Abstract
Background
While sex differences in coronary artery disease (CAD) are widely accepted with women developing more stable atherosclerosis than men, the underlying pathobiology of such differences remains largely unknown. In CAD, recent integrative systems biological studies have inferred gene regulatory networks (GRNs). Within these GRNs, key driver genes have shown great promises but have thus far been unidentified in females.
Methods
We generated sex-specific GRNs of the atherosclerotic arterial wall in 160 females and age-matched males in the Stockholm-Tartu Atherosclerosis Reverse Network Engineering Task (STARNET) study. We integrated the female GRNs with single-cell RNA-sequencing data of the human atherosclerotic plaque and single-cell RNA-sequencing of advanced atherosclerotic lesions from wildtype and Klf4 knock-out atherosclerotic SMC lineage tracing mice.
Results
By comparing sex-specific GRNs, we observed clear sex differences in network activity within the atherosclerotic tissues. Genes more active in females were associated with mesenchymal cells and endothelial cells, whereas genes more active in males were associated with the immune system. We determined that key drivers of GRNs active in female CAD were predominantly found in smooth muscle cells (SMCs) by single-cell sequencing of the human atherosclerotic plaques, as well as higher expressed in female plaque SMCs. To study the functions of these female SMC key drivers in atherosclerosis, we examined single-cell RNA-sequencing of advanced atherosclerotic lesions from wildtype and Klf4 knock-out atherosclerotic SMC lineage tracing mice. The female key drivers were found to be expressed by phenotypically modulated SMCs and affected by Klf4, suggesting that sex differences in atherosclerosis involves phenotypic switching of plaque SMCs.
Conclusions
Our systems approach provides novel insights into molecular mechanisms that underlie sex differences in atherosclerosis. To discover sex-specific therapeutic targets for atherosclerosis, an increased emphasis on sex-stratified approaches in the analysis of multi-omics datasets is warranted.
Keywords: Atherosclerosis, sex, smooth muscle cell, gene network, systems biology, transcriptome
Introduction
Coronary artery disease (CAD) is a major cause of morbidity and mortality for both men and women. Sex differences in the underlying pathogenesis of atherosclerotic disease have been described with women developing more diffuse atherosclerotic disease and more stable atherosclerotic plaques with erosion as an underlying cause of CAD, while men more often suffer from acute plaque rupture1,2. Furthermore, this discrepancy between the sexes seems to have a strong interaction with age3. Premenopausal women develop less inflamed plaques, but plaque composition changes in older women, and events ultimately exceed that of men. However, both stable and unstable plaques can lead to serious cardiovascular events.
Genome-wide association studies (GWAS) have shed light on potential treatment targets for CAD4. However, our ability to fully mitigate CAD remains limited because of an incomplete understanding of its molecular basis. Efforts to decode CAD GWAS signals into pathways and gene networks have been made through integrative systems biology approaches5. These approaches are increasingly being applied to identify coherent groups of disease genes (as opposed to individual genes) organized in gene regulatory networks (GRNs) that better mirror the inherent complexity of common disorders such as CAD. At the center of the gene networks used in systems biology lies the concept of gene activity, often probed by using gene connectivity as a proxy. Connectivity reveals the co-expression of a gene with other genes. Higher connectivity indicates that the gene is co-expressed more with other genes. We and others perceive higher connected genes as being more functional and more instrumental for disease biology6. Gene expression on the other hand is the quantifiable level at which the gene is expressed. It is the correlation of gene expression of different genes that is at the root of gene connectivity. In these GRNs, a limited number of genes termed key drivers have been shown to be essential for the activity and impact of GRNs on CAD5 suggesting that these key drivers may be suitable targets for novel therapies. So far, however, GRNs studied in CAD have been based on populations of >90% males5. This overrepresentation of male tissues combined with lack of sex-stratified analyses have uncovered GRNs and key driver genes predominantly important for male atherosclerosis. This implies that mechanisms of symptomatic female atherosclerosis is currently concealed, which warrants careful study of female atherosclerotic tissues.
To determine whether sex is important in shaping GRNs and disease mechanisms for CAD, we constructed equally powered sex-stratified GRNs in atherosclerotic tissue of patients undergoing coronary artery bypass grafting within the Stockholm-Tartu Atherosclerosis Reverse Network Engineering Task (STARNET), a human multi-tissue genetics of RNA expression study. (Fig. 1). We focused on the female-specific networks to determine potential key drivers of female-biased atherosclerosis. Single-cell RNA-sequencing (scRNA-seq) of human atherosclerotic plaques was used to determine the sex- and cell-type specific expression of female-biased key drivers. For further mechanistic understanding on the function of female-biased key drivers of atherosclerotic lesions, we used Western-diet fed smooth muscle cell (SMC) lineage tracing ApoE−/− mice and provide evidence that KLF4-dependent phenotypic switching of smooth muscle cells may be implicated in sex differences in atherosclerosis.
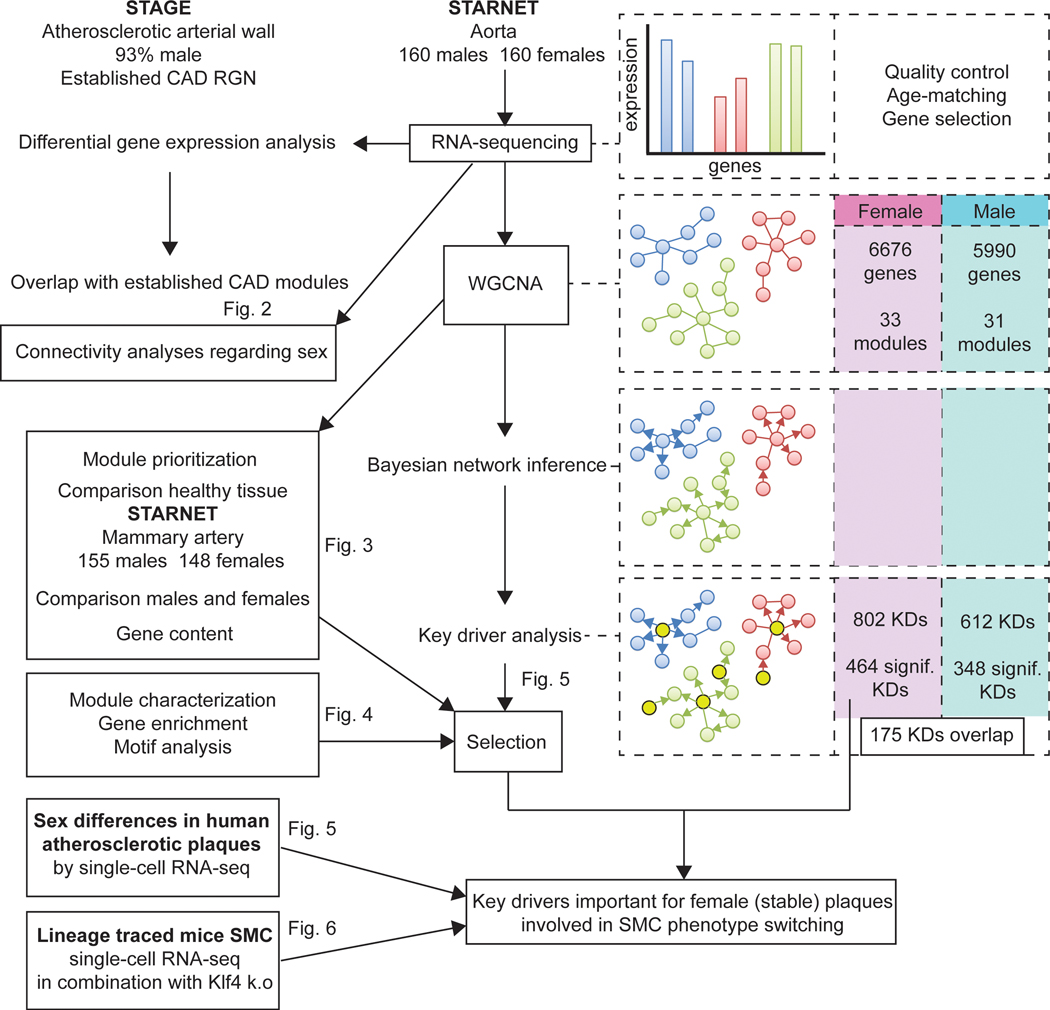
Figure 1. Workflow of sex-stratified GRN analyses.
