 orchestrates interconversion of pluripotent states by engaging Ehmt1 and Zic2. EMBO J. 39:e102591. 10.15252/embj.2019102591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacham C. E., Morrison S. J. (2013). Tumour heterogeneity and cancer cell plasticity. Nature 501 328–337. 10.1038/nature12624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles D. C., de Vries N. A., Gisler S., Lieftink C., Akhtar W., Gogola E., et al. (2017). TRIM28 is an Epigenetic Barrier to Induced Pluripotent Stem Cell Reprogramming. Stem Cells 35 147–157. 10.1002/stem.2453 [DOI] [PubMed] [Google Scholar]
- Neganova I., Cotts L., Banks P., Gassner K., Shukurov A., Armstrong L., et al. (2019). Endothelial Differentiation G Protein-Coupled Receptor 5 Plays an Important Role in Induction and Maintenance of Pluripotency. Stem Cells 37 318–331. 10.1002/stem.2954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J., Smith A. (2009). Naive and primed pluripotent states. Cell Stem Cell 4 487–492. 10.1016/j.stem.2009.05.015 [DOI] [PubMed] [Google Scholar]
- Nora E. P., Goloborodko A., Valton A. L., Gibcus J. H., Uebersohn A., Abdennur N., et al. (2017). Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell 169 930–944e922. 10.1016/j.cell.2017.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohbo K., Tomizawa S. (2015). Epigenetic regulation in stem cell development, cell fate conversion, and reprogramming. Biomol. Concepts 6 1–9. 10.1515/bmc-2014-0036 [DOI] [PubMed] [Google Scholar]
- Ohinata Y., Ohta H., Shigeta M., Yamanaka K., Wakayama T., Saitou M. (2009). A signaling principle for the specification of the germ cell lineage in mice. Cell 137 571–584. 10.1016/j.cell.2009.03.014 [DOI] [PubMed] [Google Scholar]
- Okamoto I., Otte A. P., Allis C. D., Reinberg D., Heard E. (2004). Epigenetic dynamics of imprinted X inactivation during early mouse development. Science 303 644–649. 10.1126/science.1092727 [DOI] [PubMed] [Google Scholar]
- Okamoto I., Patrat C., Thepot D., Peynot N., Fauque P., Daniel N., et al. (2011). Eutherian mammals use diverse strategies to initiate X-chromosome inactivation during development. Nature 472 370–374. 10.1038/nature09872 [DOI] [PubMed] [Google Scholar]
- Okashita N., Suwa Y., Nishimura O., Sakashita N., Kadota M., Nagamatsu G., et al. (2016). PRDM14 Drives OCT3/4 Recruitment via Active Demethylation in the Transition from Primed to Naive Pluripotency. Stem Cell Rep. 7 1072–1086. 10.1016/j.stemcr.2016.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onder T. T., Kara N., Cherry A., Sinha A. U., Zhu N., Bernt K. M., et al. (2012). Chromatin-modifying enzymes as modulators of reprogramming. Nature 483 598–602. 10.1038/nature10953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor W. A., Liu W., Chen D., Ho J., Kim R., Hunt T. J., et al. (2018). TFAP2C regulates transcription in human naive pluripotency by opening enhancers. Nat. Cell Biol. 20 553–564. 10.1038/s41556-018-0089-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrat C., Ouimette J. F., Rougeulle C. (2020). X chromosome inactivation in human development. Development 147:183095. 10.1242/dev.183095 [DOI] [PubMed] [Google Scholar]
- Peaston A. E., Evsikov A. V., Graber J. H., de Vries W. N., Holbrook A. E., Solter D., et al. (2004). Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev. Cell 7 597–606. 10.1016/j.devcel.2004.09.004 [DOI] [PubMed] [Google Scholar]
