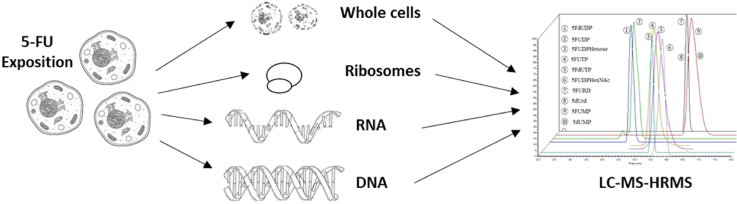
Abstract
5-Fluorouracil (5-FU) is an anticancer drug extensively used for different cancers. Intracellular metabolic activation leads to several nucleoside and nucleotide metabolites essential to exert its cytotoxic activity on multiple cellular targets such as enzymes, DNA and RNA. In this paper, we describe the development of a method based on liquid chromatography coupled with high resolution mass spectrometry suitable for the simultaneous determination of the ten anabolic metabolites (nucleoside, nucleotide and sugar nucleotide) of 5-FU. The chromatographic separation was optimized on a porous graphitic carbon column allowing the analysis of the metabolites of 5-FU as well as endogenous nucleotides. The detection was performed on an Orbitrap® tandem mass spectrometer. Linearity of the method was verified in intracellular content and in RNA extracts. The limit of detection was equal to 12 pg injected on column for nucleoside metabolites of 5-FU and 150 pg injected on column for mono- and tri-phosphate nucleotide metabolites. Matrix effect was evaluated in cellular contents, DNA and RNA extracts for nucleoside and nucleotides metabolites. The method was successfully applied to i) measure the proportion of each anabolic metabolite of 5-FU in cellular contents, ii) follow the consequence of inhibition of enzymes on the endogenous nucleotide pools, iii) study the incorporation of metabolites of 5-FU into RNA and DNA, and iv) to determine the incorporation rate of 5-FUrd into 18 S and 28 S sub-units of rRNA.
Keywords: 5-Fluorouracil, LC-MS-HRMS, Nucleotide, RNA, DNA, Incorporation rate
Graphical abstract
Highlights
-
•
The LC-MS-HRMS method allows the analysis of the ten anabolic metabolites of 5-FU.
-
•
The present method is useful to study the incorporation of 5-FU into RNA and DNA.
-
•
Method to determine the incorporation rate of 5-FU into subunit of rRNA is innovative.
1. Introduction
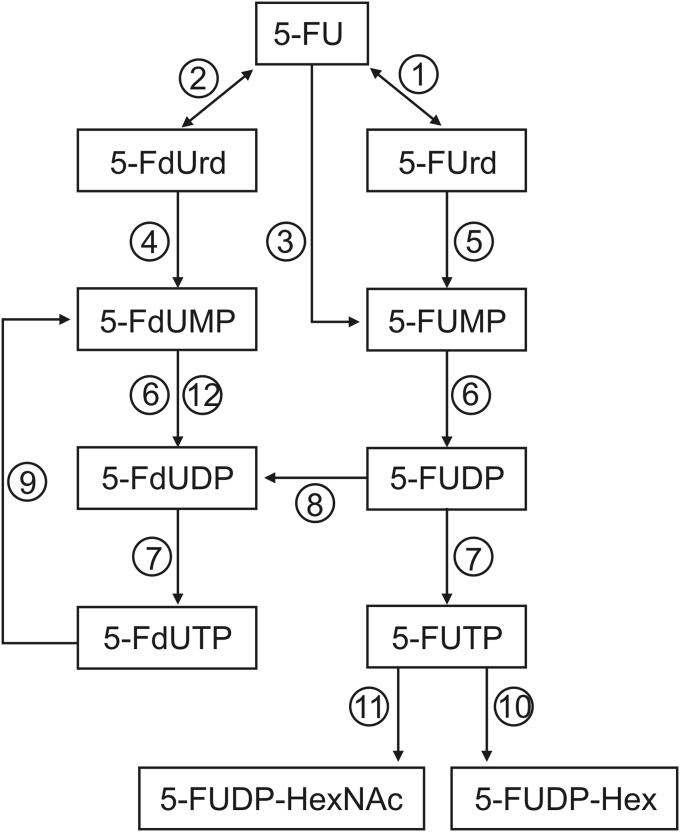
5-Fluorouracil (5-FU) is an anticancer drug which belongs to the group of antimetabolites. Its development began more than 60 years ago and 5-FU still remains extensively used throughout the world in monotherapy as well as in combination chemotherapy. 5-FU is either administered by intravenous infusion or by oral route using the prodrug capecitabine which is metabolized to 5-FU by three enzymatic steps. 5-FU is a treatment for different types of cancer including colorectal and other gastrointestinal cancers, breast cancer, and head and neck cancer. 5-FU exerts its cytotoxic action after enzymatic intracellular conversion to several nucleotide metabolites (Fig. 1). Numerous articles and reviews have been published on antitumor activity of 5-FU and here we only report briefly the main points [1,2]. Mechanism of action involved three active compounds of the drug: 5-fluoro-2′-deoxyuridine-5′-monophosphate (5-FdUMP), 5-fluorouridine-5′-triphosphate (5-FUTP) and 5-fluoro-2′-deoxyuridine-5′-triphosphate (5-FdUTP). 5-FdUMP binds to thymidylate synthase (TS), inhibiting the transformation of deoxyuridine monophosphate (dUMP) to thymidine monophosphate (TMP). The consequence is the imbalance of deoxynucleotide pool leading to DNA damage due to perturbation in synthesis and misreparation. DNA damages are also due to 5-FdUTP incorporation into DNA causing strand breaks. 5-FUTP is incorporated into different RNA species, leading to abnormal RNA function and processing and perturbation of cell growth. Moreover, Samuelsson [3] demonstrated that 5-FU inhibits pseudouridine synthases (PS), enzymes which convert uridine (Urd) to pseudouridine (pseudoUrd) in RNA. Thus, 5-FU exhibits several cellular targets related to the anabolism phase of the drug. In addition to these three active compounds, seven others are included in the anabolism phase of 5-FU: 5-fluorouridine (5-FUrd), 5-fluoro-2′-deoxyuridine (5-FdUrd), 5-fluorouridine-5′-monophosphate (5-FUMP), 5-fluorouridine-5′-diphosphate (5-FUDP), 5-fluoro-2′-deoxyuridine-5′-diphosphate (5-FdUDP), 5-FUDP-hexose (5-FUDP-Hex) and 5-FUDP-N-acetylhexosamine (5-FUDP-HexNAc) (Fig. 1) [4,5].
Fig. 1.
Anabolism phase of 5-fluorouracil (5-FU). Enzymes involved in activation: ① uridine phosphorylase, ② thymidine phosphorylase, ③ orotate phosphoribosyl transferase, ④ thymidine kinase, ⑤ uridine kinase, ⑥ UMP-CMP kinase, ⑦ nucleoside diphosphate kinase, ⑧ ribonucleotide diphosphate reductase, ⑨ dUTP hydrolase, ⑩ UDP-glucose-pyrophosphorylase, ⑪ UDP-N-acetylhexosamine-pyrophosphorylase, and ⑫ thymidylate kinase.
5-FU: 5-fluorouracil; 5-FUrd: 5-fluorouridine; 5-FdUrd: 5-fluoro-2’-deoxyuridine; 5-FUMP: 5-fluorouridine-5’-monophosphate; 5-FdUMP: 5-fluoro-2’-deoxyuridine-5’-monophosphate; 5-FUDP: 5-fluorouridine-5’-diphosphate; 5-FdUDP: 5-fluoro-2’-deoxyuridine-5’-diphosphate; 5-FUTP: 5-fluorouridine-5’-triphosphate; 5-FdUTP: 5-fluoro-2’-deoxyuridine-5’-triphosphate; 5-FUDP-Hex: 5-fluorouridine-5’-diphosphate-hexoses; 5-FUDP-HexNAc: 5-fluorouridine-5’-diphosphate-N-acetylhexosamines.
Several analytical methods reported the analysis of metabolites of 5-FU in biological matrix. While Procházková et al. [6] used capillary electrophoresis to quantify 5-FdUMP, most of other methods consist in liquid chromatography coupled with radioimmunoassay (RIA), diode-array detector (DAD) and more recently with mass spectrometry. Indeed, analysis of 5-FUrd and 5-FdUrd was reported in plasma [[7], [8], [9], [10], [11]] and in cell cultures [[12], [13], [14]]. Analysis of 5-FUMP and 5-FdUMP were described in serum [15], plasma [16], peripheral blood mononuclear cells (PBMC) [17], tissues [15] and cell cultures [12,13]. The diphosphorylated metabolites 5-FUDP and 5-FdUDP were studied in cell cultures [12] and the triphosphorylated forms 5-FUTP and 5-FdUTP were analyzed in plasma [16], PBMC [17] and cell cultures [12]. However, to the best of our knowledge, among all these methods, none allows the simultaneous analysis of the ten metabolites of the anabolism phase of 5-FU.
For the study of incorporation of metabolites of 5-FU into RNA and DNA, Benz et al. [18] and then Peters et al. [19] described a radiolabeled method applied after extraction and digestion of the nucleic acids. Keniry et al. [20] and el-Tahtawy et al. [21] reported the incorporation of metabolites of 5-FU into RNA using nuclear magnetic resonance (NMR) spectroscopy. A gas chromatography-mass spectrometry (GC-MS) method based on enzymatic degradation of 5-FUMP to 5-FU and derivatization with pentafluorobenzylbromide before analysis was also published [[22], [23], [24]]. In a study concerning the mechanisms for cytotoxicity of 5-FU, Pettersen et al. [25] used an LC-MS/MS method for the simultaneous analysis of 5-FUrd and 5-FdUrd in nucleic acids.
The aim of this work was to develop an assay based on LC-MS-HRMS, allowing the simultaneous determination of the ten nucleoside and nucleotide metabolites of the anabolism phase of 5-FU. In addition, we determined the main endogenous nucleosides and nucleotides of Urd to better illustrate the potential of this method to study i) the anabolic metabolites of 5-FU in cells, ii) the incorporation rate of 5-FU into DNA, RNA and subunit of ribosomal RNA, and iii) the consequence of TS and PS inhibition by 5-FU on endogenous nucleotide pools.