The workflow of all analyses performed in this study are shown. We started by analysing previous determined CAD GRNs in STAGE (90%+ male) by using DEG data between the sexes from AOR in STARNET. To determine whether connectivity is affected by sex, we calculated gene connectivities in a multitude of populations in which we changed the proportion of females and males. Next, we generated sex-stratified GRNs by WGCNA, Bayesian network inference and key-driver analysis. The number of sex-stratified genes, modules, and key drivers are noted in the right boxes. We prioritized modules created by WGCNA for their importance in female CAD, by comparing how these modules behaved in healthy arterial tissue (MAM) and between the sexes. Further, we correlated the expression of the gene contents with clinical parameters of CAD, measured in STARNET, among which HDL, LDL, TG, glucose, and CRP levels, as well as Syntax score (a measure for the abundance and complexity of coronary atherosclerosis). After prioritization, we annotated the modules and analysed their gene content by gene enrichment analyses and motif analyses. We then selected the key drivers for the female CAD modules and determined their sex-specifc cellular expression by single-cell RNA-sequencing of atherosclerotic plaques. Lastly, we used lineage traced mouse SMC single-cell RNA-seq data to determine female key driver functionality. AOR = atherosclerotic aortic root, CAD = coronary artery disease, GRN = gene regulatory network, MAM = mammary artery, SMC = smooth muscle cell, WGCNA = weighted gene co-expression analysis
Materials and methods
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Study populations
The Stockholm-Tartu Atherosclerosis Reverse Network Engineering Task (STARNET) is a multi-tissue genetics of RNA-expression study comprised of 600 patients that underwent a coronary artery bypass grafting procedure, of which 30% are female. Details of the study and its population have been described before7. In this study, we used RNA-sequencing data of whom atherosclerotic aortic root (AOR) tissue was available, leading to 160 females and 160 age-matched males (baseline table can be found in Supplemental Table I). We used mammary artery (MAM) of the same patients as a control tissue.
The Athero-Express Biobank, of which the study design has been published before8 is an ongoing biobank study that includes patients undergoing arterial endarterectomy. We used plaque samples of 37 patients undergoing carotid endarterectomy (11 females and 26 males) for scRNA-seq.
The Stockholm Atherosclerosis Gene Expression study (STAGE) is a multi-tissue gene expression study for determining functionally associated genes important for coronary artery disease development9. The majority of included patients in STAGE are male (over 90%). We used previously published gene regulatory networks generated in STAGE5 for comparisons with sex-specific data generated in this study in STARNET7. Ethical approval of the study was given by the respective institutional review boards of the STARNET, STAGE, and Athero-Express studies. The study was performed in accordance with the Declaration of Helsinki.
Human aortic endothelial cells
Human aortic endothelial cells (HAEC) were isolated from aortic explants of 43 female and 129 male adult heart transplant donors in the University of California, Los Angeles (UCLA) transplant program and grown to confluence in 100 mm dishes as described previously10. All protocols involving humans were approved by UCLA Institutional Review Board. Gene expression profiles of the human cells were determined using the Affymetrix HT HG-U133A microarray, which contains 18,630 probes. Intensity values were normalized with the robust multiarray average normalization method implemented in the affy package in Bioconductor. Expression data are available in Gene Expression Omnibus accession GSE30169.
Data analysis
All methods regarding data analysis of sex-stratified gene regulatory networks, human single-cell RNA sequencing and murine single-cell RNA sequencing data can be found in the Supplementary Materials. The work-flow and different analyses are shown in Figure 1.
Statistical analysis
Gene activity was determined by calculating connectivity values. K-means clustering was used to determine gene groups for the connectivity shift (Fig 2A/B). For differential gene expression, a gene was called differentially expressed if FDR < 0.05. Gene enrichment was performed using hypergeometric testing. Co-expression networks were formed by generating an adjacency matrix. The adjacency matrix was transformed into a topological overlay matrix, which takes the connectivity of neighbouring genes into account. Subsequently, modules were generated by clustering the average distance of the dissimilarity matrix. Bayesian network analysis using a Fast Greedy Equivalence Search was performed to infer directionality, upon which a key driver analysis was performed. A key driver was deemed significant if FDR < 0.05. Permutations were performed to determine enrichment of gene modules for clinical correlates and gene content. Sex differential module score from single-cell RNA sequencing data was determined by using a Welch two sample T-test.
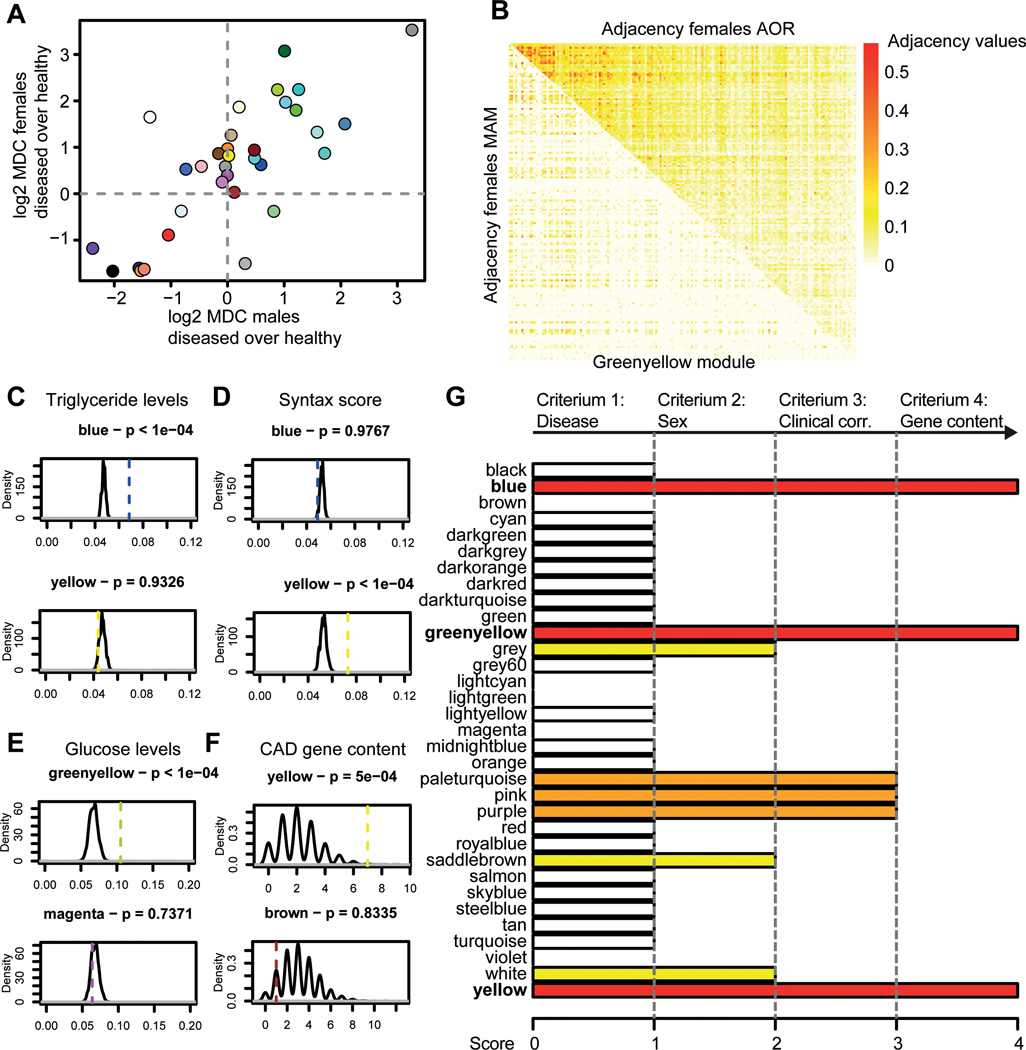
Figure 2. Sex as a biological variable in network biology.
