Summary
The Fused (Fu) kinase is a key transducer of Hedgehog signaling, but its relevant substrates have remained obscured due to the difficulty of obtaining active Fu for in vitro kinase assay. Based on the mechanism of Fu activation in vivo, we engineered a constitutively active Fu and expressed it in Sf9 cells using the baculovirus system. The kinase was affinity purified and applied for in vitro kinase assay using recombinant GST-fusion proteins as substrates to identify Fu-specific phosphorylation sites.
For complete details on the use and execution of this protocol, please refer to Han et al. (2019).
Subject areas: Molecular biology, Protein biochemistry, Protein expression and purification
Graphical Abstract
Highlights
-
•
Purification of constitutively active Fu from insect cells for in vitro kinase assay
-
•
Priming phosphorylation by Fu can allow secondary in vitro kinase assay
-
•
High-purity protein elution with Flag M2 affinity agarose and 3X Flag peptide
-
•
Sensitive phospho-protein detection via pIMAGO-biotin kit or specific antibodies
The Fused (Fu) kinase is a key transducer of Hedgehog signaling but its relevant substrates have remained obscured due to the difficulty of obtaining active Fu for in vitro kinase assay. Based on the mechanism of Fu activation in vivo, we engineered a constitutively active Fu and expressed it in Sf9 cells using the baculovirus system. The kinase was affinity purified and applied for in vitro kinase assay using recombinant GST-fusion proteins as substrates to identify Fu-specific phosphorylation sites.
Before you begin
Note: this protocol has been employed to demonstrate the direct phosphorylation of Drosophila Hedgehog pathway components, such as the transcription factor Cubitus interruptus (Ci) and the kinesin like protein Costal2 (Cos2) by Fu (Han et al., 2019). The identified phosphorylation sites fit the consensus sequence of S/T(X)5D/E, where X could be any amino acid (Han et al., 2019). The Fu phosphorylation consensus sites are also found in the Gli proteins that are mammalian homologs of Ci and are phosphorylated by the Fu-family kinase Ulk3 and STK36 (Han et al., 2019). This protocol can be applied to other protein kinases and their substrates with respect to construction of active kinases by forced-dimerization, one-step purification with Flag M2 agarose column, and in vitro kinase assay using pIMAGO-biotin phosphoprotein detection kit with the advantage of avoiding radioactive isotope.
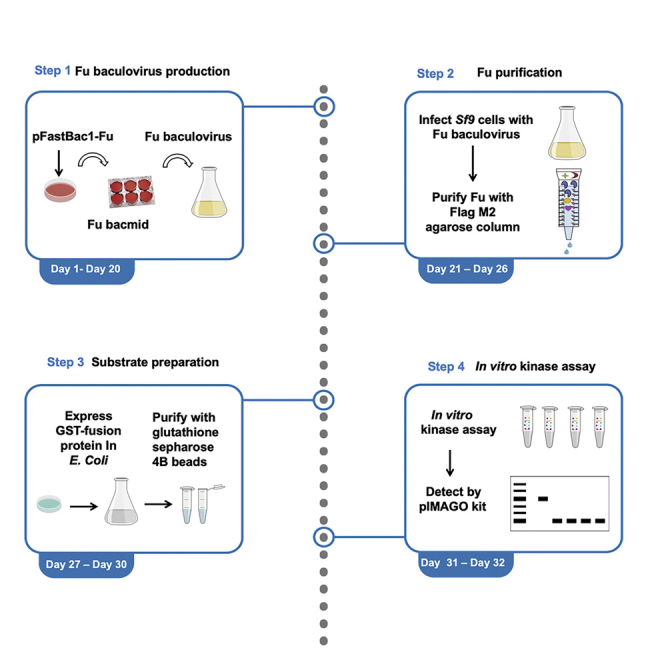
Engineering a constitutively active form of Fu for expression in Baculovirus
-
1.