- Percharde M., Lin C. J., Yin Y., Guan J., Peixoto G. A., Bulut-Karslioglu A., et al. (2018). A LINE1-Nucleolin Partnership Regulates Early Development and ESC Identity. Cell 174 391.e–405.e. 10.1016/j.cell.2018.05.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petropoulos S., Edsgard D., Reinius B., Deng Q., Panula S. P., Codeluppi S., et al. (2016). Single-Cell RNA-Seq Reveals Lineage and X Chromosome Dynamics in Human Preimplantation Embryos. Cell 167:285. 10.1016/j.cell.2016.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posfai E., Schell J. P., Janiszewski A., Rovic I., Murray A., Bradshaw B., et al. (2021). Evaluating totipotency using criteria of increasing stringency. Nat. Cell Biol. 23 49–60. 10.1038/s41556-020-00609-2 [DOI] [PubMed] [Google Scholar]
- Rao R. A., Dhele N., Cheemadan S., Ketkar A., Jayandharan G. R., Palakodeti D., et al. (2015). Ezh2 mediated H3K27me3 activity facilitates somatic transition during human pluripotent reprogramming. Sci. Rep. 5:8229. 10.1038/srep08229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes J. D. P., Feldmann A., Hernandez-Rodriguez B., Diaz N., Brown J. M., Fursova N. A., et al. (2020). Cohesin Disrupts Polycomb-Dependent Chromosome Interactions in Embryonic Stem Cells. Cell Rep. 30 820–835e810. 10.1016/j.celrep.2019.12.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossant J. (2011). The Impact of Developmental Biology on Pluripotent Stem Cell Research: Successes and Challenges. Dev. Cell 21 20–23. 10.1016/j.devcel.2011.06.010 [DOI] [PubMed] [Google Scholar]
- Rossant J., Tam P. P. L. (2017). New Insights into Early Human Development: Lessons for Stem Cell Derivation and Differentiation. Cell Stem Cell 20 18–28. 10.1016/j.stem.2016.12.004 [DOI] [PubMed] [Google Scholar]
- Rostovskaya M., Stirparo G. G., Smith A. (2019). Capacitation of human naive pluripotent stem cells for multi-lineage differentiation. Development 146:172916. 10.1242/dev.172916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe H. M., Kapopoulou A., Corsinotti A., Fasching L., Macfarlan T. S., Tarabay Y., et al. (2013). TRIM28 repression of retrotransposon-based enhancers is necessary to preserve transcriptional dynamics in embryonic stem cells. Genome Res. 23 452–461. 10.1101/gr.147678.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley M. J., Corces V. G. (2018). Organizational principles of 3D genome architecture. Nat. Rev. Genet. 19 789–800. 10.1038/s41576-018-0060-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahakyan A., Kim R., Chronis C., Sabri S., Bonora G., Theunissen T. W., et al. (2017). Human Naive Pluripotent Stem Cells Model X Chromosome Dampening and X Inactivation. Cell Stem Cell 20 87–101. 10.1016/j.stem.2016.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A., Huang X., Fidalgo M., Reimer M. H., Jr., Faiola F., Ding J., et al. (2017). The SIN3A/HDAC Corepressor Complex Functionally Cooperates with NANOG to Promote Pluripotency. Cell Rep. 18 1713–1726. 10.1016/j.celrep.2017.01.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger S., Meshorer E. (2019). Open Chromatin, Epigenetic Plasticity, and Nuclear Organization in Pluripotency. Dev. Cell 48 135–150. 10.1016/j.devcel.2019.01.003 [DOI] [PubMed] [Google Scholar]
- Schulz E. G., Meisig J., Nakamura T., Okamoto I., Sieber A., Picard C., et al. (2014). The two active X chromosomes in female ESCs block exit from the pluripotent state by modulating the ESC signaling network. Cell Stem Cell 14 203–216. 10.1016/j.stem.2013.11.022 [DOI] [PubMed] [Google Scholar]