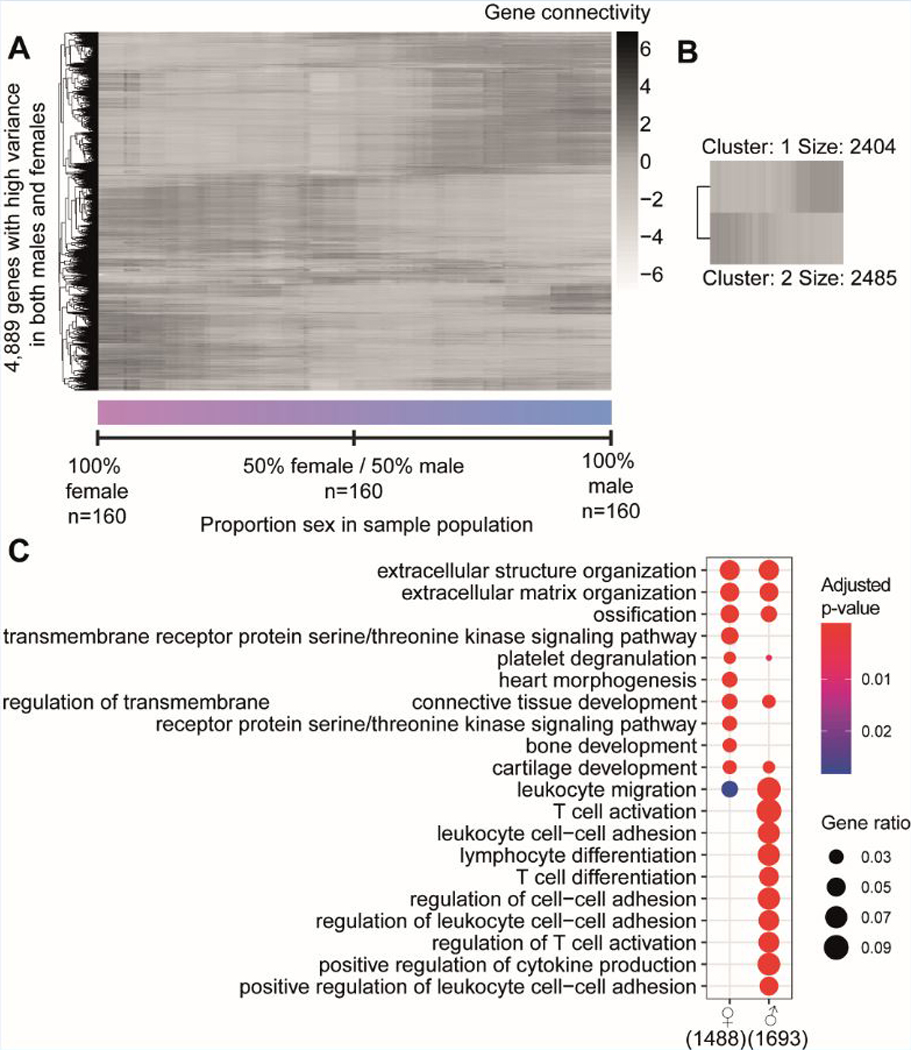
A) A median-centered heatmap of log2 - connectivity values of 4,889 genes is shown over 160 populations that differ in proportion of sex. The left-most values indicate a population that is 100% female, while the right-most values indicate 100% male. Every step in between from left to right changes the ratio by removing one female sample and adding one male sample. B) A heatmap of k-means clustering of the genes from the connectivity pattern of figure 2A (kmeans k = 2). Two clusters are formed with cluster 1 containing 2,404 genes higher connected in predominantly male populations, while cluster 2 contains 2,485 genes higher connected in female populations. C) Gene enrichment analysis of k-means clusters 1 and 2. Color indicates significance, while size of the dot shows gene ratio. Number indicates the amount of genes from the original cluster that had a use in the analysis.
Results
Sex-considerate gene co-expression in equally powered samples of atherosclerotic tissue
To investigate if sex affects gene activity in atherosclerotic tissues of 160 women and 160 age-matched men with CAD (mean age: 69 years old) obtained from STARNET, we calculated connectivities of 4,889 genes from the RNA-sequencing data (Fig. 2A). We started by calculating the connectivities in 160 females (left side of Figure 2A). Subsequently, we removed one female sample and added one male sample and recalculated the connectivity. From left to right on the x-axis of Figure 2A, females were subsequently replaced by males until the population resulted in 160 males. Gene connectivities generated from 70 to 90% males were comparable to those generated from a 100% male network. A non-stratified 50% women and 50% men composition reflects gene connectivity patterns that do not fit well for either sex. Moreover, connectivity patterns when 100% of the patients were female showed very limited overlap with connectivities when 100% of the patients were male. This indicates that sex-stratification is important for the identification of atherosclerosis-related GRNs, as a shift in connectivity for the majority of genes is evidently depending on the percentage of males and females included in a study. Upon visual inspection of the sex-driven connectivity shift (Fig. 2A), we detected 2 main groups of genes, and performed a Kmeans-clustering with 2 centres (Fig. 2B). Cluster 1 (n = 2404 genes) contains genes more connected in male atherosclerotic tissue, while cluster 2 (n = 2485 genes) contains genes more connected in atherosclerotic tissue from females. Gene enrichment analyses showed that the “male” cluster is enriched for genes mainly expressed in cells of the myeloid, immune, and hematological system (enrichment p-values 1.97E-94, 2.25E-73, 8.37E-70, respectively), whereas the “female” cluster genes are mostly expressed in endothelial and mesenchymal cells (enrichment p-values 1.54E-54, 3.59E-53 respectively, Fig. 2C for gene ontology, Supplemental Excel File I for ToppCell Atlas).
Besides gene connectivity, we also investigated differential gene expression of atherosclerotic tissues of 160 women and 160 age-matched men with CAD obtained from STARNET. This analysis revealed 3,728 differentially expressed genes (DEGs) between the sexes (FDR < 0.05), of which 1891 are higher expressed in females, and 1837 are higher expressed in males. Genes higher expressed corresponded to our gene connectivity results in males and were also enriched for immune-related processes, while genes higher expressed in females did not show a clear relation with the gene connectivity data of endothelial and mesenchymal cells (Suppl. Fig. I). The low enrichment for any process in female atherosclerotic tissue coincided with the observation that female-biased genes had fewer HGNC symbol annotations than male-biased genes (2.7x less, 379 missing in females versus 137 in males).
Comparison of previously identified male-biased GRN to expression in the equally powered male and female atherosclerotic tissues
Previous efforts have inferred GRNs for CAD from gene expression data from the STAGE study5. We checked the established STAGE CAD modules based on >90% men (and thus lacking power to investigate female GRNs) with RNA-sequencing data from the STARNET study for which we selected equally powered samples of 160 men and 160 women (Fig. 1, Suppl. Fig. IIA)7. The direction of effect of these previously identified GRN genes within these modules was male-biased, i.e. the majority of these genes was higher expressed in males as compared to females (Suppl. Fig. IIB). Gene ontology (GO) enrichment analyses on these male-driven modules indeed showed a very strong enrichment for immune response terms (Suppl. Fig. IIC). This corresponded with our observation on gene connectivity and expression in our selected population of 160 male atherosclerotic tissues. In the Supplementary Materials, we show a male GRN based on 160 men within STARNET (Supplemental Excel File I).
GRNs in female vascular tissue
The previously reported CAD modules support our view that connectivity patterns and subsequent gene expression levels in atherosclerotic tissue strongly differ between sexes. Therefore, we constructed GRNs in female tissues representing possibly unknown female atherosclerotic biology in 160 female atherosclerotic tissues.
We used 6,676 genes to build the initial female network within the atherosclerotic tissue by using weighted gene co-expression network analysis (WGCNA). 32 co-expression modules were identified harbouring all but 15 (the “grey”-module) of the 6,676 genes. Module size ranged from 26 to 1261 genes, averaged 202 genes per module and with a median size of 70 (Suppl. Fig. IIIABC). We performed Gene Ontology analysis on the modules to determine whether the generated modules match known biological pathways (Suppl. Fig. IV). Of the 32 female modules, at least 24 show enrichment for biological processes ranging from immunity to metabolism and muscle tissue development.