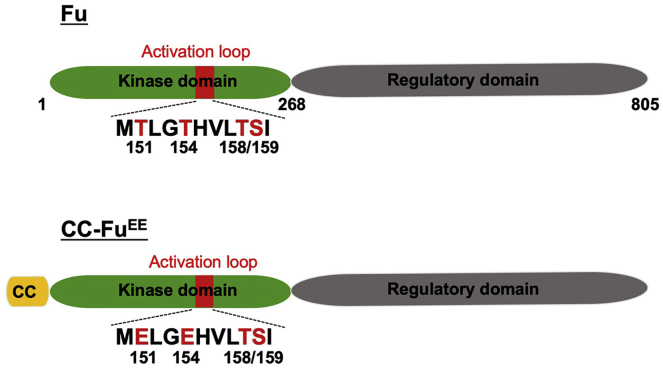
Fu kinase is activated by Hh-induced dimerization and trans-autophosphorylation (Shi et al., 2011; Zhang et al., 2011; Zhou and Kalderon, 2011). To construct a constitutively active full-length Fu kinase (amino acids (aa) 1–805), a coiled-coil (CC) dimerization motif from yeast GCN4 (aa 2–33) was fused to its N terminus and double phospho-mimetic mutations T151E/T154E were introduced to the activation loop in its kinase domain with original codon usage (Figure 1)(O'Shea et al., 1991; Shi et al., 2011).
-
2.
The resulting Fu (CC-FuEE) was subcloned into the vector pFastBac1 for expression in baculovirus using EcoRI and Not I sites.
-
3.
As a negative control, an inactive mutant of Fu (CC-FuG13V) was constructed (Liu et al., 2007).
Figure 1.
Engineering a constitutively active form of Fu
Schematic drawing of full-length wild-type Fu kinase (top) and its constitutively active form, CC-FuEE (bottom). CC, coiled-coil dimerization motif from GCN4 (aa 2–33).
Detection of in vitro phosphorylated proteins by pIMAGO or phospho-specific antibodies
-
4.
To avoid using 32P radioactive isotope in a traditional autoradiograph-based assay, the pIMAGO-biotin phosphoprotein detection kit for substrate phosphorylation recognition was employed (Iliuk et al., 2014).
-
5.
The two Fu phosphorylation sites in Ci/Gli proteins are followed by Casein kinase 1 (CK1) phosphorylation sites (Han et al., 2019). To detect these dual phosphorylation events, two antibodies that recognized these phosphorylation clusters (S218/S220 and S1230/S1233) were generated (Han et al., 2019). An in vitro kinase assay using Fu and CK1 was developed and the phosphorylation was detected by these phospho-specific antibodies.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Ci pS218/S220 | Abmart | N/A |
| Anti-Ci pS1230/S1233 | Abmart | N/A |
| IRDye 680LT donkey anti-rabbit secondary antibody | LI-COR | Cat#926-68023 |
| Bacterial and virus strains | ||
| BL21 chemically competent E. coli | Thermo Fisher Scientific | Cat#EC0114 |
| MAX efficiency DH10Bac chemically competent cells | Thermo Fisher Scientific | Cat#10361012 |
| Chemicals, peptides, and recombinant proteins | ||
| Gentamycin | Millipore Sigma | Cat#G1397 |
| Ampicillin | Millipore Sigma | Cat#A5354 |
| Kanamycin | Millipore Sigma | Cat#60615 |
| Tetracycline | Millipore Sigma | Cat#T7660 |
| IPTG | Millipore Sigma | Cat#I6758 |
| Bluo-Gal | Thermo Fisher Scientific | Cat#15519028 |
| L-Glutathione reduced | Millipore Sigma | Cat#G4251 |
| Sf-900 III SFM | Thermo Fisher Scientific | Cat#12658027 |
| 3X Flag peptide | Millipore Sigma | Cat#F4799 |
| Fetal bovine serum | Millipore Sigma | Cat#F4135 |
| Complete EDTA-free protease inhibitor cocktail | Roche | Cat#04 693 159 001 |
| PhosSTOP phosphatase inhibitor cocktail | Roche | Cat#04 906 837 001 |
| Casein kinase I | New ENGLAND BIOLABS | Cat#P6030 |
| Critical commercial assays | ||
| pIMAGO-biotin Phosphoprotein Detection Kit | Millipore Sigma | Cat#67221 |
| Anti-Flag M2 affinity gel | Millipore Sigma | Cat#A2220 |
| Glutathione Sepharose 4B | GE Healthcare | Cat#17075601 |
| SimplyBlue SafeStain | Thermo Fisher Scientific | Cat#LC6060 |
| Cellfectin II Reagent | Thermo Fisher Scientific | Cat#10362100 |
| Amicon Ultra-0.5 mL centrifugal filters | Millipore Sigma | Cat#UFC501024 |
| Experimental models: cell lines | ||
| Insect: Sf9 | Thermo Fisher Scientific | Cat#11496015 |
| Recombinant DNA | ||
| pGEX 4T-1 | GE Healthcare | Cat#28954549 |
| pFastBac1 | Thermo Fisher Scientific | Cat#10359016 |
Materials and equipment
Baculovirus production and amplification
Bluo-gal solution: prepared at the concentration of 20 mg/mL in dimethylformamide and store at −20°C with light protection.