2. Material and methods
2.1. Chemicals and reagents
5-FU, 5-FUrd, 5-FdUrd, 5-FdUMP, Urd, 13C5-uridine (Urd13C), uridine-15N2 5′-monophosphate (UMP15N), uridine-13C5,15N2 5′-monophosphate (UMP13C,15N), uridine-13C9,15N2 5′-triphosphate (UTP13C,15N), adenosine-5′-triphosphate (ATP), phosphodiesterase I from Crotalus adamanteus venom, alkaline phosphatase and benzonase nuclease were purchased from Sigma-Aldrich™ (St-Quentin-Fallavier, France). Deoxy-Nucleotide Monophosphate kinase (dNMP kinase) from bacteriophageT4 recombinant (E. Coli), 5-FdUTP and 5-FUTP were purchased at Jena Biosciences™ (Jena, Germany). 13C5-adenosine (Ad13C) came from Eurisotop™ (Saint-Aubin, France). Nuclease S1 was from Promega™ (Lyon, France) and calf 12 intestine phosphatase from New England Biolabs™ (Evry, France). PseudoUrd was from Santa Cruz Biotechnology (Heidelberg, Germany). PBS and trypsin-EDTA (0.05%) were purchased at Life technologies™ (ThermoFisher Scientific™, Les Ulis, France). Acetonitrile and methanol both of HPLC-grade, ammonia hydroxide aqueous solution (20%), acetic acid, TRIS hydrochloride, sodium chloride and magnesium chloride were obtained from Sigma-Aldrich™. Water filtered with an Elga Purelab (Flex system™, High Wycombe, United Kingdom) was used in all experiments.
2.2. Synthesis of 5-FUMP and pseudoUMP
5-FUMP was prepared by selective 5′-phosphorylation of 5-fluorouridine with POCl3 in triethylphosphate, followed by hydrolysis with triethylammonium bicarbonate (TEAB, 1 M, pH 7.5) [[26], [27], [28]]. Pseudouridine 5′-monophosphate was synthesized starting from PseudoUrd, via a 5′-H-phosphonate approach [29]. The nucleotides were purified on DEAE-Sephadex A-25 (elution: gradient of TEAB pH 7.5 from 10 mM to 0.5 M), followed by chromatography on RP18 (elution: water to 50% methanol). The triethylammonium counter ions were exchanged to sodium by passing the 5-FdUDP and 5-FUDP nucleotide solution through a DOWEX-AG 50WX2-400 column (Fluka). Yields were 10%–19%.
5-FUMP, sodium salt. 1H NMR (400 MHz, D2O): δ 8.23 (d, 3JHF = 6.3 Hz, 1H, H6), 6.01 (dd, 3JHH = 5.2 Hz and 5JHF = 1.5 Hz, 1H, H1′), 4.41 (t, 3JHH = 5.2 Hz, 1H, H2′), 4.37 (t, 3JHH = 4.1 Hz, 1H, H3′), 4.25–4.30 (m, 1H, H4′), 3.91–4.10 (m, 2H, H5′, H5′′); 31P NMR (162 MHz, D2O) δ 3.38 (s); HRMS (ESI) m/z: [M−Na]− Calcd for C9H11FN2O9P 341.0186, found: 341.0190.
Pseudouridine 5′-monophosphate, sodium salt. 1H NMR (400 MHz, D2O): δ 7.86 (s, 1H, H6), 4.76 (d, 3JHH = 4.0 Hz, 1H, H1′), 4.20–4.25 (m, 1H, H3′), 4.12–4.16 (m, 1H, H2′), 3.80–4.03 (m, 3H, H4′, H5′, H5″); 31P NMR (162 MHz, D2O) δ 3.90 (s); HRMS (ESI) m/z: [M−Na]− Calcd for C9H12N2O9P 323.0280, found: 323.0275.
2.3. Production of 5-FdUDP and 5-FUDP
The two nucleotide metabolites 5-FdUDP and 5-FUDP were produced by incubation of 50 μL of 5-FdUMP or 5-FUMP at 0.2 mM with dNMP kinase (5 μL containing 1000 units) for 30 min in a Tris-HCl buffer (pH 7.4; 100 mM) containing MgCl2 (10 mM) and ATP (2 mM). The final volume of the system was 100 μL. Then 10 μL of incubation medium were added with 100 μL of a mixture of cold methanol/water (70:30, V/V) to stop the reaction. Samples were analyzed before the reaction and at 30 min.
2.4. LC-MS-HRMS instrumentation
Liquid chromatography analysis was performed on an Ultimate 3000 system (ThermoFisher Scientific™, Bremen, Germany) equipped with two ternary pumps. The LC system was coupled with a Q-Exactive Plus Orbitrap mass spectrometer (ThermoFisher Scientific™, Bremen, Germany). The separation of the compounds was carried out with a Hypercarb® column (2.1 mm×100 mm, 5 μm; ThermoFisher Scientific™, Les Ulis, France) and was thermostated at 30 °C. The autosampler tray was maintained at 5 °C. A volume of 10 μL was injected. A stepwise gradient program with (A) NH4OH 0.25% adjusted to pH 10 with acetic acid, (B) water and (C) acetonitrile was performed. The electrospray source (ESI) operated alternatively in negative and positive modes. Spray voltage was set at 3 kV and 3.2 kV in negative and positive modes, respectively. The pressure of nitrogen sheath gas and auxiliary gas were maintained at 30 and 20 units (arbitrary units), respectively. The capillary temperature was 320 °C. A switching valve directed the eluate to waste during the first minute of the run and during reequilibration step. Mass spectrometer could operate in full scan mode (FS) and parallel reaction monitoring mode (PRM).
2.5. Samples preparation
2.5.1. Cell culture, exposure to 5-FU and isolation
HCT116 cells were grown in Dulbecco Minimum Essential Medium – GlutaMax (Invitrogen™, Villebon sur Yvette, France) supplemented with 10% fetal bovine serum (FBS). Cells were plated 48 h before 5-FU or 5-FdUrd exposure. HepaRG cells, purchased from Biopredic International™ (Rennes, France), were grown at a low density in Williams’ E medium supplemented with 10% FCS, 100 units/mL penicillin, 100 μg/mL streptomycin, 5 μg/mL insulin, 2 mM glutamine and 50 μM hydrocortisone hemisuccinate. After 2 weeks, the culture medium was supplemented with 1% DMSO and the cells were left to differentiate for 1 week (confluent DMSO-treated cells) [30].
Cells were exposed to 5-FU or 5-FdUrd for 24 h at 10 μM or 50 μM before cell isolation. For each experiment control samples consisting in non-treated cells were prepared. After exposure, cells were trypsinized after 2 washes with cold PBS. The cells were counted by adding 10 μL of trypan blue with 10 μL of cells, and then the mixture was introduced in automated cell counter (Countess™, Invitrogen, ThermoFisher, France). Cell pellet was washed once with cold PBS and then stored at −80 °C after removing the supernatant.
2.5.2. Extraction of intracellular metabolites
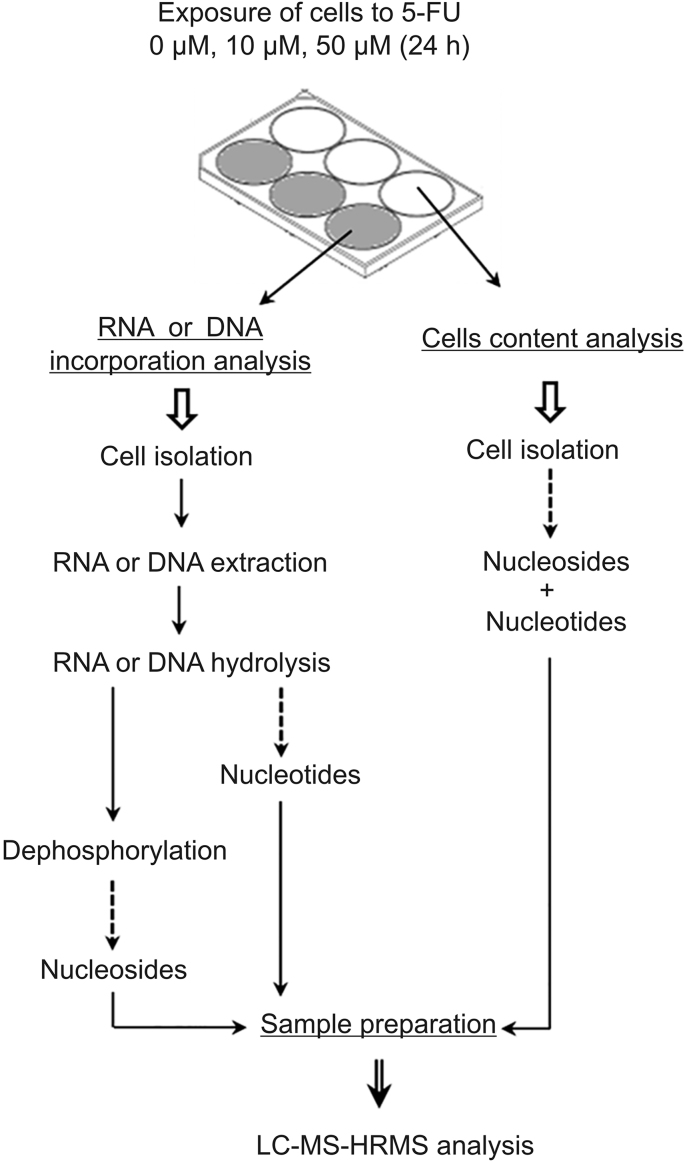
Intracellular content was extracted from cell pellet by lysis with a mixture of cold methanol/water (70:30, V/V) (Fig. 2). The following internal standards were added at 0.02 mM: Urd13C, UMP13C,15N and UTP13C,15N. Then samples were vigorously vortexed and centrifuged for 10 min at 13,000 g. Supernatant was evaporated to dryness under nitrogen at 37 °C. The residue was resuspended with 100 μL of water and then transferred to a vial for injection.
Fig. 2.
Sample workflow for the study of intracellular anabolism of 5-FU and the incorporation into RNA and DNA.
