Abstract
Natural products (NPs) are critical sources of drug molecules for decades. About two-thirds of natural antibiotics are produced by Streptomyces. Streptomyces have a large number of secondary metabolite biosynthetic gene clusters (SM-BGCs) that may encode NPs. However, most of these BGCs are silent under standard laboratory conditions. Hence, activation of these silent BGCs is essential to current natural products discovery research. In this review, we described the commonly used strategies for silent BGC activation in Streptomyces from two aspects. One focused on the strategies applied in heterologous host, including methods to clone and reconstruct BGCs along with advances in chassis engineering; the other focused on methods applied in native host which includes engineering of promoters, regulatory factors, and ribosomes. With the metabolic network being elucidated more comprehensively and methods optimized more high-thoroughly, the discovery of NPs will be greatly accelerated.
Keywords: Streptomyces, natural products, biosynthetic gene cluster, heterologous expression, in situ activation, synthetic biology
Introduction
Natural products (NPs) are major sources of drug molecules, including antibiotic, anticancer, antifungal, antiparasitic, and immunosuppressive compounds. Streptomyces plays a central role in the discovery of NPs, and the genes responsible for NPs biosynthesis are generally clustered in a continuous region of the genome termed as biosynthetic gene clusters (BGCs). With the rapid development of sequencing technologies, especially the third generation sequencing technology (Loman and Pallen, 2015), more and more genomic information of Streptomyces was clarified. Analysis of sequenced Streptomyces genome data revealed that a single Streptomyces’ genome generally encodes 25–50 BGCs, ∼90% of which are silent or cryptic under standard laboratory growth conditions (Walsh and Fischbach, 2010; Rutledge and Challis, 2015; Mao et al., 2018). Therefore, to increase the production of the encoded natural product, methods to unlock or up-regulate these so called “silent” gene clusters have become the interest of research in recent years.
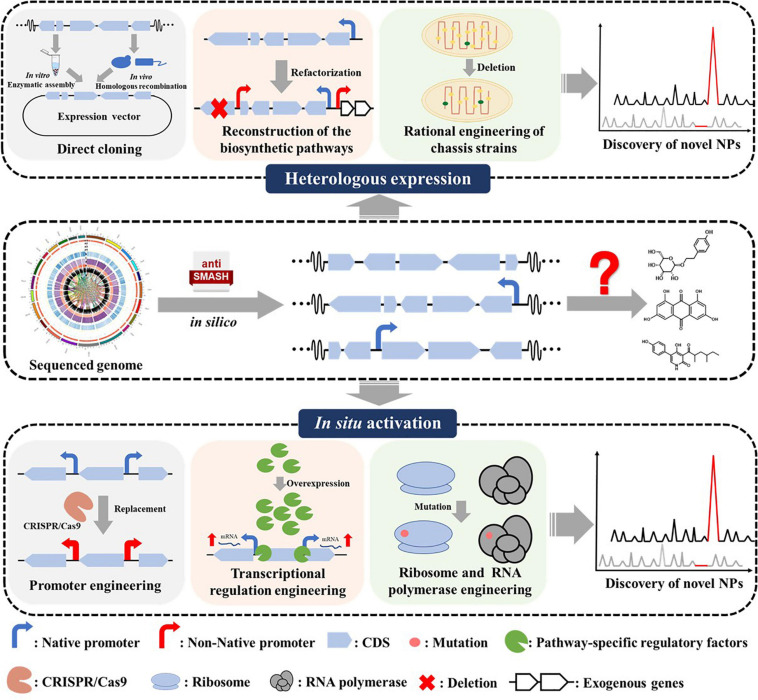
Numbers of methods have been developed to activate silent BGCs in recent years (Rutledge and Challis, 2015; Onaka, 2017; Mao et al., 2018; Lewis, 2020). Powerful bioinformatics approaches for genome mining and identification of NPs BGCs are well summarized in some recent reviews (Lee et al., 2020; Ren et al., 2020; Van Santen et al., 2020; Kenshole et al., 2021). Herein, we provide a concise overview as an introductory guide to the recent advances in silent BGCs activation in Streptomyces from two aspects, involving heterologous reconstruction and in situ activation (Figure 1). For heterologous reconstruction, we discussed different cloning strategies, biosynthetic pathways reconstruction methods, and chassis strain engineering approaches. For in situ activation, we summarized the methods including promoter engineering, transcription factors operating, and ribosome engineering (Table 1).