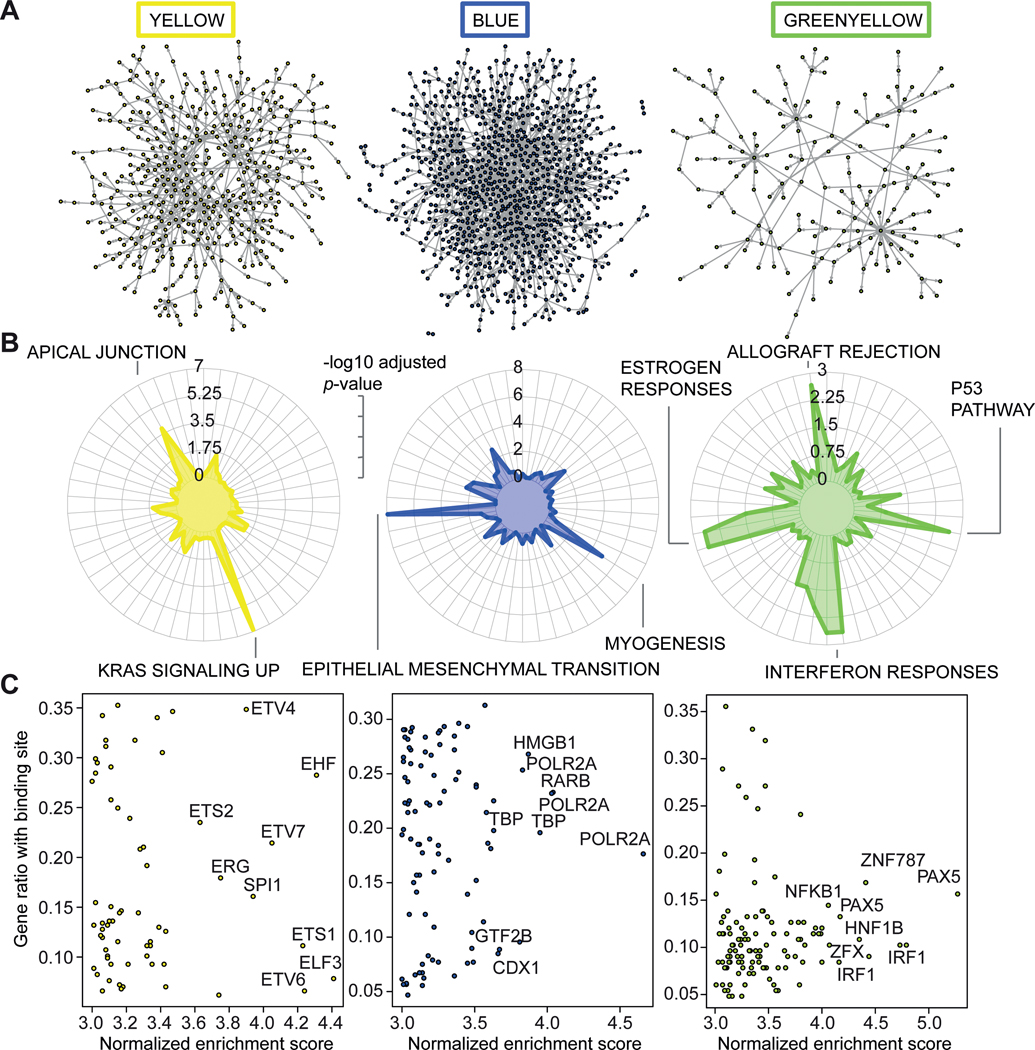
Module prioritization and characterization of female GRNs
To prioritize female modules in atherosclerotic tissue that are clinically relevant, we required that 1) the gene activity was higher in the diseased tissue (Fig. 3A/B, Suppl. Fig. VA); 2) the gene activity was higher in females as compared to males (Fig. 3A, Suppl. Fig. VB); 3) the gene content of the module was associated more to the degree of atherosclerosis and risk factor profiles than random genes (Fig. 3C-E, Suppl. Fig. VI-XII); 4) the gene content of the module was more enriched for genes linked to CAD (Fig. 3F, Suppl. Fig. XIII-XV). This led to the prioritization of three clinically relevant female modules for atherosclerosis (Fig. 3G). For detailed description of the selection, we refer to the Supplementary Methods section. We continued with these three (yellow, blue, and green-yellow) female modules for further investigation. Since the three prioritized female modules contain hundreds of genes (yellow: 485, blue: 1026, green-yellow: 166, structure depicted in Fig. 4A), we first determined what types of processes the modules were most likely representing. We used the Hallmark gene sets11 to determine overrepresented processes from 50 well-curated lists (Fig. 4B). The top 2 enrichments for the female yellow module were “KRAS signalling up” (p=1.35*10−7, 21 genes out of 200 in the set) and “Apical Junction” (p=1.49*10−4, 16 genes out of 200). The top two enrichments for blue were enriched for “Epithelial-to-mesenchymal transition” (p=1.20*10−8, 33 genes out of 202) and “myogenesis” (p=1.26*10−5, 27 genes out of 200). Lastly, green-yellow was enriched for interferon responses (p=0.002 and 0.002, 5 genes out of 97 and 8 genes out of 200), estrogen responses (p=0.002, 7 genes out of 200 for both) and allograft rejection (p=0.002, 7 out of 200 genes). We hypothesized that cell type was a major determinant for these three modules; endothelial cells for yellow, SMCs for blue, and immune cells for green-yellow. Motif analysis further underscored this notion (fig. 4C). The top 10 transcription factors with the highest normalized enrichment score in yellow were all ETS-family members, whereas for green-yellow this pertained to factors involved in inflammation and immune regulation, such as NFKB1, PAX5 and IRF1. Blue was enriched for factors involved in the generic transcriptional machinery, such as TBP and POLR2A. We further showed that gene connectivity of the yellow module was indeed higher in human aortic endothelial cells of females as compared to males (p<2.2e-16, Suppl. Fig. XVI).
Figure 3. Module prioritization.
A) A scatterplot of module differential connectivity (MDC) values is shown, colors indicate the different WGCNA-generated modules. The x-axis shows log2 MDC values of AOR/MAM in males, while the y-axis shows log2 MDC values of AOR/MAM in females. For example, yellow is higher connected in females in disease (MDC > 1.5), but equally connected in diseased and healthy tissue in males (MDC ~ 0). B) An example of a differentially connected module (greenyellow) is shown in a differential adjacency matrix. Upper triangle of the matrix shows adjacency values of greenyellow in female AOR, while the lower triangle shows female MAM. Color code indicates adjacency, a measure of connectivity, from white (not adjacent) to red (strongly adjacent). C-F) Density plots are shown indicating the distribution of permuted median absolute correlation values to clinical traits (C-E) and overlap with CAD gene content (F). Dashed lines indicate the observed comparator value. Permutation p-values and modules are indicated in the title of the panel. Two examples are given per panel, all comparison can be found in the Suppl. Fig. VI-XV. G) A barplot indicating score for prioritization of modules is shown. We selected 4 criteria to test the WGCNA-generated modules, of which differential connectivity in health and disease, sex-differential connectivity, association to CAD-related clinical parameters, and enrichment for CAD-related gene content. Color indicates score (white = 1, yellow = 2, orange = 3, red (selected) = 4). For example, the yellow module is differentially connected in healthy and diseased tissue, while also being differentially connected between males and females. Yellow is also more associated to clinical parameters for CAD than random genes, and contains more genes relevant to CAD than random genes. AOR = atherosclerotic aortic root, CAD = coronary artery disease, MAM = mammary artery, MDC = module differential connectivity, WGCNA = weighted gene co-expression analysis.
Figure 4. Module characterization.