2.5.3. Extraction, hydrolysis and dephosphorylation of RNA
RNA extraction was performed using RNeasy Mini Kit (Qiagen™, Les Ulis, France) after lysis of cell pellet with the lysis buffer provided by the kit. Quantity of RNA extracted was measured using a spectrophotometer NanoDrop 2000® (Ozyme™, Saint-Cyr-L’école, France). Samples were then divided in aliquots containing a maximum of 3 μg of RNA before hydrolysis. Hydrolysis of RNA extracts was performed overnight at 37 °C by incubation with 270 units of Nuclease S1 using the supplied buffer. For analysis of RNA extracts under dephosphophyraleted form a dephosphorylation step was achieved by adding 5 U of calf intestine phosphatase in 100 mM Tris-HCl, 50 mM NaCl, 10 mM MgCl2, and 0.025% Triton® X-100. Digested extracts were stored at −80 °C before analysis.
Before dephosphorylation, samples were spiked with 20 μL of UMP15N to be used as a standard for this step. Thus, UMP15N was dephosphorylated in Urd15N and Urd15N was followed during analysis in samples which underwent dephosphorylation.
A volume of 300 μL of pure methanol was added to RNA extracts. Then, 10 μL of the following internal standards at 0.02 mM were added: Urd13C, Ad13C and UMP13C,15N. Then samples were vigorously vortexed and centrifuged for 10 min at 13,000 g. Supernatant was evaporated to dryness under nitrogen at 37 °C. The residue was resuspended with 100 μL of water and then transferred to a vial for injection.
2.5.4. Extraction, hydrolysis and dephosphorylation of DNA
DNA extraction was performed using QIAamp DNA Mini Kit® (Qiagen™, Les Ulis, France) after lysis of cell pellet with the lysis buffer provided by the kit. Quantity of DNA extracted was measured using a spectrophotometer NanoDrop 2000® (Ozyme™, Saint-Cyr-L’école, France). The procedure of digestion of DNA to deoxyribonucleoside was derived from a previous work published by Quinlivan and Gregory [31]. Briefly, DNA extracts were divided in aliquots containing 3 μg of DNA. Then 150 μL of digest mix was added. The composition of the digest mix was 250 U Benzonase® nuclease, 300 mU phosphodiesterase I (Sigma-Aldrich, Lyon, France) and 200 U alkaline phosphatase (New England Biolabs™, Evry, France) to 5 mL Tris-HCl buffer (20 mM, pH 7.9) containing 100 mM NaCl and 20 mM MgCl2. DNA extracts were then incubated overnight at 37 °C. In the end, samples were stored at −80 °C until analysis.
To analyze samples representing a higher quantity of DNA, a supplementary step of concentration was performed. Briefly, five DNA digested extracts were pooled after resuspension with 300 μL of pure methanol. Then, 10 μL of Urd13C and Ad13C at 0.02 mM were added as internal standard. The total volume was evaporated under nitrogen at 37 °C and the residue was resuspended with 100 μL of water and transferred to a vial for injection. Finally, 20 μL was injected. Thus, this concentration step allowed the analysis of samples containing 10-fold higher DNA than the procedure described for RNA extracts.
2.5.5. 18 S and 28 S ribosomal RNA purification
Ribosomes were purified from HCT116 cells as previously described by Belin et al. [32] by sedimentation through sucrose cushion. Ribosomal RNAs (rRNAs) were purified using RNeasy Mini Kit as described above (Section 2.5.3). For 18 S and 28 S rRNAs purification, total rRNAs were separated by electrophoresis in a 1% agarose gel and the bands corresponding to 18 S and 28 S rRNA were visualized under UV fluorescence and cut out of the agarose gel. Each rRNA was extracted from the agarose gel piece using the NucleoSpin RNA kit and NTC buffer (Macherey-Nagel, Hoerdt, France). Then, 18 S and 28 S rRNAs were quantified and digested and stored at −80 °C as described in Section 2.5.3. The rest of preparation was identical to the one described above for DNA and RNA with the addition of 300 μL of pure methanol and 10 μL of Urd13C and Ad13C at 0.02 mM.
2.6. Method evaluation
2.6.1. Linearity of the method
To ensure the linearity of the analytical method for cellular content, samples containing increased intracellular contents of cells exposed to 50 μM 5-FU during 24 h were analyzed. Thus, lysates of HCT116 cells representing 0.5×106, 1×106, 2×106 and 3×106 cells were prepared. Linearity was verified by plotting the peak area ratio (metabolites of 5-FU/internal standard) as function of the cell number. Linearity was also tested by spiking intracellular extracts of 1×106 cells with increasing concentrations of 6 metabolites available in pure form. The range of the quantities injected was from 0.1 to 3.75 ng for 5-FUrd and 5-FdUrd, from 0.5 to 10 ng for 5-FUMP and 5-FdUMP and from 1 to 10 ng for 5-FUTP and 5-FdUTP.
To ensure the linearity of the analytical method for RNA extracts, samples containing increasing quantity of RNA extracted from cells exposed to 50 μM 5-FU during 24 h were analyzed. Thus, quantities of extracted RNA of 0.2, 0.5, 1 and 3 μg were hydrolyzed and dephosphorylated. Linearity was verified by plotting the peak area ratio (5-FUrd/internal standard) as function of the quantity of RNA.
2.6.2. Matrix effect
For cellular content, matrix effect of 5-FUrd, 5-FdUrd, 5-FUMP, 5-FdUMP, 5-FUTP and 5-FdUTP was studied by spiking non-treated cells extracts with 30 ng of each metabolite. Results were compared with the signal obtained by injecting on the chromatographic column the same quantity of the metabolites from a pure solution.
For nucleic acids, matrix effect was studied by spiking non-treated phosphorylated and dephosphorylated RNA with 5-FUMP and 5-FUrd, respectively and non-treated phosphorylated and dephosphorylated DNA with 5-FdUMP and 5-FdUrd, respectively. For the four metabolites, a quantity of 3 ng was added to DNA or RNA extracts of HCT116 cells. Samples were then prepared as explained previously. Results were compared with the signal obtained by injecting on the chromatographic column the same quantity of the metabolites from a pure solution. Experiments were performed three times. The ion suppression was also investigated by a post-column infusion system. Extract samples from non-treated DNA or RNA were injected with LC device, and post-column infusion of 5-FUrd, 5-FdUrd, 5-FUMP or 5-FdUMP was performed during the chromatographic run.
2.6.3. Determination of the limit of detection
Limits of detection (LOD) were determined for 5-FUrd, 5-FdUrd, 5-FUMP, 5-FdUMP, 5-FUTP and 5-FdUTP. For this work, we defined the LOD as the lowest injected quantity exhibited a coefficient of variation lower than 20% after analysis of three samples.
2.7. Applications
The study of the intracellular metabolites of the anabolism phase of 5-FU was performed on cell extracts after exposure to 5-FU or 5-FdUrd at 50 μM during 24 h. To observe the effect of exposure to 5-FU or 5-FdUrd on TS dUMP, dUDP, dUTP, TMP, TDP and TTP were quantified in these cell extracts. Incorporation of metabolites of 5-FU into DNA and RNA and determination of the incorporation rate into RNA were studied on RNA, DNA and ribosome extracts of HCT116 cells exposed to 5-FU or 5-FdUrd at 10 or 50 μM during 24 h.
The quantification of 5-FUrd, adenosine, guanosine, cytidine, Urd and pseudoUrd in RNA extracts was performed using calibration curves. Concentrations of the lowest standard and the highest standard were equal to 0.03 and 5.00 μg/mL for Urd, adenosine, guanosine and cytidine, 1.67 and 250 ng/mL for pseudoUrd, and 0.21 and 31.25 ng/mL for 5-FUrd. Urd13C was used as internal standard for 5-FUrd, Urd, pseudoUrd and cytidine, and Ad13C was the internal standard of adenosine and guanosine. Calibration curves were constructed by plotting the ion abundance peak area ratio (5-FUrd or endogenous nucleosides/internal standard) as function of metabolite concentration. Data were fitted by weighted (1/concentration) for least-squares regression, and standard curves were determined using linear regression analysis.
3. Results
3.1. Analytical method for the identification of metabolites of 5-FU
3.1.1. LC-MS-HRMS method
A liquid chromatography method was developed for the simultaneous determination of the ten nucleoside and nucleotide metabolites of the anabolism phase of 5-FU. Six metabolites (5-FUrd, 5-FdUrd, 5-FUMP, 5-FdUMP, 5-FUTP and 5-FdUTP) available as pure compounds were used for the development of the method. 5-FUMP was synthetized since this compound was not commercially available. The study of the chromatographic behavior and mass spectrometry characteristics of nucleoside and nucleotide metabolites was performed. The method was adapted from a previously validated assay for the quantification of endogenous nucleotides [33]. A stepwise gradient program with (A) 0.25% NH4OH adjusted to pH 10 with acetic acid, (B) water and (C) acetonitrile was performed (Table 1). The percentage of (A) was maintained to 10%, while the percentage of (C) was increased from 12% to 53%. The column was equilibrated with 10% of (A) and 90% of (B). Equilibration of the column was performed without acetonitrile to allow the retention of nucleosides on the Hypercarb® column. Metabolites of 5-FU were eluted between 4.5 and 6.1 min (Table 1 and Fig. 3).
Table 1.