FIGURE 1.
Overview of strategies for silent BGCs activation. BGC, biosynthetic gene cluster; CDS, coding sequences; RBS, ribosome binding site.
TABLE 1.
Examples of BGCs activation in Streptomyces.
| Hosts | Targets | Approaches | Expression strains | Effect | References |
| Heterologous host | Cloning BGC | Environmental DNA (eDNA) cosmid libraries | S. albus J1074 | To clone the complete malacidin BGC from environmental metagenome samples | Hover et al., 2018 |
| Streptomyces bacterial artificial chromosome system (pSBAC) | S. lividans and S. coelicolor | To clone a 60 kb pikromycin BGC | Pyeon et al., 2017 | ||
| Exonuclease Combined with RecET recombination (ExoCET) | To clone a 106 kb salinomycin BGC | Wang et al., 2018 | |||
| Cas9-Assisted Targeting of chromosome segments (CATCH) | To clone a 36 kb jadomycin BGC | Jiang et al., 2015 | |||
| Combining CRISPR/Cas9 system with in vitro λ packaging system | S. avermitilis MA-4680 | To clone 27.4 kb Tü3010 and 40.7 kb sisomicin BGC | Tao et al., 2019 | ||
| Reconstruction | Regulatory sequences cassettes | S. albus J1074 | To activate the actinorhodin BGC. | Ji et al., 2018 | |
| RedEx | S. albus J1074 | To activate the spinosyn BGC and butenyl-spinosyn A production is 2.36 mg/l, spinosyn Jproduction is 7.34 mg/l | Song et al., 2020 | ||
| Chassis strain | Deletion phosphofructokinases (encoded by pfk) | S. albus J1074 | To increase the precursor level of NADPH and heterologous expression of actinorhodin | Kallifidas et al., 2018 | |
| Deletion genomic regions | S. Chattanoogensis L320 | To delete 0.7 Mb non-essential genomic regions | Bu et al., 2019 | ||
| Native host | Promoter | Identification strong promoters | S. griseus | To activate a PTM BGC. | Luo et al., 2015 |
| Characterization native or synthetic promoters and Ribosomal binding sites (RBSs) | S. avermitilis | To activate and overproduce the lycopene BGC. | Bai et al., 2015 | ||
| Identification a strong promoter groESp | S. chattanoogensis L10 | To activate the natamycin BGC and to increase yield by 20%. | Wang K. et al., 2019 | ||
| Promoter engineering of the PAS-LuxR (pimM) | Streptomyces | To activate the polyene BGC. | Barreales et al., 2018 | ||
| CRISPR-Cas9 knock-in strategy | Streptomyces | To activate multiple BGCs of Streptomyces and to trigger the production of a pentangular polyketide. | Zhang et al., 2017 | ||
| Regulator | Expression of bldA | S. coelicolor | To activate the actinorhodin, undecylprodigiosin and methylenomycin BGCs. | Hackl and Bechthold, 2015) | |
| Overexpression of slnR | S. albus | To activate the salinomycin BGC. | Zhu et al., 2017) | ||
| Heterologous expression of papR2 | S. lividans | To activate the undecylprodigiosin BGC. | Krause et al., 2020) | ||
| Expression of gdmRIII | S. autolyticus | To positively control the biosynthesis of geldanamycin. | Jiang et al., 2017 | ||
| Overexpression of toyA | S. diastatochromogenes | To activate the toyocamycin BGC and toyocamycin highest titer is 456.3 mg/l. | Xu et al., 2019 | ||
| Expression of aveI | S. avermitilis | To activate the melanin BGC. | Liu et al., 2019 | ||
| Ribosome and RNA polymerase | Overexpression of exogenous rpsL and rpoB genes containing beneficial mutations | Marine Streptomycete | To activate the piloquinone and homopiloquinone BGCs | Zhang Q. et al., 2020 | |
| Mutation RNA polymerase: rpoB (H437Y) | S. chattanoogensis L10 (CGMCC 2644) | To activate the anthrachamycin BGC. | Li Z.Y. et al., 2019 | ||
| Mutation RNA polymerase:guanosine-tetraphosphate (ppGpp) | S.sp. SoC090715LN-16 S55-50-5 | To identify/overproduce a novel isoindolinomycin. | Thong et al., 2018 |
Heterologous Expression of Target BGCs
Heterologous expression is an efficient and established approach to unlock silent or cryptic gene clusters that have been identified by genome mining. Compared to expressions in native hosts, heterologous expression owns several advantages. (1) It can express BGCs whose native host is uncultivable or grows slowly under laboratory growth conditions; (2) heterologous host usually holds mature genetic manipulation tools; (3) the background information of the heterologous host is clearer (Xu and Wright, 2019). Generally, heterologous expression includes three steps-cloning of the target BGCs; engineering of the target BGCs; and transformation to the selected heterologous host. In this section, we will briefly update on the progress of heterologous expression from these three aspects.