A) Graphical representations of the selected modules (yellow, blue and greenyellow) are shown in their respective colors. Edges (lines) show connections, whereas nodes (dots) show genes. Gene names have been omitted for clarity. B) Hallmark gene enrichments are shown in radar plots for the selected modules. The color highlights the significance of the gene enrichment of genes in their respective modules. The length of the radius measures significance (-log10 adjusted p-value). The most significant terms are highlighted by name, such as KRAS signalling up for yellow, and epithelial mesenchymal transition for blue. C) Promoter motif enrichment scores are shown in scatterplots for the selected modules. X-axis indicates the normalized enrichment score (NES) for the factor, while the y-axis shows the ratio of genes in the module with a binding site for that factor. The factors with the highest NES are highlighted with their gene name.
Bayesian network inference and key driver analysis of female gene regulatory networks, and expression patterns in single cells from human atherosclerotic plaques
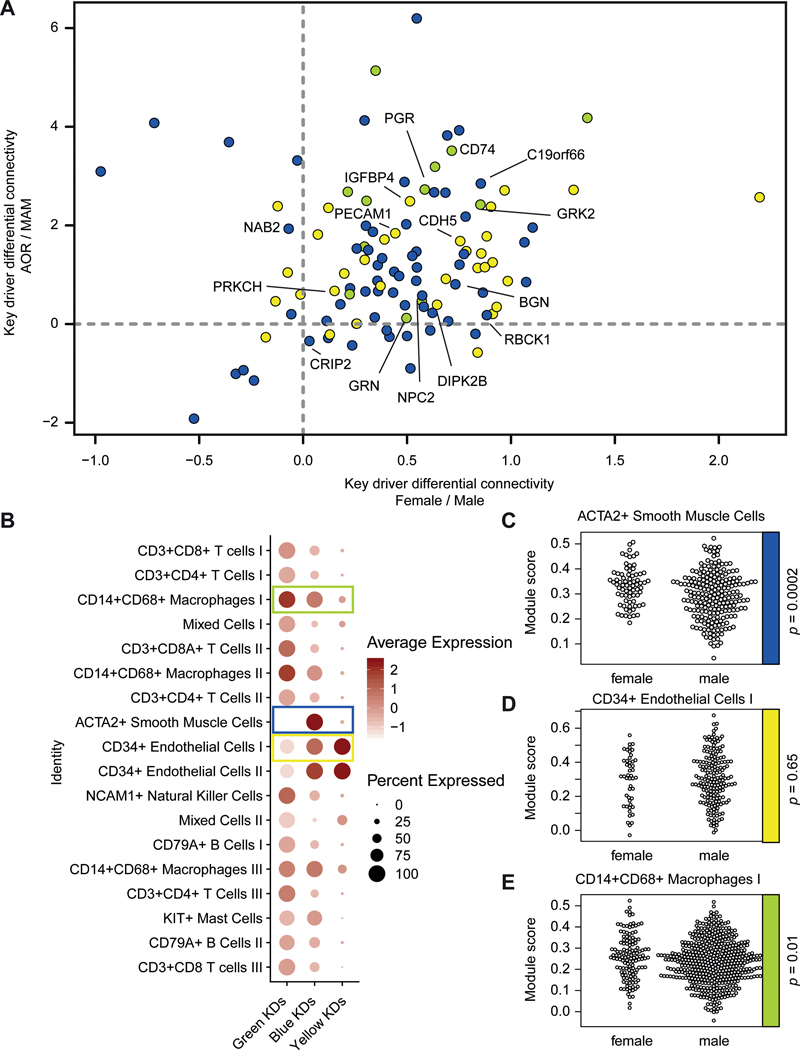
As WGCNA cannot infer directionality of nodes within a network, we applied Bayesian network algorithms to infer directed networks (i.e. gene regulatory networks (GRNs)) from the WGCNA-generated co-expression modules. Next, we performed key driver analysis12 to identify the top-hierarchical regulatory genes of each GRN governing the gene activity in each GRN. Key driver analysis identified 51 genes in the yellow GRN, 109 key drivers in the blue GRN, and 19 key drivers in the green yellow GRN, of which 33, 63, and 12 were significant, respectively. By calculating the differential gene connectivity of the significant female key drivers in atherosclerotic tissues, we found that the majority of key drivers of the female modules were more highly connected in disease than in healthy tissue, and more highly connected in females as compared to males (Fig. 5A).
Figure 5. Female CAD key drivers.
A) Gene differential connectivity of the key drivers of the female CAD modules is shown in a scatterplot. The x-axis shows differential connectivity in females over males, whereas the y-axis shows differential connectivity in AOR over MAM in females. Color indicates to which module the key driver belongs (yellow, blue, or greenyellow). The majority of the key drivers are grouped together in the upper right quadrant, indicating that they are more connected in females and in disease. The most significant key drivers of the three modules are highlighted with their gene name. B) Single cell RNA-sequencing expression in human carotid atherosclerotic plaques of the female CAD key drivers is shown in a dotplot. The columns show the module, whereas the rows show cell clusters found within the single cell RNA-sequencing of human atherosclerotic plaques. Each data point show the averaged expression of the key drivers in a module in that particular cell cluster. Color indicates expression, size of the dot indicate the percentage of expression. E.g. the key drivers of the yellow module are mostly expressed in endothelial cells of the human plaque. Key drivers selected for panel C-E are boxed in their respective color. C-E) Sex-stratified module scores of selected modules are shown in their respective cell-type. From top to bottom: blue key drivers in ACTA2+ SMCs, yellow key drivers in CD34+ endothelial cells I, green key drivers in CD14+CD68+ macrophages I. The p-value of the sex difference is shown on the right side (Welch Two Sample T-Test). AOR = atherosclerotic aortic root, CAD = coronary artery disease, MAM = mammary artery, SMC = smooth muscle cell.
Sex-considerate expression patterns in single cells from human atherosclerotic plaques
To characterize the cell-specificity of the modules, we analysed their key driver gene expression using scRNA-seq data of human atherosclerotic plaques obtained from carotid endarterectomies on 37 patients (26 males, 11 females)13 (Supplemental Fig. XVIIAB). The scRNA-seq data revealed 18 distinct cell populations, including inflammatory and non-inflammatory cell-types (Suppl. Fig. XVIIC). We calculated the average gene expression of the female CAD key drivers over these cell populations (Fig. 5B). The scRNA-seq data confirmed the cell-specificity, i.e. key drivers from yellow are mostly expressed in endothelial cells from atherosclerotic plaques, key drivers from blue are mostly expressed in endothelial cells and SMCs, while green-yellow is most prominent in macrophages. Next, we calculated a module score for the key-driver expression in the cell clusters and found significant sex differences in gene expression for the blue and green-yellow module. The key drivers from both the blue and the green-yellow module were expressed at a higher level in their respective cell-types derived from female plaques (p = 0.0002 and p = 0.01, respectively, Fig. 5C-E) as compared to male plaques. To see whether key drivers were differentially expressed between the sexes in their respective cell-type, we performed sex differential gene expression analysis in the scRNA-seq data. Overlap between blue module (SMC) key-drivers and differentially expressed genes between the sexes in plaque SMCs highlighted GAS6 and SERPING1 as potential female key drivers of CAD being more highly expressed in female SMCs.