Stepwise gradient program for the analysis of metabolites of 5-FU. Mobile phase A: 0.25% NH4OH adjusted to pH 10 with acetic acid, mobile phase B: water, and mobile phase C: acetonitrile.
| Time (min) | Mobile phase A (%) | Mobile phase B (%) | Mobile phase C (%) | Flow rate (μL/min) |
|---|---|---|---|---|
| 0.0 | 10 | 78 | 12 | 200 |
| 15.0 | 10 | 37 | 53 | 200 |
| 15.1 | 10 | 90 | 0 | 400 |
| 20.0 | 10 | 90 | 0 | 400 |
| 20.1 | 10 | 90 | 0 | 200 |
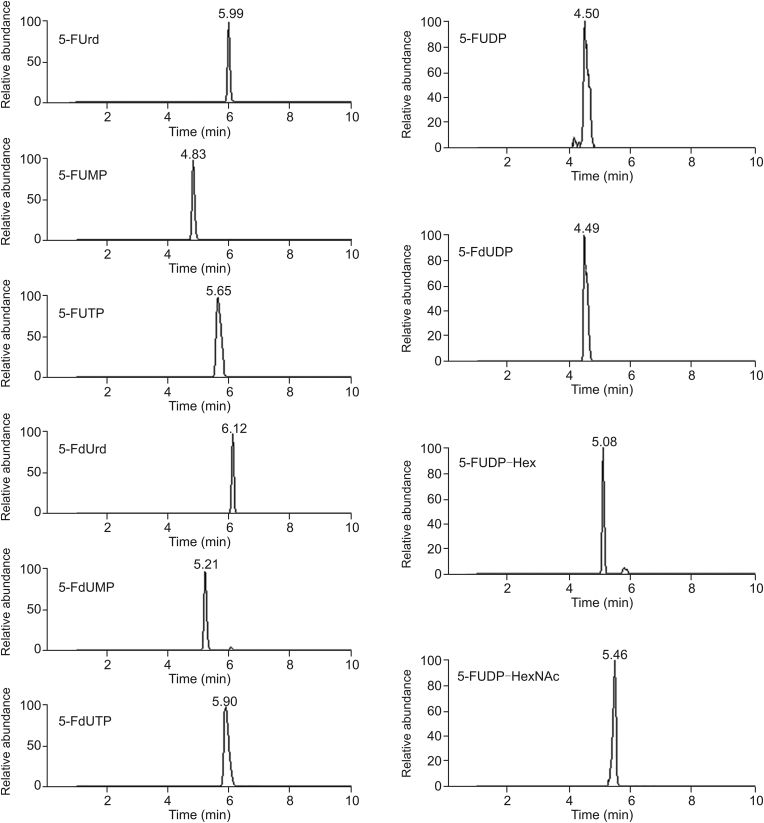
Fig. 3.
Extracted chromatograms.
5-fluorouridine (5-FUrd), 5-fluoro-2′-deoxyuridine (5-FdUrd), 5-fluorouridine-5′-monophosphate (5-FUMP), 5-fluoro-2′-deoxyuridine-5′-monophosphate (5-FdUMP), 5 fluorouridine-5′-triphosphate (5-FUTP) and 5-fluoro-2′-deoxyuridine-5′-truphosphate (5-FdUTP) in pure solutions, 5-fluorouridine-5′-diphosphate (5-FUDP) and 5-fluoro-2′-deoxyuridine-5′-diphosphate (5-FdUDP) after production by enzymatic reaction, 5-FUDP-hexose (5-FUDP-Hex) and 5-FUDP-N-acetylhexosamine (5-FUDP-HexNAc) in a cellular extract after negative ionization.
On the Q-Exactive Plus mass spectrometer, full scan mode from m/z 100 to m/z 750 with a resolution of 70,000 was applied after positive and negative ionization. AGC target was programmed to 1×106 and the max IT was 100 ms. For all analytes, response after positive and negative ionization were compared. The ionization mode inducing the better signal to noise ratio was selected. Thus, negative ionization mode were selected for 5-FUrd, 5-FdUrd, 5-FUMP, 5-FdUMP, 5-FUTP and 5-FdUTP (Table 2). From pure solutions diluted in water at the same concentration, the signal (area of chromatographic peak) was identical for 5-FdUrd and 5-FUrd in one hand and for 5-FdUMP, 5-FUMP, 5-FdUTP and 5-FUTP in another hand. However, the signal was 3 times higher for metabolites as nucleoside form than as nucleotide form.
Table 2.
Retention times and mass spectrometry parameters of anabolic metabolites of 5-FU.
| Compounds | Chemical formula | Retention time (min) | HRMS |
MS-HRMS |
||
|---|---|---|---|---|---|---|
| m/z negative mode | m/z positive mode | Ionization mode: ratio (-)/(+) | Main fragments after PRM (NCE = 35)∗ | |||
| 5-FUrd | C9H11FN2O6 | 6.0 | 261.05284 | 263.06739 | 460 | (-) 171.02112 (100%), 129.01054 (90%), 108.00913 (80%), 128.01536 (50%) |
| 5-FUMP | C9H12FN2O9P | 4.8 | 341.01917 | 343.03372 | 2 | (-) 96.96958 (100%), 78.95902 (85%), 129.01053 (55%), 211.00147 (30%) |
| 5-FUDP | C9H13FN2O12P2 | 4.5 | 420.98550 | 423.00005 | No signal in (+) | (-) 78.95906 (100%), 129.01068 (90%), 159.92543 (85%), 96.96955 (35%) |
| 5-FUTP | C9H14FN2O15P3 | 5.7 | 500.95183 | 502.96638 | 35 | (-) 158.92543 (100%), 272.95746 (20%), 78.95907 (15%), 129.01058 (12%) |
| 5-FdUrd | C9H11FN2O5 | 6.1 | 245.05792 | 247.07248 | 570 | (-) 155.02619 (100%), 129.01057 (15%) |
| 5-FdUMP | C9H12FN2O8P | 5.2 | 325.02425 | 327.03881 | 3 | (-) 129.01057 (100%), 78.95902 (45%), 195.00653 (45%), 96.96957 (15%) |
| 5-FdUDP | C9H13FN2O11P2 | 4.5 | 404.99058 | 407.00514 | 47 | (-) 78.95904 (100%), 158.92541 (45%), 96.96953 (40%), 129.01057 (35%) |
| 5-FdUTP | C9H14FN2O14P3 | 5.9 | 484.95691 | 486.97147 | 48 | (-) 158.92532 (100%), 256.96249 (30%), 78.95904 (20%), 176.93602 (15%) |
| 5-FUDP-Hex | C15H23FN2O17P2 | 5.1 | 583.03832 | 585.05287 | No signal in (+) | (-) 78.95909 (100%), 96.96962 (95%), 211.00153 (55%), 129.01060 (35%), 341.01965 (35%) |
| 5-FUDP-HexNAc | C17H26FN3O17P2 | 5.5 | 624.06487 | 626.07942 | 3.5 | (-) 78.95911 (100%), 158.92535 (85%), 272.95718 (60%), 96.96967 (45%), 129.01064 (40%) |
Ratio (-)/(+): response in negative mode/response in positive mode; NCE: normalized collision energy; ∗: fragments containing fluorine are in bold. In brackets: relative abundance of each fragment.
3.1.2. Identification of 5-FUDP, 5-FdUDP, 5-FUDP-Hex and 5-FUDP-HexNAc
Ten metabolites of 5-FU have been previously described in the anabolism phase (Fig. 1). However, only six of them were available as pure compounds. Thus, to test the ability of our method to detect all the metabolites, 5-FUDP-Hex and 5-FUDP-HexNAc were produced from cell lines, and 5-FUDP and 5-FdUDP came from an enzymatic reaction.
In order to produce diphosphorylated metabolites of 5-FU, 5-FdUMP and 5-FUMP were incubated separately with dNMP kinase. Although the dNMP kinase exhibits a high selectivity for dNMP substrate, NMP substrate can also be metabolized in a limited extend [34]. Incubation of 5-FdUMP or 5-FUMP with dNMP kinase showed a decrease of both substrates over time. With 5-FdUMP, signal intensity increased at 4.5 min with m/z at 407.00514 in positive mode and 404.99058 in negative mode and corresponding to the chemical formula C9H13FN2O11P2. The isotopic abundance was also in agreement (5 ppm) with this chemical formula. The signal intensity was about 50 times higher in negative mode than in positive mode. The fragmentation using PRM mode (Rs = 17,500) exhibited main fragments at m/z 78.95904, 158.92541, 96.96953 and 129.01057 (Table 2). The fragment at m/z 129.01057 corresponded to the nucleobase 5-FU. Other main fragments corresponded to phosphorylated moiety. All these data strongly suggested that 5-FdUDP was produced by the enzymatic reaction. With 5-FUMP, signal intensity increased at 4.5 min only in negative mode with m/z at 420.98550 and corresponding to the chemical formula C9H13FN2O12P2. The isotopic abundance was also in agreement (5 ppm) with this chemical formula. The fragmentation using PRM mode (Rs = 17,500) exhibited main fragments at m/z 78.95906, 129.01068, 159.92543 and 96.96955 (Table 2). The fragment at m/z 129.01068 corresponds to the nucleobase 5-FU. All these data strongly suggested that 5-FUDP was produced by the enzymatic reaction. Thus, our method allowed the analysis of the diphosphorylated metabolites of 5-FU.
After cell exposure to 5-FU, sugar nucleotide metabolites were also identified. The metabolite 5-FUDP-Hex was detected with a retention time at 5.1 min. This compound was only detected in negative mode at m/z 583.03832 with an isotopic abundance in agreement (5 ppm) with the chemical formula C15H23FN2O17P2. The fragmentation using PRM mode (Rs = 17,500) exhibited main fragments at m/z 78.95909, 96.96962, 211.00153, 129.01060 and 341.01965 (Table 2). The fragments at m/z 129.01060 and 341.01965 corresponded to the nucleobase 5-FU and the nucleoside 5-FUMP, respectively. The metabolite 5-FUDP-HexNAc was identified with a retention time at 5.5 min. This compound was detected in negative and positive modes at m/z 624.06487 and 626.07942, respectively. The signal intensity was about 3.5 times higher in negative mode. The exact mass and the isotopic abundance were in agreement (5 ppm) with the chemical formula C17H26FN3O17P2. In negative ionization mode, the fragmentation using PRM mode (Rs = 17,500) exhibited main fragments at m/z 78.95911, 158.92535, 272.95718, 96.96967 and 129.01064 (Table 2). The fragment at m/z 129.01064 corresponds to the nucleobase 5-FU. These two metabolites are assigned as hexose (or hexosamine) because the mass spectrometer could not decipher if the sugar is glucose, mannose or galactose (or glucosamine, mannosamine or galactosamine) [35]. To conclude, the present LC-MS-HRMS method allows the analysis in a single run of all metabolites of 5-FU described in the anabolism phase of this drug. An extracted chromatogram of each analyte is presented in Fig. 3.