Cloning of Large BGCs
Traditional methods for cloning large BGCs generally employ genomic library constructed by cosmid, fosmid, BAC (Bacterial Artificial Chromosomal), and PAC (P1-derived Artificial Chromosome) vectors (Blodgett et al., 2005; Jones et al., 2013; Xu et al., 2016). In a recent research, 90 Actinomycetes NP BGCs have been successfully heterologous expressed, and about 83% of them were constructed via the cosmid/fosmid library method (Nah et al., 2017). However, these techniques are often time-consuming as well as laborious.
TAR (Transformation-Associated Recombination) cloning is a powerful and reliable system to directly clone large size BGCs (Orr-Weaver et al., 1981). The ends of the linearized TAR cloning vector contain specific homologous sequences of target BGCs as hooks to stimulate homologous recombination (Kouprina and Larionov, 2016, 2019). Bonet et al. (2015) reported the first case of heterologous expression of a natural product BGC from the marine Streptomyces Salinispora via the TAR-mediate pCAP01 vector. Later, Kang et al. (2016) built a mCRISTAR platform that combines CRISPR/Cas9 with TAR to simultaneously replace multiple promoters in the tetarimycin BGC. The system was further improved as mpCRISTAR (Multiple Plasmids-based CRISPR/Cas9 and TAR) by employing multiple plasmids, each harboring one or two unique guide RNAs. Based on mpCRISTAR, six or eight promoters can be simultaneously replaced with an efficiency of 68 and 32%, respectively (Kim et al., 2020).
Meanwhile, approaches based on site-specific recombinase systems have also been developed to directly clone BGCs. The integrase-mediated recombination (IR) system employs phage ΦBT1 attP-attB-int system to induce site-specific recombination (Du et al., 2015). Liu et al. (2009) described a versatile E. coli-Streptomyces shuttle vector system, pSBAC, employing the ΦBT1 IR system. Pyeon et al. further optimized the above system with additional restriction recognition sites on pSBAC to simplify the cloning procedure. They successfully cloned the 80 kb tautomycetin BGC and 60 kb pikromycin BGC for heterologous expression with the modified system (Pyeon et al., 2017). Zhang et al. (1998, 2000) developed a powerful Red/ET recombineering tool to assemble large DNA fragments using homologous recombination in E. coli. Later, Wang et al. (2018) upgraded this system by employing T4 polymerase to facilitate annealing between the linear target DNA and vector in vitro, and they termed the system as ExoCET. They used ExoCET to successfully cloned the intact 106 kb salinomycin BGC from S. albus.