LB Agar plates (DH10Bac): containing 50 μg/mL kanamycin, 7 μg/mL gentamycin, 10 μg/mL tetracycline and 40 μg/mL IPTG. 50–100 μL Bluo-gal (for 10 cm plate) was spread over the surface of the LB Agar plates and air dried in a ventilated hood for 5–10 min.
DH10Bac cell growth medium: LB medium containing 50 μg/mL kanamycin, 7 μg/mL gentamycin, 10 μg/mL tetracycline.
P1 buffer: 50 mM Tris-HCl pH 8.0, 10mM EDTA, 100 μg/mL RNase A (4°C, 6 months).
P2 buffer: 200 mM NaOH, 1% SDS (25°C, 6 months).
P3 buffer: 3 M potassium acetate pH 5.5 (25°C, 6 months).
TE buffer: 10 mM Tris-HCl pH 8.0, 1 mM EDTA (25°C, 12 months).
Sf9 cell growth medium: Sf-900 III SFM with 10% FBS and 50 μg/mL Gentamycin.
Fu kinase expression and purification
Lysis buffer: 20 mM Tris-HCl pH 8.0, 150 mM NaCl, 5 mM EDTA, 1% IGEPAL CA-630, 10% Glycerol, 1× Complete protease inhibitors cocktail, 1× phosSTOP phosphatase inhibitors cocktail (4°C, 1 month).
Storage buffer: 20 mM Tris-HCl pH 7.0, 100 mM NaCl, 1 mM DTT, 1 mM EDTA, 1 mM EGTA, 0.1% Triton X-100 (−20°C, 12 months).
GST-fusion substrate expression and purification
Extraction/binding buffer: 20 mM Tris-HCl pH 8.0, 100 mM NaCl, 5 mM EDTA (25°C, 6 months).
Elution buffer: 20 mM Tris-HCl pH 8.0, 100 mM NaCl, 5 mM EDTA, 10 mM GSH (4°C, 1 month).
Q700 Sonicator (Qsonica)
In vitro kinase assay
Reaction buffer (1×): 15 mM Tris-HCl pH 7.5 (25°C), 0.2 mM Mg2+/ATP
(−20°C, 12 months)
Detection of protein phosphorylation by pIMAGO-biotin phosphoprotein detection kit or standard immunoblot with phospho-specific antibodies
Phospho-specific polyclonal antibodies were made by Abmart (Shanghai, China) using the following phospho-peptides as antigens: RKRALS(p)SS(p)PYSDS for pS218/220 (CiN), and QIIDSS(p)MTS(p)LPEL for pS1230/1233 (CiC), respectively. The corresponding non-phosphorylated peptides were synthesized to serve as a negative control for affinity purification. The crude serum samples from immunized rabbits were loaded onto columns with immobilized non-phosphorylated antigen peptides to absorb antibodies that recognize non-phosphorylated antigens. The flow-throughs were subsequently loaded onto columns with immobilized phosphorylated antigen peptides to enrich the pool of antibodies recognizing the phosphorylated antigens. The resultant antibody fractions were collected and tested for specificity using substrates with phosphorylated sites mutated as negative controls (Han et al., 2019).
Loading buffer (4×): 0.25 M Tris-HCl (pH 6.8), 8% SDS, 20% 2-mercaptoethanol, 40% glycerol, and 0.008% bromophenol blue (−20°C, 12 months) .
Running buffer: 25 mM Tris base, 190 mM glycine, 0.1% SDS (25°C, 3 months).
Transfer buffer: 25 mM Tris base, 190 mM glycine, 20% methanol (25°C, 3 months).
TBST solution: 20 mM Tris-HCl (pH 7.6), 150 mM NaCl, and 0.1% Tween 20 (25°C, 3 months).
Blocking buffer: 5% nonfat dried milk or bovine serum albumin (BSA) in TBST (4°C, 2 weeks).
Odyssey CLx Imaging System (LI-COR).