3.2. Evaluation of the method
3.2.1. Linearity
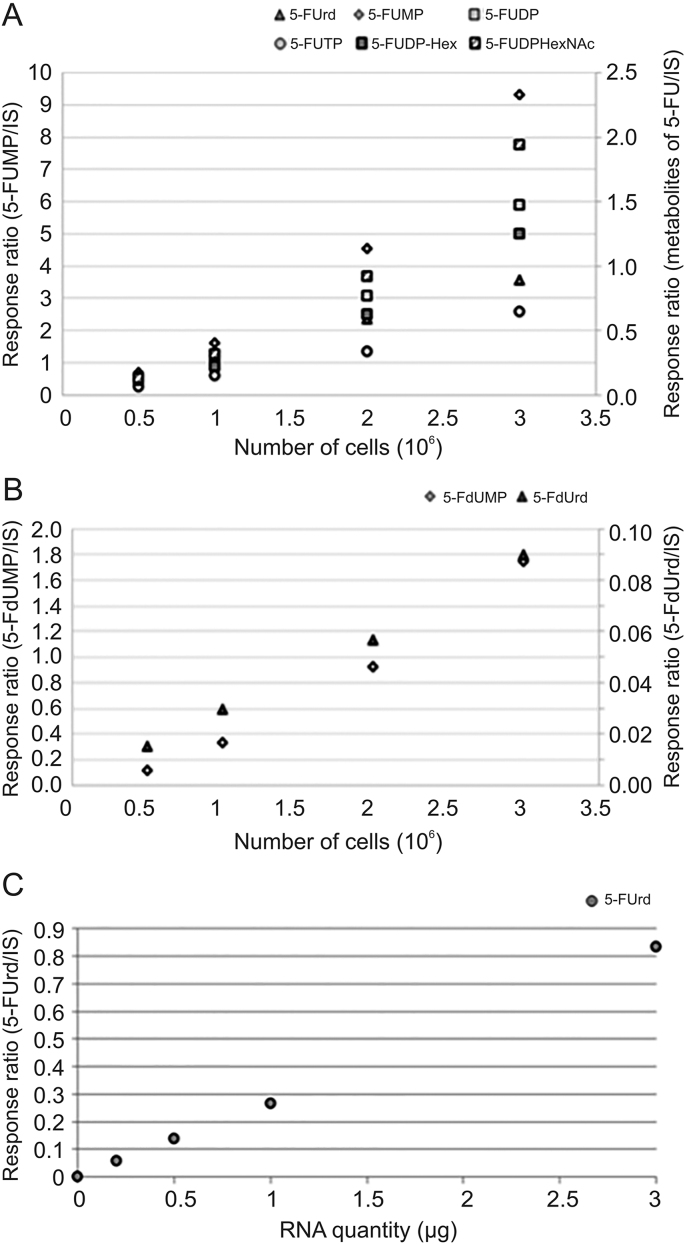
Analysis of intracellular content of HCT116 cells exposed to 5-FU revealed the presence of 5-FUrd, 5-FUMP, 5-FUDP, 5-FUTP, 5-FUDP-Hex, 5-FUDP-HexNAc, 5-FdUrd and 5-FdUMP. Cellular contents corresponding to 0.5 × 106, 1 × 106, 2 × 106 and 3 × 106 cells previously exposed to 50 μM 5-FU were prepared. As shown in Figs. 4A and B, the present method is linear since responses of each metabolite of 5-FU increased proportionally with the number of cells. The linearity of the method was also observed with the proportionality of the response as a function of the increasing quantities injected after having spiked a fixed number of cells with increasing concentrations of metabolites (Fig. S1). Moreover, RNA extracts of 0.2, 0.5, 1 and 3 μg were prepared and response of 5-FUrd was linear since it increased proportionally with the quantity of RNA (Fig. 4C).
Fig. 4.
Linearity of the method according to the number of cells and the quantity of RNA extract. (A) Response ratio of 5-FUrd, 5-FUMP, 5-FUDP, 5-FUDP-Hex and 5-FUDP-HexNAc as function of the number of cells, (B) response ratio of 5-FdUrd and 5-FdUMP as function of the number of cells, and (C) response ratio of 5-FUrd as function of the quantity of RNA extract. IS: internal standard.
3.2.2. LOD and matrix effect
To determine the analytical performances of our method and its interest for various applications around 5-FU, we determined the LOD of metabolites available as pure compounds. The LOD was equal to 12 pg injected on column for 5-FdUrd and 5-FUrd, 150 pg injected on column for 5-FUMP, 5-FdUMP, 5-FUTP and 5-FdUTP.
The matrix effect was evaluated for cellular contents, DNA and RNA extracts (Table 3). Depending on the compounds and biological matrices, the matrix effect is variable but is overall more important for nucleotides. No ion suppression due to RNA or DNA extracts constituents was observed by post-column infusion for 5-FUrd and 5-FdUrd. An ion suppression equal to 10% was observed for 5-FUMP and 5-FdUMP (Fig. S2).
Table 3.
Evaluation of the matrix effect of metabolites of 5-FU from cellular content, DNA and RNA extracts.
| Matrix | Metabolites | Matrix effect (%) |
|---|---|---|
| Cellular content | 5-FUrd | +20 |
| 5-FUMP | −24 | |
| 5-FUTP | +20 | |
| 5-FdUrd | +20 | |
| 5-FdUMP | +50 | |
| 5-FdUTP | −39 | |
| RNA extract | 5-FUrd | <10 |
| 5-FUMP | −56 | |
| DNA extract | 5-FdUrd | <10 |
| 5-FdUMP | −35 |
3.3. Applications
3.3.1. Intracellular anabolism of 5-FU
In cellular content of HCT116 cells exposed to 10 or 50 μM 5-FU or 5-FdUrd, the 8 following metabolites of the anabolism phase were observed: 5-FUrd, 5-FUMP, 5-FUDP, 5-FUTP, 5-FdUrd, 5-FdUMP, 5-FUDP-Hex and 5-FUDP-HexNAc. The two metabolites 5-FdUDP and 5-FdUTP were not detected in cellular contents, neither after 5-FU nor 5-FdUrd exposure. This absence of response was confirmed by using another cell line, HepaRG cells. In HCT116 and HepaRG cells, nucleotide metabolites of 5-FU were clearly predominant compared to nucleoside forms (nucleotides/nucleosides ratio > 10). Under the same exposure conditions, the proportion of each nucleotide metabolite of 5-FU depended on cell lines. The metabolite 5-FUTP was the most abundant in HCT116 cells, followed by 5-FUDP and then 5-FUMP while in HepaRG cells the distribution was reversed with the predominance of 5-FUMP. After cell exposure to 50 μM 5-FdUrd, all the ribonucleotide metabolites were found at the same amount than after 5-FU exposure, indicating that thymidine phosphorylase catalyzes a high proportion of 5-FdUrd to 5-FU (Fig. 1). After cells exposure to 50 μM 5-FU for 24 h, the production of nucleotide metabolites of 5-FU was very important in HCT116 cells since the sum of the nucleotide metabolites (5-FUMP + 5-FUDP + 5-FUTP) was equivalent to that the endogenous nucleotides of uridine (UMP + UDP + UTP). In HepaRG cells the endogenous nucleotides remained in the majority (ratio 5/1).
3.3.2. Inhibition of TS by 5-FdUMP
TS is an enzyme that catalyzes the transformation of dUMP to TMP. Inhibition of TS by 5-FdUMP is one of the main mechanisms of action of 5-FU [1]. After exposure to 50 μM 5-FU, an increase of dUMP (about 5000 fold) and dUDP (about 10 fold) was shown. It was associated with a decrease of TMP (about a factor 5) and TTP (about a factor 7).
3.3.3. Analysis of 5-FU metabolites in RNA and DNA
The study of the incorporation of 5-FU into DNA and RNA can be performed by either (deoxy-)nucleoside or (deoxy-)nucleotide metabolites analysis. As described above in Section 3.1.3, it appears more relevant, according to the LOD and matrix effect, to analyze metabolites of 5-FU as nucleoside forms rather than nucleotide forms. In this case, in order to verify the completeness of the dephosphorylation step, UMP15N was added before addition of phosphatase. After incubation, it was only observed Urd15N whereas no UMP15N was detected.
RNA (3 μg) from non-treated cells or cells exposed to 10 or 50 μM 5-FU or 5-FdUrd was analyzed. Incorporation of 5-FU into RNA was detected both from dephosphorylated (5-FUrd) and phosphorylated (5-FUMP) extracts. The rate of incorporation of 5-FUrd or 5-FUMP into RNA was similar when cells were exposed either to 5-FU or to 5-FdUrd at the same concentration. The amount of 5-FUrd incorporated in RNA increased with the dose of 5-FU. This rate of incorporation was determined to 6–8 pmol/nmol of Urd and 15–17 pmol/nmol of Urd after treatment with 10 or 50 μM 5-FU, respectively. Thus, a factor of 2.5 was observed according to the cell exposure to 10 μM and 50 μM 5-FU.
DNA (3 μg) from untreated cells or cells treated with 10 or 50 μM 5-FU was also analyzed. Since a lower incorporation of 5-FU metabolite was previously described into DNA in comparison to RNA and a lower analytical response of nucleotides in comparison to nucleosides, only dephosphorylated DNA extracts were analyzed [25]. In this condition, no signal was observed for 5-FdUrd, indicating a lower incorporation of 5-FU into DNA than into RNA. Exposure of the cells to 50 μM of 5-FdUrd also failed to result in 5-FU detection from 3 μg of DNA extracts. In the present study, incorporation of 5-FU into DNA was only observed after exposure of cells to 50 μM for 24 h and when 30 μg DNA was analyzed. The rate of incorporation of 5-FU into DNA was assessed to 0.10–0.20 pmol/nmol of thymidine. Thus, the rate of incorporation of metabolites of 5-FU into RNA was 50–100 fold higher than the rate of incorporation into DNA.