Apart from in vivo cloning technologies, there are various in vitro cloning strategies. Gibson assembly has been well applied in multi-segment assembly in vitro (Gibson et al., 2009). For example, the 41 kb conglobatin BGC from S. conglobatus was cloned through Gibson assembly (Zhou et al., 2015). However, Gibson assembly is inefficient for large DNA fragments with high G + C content (Casini et al., 2014; Li et al., 2015). Therefore, Jiang et al. (2015) combined Gibson assembly with CRISPR-Cas9, termed as CATCH (Cas9-Assisted Targeting of CHromosome segments). They successfully cloned the 36 kb jad gene cluster from S.venezuelae and the 32 kb ctc gene cluster from S. aureofaciens into the p15A vector via CATCH. Similarly, Tao et al. (2019) illustrated an in vitro one-step targeted cloning approach combining CRISPR/Cas9 system with in vitro λ packaging system, and the pathways of Tü3010 (27.4 kb) and sisomicin (40.7 kb) were successfully cloned, respectively.
In summary, each strategy for cloning large BGCs has pros and cons. The methods of genomic library construction are random, but they are beneficial for metagenome-driven natural product discovery (Katz et al., 2016; Hover et al., 2018). The pSBAC is suitable for cloning large DNA fragments with specific restriction digestion sites, which are not generally available at both ends of target BGCs. As for tools based on homologous recombination, like TAR and Red/ET, both are commonly used in cloning large DNA fragments but may introduce some undesired recombination. Although CRISPR tools solve the limitation of insufficient restriction sites, it still faces the bottleneck of isolating targeted BGCs from the genomic DNA. With the increasing number of sequenced genomes, developing high-throughput cloning tools becomes imminent, such as combining current tools with automated platforms (Burger et al., 2020).
Reconstruction of the Biosynthetic Pathways
It has been reported that the complexity of the regulatory network in host cells was a major challenge for metabolic engineering (Shao et al., 2013). Therefore, reconstruction and heterologous expression of the biosynthetic pathways can release them from the complex metabolic network. Nevertheless, BGCs controlled by promoters of different strengths increase the complexity of the reconstruction (Horbal et al., 2018). At present, the reconstruction process mainly includes: (1) gene substitution, (2) enzyme evolution, (3) promoter replacement, (4) transcriptional repressor knockout (Li L. et al., 2019). For example, Alberti et al. (2019) successfully activated the scl BGC by inactivating the transcriptional repressors via CRISPR/Cas9. AGOS (Artificial Gene Operon assembly System) is a plug and play method designed for the construction of artificial gene operons through Red/ET mediated recombination. Four gene operons of novobiocin BGCs were heterologously integrated into the genome of S. coelicolor M1146 via AGOS, leading to the production of novobiocin and novobiocin precursors (Basitta et al., 2017). Marín et al. cloned the synthetic genes encoding tyrosine ammonia lyase, 4-coumaroyl CoA ligase, chalcone synthase, chalcone isomerase and flavone synthase into a high copy number shuttle vector including a strong promoter ermE∗p. The final plasmid pAPI was transformed into the heterologous host S. albus and successfully produced apigenin at 0.08 mg/L (Marín et al., 2017). In another case, Song et al. (2020) refactored the spinosyn BGCs via RedEx to test whether the ethyl group at C-21 of spinosyn A can be replaced by butene group.
Among the above mentioned methods, promoter replacement is the most effective and well established method to activate silent BGCs, especially in Streptomyces (Luo et al., 2013). For example, Luo et al. identified strong promoters from S. albus J1074, whose strength is 200–1,300% the strength of the well-known strong promoter ermE∗p. They used a plug-and-play scaffold to successfully activate the silent PTM BGC of S. griseus in three widely used Streptomyces chassis strains (Luo et al., 2015). Ji et al. (2018) used synthetic regulatory sequences cassettes to successfully activate actinorhodin BGCs. In another case, the silent streptophenazine BGC in marine Streptomyces S.sp is non-transcriptional active in heterologous environment. After introducing four constitutive promoters (ermE∗p/actIp/sp44/p21) at different positions in the BGC, the production of streptophenazine was detected (Bauman et al., 2019).
All in all, thorough reconstruction of BGCs often leads to the activation of silent BGCs and the discovery of new NPs (Luo et al., 2016; Zhao et al., 2019). However, compared with E. coli and other model strains, the genetic manipulation tools in Streptomyces are still limited. Therefore, new methods are urgently needed and some new developments are well summarized in other reviews (Tan and Liu, 2017; Tao et al., 2018; Zhao Y. et al., 2020).