Female key-driver genes point to SMC phenotype switching
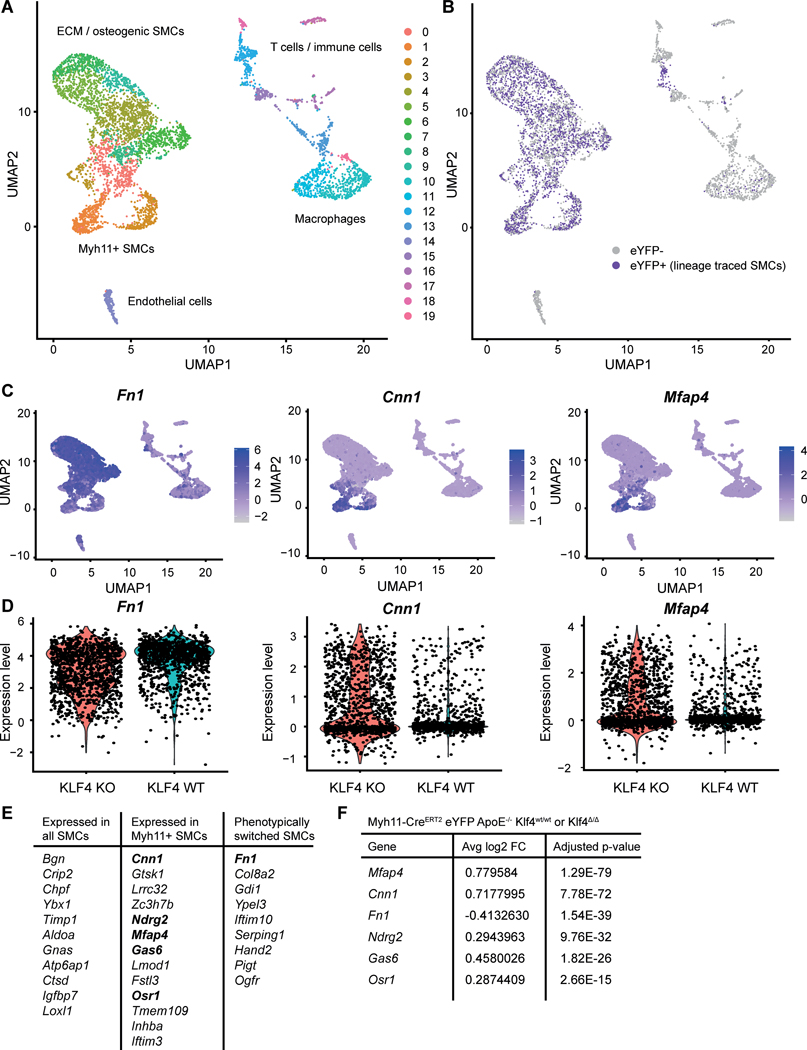
Since the blue module was enriched for myogenesis and epithelial to mesenchymal transition, and contained key drivers differentially expressed between the sexes in SMCs in plaques, we focused on the function of the key driver genes of the blue module in SMC transitions. Recent single cell SMC lineage tracing data14 was used to determine whether or not the key drivers in the blue module are involved in SMC phenotypic transitions important in atherosclerotic lesion development. scRNA-seq data from microdissected, late-stage atherosclerotic lesions from Myh11-CreERT2 eYFP ApoE−/− Klf4Δ/Δ and Klf4wt/wt mice was queried for expression of key driver genes from the yellow, green, and blue modules in SMC lineage-traced libraries. Remarkably, 33/57 key driver genes for the blue module (58%) were expressed in SMC-lineage traced cells, compared to 7/33 (21%) key driver genes for the yellow module and 2/12 (17%) key driver genes for the green module (Supplemental Excel File I). Thirteen blue module key driver genes were found in Myh11-positive clusters representing traditional SMC, 9 blue module key driver genes were found specifically in Myh11-negative clusters representing phenotypically switched SMC, and the remaining 11 key drivers were expressed in all SMC-derived clusters (Figure 6A-B, D). SMC-specific knockout of Klf4, which results in smaller lesions with evidence of increased stability15, affected expression of 6 female-biased genes in SMC-derived cells (Mfap4, Cnn1, Fn1, Ndrg2, Gas6, and Osr1). Fn1 (expressed in clusters 0, 1, 7 and 12, including collagen-rich and osteogenic SMC) was downregulated in SMC-specific Klf4 knockout, while the other 5 genes (expressed in clusters 4 and 6, including Acta2+ SMC) were upregulated (Figure 6C, E). Of note, Col8a2, which is also present in ECM-rich and osteogenic clusters of SMC and implicated in SMC phenotypic switching16, was downregulated with SMC-Klf4 knockout, but did not achieve statistical significance.
Figure 6. Female key drivers and SMC phenotypic switching.
Genes enriched in female plaques reflect Klf4-dependent SMC phenotypic switching. A) UMAP of scRNAseq data from advanced BCA plaques of SMC lineage tracing Myh11-CreERT2 eYFP ApoE−/− Klf4wt/wt or Klf4△/△ mice, organized by UMAP cluster (annotations refer to markers in Supplement). B) Clusters 0–7 represent SMC lineage traced cells, as shown by eYFP status. C) Feature plots of representative blue module driver genes. D) Violin plots of representative blue module driver genes with and without knockout of Klf4. E) Table of blue module driver genes expressed by lineage traced SMC. Boldly printed genes are expanded upon in F. F) Differential expression analysis of data in A revealed six blue module drivers are significantly regulated by Klf4 in SMCs (Avg_log2FC expressed relative to wild-type). SMC = smooth muscle cell.
Discussion
In most CAD cohort studies, women are underrepresented and the data is often pooled which subsequently leads to outcomes that may be male-biased. Our results demonstrate that this male overrepresentation in studies undermines the sex-diversity in known mechanisms of atherosclerotic disease. This knowledge is crucial in understanding why women and men show different features in atherosclerotic tissues.
We used male and female atherosclerotic samples of similar age, since we age-matched the women in the STARNET study to the men. This was also done to obtain similar powered analyses within the women and men, which is often a problem in cardiovascular studies. Our included population is on average 69 years of age, indicating that the male and female biology of atherosclerosis around this age is quite different. This in spite of the fact that these women are postmenopausal for years. Sex differences observed in our study may even be larger in younger populations as autopsy studies revealed that the plaque erosion pathology in women is especially pronounced around menopause17,18.
One of the strengths of our study is the extensive well powered data sources we used to study sex differences in atherosclerosis, both in human and mice, uncovering important female atherosclerotic biology in SMCs.
There may be multiple ways in which sex can affect SMC biology. First, sex chromosomes are different between males and females from the beginning of conception. Female cells enjoy an XX-sex chromosome complement, while males instead have an XY complement. Sex chromosome complements have been shown to affect traits and cardiovascular diseases, e.g. an XX complement promotes atherosclerosis in mice by increasing the bioavailability of dietary fat19. A second way can be by gonadal hormones that affect a plethora of processes. Activated estrogen or androgen receptors may regulate gene expression of multiple downstream targets and thereby influence the activity of GRNs. Interestingly, GAS6, one of the female key drivers in SMCs, has been shown to be induced by estrogen in epithelial cells20. Estrogen might similarly play a role in GAS6 activity in plaque SMCs. Of note, GAS6-Axl induced migration of cultured SMC is increased by hyperglycemia and postulated to contribute to exacerbation of atherosclerosis associated with diabetes21. Moreover, PI3K induced Akt1 activation, which is downstream of the GAS6-Axl axis, has been shown to be important in plaque stabilization22. Gas6 has also been shown to be secreted by SMCs in atherosclerotic plaques, with its expression being higher in stable plaques as compared to unstable plaques23. Smooth muscle cells are also involved in the process of arterial calcification by means of phenotype switching24, and sex differences within arterial calcifications, either intimal or medial, are well established with a higher incidence of calcifications in males25,26. Interestingly, GAS6 has been implicated in vascular calcifications27. On top of that, both estrogen and testosterone are mediators of the function described for GAS6 in calcifications28,29. This highlights a strong possible sex difference in vascular calcifications mediated by GAS6 to be followed up in other studies.
We showed in multiple ways that male-biased genes in our atherosclerotic lesions, albeit in activity as measured by connectivity or gene expression, are involved in immune regulation. These findings coincide with histological and imaging studies that show that male plaques are more inflammatory. Interestingly, the importance of the immune system in males and cardiovascular risk has also been highlighted by differential expression of Y chromosomal genes in macrophages of men with a Y chromosomal haplotype that harbors an increased cardiovascular risk30. Another striking finding was that genes higher expressed in females contain fewer HGNC symbol annotations than those higher expressed in males. One may speculate that this is because most studies have been performed on male tissues31.