3.3.4. Determination of incorporation rate of 5-FU into sub-units of rRNA
It was previously demonstrated that 5-FU is incorporated into different RNA types [4]. Kanamaru et al. [36] previously showed, using L1210 cells and radiolabelled 3H-5FU, that the drug is incorporated into 18 S and 28 S sub-units of rRNA. With our method, we confirmed the incorporation of 5-FU into 18 S and 28 S rRNAs. In humans, the 18 S and 28 S rRNA molecules are composed of 1869 and 5035 nucleotides, respectively [37]. In 18 S and 28 S rRNA extracts, endogenous nucleosides (pseudoUrd, Urd, A, C, G) and 5-FUrd were quantified (between-day repeatability and accuracy results are presented in Table S1). When cells were exposed to 50 μM 5-FU for 24 h, 5-FUrd represented 0.40% and 0.33% of endogenous nucleosides corresponding to 7.5 ± 0.4 and 16.7 ± 0.2 molecules of 5-FUrd into 18 S and 28 S rRNA molecules, respectively (n = 3).
3.3.5. Inhibition of PS by 5-FU in rRNA extracts
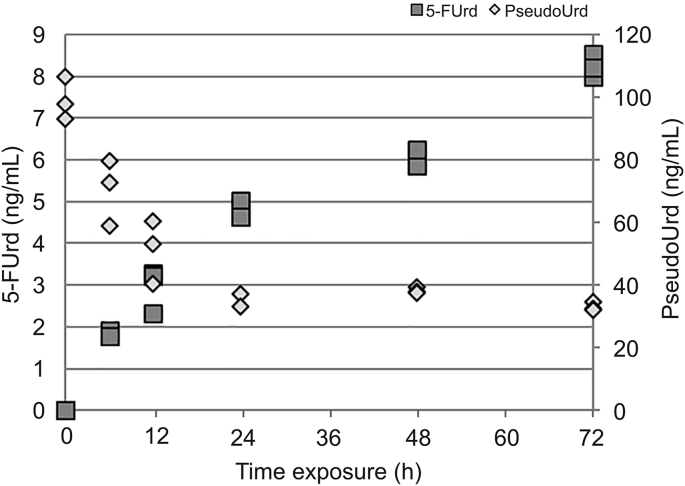
PseudoUrd is the most abundant post-transcriptional modified nucleoside present in RNAs and plays an important biological role in different classes of RNAs [38]. The isomerization of Urd to PseudoUrd by PS leads to a C-C bond between the ribose and the Urd. The present method allows the analysis of pseudoUrd as well as pseudoUMP in dephosphorylated and phosphorylated RNA extracts, respectively. As described above for metabolites of 5-FU, the analytical response is higher for pseudoUrd than for pseudoUMP with a matrix effect clearly lower for nucleoside compared to nucleotide form. Another analytical interest in measuring pseudoUrd rather than pseudoUMP is that Urd and pseudoUrd were well separated chromatographically (6.0 min for pseudoUrd and 7.0 min for Urd). On the contrary, UMP and pseudoUMP were co-eluted (4.3 min), thus requiring the fragmentation of the two compounds to distinguish them (specific ion after positive ionization at m/z 97,02837 and m/z 125,03454 for UMP and pseudoUMP, respectively). Hengesbach et al. [39] demonstrated that 5-FU is a potent inhibitor of PS activity. As shown in a kinetic assay, the levels of pseudoUrd in RNA extracts decreased while incorporation of 5-FUrd increased when cells were exposed to 5-FU (Fig. 5).
Fig. 5.
Kinetic of 5-FUrd and pseudouridine (pseudoUrd) in RNA extracted from cells exposed to 5-FU at 50 μM (n = 3).
4. Discussion
5-FU is an “old” drug that was developed in the 1960s. However, it is still the gold standard for first-line treatment of colorectal cancer and widely used in other cancers. The medical and scientific interest remains present for this compound and many recent works focused on the study of new targets or mechanisms of chemoresistance [[40], [41], [42], [43]]. Thus, it is relevant to propose a new sensitive and selective LC-MS-HRMS method, allowing the analysis of the anabolic metabolites of 5-FU and their incorporation into different cellular organelles.
4.1. Analytical method
Numerous methods reported the analysis of anabolic metabolites of 5-FU. However, the majority of them only focused on nucleoside metabolites of 5-FU [[7], [8], [9],11,14,44,45]. Chromatographic analysis of nucleoside forms is easier by using, for example, reversed stationary phase with mobile phase based on methanol, acetonitrile and water [11,44]. Analysis of nucleotide forms is more challenging due to the poor retention of compounds on reversed stationary phase caused by the polarity of the phosphate moiety [33]. To overcome this issue, our separation was performed with a porous graphitic carbon (PGC) column. Four interactions could occur between the PGC stationary phase, mobile phase and compounds: hydrophobic or dispersive, electronic, steric and redox interactions [46]. This stationary phase has been already successfully used for analysis of endogenous nucleotides and nucleoside analogues [33,47]. A reverse phase column could be used, but an ion pairing agent has to be added in the mobile phase to ensure retention of di- and tri-phosphorylated forms [12]. Anion exchange columns also allowed a good separation of metabolites of 5-FU even nucleotide forms [16,17]. Unlike Ciccolini et al. [12] and Derissen et al. [17], who had used high levels of salts or ion-pairing agents, the chromatographic separation in our method was performed without these additives which are known to hamper mass spectrometric detection. Thus our method is able to separate nucleoside forms and nucleotide forms of metabolites of 5-FU without any inconvenience for mass spectrometric detection.
In our conditions, the response with ESI in negative mode was higher for the ten metabolites of 5-FU than in positive mode. This response gap is much more pronounced with nucleoside metabolites than with nucleotides. In addition, it was observed that, with the same composition of mobile phase, endogenous nucleotides have a higher response in positive mode [33]. In a mobile phase containing ammonium acetate, acetonitrile and water, Jansen et al. [47] showed a higher ionization in negative mode for 2′,2′-difluorodeoxyuridine (dFdU) and in positive mode for its phosphorylated metabolites (dFdUMP, dFdUDP and dFdUTP). In an acidic mobile phase, 5-fluoro-2′-deoxycytidine was ionized in positive mode [48]. We expected that our mobile phase at pH 10 and the presence of fluorine on the nucleobase enhanced ESI negative ionization. Mass fragmentation after negative ionization exhibited one or more fragments containing the fluorine for all of the metabolites, thus ensuring the selectivity of the detection with endogenous nucleotides. The cleavage of the glycosidic bond leading to a fragment at m/z 129.0106 corresponding [M−H]− 5-FU ion was observed for all the metabolites except 5-FdUTP. The cleavage of the ribose ring leading to a fragment at m/z 171.0211 and 155.0262 was shown for 5-FUrd and 5-FdUrd, respectively, as previously observed [13]. Mass fragmentation spectrum of 5-FdUTP exhibited a fragment at m/z 404.9905, with a relative abundance of 5%, corresponding to 5-FdUDP. Fragments at m/z 78.9591, 96.9696 and 158.9254 corresponding to the phosphate moiety were observed for nucleotide and sugar forms. Mass spectra after positive ionization are much less informative and specific to 5-FU since only fragments at m/z 97.0284 and 81.0336 corresponding to the ribose and deoxyribose moiety respectively were observed (data not shown). The study of matrix effect has shown an enhancement of ionization for some metabolites of 5-FU in intracellular content while the same metabolites encountered no matrix effect or an ion suppression in DNA or RNA matrix. Matrix effect may be due to additives present in the mobile phase, type of ionization source, but also to co-eluted compounds and depends on compounds analyzed. In nucleic acids a lower LOD was obtained for 5-FUrd and 5-FdUrd in comparison to 5-FUMP and 5-FdUMP. Firstly, this is due to a better response with nucleosides than with nucleotides. Secondly, no inhibitory matrix effect was measured in nucleic acids extracts after dephosphorylation while a matrix effect greater than 30% was measured with nucleotides. Altogether these results encouraged to analyze metabolites as nucleoside forms (5-FUrd and 5-FdUrd) than nucleotide forms (5-FUMP and 5-FdUMP) for the incorporation into nucleic acids. Although dephosphorylation using phosphodiesterase requires an additional step, the impact on samples preparation time is very limited since this step can be coupled with hydrolysis step.
Numerous publications reported the analyses of metabolites of the anabolism phase of 5-FU in biological matrix. The most analyzed metabolites are 5-FUrd and 5-FdUrd in plasma and cell lines [[7], [8], [9], [10], [11],13,14,18]. Few studies have focused on the analysis of nucleotide metabolites of 5-FU. Procházková et al. [6], Carli et al. [13], Wrightson et al. [15] and Kamm et al. [23] studied 5-FdUMP in plasma, cell lines and tissues. Derissen et al. [16,17] described the analysis of 5-FdUMP, 5-FdUDP, 5-FdUTP and 5-FUTP in plasma and PBMC. Ciccolini et al. [12] analyzed 8 metabolites of the anabolism phase of 5-FU in a single run: 5-FUrd, 5-FdUrd, 5-FUMP, 5-FdUMP, 5-FUDP, 5-FdUDP, 5-FUTP and 5-FdUTP in cell lines. Thus to our knowledge, here we present the first LC-MS-HRMS method able to analyze the ten metabolites of the anabolism phase of 5-FU.
For the study of the incorporation of 5-FU into nucleic acids, Benz et al. [18] and then Peters et al. [19] reported a radiolabeled method by using [14C-5-FU] or [3H-5-FU] for cell exposure followed by extraction and digestion of the nucleic acids. More recently, a GC-MS method requiring an enzymatic degradation of 5-FUMP or 5-FdUMP in 5-FU in RNA or in DNA extracts followed by a derivatization with pentafluorobenzylbromide was published [[22], [23], [24]]. Both methods are time-consuming and not easy to use, especially the radiolabeled methods. Furthermore, the selectivity of these methods depends only on the chromatographic separation if there is one. Thanks to HRMS and fragmentation, the selectivity of our method depends on both the chromatographic separation and the mass detection. Thus our method is highly selective. For the GC-MS method, the disadvantage of analyzing metabolites as 5-FU and not as 5-FUrd or as 5-FdUrd is that it requires a DNA extract without any contamination with RNA and vice versa, which is difficult to obtain. The contamination of RNA by DNA, and inversely, will contribute to an overestimation of the incorporation in each nucleic acid. The analysis of 5-FUrd and 5-FdUrd as themselves as reported in our method avoids this risk of overestimation. An LC-MS/MS method for the determination of incorporation metabolites into RNA and DNA was recently published [25]. This method coupled a separation on a reversed phase stationary phase and a detection with a triple quadrupole mass spectrometer. This method was specific to 5-FUrd and 5-FdUrd. However, the quantification of endogenous nucleosides of RNA is essential for the determination of the incorporation rate of metabolites of 5-FU into RNA. Thus Pettersen et al. [25] had to use another LC-MS/MS method for the quantification of endogenous nucleosides. Samples were therefore separated into two portions: one portion per method. As our method is applicable to the analysis of metabolites of 5-FU as well as endogenous nucleosides in a single run, all analytes are quantified in the same sample and the determination of the incorporation rate of metabolites of 5-FU into RNA is less time-consuming. The LC-MS-HRMS method described in this paper is, to the best of our knowledge, the first one allowing the simultaneous analysis of the ten metabolites of the anabolism phase of 5-FU and is suitable for the analysis of 5-FU incorporation into nucleic acids.