Step-by-step method details
DH10Bac cell transformation
Timing: 52–54 h
-
1.
Thaw DH10Bac chemically competent cells on ice.
-
2.
Add 1–10 ng of pFastBac1 vector containing the coding sequence of N-terminally Flag-tagged CC-FuEE and CC-FuGV (G13V mutant, a kinase dead form) into 100 μL DH10Bac chemically competent cells and incubate on ice for 30 min.
-
3.
Heat shock the cells at 42°C for 45 s.
-
4.
Cool down the cells on ice for 2 min.
-
5.
Add 900 μL SOC medium and incubate the cells at 37°C for 4 h with shaking.
-
6.
Dilute the mixture with SOC medium at 1:10 and 1:100 (namely, 1 μL mixture with 99 μL SOC medium and 10 μL mixture with 90 μL SOC medium), for each plasmid, spread 100 μL of undiluted mixture along with the above two dilutions (100 μL each) onto one LB Agar plate (DH10Bac).
CRITICAL: Different amount of mixture were spread on plates to ensure that at least there would be one plate with appropriate number of colonies.
-
7.
Grow the cells at 37°C protected from light for at least 48 h.
Bacmid preparation
Timing: 24 h
-
8.
Pick up a white colony from the above plate and inoculate in 1.5 mL DH10Bac cell growth medium at 37°C for 16 h.
-
9.
The next day, collect the cells by centrifugation and resuspend in 300 μL P1 buffer.
-
10.
Mix with 300 μL P2 buffer gently and incubate at 25°C for 2–3 min.
-
11.
Neutralize pH by adding 300 μL of P3 buffer and gently invert the tubes 4–6 times.
-
12.
Sediment the precipitate by centrifugation at 14,000 × g for 10 min at 4°C.
-
13.
Carefully withdraw around 800 μL of supernatant (avoid disturbing the white pellet) and mix gently with 560 μL (0.7 V) of isopropanol. Let the mixture sit on ice for 10–20 min or at −18°C for 12 h.
-
14.
Collect the Bacmid DNA by centrifugation at 14,000 × g for 10–15 min and wash with 500 μL of 70% ethanol.
-
15.
Desiccate the Bacmid DNA at 25°C in open air for 10 min (can be done alternatively at a 37°C incubator).
-
16.
Dissolve the Bacmid DNA in 40 μL TE buffer.
Pause point: Bacmid DNA could be stored at −20°C for at least 6 months.
Baculovirus production and amplification
Timing: 2–3 weeks
-
17.
Determine the DNA concentration of recombinant Bacmid.
-
18.
Seed 1 million of Sf9 cells into one well of a 6-well plate.
CRITICAL: At least 90% of cells should settle down within 5–10 min.
-
19.
Allow cells grow at 27°C incubator for at least 1–2 h.
-
20.
Replace the growth medium by 2 mL SF900-III medium with 1.5% FBS.
-
21.
For each transfection, dilute 8 μL Cellfectin II reagent (pre-warm to 25°C and gently vortex 1–2 S) in 100 μL SF900-III medium without serum or antibiotics.
-
22.
For each transfection, dilute 1–3 μg Bacmid DNA (usually 2–3 μL/40 μL Bacmid DNA solution) in 100 μL SF900-III medium without serum or antibiotics.
-
23.
Combine diluted DNA and Cellfectin II and incubate at 25°C for 15 min.
-
24.
Add the above mixture to Sf9 cells dropwise.
-
25.
Let the cells sit at 27°C for 3 h.
-
26.
Change medium to regular Sf9 growth medium.
-
27.
Incubate cells at 27°C for 72 h before collecting medium containing P1 virus. Store the P1 virus at 4°C with protection from light.
-
28.
Seed 1–2 million Sf9 cells into one well of 6-well plate and let the cells grow 1–2 h at 27°C.
-
29.
Add 1–2 μL P1 virus into each well and let the cells grow for 2–3 days. Collect the medium containing P2 virus and verify the expression of intended protein in the cells with standard western blot procedure.
-
30.
For large-scale virus production (P3 virus), seed 50–100 mL of Sf9 cells at the density of 1 million cells per mL in 200 mL flask. The next day, add 1/2000 volume of P2 virus and incubate 3 days at 27°C.
-
31.
Collect P3 virus and store at 4°C for future infection.
Pause point: Baculovirus (P1, P2 and P3) could be stored at 4°C with light protection at least 3–6 months.