- Shan Y., Liang Z., Xing Q., Zhang T., Wang B., Tian S., et al. (2017). PRC2 specifies ectoderm lineages and maintains pluripotency in primed but not naive ESCs. Nat. Commun. 8:672. 10.1038/s41467-017-00668-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N., Ng H. H. (2017). The metabolic programming of stem cells. Genes Dev. 31 336–346. 10.1101/gad.293167.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J., Nichols J., Theunissen T. W., Guo G., van Oosten A. L., Barrandon O., et al. (2009). Nanog is the gateway to the pluripotent ground state. Cell 138 722–737. 10.1016/j.cell.2009.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim Y. J., Kim M. S., Nayfeh A., Yun Y. J., Kim S. J., Park K. T., et al. (2017). 2i Maintains a Naive Ground State in ESCs through Two Distinct Epigenetic Mechanisms. Stem Cell Rep. 8 1312–1328. 10.1016/j.stemcr.2017.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. (2017). Formative pluripotency: the executive phase in a developmental continuum. Development 144 365–373. 10.1242/dev.142679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. G., Heath J. K., Donaldson D. D., Wong G. G., Moreau J., Stahl M., et al. (1988). Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature 336 688–690. 10.1038/336688a0 [DOI] [PubMed] [Google Scholar]
- Smith D. G., Sturmey R. G. (2013). Parallels between embryo and cancer cell metabolism. Biochem. Soc. Trans. 41 664–669. 10.1042/BST20120352 [DOI] [PubMed] [Google Scholar]
- Smith Z. D., Chan M. M., Humm K. C., Karnik R., Mekhoubad S., Regev A., et al. (2014). DNA methylation dynamics of the human preimplantation embryo. Nature 511 611–615. 10.1038/nature13581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperber H., Mathieu J., Wang Y., Ferreccio A., Hesson J., Xu Z., et al. (2015). The metabolome regulates the epigenetic landscape during naive-to-primed human embryonic stem cell transition. Nat. Cell Biol. 17 1523–1535. 10.1038/ncb3264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadhouders R., Vidal E., Serra F., Di Stefano B., Le Dily F., Quilez J., et al. (2018). Transcription factors orchestrate dynamic interplay between genome topology and gene regulation during cell reprogramming. Nat. Genet. 50 238–249. 10.1038/s41588-017-0030-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart H. T., Stirparo G. G., Lohoff T., Bates L. E., Kinoshita M., Lim C. Y., et al. (2019). Distinct Molecular Trajectories Converge to Induce Naive Pluripotency. Cell Stem Cell 25 388–406e388. 10.1016/j.stem.2019.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126 663–676. 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Takashima Y., Guo G., Loos R., Nichols J., Ficz G., Krueger F., et al. (2014). Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell 158 1254–1269. 10.1016/j.cell.2014.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima Y., Guo G., Loos R., Nichols J., Ficz G., Krueger F., et al. (2015). Resetting Transcription Factor Control Circuitry toward Ground-State Pluripotency in Human. Cell 162 452–453. 10.1016/j.cell.2015.06.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Berge D., Kurek D., Blauwkamp T., Koole W., Maas A., Eroglu E., et al. (2011). Embryonic stem cells require Wnt proteins to prevent differentiation to epiblast stem cells. Nat. Cell Biol. 13 1070–1075. 10.1038/ncb2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesar P. J., Chenoweth J. G., Brook F. A., Davies T. J., Evans E. P., Mack D. L., et al. (2007). New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448 196–199. 10.1038/nature05972 [DOI] [PubMed] [Google Scholar]