4.2. Applications
After exposure of 3×10 6 cells to 10 μM 5-FU for 24 h, 8 metabolites of the anabolism phase were identified in the cellular content. Nucleotide metabolites were produced in a large extent in comparison to nucleoside metabolites. The predominance of nucleotide forms was previously described by Benz et al. in HCT-8 cells which reported a proportion of 85% for nucleotide forms [18]. We did not detect the two metabolites 5-FdUDP and 5-FdUTP in cellular contents, neither after 5-FU nor 5-FdUrd exposure. Ciccolini et al. [12], with HT29 cells, have observed these two metabolites only after addition of d-Inosine in the cell medium in order to stimulate thymidine phosphorylase. In mammalian cells, dUMP is phosphorylated by thymidylate kinase and by UMP-CMP kinase whereas UMP is only phosphorylated by UMP-CMP kinase. The activation of anticancer nucleoside analogues such as 5-FU is also dependent on these enzymes [34]. It was previously demonstrated that UMP is the best enzyme substrate (of the order of 50 times) than dUMP [34,49]. Thus, these data may explain the high production of 5-FUDP rather than 5-FdUDP. By analyzing the cellular content, the imbalance in deoxynucleotide pool according to the inhibition of TS by 5-FdUMP can clearly be observed with the present method. A stable ternary complex between TS, 5-FdUMP and 5,10-methylene-tetrahydrofolate leads to preventing the fixation of the natural substrate dUMP [1]. The consequence is the accumulation of dUMP and the decrease in synthesis of TMP as previously observed [13].
Our method has shown the incorporation of metabolites of 5-FU both into RNA and DNA. These anabolism pathways are known as a part of the mechanism of action of 5-FU (Fig. 1). Incorporation of metabolites of 5-FU into RNA has been reported since the 1980s by Benz et al. [18] using a radiolabeled method, by Keniry et al. [20] and el-Tahtawy et al. [21] using NMR spectroscopy. With a radiolabel method, Peters et al. [19] demonstrated the incorporation of metabolites of 5-FU both into RNA and DNA. Using a GC-MS method the incorporation was also demonstrated in both nucleic acids [22]. Incorporation of 5-FU into DNA is more challenging to measure than incorporation into RNA. Indeed, the incorporation rate of metabolites of 5-FU into RNA was estimated to be 50 to 100-fold higher than into DNA in our conditions requiring a higher quantity of DNA than RNA for analysis. From RNA and DNA extracted from tumor and tissues, Noordhuis et al. [24] found a 2 to 30-fold higher incorporation rate into RNA than into DNA. From 5-FU-treated cells, Pettersen et al. [25] observed an accumulation 3000 to 15,000-fold more of metabolites of 5-FU into RNA than into DNA, depending on cell lines. As mentioned above, 5-FdUDP and 5-FdUTP were not detected in cellular extracts after 24 h incubation of 5-FU or 5-FdUrd. The very weak production of 5-FdUDP from 5-FdUMP seems to be the rate-limiting step in the phosphorylation pathway since 5-FU is incorporated into DNA by the DNA polymerase following its conversion to 5-FdUTP. This may explain the greater incorporation of 5-FU metabolites into RNA rather than into DNA. Level of incorporation of metabolites of 5-FU into RNA was similar after an exposure of cells to 5-FU or 5-FdUrd. This observation correlated with our results obtained for intracellular metabolites since exposure to 5-FU or 5-FdUrd led to the same level of 5-FUTP. In the present work, incorporation rate of 5-FUrd into 18 S and 28 S sub-units of rRNA was determined. This measurement is also applicable to other RNAs since the number of total endogenous nucleotides is known. To the best of our knowledge, it is the first report on the determination of a such rate. This could be useful to explore the consequence of the synthesis of fluorinated rRNA as a potential cytotoxic or resistance mechanism of 5-FU.
5. Conclusion
We developed an LC-MS-HRMS method for the simultaneous determination of the ten metabolites (nucleoside, nucleotide and sugar nucleotide) of the anabolism phase of 5-FU. The analytical conditions were optimized to obtain an excellent sensitivity and selectivity. The presented method is suitable for determining the metabolites of 5-FU in cell extracts and to study the incorporation of 5-FU metabolite into DNA and RNA. Moreover, as the method allows in the same run the analysis of endogenous nucleosides and nucleotides, we were able to accurately determine the incorporation rate of metabolites of 5-FU into sub-units rRNA and the consequence of thymidylate synthase and pseudouridine synthase inhibition by 5-FU. In view of these results, the LC-MS-HRMS method could also be used to determine metabolites and incorporation of 5-FU into DNA and RNA in target tissues such as tumor.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgments
This work was supported by Institut National Du Cancer (grant number 2018-131).
Footnotes
Peer review under responsibility of Xi’an Jiaotong University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpha.2020.04.001.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Longley D.B., Harkin D.P., Johnston P.G. 5-fluorouracil: mechanisms of action and clinical strategies. Nat. Rev. Canc. 2003;3:330–338. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 2.Tsesmetzis N., Paulin C.B.J., Rudd S.G. Nucleobase and nucleoside analogues: resistance and Re-sensitisation at the level of pharmacokinetics, pharmacodynamics and metabolism. Cancers (Basel) 2018;10 doi: 10.3390/cancers10070240. 240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Samuelsson T. Interactions of transfer RNA pseudouridine synthases with RNAs substituted with fluorouracil. Nucleic Acids Res. 1991;19:6139–6144. doi: 10.1093/nar/19.22.6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parker W.B., Cheng Y.C. Metabolism and mechanism of action of 5-fluorouracil. Pharmacol. Ther. 1990;48:381–395. doi: 10.1016/0163-7258(90)90056-8. [DOI] [PubMed] [Google Scholar]
- 5.Weckbecker G., Keppler D.O. Substrate properties of 5-fluorouridine diphospho sugars detected in hepatoma cells. Biochem. Pharmacol. 1984;33:2291–2298. doi: 10.1016/0006-2952(84)90669-5. [DOI] [PubMed] [Google Scholar]
- 6.Procházková A., Liu S., Friess H. Determination of 5-fluorouracil and 5-fluoro-2’-deoxyuridine-5’-monophosphate in pancreatic cancer cell line and other biological materials using capillary electrophoresis. J. Chromatogr. A. 2001;916:215–224. doi: 10.1016/s0021-9673(00)01171-7. [DOI] [PubMed] [Google Scholar]
- 7.Ishii H., Shimada M., Yamaguchi H. A simultaneous determination method for 5-fluorouracil and its metabolites in human plasma with linear range adjusted by in-source collision-induced dissociation using hydrophilic interaction liquid chromatography-electrospray ionization-tandem mass spectrometry. Biomed. Chromatogr. 2016;30:1882–1886. doi: 10.1002/bmc.3743. [DOI] [PubMed] [Google Scholar]
- 8.Deenen M.J., Rosing H., Hillebrand M.J. Quantitative determination of capecitabine and its six metabolites in human plasma using liquid chromatography coupled to electrospray tandem mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 913–914. 2013:30–40. doi: 10.1016/j.jchromb.2012.11.033. [DOI] [PubMed] [Google Scholar]
- 9.Casale F., Canaparo R., Serpe L. Plasma concentrations of 5-fluorouracil and its metabolites in colon cancer patients. Pharmacol. Res. 2004;50:173–179. doi: 10.1016/j.phrs.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 10.Deng P., Ji C., Dai X. Simultaneous determination of capecitabine and its three nucleoside metabolites in human plasma by high performance liquid chromatography-tandem mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2015;989:71–79. doi: 10.1016/j.jchromb.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 11.Holleran J.L., Eiseman J.L., Parise R.A. LC-MS/MS assay for the quantitation of FdCyd and its metabolites FdUrd and FU in human plasma. J. Pharmaceut. Biomed. 2016;129:359–366. doi: 10.1016/j.jpba.2016.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ciccolini J., Peillard L., Aubert C. Monitoring of the intracellular activation of 5-fluorouracil to deoxyribonucleotides in HT29 human colon cell line: application to modulation of metabolism and cytotoxicity study. Fund. Clin. Pharmacol. 2000;14:147–154. doi: 10.1111/j.1472-8206.2000.tb00403.x. [DOI] [PubMed] [Google Scholar]
- 13.Carli D., Honorat M., Cohen S. Simultaneous quantification of 5-FU, 5-FUrd, 5-FdUrd, 5-FdUMP, dUMP and TMP in cultured cell models by LC-MS/MS. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2009;877:2937–2944. doi: 10.1016/j.jchromb.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 14.Tsume Y., Provoda C.J., Amidon G.L. The achievement of mass balance by simultaneous quantification of floxuridine prodrug, floxuridine, 5-fluorouracil, 5-dihydrouracil, α-fluoro-β-ureidopropionate, α-fluoro-β-alanine using LC-MS. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2011;879:915–920. doi: 10.1016/j.jchromb.2011.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wrightson W.R., Myers S.R., Galandiuk S. HPLC analysis of 5-FU and FdUMP in tissue and serum. Biochem. Biophys. Res. Commun. 1995;216:808–813. doi: 10.1006/bbrc.1995.2693. [DOI] [PubMed] [Google Scholar]