Fu kinase expression and purification
Timing: 5–7 days
-
32.
Seed 600 mL Sf9 cells into 2L flask at the density of 1 million cells per mL and infect the cells with 6 mL Flag-CC-FuEE or CC-FuGV P3 virus on the next day.
-
33.
48 h after infection, collect the cells by centrifugation at 5,000 × g for 20 min.
-
34.
Resuspend the cell pellet with 40 mL lysis buffer and incubate at 4°C for 10 min with agitation.
-
35.
Remove the cell debris by centrifugation at 12,000 × g for 20 min.
-
36.
Incubate the resultant cell lysate with 100 μL (bed volume) Flag M2 affinity agarose at 4°C for 2 h with shaking.
-
37.
Wash the beads with 3 changes of fresh cold lysis buffer at 4°C for 5 min each.
-
38.
Elute the bound protein by incubation with 2× Kinase storage buffer containing 200 μg/mL 3X Flag peptide at 4°C for 2 h with shaking.
-
39.
Load a small aliquot of eluted protein onto SDS-PAGE gel and stain the gel with Coomassie blue to determine the purity and yield of Fu kinase.
-
40.
Concentrate the kinase to 0.2 μg/μL in 2× kinase storage buffer using Amicon Ultra centrifugal filter with 10K cut-off simultaneously reducing the concentration of Flag peptide in the sample to minimal level.
-
41.
Dilute the kinase with equal volume of glycerol; aliquot and flash-freeze with liquid nitrogen and store at −80°C.
GST-fusion substrate expression and purification
Timing: 2–3 days
-
42.
Transform BL21 chemically competent cells with pGEX 4T-1 and pGEX 4T-1 vectors with expression constructs for Ci/Cos2 polypeptides (CiN: aa 213–234; CiC: aa 1,222–1,240; Cos2: aa 564–585).
-
43.
Inoculate single colony into 3 mL LB medium with ampicillin and incubate at 37°C on a shaker at 250 rpm for 16 h.
-
44.
Dilute with the fresh LB medium at the ratio 1:200 (total 600 mL) and continue the incubation until OD600 reaches around 0.6.
-
45.
After cooling down on ice, induce the GST/GST-fusion protein expression with 0.1 M IPTG and grow the cells at 30°C for 3–4 h.
-
46.
Collect the cells by centrifugation at 5,000 × g for 15 min.
-
47.
Resuspend the bacteria in 40 mL lysis/binding buffer and break the cells with a sonicator on ice in short burst (15 s on, 15 s off, 20 min in total, 30%–40% output).
-
48.
Centrifuge at 12,000 × g for 15 min at 4°C and collect the supernatant.
-
49.
Incubate the supernatant with 0.5 mL glutathione Sepharose at 4°C for 16 h with shaking.
-
50.
Wash the beads thrice with 1 mL of fresh lysis/binding buffer for 5 min at 4°C with agitation.
-
51.
Elute GST/GST-fusion proteins by addition of 1 mL of elution buffer and incubate 1–2 h at 4°C. Collect the eluted proteins by 2,000 × g centrifugation at 4°C for 2 min.
-
52.
Determine the protein yield and purity by on a Coomassie Blue stained SDS-PAGE gel against BSA standard applied in the amount of 0.1 μg, 0.2 μg, 0.4 μg, 0.6 μg, 0.8 μg, and 1 μg per lane. Assess the protein amount by scanning of the dry gel using Odyssey system at 700 nm wavelength.
-
53.
Concentrate the protein by ultracentrifugation to the final concentration to 0.5–1 mg/mL with Amicon Ultra-0.5 mL centrifugal filters with 10K cut-off with simultaneous buffer exchange to 30 mM Tris-HCl pH 7.5 100 mM NaCl. Dilute concentrated proteins with equal volume of glycerol, flash-freeze with liquid nitrogen, and store at −80°C.
In vitro kinase assay with Fu kinase alone
-
54.
For each experimental point, set up a 25 μL reaction mixture with 1× in vitro kinase buffer, using 0.1 ug of catalytically active/inactive protein of Fu kinase purified from Sf9 cells. and 1 μg of GST Ci/Cos2 fragment fusion protein as substrate.
-
55.
Incubate the reaction mixture at 30°C for 15–30 min.
-
56.
Stop the reaction by adding equal volume of 2× SDS loading buffer and boil for 5 min.