- Teslaa T., Teitell M. A. (2015). Pluripotent stem cell energy metabolism: an update. EMBO J. 34 138–153. 10.15252/embj.201490446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen T. W., Friedli M., He Y., Planet E., O’Neil R. C., Markoulaki S., et al. (2016). Molecular Criteria for Defining the Naive Human Pluripotent State. Cell Stem Cell 19 502–515. 10.1016/j.stem.2016.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson J. A., Itskovitz-Eldor J., Shapiro S. S., Waknitz M. A., Swiergiel J. J., Marshall V. S., et al. (1998). Embryonic stem cell lines derived from human blastocysts. Science 282 1145–1147. 10.1126/science.282.5391.1145 [DOI] [PubMed] [Google Scholar]
- Toh C. X., Chan J. W., Chong Z. S., Wang H. F., Guo H. C., Satapathy S., et al. (2016). RNAi Reveals Phase-Specific Global Regulators of Human Somatic Cell Reprogramming. Cell Rep. 15 2597–2607. 10.1016/j.celrep.2016.05.049 [DOI] [PubMed] [Google Scholar]
- Tosolini M., Jouneau A. (2016). From Naive to Primed Pluripotency: In Vitro Conversion of Mouse Embryonic Stem Cells in Epiblast Stem Cells. Methods Mol. Biol. 1341 209–216. 10.1007/7651_2015_208 [DOI] [PubMed] [Google Scholar]
- van Mierlo G., Dirks R. A. M., De Clerck L., Brinkman A. B., Huth M., Kloet S. L., et al. (2019). Integrative Proteomic Profiling Reveals PRC2-Dependent Epigenetic Crosstalk Maintains Ground-State Pluripotency. Cell Stem Cell 24 123–137e128. 10.1016/j.stem.2018.10.017 [DOI] [PubMed] [Google Scholar]
- von Meyenn F., Iurlaro M., Habibi E., Liu N. Q., Salehzadeh-Yazdi A., Santos F., et al. (2016). Impairment of DNA Methylation Maintenance Is the Main Cause of Global Demethylation in Naive Embryonic Stem Cells. Mol. Cell 62:983. 10.1016/j.molcel.2016.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M., Teissandier A., Perez-Palacios R., Bourc’his D. (2016). An epigenetic switch ensures transposon repression upon dynamic loss of DNA methylation in embryonic stem cells. Elife 5:11418. 10.7554/eLife.11418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Liu X., Gao Y., Yang L., Li C., Liu W., et al. (2018). Reprogramming of H3K9me3-dependent heterochromatin during mammalian embryo development. Nat. Cell Biol. 20 620–631. 10.1038/s41556-018-0093-4 [DOI] [PubMed] [Google Scholar]
- Wang J., Huang J., Shi G. (2020). Retrotransposons in pluripotent stem cells. Cell Regen. 9:4. 10.1186/s13619-020-00046-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Xie G., Singh M., Ghanbarian A. T., Rasko T., Szvetnik A., et al. (2014). Primate-specific endogenous retrovirus-driven transcription defines naive-like stem cells. Nature 516 405–409. 10.1038/nature13804 [DOI] [PubMed] [Google Scholar]
- Wang T., Chen K., Zeng X., Yang J., Wu Y., Shi X., et al. (2011). The histone demethylases Jhdm1a/1b enhance somatic cell reprogramming in a vitamin-C-dependent manner. Cell Stem Cell 9 575–587. 10.1016/j.stem.2011.10.005 [DOI] [PubMed] [Google Scholar]
- Wang Y., Bi Y., Gao S. (2017). Epigenetic regulation of somatic cell reprogramming. Curr. Opin. Genet. Dev. 46 156–163. 10.1016/j.gde.2017.07.002 [DOI] [PubMed] [Google Scholar]
- Ware C. B., Nelson A. M., Mecham B., Hesson J., Zhou W., Jonlin E. C., et al. (2014). Derivation of naive human embryonic stem cells. Proc. Natl. Acad. Sci. U S A. 111 4484–4489. 10.1073/pnas.1319738111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z., Gao F., Kim S., Yang H., Lyu J., An W., et al. (2013). Klf4 organizes long-range chromosomal interactions with the oct4 locus in reprogramming and pluripotency. Cell Stem Cell 13 36–47. 10.1016/j.stem.2013.05.010 [DOI] [PubMed] [Google Scholar]