One mechanism that may contribute to female plaque pathogenesis is SMC phenotypic switching. This phenomenon has been documented through recent lineage tracing and scRNA-seq analyses of mouse and human lesions, with different SMC phenotypes associated with atherosclerosis-promoting changes dependent on Klf4 or atherosclerosis-protective changes dependent on Tcf21 and Oct432,33. We observed that several key driver genes in the blue (SMC) module are expressed in SMCs negative for traditional markers, identified by lineage tracing, and that many traditional SMC markers enriched in females are regulated by Klf4. Two examples are GAS6 and FN1, which have both been implicated in the process of vascular remodelling22,23,34,35.
Studies of SMC phenotypic switching in females have been limited by the fact that existing lineage tracing mice carry the Myh11-CreERT2 transgene on the Y chromosome. While our analysis demonstrated that sex-stratified GRN key drivers in humans may function by affecting SMC phenotypic switching, future studies with novel transgenic mice not reliant on the Y chromosome transgene are needed to directly examine sex differences in SMC phenotypic switching and atherogenesis in vivo.
Limitations
We used atherosclerotic aortic tissue as a proxy for atherosclerosis in the coronary arteries, since it is difficult to obtain human atherosclerotic coronary artery tissue. Equally so, atherosclerosis is viewed as a systemic disease, and aortic atherosclerotic tissue may reflect disease patterns in a multitude of vascular beds.
Sex differences in tissue composition of the atherosclerotic plaque have been described before and may contribute to sex differences found in network connectivity. Hence, we might have detected our female key drivers because of differences in cellular composition, instead of differences on a cellular level. However, the sex-biased activity of the yellow (endothelial cell) module could be confirmed in human aortic endothelial cells, pointing towards differences intrinsic to the endothelium. This indicates that sex differences exist on a cellular level as well. The different genes of the modules are not necessarily cell-specific, since they may be expressed in other cell-types as well. However, the conglomerate module and its genes, and thus its biological function, is likely most important together as detected in the cell-types that we observed. Lastly, we were not able to distinguish potential effects driven by gender, since gender encompasses the cultural and societal differences between men and women, whereas sex differences stem from biological sex.
Concluding remarks
Sex should be treated as a biological variable in systems biology, since gene expression and network connectivity are heavily influenced by sex. We highlight disease mechanisms important for understanding why men and women develop atherosclerotic lesions with different compositions. Since most studies focus on pooled data or lack sex stratification, we argue that sex-specific research may provide additional important mechanistic insights in atherosclerotic disease.
Supplementary Material
Clinical perspective.
What Is New?
We show that biological sex affects gene expression and network connectivity in atherosclerotic tissues.
By generating sex-stratified gene regulatory networks, we highlight smooth muscle cell biology as a mechanism of pathophysiological differences in atherosclerosis between females and males.
What Are the Clinical Implications?
When systematically done, sex-stratified multi-omics data have the potential to lead to the discovery of new sex-specific therapeutic targets for atherosclerosis.
These results highlight novel smooth muscle biology in female atherosclerosis.
Acknowledgements
We acknowledge the service of Single Cell Discoveries for single-cell RNA-sequencing of human plaque material.
Funding
This study was funded by the Dutch Heart Foundation (Queen of Hearts 2013T084), EU project ERC consolidator grant 866478 (UCARE), and ERA-CVD 2017T099 ENDLESS.
Johan LM Björkegren (J.L.M.B.) acknowledges research support from NIH R01HL125863, American Heart Association (A14SFRN20840000), the Swedish Research Council (2018-02529), the Heart Lung Foundation (20170265) by Astra-Zeneca through ICMC, Karolinska Institutet, Sweden and from the Foundation Leducq(PlaqueOmics: Novel Roles of Smooth Muscle and Other Matrix Producing Cells in Atherosclerotic Plaque Stability and Rupture, 18CVD02.
List of abbreviations
- AOR
Atherosclerotic aortic root
- CAD
Coronary artery disease
- DEG
Differentially expressed gene
- GRN
Gene regulatory network
- GWAS
Genome-wide association study
- KDA
Key driver analysis
- MAM
Mammary artery
- scRNA-seq
Single-cell RNA-sequencing
- SMC
Smooth muscle cell
- STAGE
Stockholm Atherosclerosis Gene Expression study
- STARNET
Stockholm-Tartu Atherosclerosis Reverse Network Engineering Task
- WGCNA
Weighted gene co-expression network analysis
Footnotes
Conflict of interest:
There are no conflicts of interest to disclose.
References
- 1.Vrijenhoek JEP, Haitjema S, De Borst GJ, De Vries JPPM, Vaartjes I, Moll FL, Pasterkamp G, Den Ruijter HM. The impact of female sex on long-term survival of patients with severe atherosclerosis undergoing endarterectomy. Atherosclerosis. 2014;237:521–527. [DOI] [PubMed] [Google Scholar]
- 2.Vrijenhoek JEP, Den Ruijter HM, De Borst GJ, De Kleijn DP V, De Vries JPPM, Bots ML, Van De Weg SM, Vink A, Moll FL, Pasterkamp G. Sex is associated with the presence of atherosclerotic plaque hemorrhage and modifies the relation between plaque hemorrhage and cardiovascular outcome. Stroke. 2013;44:3318–3323. [DOI] [PubMed] [Google Scholar]
- 3.Man JJ, Beckman JA, Jaffe IZ. Sex as a Biological Variable in Atherosclerosis. Circ Res. 2020; 123: 1297–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pim V der H, Niek V. Identification of 64 Novel Genetic Loci Provides an Expanded View on the Genetic Architecture of Coronary Artery Disease. Circ Res. 2018;122:433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Talukdar HA, Foroughi Asl H, Jain RK, Ermel R, Ruusalepp A, Franzén O, Kidd BA, Readhead B, Giannarelli C, Kovacic JC et al. Cross-Tissue Regulatory Gene Networks in Coronary Artery Disease. Cell Syst. 2016;2:196–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeong H, Mason SP, Barabási AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. [DOI] [PubMed] [Google Scholar]
- 7.Franzén O, Ermel R, Cohain A, Akers NK, Di Narzo A, Talukdar HA, Foroughi-Asl H, Giambartolomei C, Fullard JF, Sukhavasi K et al. Cardiometabolic risk loci share downstream cis- and trans-gene regulation across tissues and diseases. Science. 2016;353:827–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verhoeven BAN, Velema E, Schoneveld AH, De Vries JPPM, De Bruin P, Seldenrijk CA, De Kleijn DPV, Busser E, Van Der Graaf Y, Moll F et al. Athero-express: Differential atherosclerotic plaque expression of mRNA and protein in relation to cardiovascular events and patient characteristics. Rationale and design. Eur J Epidemiol. 2004;19:1127–1133. [DOI] [PubMed] [Google Scholar]