In vitro kinase assay with sequential phosphorylation by Fu and CK1
-
57.
After the first step of in vitro kinase assay with Fu, instead of termination of the reaction by addition of 2× SDS loading buffer, add 20 μL of glutathione beads (previously incubated with 2 mg/mL BSA at 4°C for 1 h with agitation to minimize the non-specific binding) and incubate at 4°C with shaking for at least 1 h to pull down the GST-fusion substrates.
-
58.
Wash the beads with thrice 1× kinase buffer.
-
59.
Use washed beads as the substrate in final kinase assay with CK1. All other steps are similar to that described for Fu kinase assay except for using CK1 as a kinase.
-
60.
Stop the reaction by adding equal volume of 2× SDS loading buffer and boil for 5 min.
Detection of protein phosphorylation by pIMAGO-biotin phosphoprotein detection kit
-
61.
Add 5× IAA solution (10 μL) to the reaction mixture (40 μL) and incubated at dark for 15 min.
-
62.
Load a small aliquot of reaction mixture (5–10 μL) onto an SDS-PAGE gel and transfer proteins to a nitrocellulose membrane.
-
63.
Block the membrane with 10 mL 1× blocking buffer at 25°C for 1 h with constant shaking.
-
64.
Incubate the membrane with 1× pIMAGO buffer supplemented with 1:1,000 pIMAGO reagent at 25°C for 1 h.
-
65.
Wash the membrane with 1× wash buffer three times for 5 min each.
-
66.
Wash the membrane with TBST for 5 min.
-
67.
Incubate the membrane with 1× blocking buffer supplemented with 1:1,000 Avidin-Fluor at 25°C for 1 h.
-
68.
Wash the membrane with TBST three times for 5 min each.
-
69.
Scan the membrane with an Odyssey CLx imaging system.
Immunoblot with Ci phospho-specific antibodies
-
70.
Load 1–5 μL of the reaction mixture from an in vitro sequential kinase assay onto an SDS-PAGE and transfer proteins to a nitrocellulose membrane using standard Western blot procedure.
-
71.
Block the membrane with 5% nonfat milk for 1 h at 25°C.
-
72.
Incubate the membrane with Ci phospho-specific antibodies (primary, for CiN phospho antibody pS218/S220, 1:1,000; CiC phospho antibody pS1230/S1233, 1:4,000) at 25°C for 2 h with agitation followed by wash with TBST solution for 3 times with agitation (5 min each time) , and incubation with fluorogenic donkey anti-rabbit antibody (secondary, 1:10,000) for 1 h at 25°C with shaking.
-
73.
Scan the membrane by an Odyssey CLx imaging system at 700 nm wavelength.
Expected outcomes
Sf9/Baculovirus system was applied to expressing recombinant Fu kinase. Significant amount of Fu protein with high purity could be acquired using Flag M2 affinity agarose/3X Flag peptide purification system as demonstrated in Figure 2.
Figure 2.
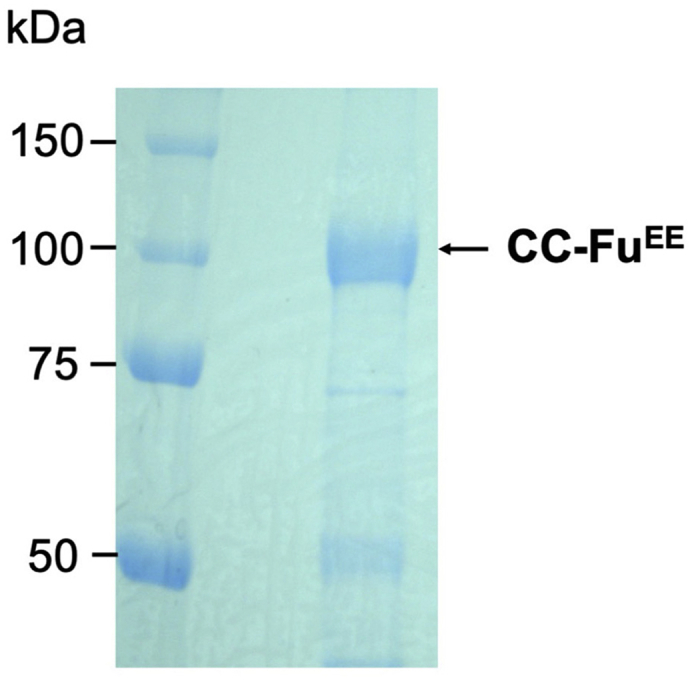
Purification of Fu kinase
Coomassie blue staining of Flag-CC-FuEE purified from Sf9 cells.