- Weinberger L., Ayyash M., Novershtern N., Hanna J. H. (2016). Dynamic stem cell states: naive to primed pluripotency in rodents and humans. Nat. Rev. Mol. Cell Biol. 17 155–169. 10.1038/nrm.2015.28 [DOI] [PubMed] [Google Scholar]
- Whyte W. A., Orlando D. A., Hnisz D., Abraham B. J., Lin C. Y., Kagey M. H., et al. (2013). Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153 307–319. 10.1016/j.cell.2013.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R. L., Hilton D. J., Pease S., Willson T. A., Stewart C. L., Gearing D. P., et al. (1988). Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature 336 684–687. 10.1038/336684a0 [DOI] [PubMed] [Google Scholar]
- Wu K., Liu H., Wang Y., He J., Xu S., Chen Y., et al. (2020). SETDB1-Mediated Cell Fate Transition between 2C-Like and Pluripotent States. Cell Rep. 30 25–36e26. 10.1016/j.celrep.2019.12.010 [DOI] [PubMed] [Google Scholar]
- Xia W., Xie W. (2020). Rebooting the Epigenomes during Mammalian Early Embryogenesis. Stem Cell Rep. 2020:5. 10.1016/j.stemcr.2020.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S., Lu J., Sridhar B., Cao X., Yu P., Zhao T., et al. (2017). SMARCAD1 Contributes to the Regulation of Naive Pluripotency by Interacting with Histone Citrullination. Cell Rep. 18 3117–3128. 10.1016/j.celrep.2017.02.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R. H., Peck R. M., Li D. S., Feng X., Ludwig T., Thomson J. A. (2005). Basic FGF and suppression of BMP signaling sustain undifferentiated proliferation of human ES cells. Nat. Methods 2 185–190. 10.1038/nmeth744 [DOI] [PubMed] [Google Scholar]
- Xu Y., Zhang M., Li W., Zhu X., Bao X., Qin B., et al. (2016). Transcriptional Control of Somatic Cell Reprogramming. Trends Cell Biol. 26 272–288. 10.1016/j.tcb.2015.12.003 [DOI] [PubMed] [Google Scholar]
- Yang B. X., El Farran C. A., Guo H. C., Yu T., Fang H. T., Wang H. F., et al. (2015). Systematic Identification of Factors for Provirus Silencing in Embryonic Stem Cells. Cell 163 230–245. 10.1016/j.cell.2015.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Huang X., Zang R., Chen J., Fidalgo M., Sanchez-Priego C., et al. (2020). DUX-miR-344-ZMYM2-Mediated Activation of MERVL LTRs Induces a Totipotent 2C-like State. Cell Stem Cell 26 234–250e237. 10.1016/j.stem.2020.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Rajan S. S., Friedrich M. J., Lan G., Zou X., Ponstingl H., et al. (2019a). Genome-Scale CRISPRa Screen Identifies Novel Factors for Cellular Reprogramming. Stem Cell Rep. 12 757–771. 10.1016/j.stemcr.2019.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Ryan D. J., Wang W., Tsang J. C., Lan G., Masaki H., et al. (2017a). Establishment of mouse expanded potential stem cells. Nature 550 393–397. 10.1038/nature24052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P., Humphrey S. J., Cinghu S., Pathania R., Oldfield A. J., Kumar D., et al. (2019b). Multi-omic Profiling Reveals Dynamics of the Phased Progression of Pluripotency. Cell Syst. 8 427–445e410. 10.1016/j.cels.2019.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S. H., Andrabi M., Biss R., Murtuza Baker S., Iqbal M., Sharrocks A. D. (2019c). ZIC3 Controls the Transition from Naive to Primed Pluripotency. Cell Rep. 27 3215–3227e3216. 10.1016/j.celrep.2019.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Liu B., Xu J., Wang J., Wu J., Shi C., et al. (2017b). Derivation of Pluripotent Stem Cells with In Vivo Embryonic and Extraembryonic Potency. Cell 169 243–257e225. 10.1016/j.cell.2017.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeom Y. I., Fuhrmann G., Ovitt C. E., Brehm A., Ohbo K., Gross M., et al. (1996). Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development 122 881–894. [DOI] [PubMed] [Google Scholar]