- 9.Hägg S, Skogsberg J, Lundström J, Noori P, Nilsson R, Zhong H, Maleki S, Shang M-M, Brinne B, Bradshaw M et al. Multi-Organ Expression Profiling Uncovers a Gene Module in Coronary Artery Disease Involving Transendothelial Migration of Leukocytes and LIM Domain Binding 2: The Stockholm Atherosclerosis Gene Expression (STAGE) Study. PLoS Genet. 2009;5:e1000754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erbilgin A, Civelek M, Romanoski CE, Pan C, Hagopian R, Berliner JA, Lusis AJ. Identification of CAD candidate genes in GWAS loci and their expression in vascular cells. J Lipid Res. 2013;54:1894–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database Hallmark Gene Set Collection. Cell Syst. 2015;1:417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shu L, Zhao Y, Kurt Z, Byars SG, Tukiainen T, Kettunen J, Orozco LD, Pellegrini M, Lusis AJ, Ripatti S et al. Mergeomics: Multidimensional data integration to identify pathogenic perturbations to biological systems. BMC Genomics. 2016; 17:874. doi: 10.1186/s12864-016-3198-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Depuydt MA, Prange KH, Slenders L, Örd T, Elbersen D, Boltjes A, de Jager SC, Asselbergs FW, de Borst GJ, Aavik E et al. Microanatomy of the Human Atherosclerotic Plaque by Single-Cell Transcriptomics. Circ Res. 2020; 127: 1437–1455. doi: 10.1161/CIRCRESAHA.120.316770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alencar GF, Owsiany KM, K S, Sukhavasi K, Mocci G, Nguyen A, Williams CM, Shamsuzzaman S, Mokry M, Henderson CA et al. Stem Cell Pluripotency Genes Klf4 and Oct4 Regulate Complex SMC Phenotypic Changes Critical in Late-Stage Atherosclerotic Lesion Pathogenesis. Circulation. 2020;142: 2045–2059. Doi: 10.1161/CIRCULATIONAHA.120.046672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shankman LS, Gomez D, Cherepanova OA, Salmon M, Alencar GF, Haskins RM, Swiatlowska P, Newman AAC, Greene ES, Straub AC et al. KLF4-dependent phenotypic modulation of smooth muscle cells has a key role in atherosclerotic plaque pathogenesis. Nat Med. 2015;21:628–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cherepanova OA, Pidkovka NA, Sarmento OF, Yoshida T, Gan Q, Adiguzel E, Bendeck MP, Berliner J, Leitinger N, Owens GK. Oxidized Phospholipids Induce Type VIII Collagen Expression and Vascular Smooth Muscle Cell Migration. Circ Res. 2009;104:609–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burke AP, Farb A, Malcom G, Virmani R. Effect of menopause on plaque morphologic characteristics in coronary atherosclerosis. Am Heart J. 2001;141:S58–62. [DOI] [PubMed] [Google Scholar]
- 18.Yahagi K, Davis HR, Arbustini E, Virmani R. Sex differences in coronary artery disease: Pathological observations. Atherosclerosis. 2015;239:260–267. [DOI] [PubMed] [Google Scholar]
- 19.AlSiraj Y, Chen X, Thatcher SE, Temel RE, Cai L, Blalock E, Katz W, Ali HM, Petriello M, Deng P et al. XX sex chromosome complement promotes atherosclerosis in mice. Nat Commun. 2019;10:2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mo R, Tony Zhu Y, Zhang Z, Rao SM, Zhu Y-J. GAS6 is an estrogen-inducible gene in mammary epithelial cells. Biochem Biophys Res Commun. 2007;353:189–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cavet ME, Smolock EM, Ozturk OH, World C, Pang J, Konishi A, Berk BC. Gas6–Axl Receptor Signaling Is Regulated by Glucose in Vascular Smooth Muscle Cells. Arterioscler Thromb Vasc Biol. 2008;28:886–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernández-Hernando C, József L, Jenkins D, Di Lorenzo A, Sessa WC. Absence of Akt1 Reduces Vascular Smooth Muscle Cell Migration and Survival and Induces Features of Plaque Vulnerability and Cardiac Dysfunction During Atherosclerosis. Arterioscler Thromb Vasc Biol. 2009;29:2033–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clauser S, Meilhac O, Bièche I, Raynal P, Bruneval P, Michel JB, Borgel D. Increased secretion of gas6 by smooth muscle cells in human atherosclerotic carotid plaques. Thromb Haemost. 2012;107:140–149. [DOI] [PubMed] [Google Scholar]
- 24.Durham AL, Speer MY, Scatena M, Giachelli CM, Shanahan CM. Role of smooth muscle cells in vascular calcification: Implications in atherosclerosis and arterial stiffness. Cardiovasc Res. 2018;114:590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zwakenberg SR, de Jong PA, Hendriks EJ, Westerink J, Spiering W, de Borst GJ, Cramer MJ, Bartstra JW, Doesburg T, Rutters F et al. Intimal and medial calcification in relation to cardiovascular risk factors. PLoS One. 2020;15:e0235228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang B, Miller VM, Miller JD. Influences of Sex and Estrogen in Arterial and Valvular Calcification. Front Endocrinol (Lausanne). 2019;10:622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qiu C, Zheng H, Tao H, Yu W, Jiang X, Li A, Jin H, Lv A, Li H. Vitamin K2 inhibits rat vascular smooth muscle cell calcification by restoring the Gas6/Axl/Akt anti-apoptotic pathway. Mol Cell Biochem. 2017;433:149–159. [DOI] [PubMed] [Google Scholar]
- 28.Nanao-Hamai M, Son B-K, Hashizume T, Ogawa S, Akishita M. Protective effects of estrogen against vascular calcification via estrogen receptor α-dependent growth arrest-specific gene 6 transactivation. Biochem Biophys Res Commun. 2016;480:429–435. [DOI] [PubMed] [Google Scholar]
- 29.Son B-K, Akishita M, Iijima K, Ogawa S, Maemura K, Yu J, Takeyama K, Kato S, Eto M, Ouchi Y. Androgen receptor-dependent transactivation of growth arrest-specific gene 6 mediates inhibitory effects of testosterone on vascular calcification. J Biol Chem. 2010;285:7537–7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bloomer LDS, Nelson CP, Eales J, Denniff M, Christofidou P, Debiec R, Moore J, Consortium C, Zukowska-Szczechowska E, Goodall AH et al. Male-Specific Region of the Y Chromosome and Cardiovascular Risk. Arterioscler Thromb Vasc Biol. 2013;33:1722–1727. [DOI] [PubMed] [Google Scholar]
- 31.Ventura-Clapier R, Dworatzek E, Seeland U, Kararigas G, Arnal JF, Brunelleschi S, Carpenter TC, Erdmann J, Franconi F, Giannetta E et al. Sex in basic research: Concepts in the cardiovascular field. Cardiovasc Res. 2017;113:711–724. [DOI] [PubMed] [Google Scholar]
- 32.Wirka RC, Wagh D, Paik DT, Pjanic M, Nguyen T, Miller CL, Kundu R, Nagao M, Coller J, Koyano TK et al. Atheroprotective roles of smooth muscle cell phenotypic modulation and the TCF21 disease gene as revealed by single-cell analysis. Nat Med. 2019;25:1280–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cherepanova OA, Gomez D, Shankman LS, Swiatlowska P, Williams J, Sarmento OF, Alencar GF, Hess DL, Bevard MH, Greene ES et al. Activation of the pluripotency factor OCT4 in smooth muscle cells is atheroprotective. Nat Med. 2016;22:657–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doddapattar P, Dev R, Jain M, Dhanesha N, Chauhan AK. Differential Roles of Endothelial Cell-Derived and Smooth Muscle Cell-Derived Fibronectin Containing Extra Domain A in Early and Late Atherosclerosis. Arterioscler Thromb Vasc Biol. 2020;40:1738–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Budatha M, Zhang J, Zhuang ZW, Yun S, Dahlman JE, Anderson DG, Schwartz MA. Inhibiting Integrin α5 Cytoplasmic Domain Signaling Reduces Atherosclerosis and Promotes Arteriogenesis. J Am Heart Assoc. 2018;7:e007501. doi: 10.1161/JAHA.117.007501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R Package for Comparing Biological Themes Among Gene Clusters. Omi A J Integr Biol. 2012;16:284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:305–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The SVA package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khera A V, Kathiresan S. Genetics of coronary artery disease: Discovery, biology and clinical translation. Nat Rev Genet. 2017;18:331–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hartman RJG, Korporaal SJA, Mokry M, de Jager SCA, Meeuwsen JAL, van der Laan SW, Lansu NR, Zoet GA, Pasterkamp G, Urbanus RT et al. Platelet RNA modules point to coronary calcification in asymptomatic women with former preeclampsia. Atherosclerosis. 2019;291:114–121. doi: 10.1016/j.atherosclerosis.2019.10.009. [DOI] [PubMed] [Google Scholar]
- 43.Aibar S, González-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, Rambow F, Marine J, Geurts P, Aerts J et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14:1083–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muraro MJ, Dharmadhikari G, Grün D, Groen N, Dielen T, Jansen E, van Gurp L, Engelse MA, Carlotti F, de Koning EJP et al. A Single-Cell Transcriptome Atlas of the Human Pancreas. Cell Syst. 2016;3:385–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hashimshony T, Senderovich N, Avital G, Klochendler A, de Leeuw Y, Anavy L, Gennert D, Li S, Livak KJ, Rozenblatt-Rosen O et al. CEL-Seq2: Sensitive highly-multiplexed single-cell RNA-Seq. Genome Biol. 2016;17:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.