The constitutively active form of Fu kinase, CC-FuEE, exhibits robust activity toward the GST-fusion protein substrates in in vitro kinase assay. Utilization of pIMAGO-biotin Kit enables convenient and specific detection of phosphorylated substrates as shown in Figure 3, while prevents usage of traditional 32P radioactive isotope.
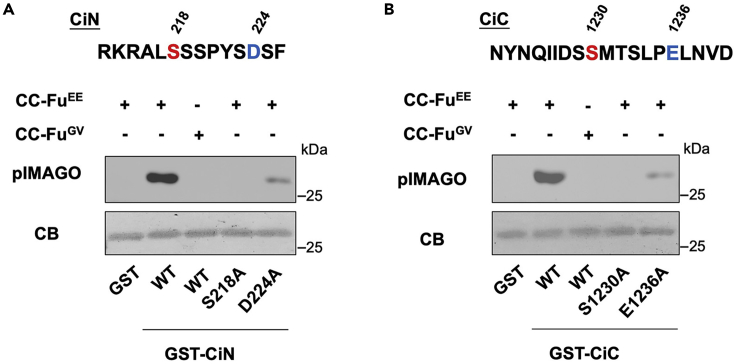
Figure 3.
In vitro kinase assay with purified Fu and detection with pIMAGO
In vitro kinase assay using purified CC-FuEE or CC-FuGV as source of kinase and GST-fusion proteins containing either CiN (A) or CiC (B) fragment with the wild-type sequence or the indicated amino acid substitutions. Phosphorylation was detected by the pIMAGO kit. GST-fusion proteins were visualized by Coomassie blue gel staining (CB). The Fu phosphorylation sites are colored in red and the critical acid residues at +6 position are colored in blue. Figure reprinted with permission from Han et al. (2019).
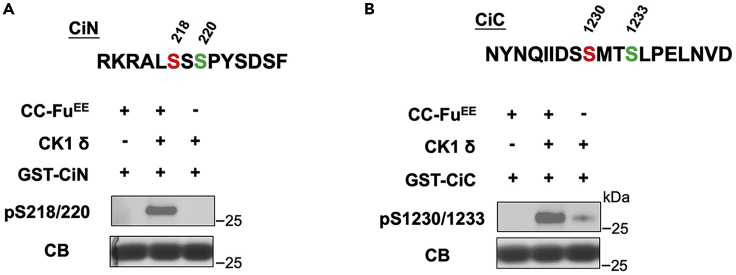
Both Fu phosphorylation sites in Ci protein have a secondary CK1 site in proximity. To verify the involvement of CK1 in phosphorylation of these two clusters after priming phosphorylation by Fu, we developed two phospho-antibodies against them. The immunoblot data shown in Figure 4 provide strong evidence that sequential phosphorylation by Fu and CK1 occurs at these two clusters.
Figure 4.
In vitro kinase assay with Fu and CK1 followed by detection with phospho-specific antibodies
In vitro kinase assay using purified CC-FuEE and/or recombinant CK1 as kinases and GST-fusion substrates containing either CiN (A) or CiC (B) fragment with the indicated amino acid sequence. Phosphorylation was detected by western blot using the indicated phospho-specific antibodies. GST-fusion proteins were visualized by Coomassie blue gel staining (CB). The Fu and CK1 phosphorylation sites are colored in red and green, respectively. Figure reprinted with permission from Han et al. (2019).
Limitations
The in vitro kinase assay carried out using current protocol could only demonstrate that Fu kinase alone or combined with CK1 are able to phosphorylate certain sites within the substrates under optimized in vitro conditions. However, other experiments such as cell-based phosphorylation assay and in vivo functional study are needed to demonstrate the physiological significance of these phosphorylation events by the Fu kinase.
Troubleshooting
Problem 1
It is difficult to discern white colonies of DH10Bac cells after transformation (step 8).
Potential solution
DH10Bac cells grow quite slowly on these LB Agar plates containing multiple antibiotics, it is much easier for pick up white colonies after 48 h incubation at 37°C. Consider picking up big white colonies surrounded by multiple blue colonies.
Problem 2
Bacmid transfection is not successful (step 25).