- 16.Derissen E.J.B., Jacobs B.A.W., Huitema A.D.R. Exploring the intracellular pharmacokinetics of the 5-fluorouracil nucleotides during capecitabine treatment, Brit. J. Clin. Pharmacol. 2016;81:949–957. doi: 10.1111/bcp.12877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Derissen E.J.B., Hillebrand M.J.X., Rosing H. Development of an LC-MS/MS assay for the quantitative determination of the intracellular 5-fluorouracil nucleotides responsible for the anticancer effect of 5-fluorouracil. J. Pharmaceut. Biomed. 2015;110:58–66. doi: 10.1016/j.jpba.2015.02.051. [DOI] [PubMed] [Google Scholar]
- 18.Benz C., Cadman E. Modulation of 5-fluorouracil metabolism and cytotoxicity by antimetabolite pretreatment in human colorectal adenocarcinoma HCT-8. Canc. Res. 1981;41:994–999. [PubMed] [Google Scholar]
- 19.Peters G.J., Laurensse E., Leyva A. Sensitivity of human, murine, and rat cells to 5-fluorouracil and 5’-deoxy-5-fluorouridine in relation to drug-metabolizing enzymes. Canc. Res. 1986;46:20–28. [PubMed] [Google Scholar]
- 20.Keniry M., Benz C., Shafer R.H. Noninvasive spectroscopic analysis of fluoropyrimidine metabolism in cultured tumor cells. Canc. Res. 1986;46:1754–1758. [PubMed] [Google Scholar]
- 21.el-Tahtawy A., Wolf W. In vivo measurements of intratumoral metabolism, modulation, and pharmacokinetics of 5-fluorouracil, using 19F nuclear magnetic resonance spectroscopy. Canc. Res. 1991;51:5806–5812. [PubMed] [Google Scholar]
- 22.Peters G.J., Noordhuis P., Komissarov A. Quantification of 5-fluorouracil incorporation into RNA of human and murine tumors as measured with a sensitive gas chromatography-mass spectrometry assay. Anal. Biochem. 1995;231:157–163. doi: 10.1006/abio.1995.1515. [DOI] [PubMed] [Google Scholar]
- 23.Kamm Y.J.L., Peters G.J., Hull W.E. Correlation between 5-fluorouracil metabolism and treatment response in two variants of C26 murine colon carcinoma. Br. J. Canc. 2003;89:754–762. doi: 10.1038/sj.bjc.6601162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noordhuis P., Holwerda U., Van der Wilt C.L. 5-Fluorouracil incorporation into RNA and DNA in relation to thymidylate synthase inhibition of human colorectal cancers. Ann. Oncol. 2004;15:1025–1032. doi: 10.1093/annonc/mdh264. [DOI] [PubMed] [Google Scholar]
- 25.Pettersen H.S., Visnes T., Vågbø C.B. UNG-initiated base excision repair is the major repair route for 5-fluorouracil in DNA, but 5-fluorouracil cytotoxicity depends mainly on RNA incorporation. Nucleic Acids Res. 2011;39:8430–8444. doi: 10.1093/nar/gkr563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ginsburg-Shmuel T., Haas M., Schumann M. 5-OMe-UDP is a potent and selective P2Y(6)-receptor agonist. J. Med. Chem. 2010;53:1673–1685. doi: 10.1021/jm901450d. [DOI] [PubMed] [Google Scholar]
- 27.Tsang W.-Y., Wood B.M., Wong F.M. Proton transfer from C-6 of uridine 5’-monophosphate catalyzed by orotidine 5’-monophosphate decarboxylase: formation and stability of a vinyl carbanion intermediate and the effect of a 5-fluoro substituent. J. Am. Chem. Soc. 2012;134:14580–14594. doi: 10.1021/ja3058474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Béres J., Bentrude W.G., Kálmán A. Synthesis, structure, and antitumor and antiviral activities of a series of 5-halouridine cyclic 3’,5’-monophosphates. J. Med. Chem. 1986;29:488–493. doi: 10.1021/jm00154a011. [DOI] [PubMed] [Google Scholar]
- 29.Roy B., Lefebvre I., Puy J.-Y. A facile and effective synthesis of lamivudine 5′-diphosphate. Tetrahedron Lett. 2011;52:1250–1252. [Google Scholar]
- 30.Gripon P., Rumin S., Urban S. Infection of a human hepatoma cell line by hepatitis B virus. Proc. Natl. Acad. Sci. U.S.A. 2002;99:15655–15660. doi: 10.1073/pnas.232137699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quinlivan E.P., Gregory J.F. DNA digestion to deoxyribonucleoside: a simplified one-step procedure. Anal. Biochem. 2008;373:383–385. doi: 10.1016/j.ab.2007.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Belin S., Beghin A., Solano-Gonzàlez E. Dysregulation of ribosome biogenesis and translational capacity is associated with tumor progression of human breast cancer cells. PloS One. 2009;4 doi: 10.1371/journal.pone.0007147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Machon C., Jordheim L.P., Puy J.-Y. Fully validated assay for the quantification of endogenous nucleoside mono- and triphosphates using online extraction coupled with liquid chromatography-tandem mass spectrometry. Anal. Bioanal. Chem. 2014;406:2925–2941. doi: 10.1007/s00216-014-7711-1. [DOI] [PubMed] [Google Scholar]
- 34.Van Rompay A.R., Johansson M., Karlsson A. Phosphorylation of deoxycytidine analog monophosphates by UMP-CMP kinase: molecular characterization of the human enzyme. Mol. Pharmacol. 1999;56:562–569. doi: 10.1124/mol.56.3.562. [DOI] [PubMed] [Google Scholar]
- 35.Günther Sillero M.A., Pérez-Zúñiga F., Gomes J. Synthesis of FUDP-N-acetylglucosamine and FUDP-glucose in Saccharomyces cerevisiae cells treated with 5-fluorouracil. FEMS Yeast Res. 2008;8:257–265. doi: 10.1111/j.1567-1364.2007.00321.x. [DOI] [PubMed] [Google Scholar]
- 36.Kanamaru R., Kakuta H., Sato T. The inhibitory effects of 5-fluorouracil on the metabolism of preribosomal and ribosomal RNA in L-1210 cells in vitro, Cancer Chemother. Pharmacology. 1986;17:43–46. doi: 10.1007/BF00299864. [DOI] [PubMed] [Google Scholar]
- 37.Eichler D.C., Craig N. Processing of eukaryotic ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol. 1994;49:197–239. doi: 10.1016/s0079-6603(08)60051-3. [DOI] [PubMed] [Google Scholar]
- 38.Charette M., Gray M.W. Pseudouridine in RNA: what, where, how, and why. IUBMB Life. 2000;49:341–351. doi: 10.1080/152165400410182. [DOI] [PubMed] [Google Scholar]
- 39.Hengesbach M., Voigts-Hoffmann F., Hofmann B. Formation of a stalled early intermediate of pseudouridine synthesis monitored by real-time FRET. RNA. 2010;16:610–620. doi: 10.1261/rna.1832510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li M., Cui Z.-G., Zakki S.A. Aluminum chloride causes 5-fluorouracil resistance in hepatocellular carcinoma HepG2 cells. J. Cell. Physiol. 2019;234:20249–20265. doi: 10.1002/jcp.28625. [DOI] [PubMed] [Google Scholar]
- 41.Francipane M.G., Bulanin D., Lagasse E. Establishment and characterization of 5-fluorouracil-resistant human colorectal cancer stem-like cells: tumor dynamics under selection pressure. Int. J. Mol. Sci. 2019;20:E1817. doi: 10.3390/ijms20081817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sakatani A., Sonohara F., Goel A. Melatonin-mediated downregulation of thymidylate synthase as a novel mechanism for overcoming 5-fluorouracil associated chemoresistance in colorectal cancer cells. Carcinogenesis. 2019;40:422–431. doi: 10.1093/carcin/bgy186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Humphrey T.C. Identifying new targets for cancer drug 5’-fluorouracil. Cell Cycle. 2015;14:1353. doi: 10.1080/15384101.2015.1022062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jung M., Berger G., Pohlen U. Simultaneous determination of 5-fluorouracil and its active metabolites in serum and tissue by high-performance liquid chromatography. J. Chromatogr. B Biomed. Sci. Appl. 1997;702:193–202. doi: 10.1016/s0378-4347(97)00368-x. [DOI] [PubMed] [Google Scholar]
- 45.Joulia J.M., Pinguet F., Grosse P.Y. Determination of 5-fluorouracil and its main metabolites in plasma by high-performance liquid chromatography: application to a pharmacokinetic study. J. Chromatogr. B Biomed. Sci. Appl. 1997;692:427–435. doi: 10.1016/s0378-4347(96)00518-x. [DOI] [PubMed] [Google Scholar]
- 46.Jansen R.S., Rosing H., Schellens J.H.M. Retention studies of 2’-2’-difluorodeoxycytidine and 2’-2’-difluorodeoxyuridine nucleosides and nucleotides on porous graphitic carbon: development of a liquid chromatography-tandem mass spectrometry method. J. Chromatogr. A. 2009;1216:3168–3174. doi: 10.1016/j.chroma.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 47.Jansen R.S., Rosing H., Schellens J.H.M. Simultaneous quantification of 2’,2’-difluorodeoxycytidine and 2’,2’-difluorodeoxyuridine nucleosides and nucleotides in white blood cells using porous graphitic carbon chromatography coupled with tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2009;23:3040–3050. doi: 10.1002/rcm.4212. [DOI] [PubMed] [Google Scholar]
- 48.Byun J., Henderson J.P., Heinecke J.W. Identification and quantification of mutagenic halogenated cytosines by gas chromatography, fast atom bombardment, and electrospray ionization tandem mass spectrometry. Anal. Biochem. 2003;317:201–209. doi: 10.1016/s0003-2697(03)00093-9. [DOI] [PubMed] [Google Scholar]
- 49.Hsu C.-H., Liou J.-Y., Dutschman G.E. Phosphorylation of cytidine, deoxycytidine, and their analog monophosphates by human UMP/CMP kinase is differentially regulated by ATP and magnesium. Mol. Pharmacol. 2005;67:806–814. doi: 10.1124/mol.104.006098. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.