- Yilmaz A., Benvenisty N. (2019). Defining Human Pluripotency. Cell Stem Cell 25 9–22. 10.1016/j.stem.2019.06.010 [DOI] [PubMed] [Google Scholar]
- Ying Q. L., Nichols J., Chambers I., Smith A. (2003). BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 115 281–292. 10.1016/s0092-8674(03)00847-x [DOI] [PubMed] [Google Scholar]
- Ying Q. L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., et al. (2008). The ground state of embryonic stem cell self-renewal. Nature 453 519–523. 10.1038/nature06968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Wei Y., Sun H. X., Mahdi A. K., Pinzon Arteaga C. A., Sakurai M., et al. (2020a). Derivation of Intermediate Pluripotent Stem Cells Amenable to Primordial Germ Cell Specification. Cell Stem Cell 2020:3. 10.1016/j.stem.2020.11.003 [DOI] [PubMed] [Google Scholar]
- Yu S., Zhou C., Cao S., He J., Cai B., Wu K., et al. (2020b). BMP4 resets mouse epiblast stem cells to naive pluripotency through ZBTB7A/B-mediated chromatin remodelling. Nat. Cell Biol. 22 651–662. 10.1038/s41556-020-0516-x [DOI] [PubMed] [Google Scholar]
- Zapatka M., Borozan I., Brewer D. S., Iskar M., Grundhoff A., Alawi M., et al. (2020). The landscape of viral associations in human cancers. Nat. Genet. 52 320–330. 10.1038/s41588-019-0558-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Gayen S., Xiong J., Zhou B., Shanmugam A. K., Sun Y., et al. (2016). MLL1 Inhibition Reprograms Epiblast Stem Cells to Naive Pluripotency. Cell Stem Cell 18 481–494. 10.1016/j.stem.2016.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Wu D. Y., Zheng H., Wang Y., Sun Q. R., Liu X., et al. (2020a). Analysis of Genome Architecture during SCNT Reveals a Role of Cohesin in Impeding Minor ZGA. Mol. Cell 79 234–250e239. 10.1016/j.molcel.2020.06.001 [DOI] [PubMed] [Google Scholar]
- Zhang M., Lai Y., Krupalnik V., Guo P., Guo X., Zhou J., et al. (2020b). beta-Catenin safeguards the ground state of mouse pluripotency by strengthening the robustness of the transcriptional apparatus. Sci. Adv. 6:eaba1593. 10.1126/sciadv.aba1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Chen F., Chen R., Xie D., Yang J., Zhao X., et al. (2019). Zscan4c activates endogenous retrovirus MERVL and cleavage embryo genes. Nucleic Acids Res. 47 8485–8501. 10.1093/nar/gkz594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Li W., Zhu S., Joo J. Y., Do J. T., Xiong W., et al. (2010). Conversion of mouse epiblast stem cells to an earlier pluripotency state by small molecules. J. Biol. Chem. 285 29676–29680. 10.1074/jbc.C110.150599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Choi M., Margineantu D., Margaretha L., Hesson J., Cavanaugh C., et al. (2012). HIF1α induced switch from bivalent to exclusively glycolytic metabolism during ESC-to-EpiSC/hESC transition. EMBO J. 31 2103–2116. 10.1038/emboj.2012.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Q., Li W., Benda C., Huang Z., Ahmed T., Liu P., et al. (2018). NCoR/SMRT co-repressors cooperate with c-MYC to create an epigenetic barrier to somatic cell reprogramming. Nat. Cell Biol. 20 400–412. 10.1038/s41556-018-0047-x [DOI] [PubMed] [Google Scholar]
- Zimmerlin L., Zambidis E. T. (2020). Pleiotropic roles of tankyrase/PARP proteins in the establishment and maintenance of human naive pluripotency. Exp. Cell Res. 390:111935. 10.1016/j.yexcr.2020.111935 [DOI] [PMC free article] [PubMed] [Google Scholar]