Potential solution
Firstly, do not over-transfect Sf9 cells. Usually 1–2 μg of Bacmid is enough and too much of that will kill Sf9 cells. Secondly, make sure only healthy Sf9 cells are seeded for transfection. Usually, Sf9 cells in suspension culture are seeded onto 6-well plates before transfection and at least 90% cells should attach to the bottom within 5–10 min if Sf9 are healthy enough for further manipulations.
Problem 3
During the Bacmid transfection, a lot of Sf9 cells died (step 27).
Potential solution
Please be aware that Sf9 is extremely sensitive to desiccation. Without any medium, they will die as short as 40–50 s.
Problem 4
The protein expression level of baculovirus is low (step 29).
Potential solution
Make sure that only healthy Sf9 cells are taken for infection. Keep your baculovirus at 4°C and protect it from light. It is also critical to always amplify baculovirus using P1 or P2 virus, not P3 virus, and keep the MOI as low as 0.01–0.1.
Problem 5
The yield and purity of the GST-fusion protein are poor (step 52).
Potential solution
Try to induce the expression with low concentrations of IPTG (such as 0.1 mM) and decrease the growth temperature of culture to 18°C–20°C with longer incubation (could be extended to 12 h). For GST-fusion proteins with poor yield, use high volume of cell lysate versus low amount of GST beads that could improve both yield and purity.
Problem 6
The sonication of bacterial cells is not successful (step 47).
Potential solution
Increase the volume of lysis buffer used in the sonication.
Problem 7
The phospho-specific antibodies recognize non-phosphorylated substrates (step 73).
Potential solution
The two-step purification scheme described in Materials and equipment section should largely remove antibodies that recognize non-phosphorylated substrates and enrich the content of antibodies that recognize phosphorylated substrates so that the antibodies produce good signal to noise ratio. One approach to further reduce the noise is to dilute the working concentration of the antibodies. If this does not work, try one more round of purification that may reduce non-specific signals.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Jin Jiang, E-mail: jin.jiang@utsouthwestern.edu.
Materials availability
All unique/stable reagents generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
Data and code availability
This study did not generate/analyze datasets and codes.
Acknowledgments
This work was supported by grants from the National Institutes of Health (R35GM118063) and Welch Foundation (I-1603).
Author contributions
Conceptualization, Y.H. and J.J.; Investigation, Y.H.; Writing – Original Draft, Y.H.; Writing – Review & Editing, J.J.; Funding Acquisition, J.J.; Supervision, J.J.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Yuhong Han, Email: yuhong.han@utsouthwestern.edu.
Jin Jiang, Email: jin.jiang@utsouthwestern.edu.
References
- Iliuk A., Li L., Pan L., Tao W.A. Vol. 74. AACR; Cancer Res 2014; 2014. Development of pIMAGO for universal detection of protein phosphorylation. [abstract] (Proceedings of the 105th Annual Meeting of the American Association for Cancer Research Apr 5-9; San Diego, CA. Philadelphia (PA)). [DOI] [Google Scholar]
- Han Y., Wang B., Cho Y.S., Zhu J., Wu J., Chen Y., Jiang J. Phosphorylation of Ci/Gli by fused family kinases promotes Hedgehog signaling. Dev. Cell. 2019;50:610–626.e614. doi: 10.1016/j.devcel.2019.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Cao X., Jiang J., Jia J. Fused-Costal2 protein complex regulates Hedgehog-induced Smo phosphorylation and cell-surface accumulation. Genes Dev. 2007;21:1949–1963. doi: 10.1101/gad.1557407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shea E.K., Klemm J.D., Kim P.S., Alber T. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science. 1991;254:539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- Shi Q., Li S., Jia J., Jiang J. The Hedgehog-induced Smoothened conformational switch assembles a signaling complex that activates Fused by promoting its dimerization and phosphorylation. Development. 2011;138:4219–4231. doi: 10.1242/dev.067959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Mao F., Lu Y., Wu W., Zhang L., Zhao Y. Transduction of the Hedgehog signal through the dimerization of Fused and the nuclear translocation of Cubitus interruptus. Cell Res. 2011;21:1436–1451. doi: 10.1038/cr.2011.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Kalderon D. Hedgehog activates Fused through phosphorylation to elicit a full spectrum of pathway responses. Dev. Cell. 2011;20:802–814. doi: 10.1016/j.devcel.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate/analyze datasets and codes.