Rational Engineering of Chassis Strains
Streptomyces are rich in inherently valuable secondary metabolites. Therefore, series of Streptomyces species have been developed as chassis to express heterologous BGCs, such as S. coelicolor, S. lividans, and S. albus (Ziermann and Betlach, 1999; Zhou et al., 2012; Myronovskyi et al., 2018). A suitable surrogate expression host should contain several essential features: (1) a variety of natural product precursors which are conducive to construct abundant complex molecules; (2) a simplified secondary metabolite background; (3) an efficient transportation system to transfer various bioactive compounds; (4) a known regulatory network; (5) a mature fermentation and upscaling process; (6) powerful genetic manipulation tools (Baltz, 2010, 2016; Myronovskyi and Luzhetskyy, 2019; Xu and Wright, 2019). Researchers did a comprehensive and detailed introduction of Streptomyces species used as heterologous hosts from 2010 to 2018 in another excellent review (Myronovskyi and Luzhetskyy, 2019).
The commonly used chassis engineering strategy is to reduce the background of secondary metabolism (Lee et al., 2019). Non-essential genomic regions and secondary metabolic genes mainly appearing in the end region of the chromosome. They are not stable and prone to chromosomal rearrangements, hence knocking out of them may generate clean background chassis strains. Ahmed et al. (2020) developed a set of S. lividans chassis strains. The S. lividans ΔYA11 was obtained by deleting 11 gene clusters (228.5 kb) and inserting two attB sites. Bu et al. (2019) rational constructed two genome-reduced Streptomyces chassis strains, the S. chattanoogensis L320 and L321, through multiple computational approaches and site-specific recombination systems, with non-essential genomic regions deletion of 1.3 and 0.7 Mb, respectively. Sometimes, the low yield of heterologous produced NPs may be due to insufficient precursors in the expression hosts. Therefore, increasing the supply of the precursors is a promising strategy. Borodina et al. (2008) rationally engineered the S. coelicolor A3(2) strain by deleting the phosphofructokinases (encode by pfkA2) gene, thus the precursor level of NADPH was increased and the production of actinorhodin and undecylprodigiosin were upregulated correspondingly. Dang et al. (2017) knocked out pfk in S. hygroscopicus ATCC 29253. The titer of rapamycin increased by 30.8% in the engineered strain. Kallifidas et al. (2018) successfully heterologous expressed actinorhodin in S. albus, and then further increased its yield by knocking out pfkSA.
Currently, a set of powerful bioinformatics approaches are developed to design chassis strains rationally (Ren et al., 2020). Meanwhile, the powerful genetic editing tool CRISPR has been applied in Streptomyces for genome engineering (Tong et al., 2019a,b; Zhang J. et al., 2020; Zhao Y.W. et al., 2020). These techniques are expected to accelerate the development of Streptomyces chassis strains.
Heterologous expression has numerous advantages, but some limitations still exist. (1) The size of SM-BGCs is highly variable (1–100 kb), and most are more than 10 kb. Currently, there is no certain method that is universal, large-size endurable, efficient, and high-throughput; (2) Because of the complicated metabolic networks of Streptomyces, clarify the interaction between the host strain and the heterologous BGCs is hard; (3) At present, Streptomyces chassis compatible with all NPs’ production has not been reported; (4) Current genetic manipulation tools of Streptomyces are not applicable in all species, thus more powerful and universal genetic tools are needed.
In situ Activation of Target BGCs
The expression of NPs BGCs in Streptomyces is governed by a complex metabolic regulatory network. The production of antibiotics can be greatly enhanced by rewiring the regulatory network (Xia et al., 2020). Therefore, a better understanding and manipulation of the regulatory network in these silent BGCs could help to activate BGCs. In this section, we described different strategies to manipulate the regulatory modules in the native hosts for silent BGCs activation.
Promoter Engineering
With regard to cluster activation, promoter elements are of indisputable importance as they are responsible for efficient transcription, which is the first stage of gene expression (Myronovskyi and Luzhetskyy, 2016). Promoter engineering employs a set of regulatory sequences with known functions, to release the following gene expression from the native complex regulations. Constitutive promoters commonly used to activate gene expression include: the promoter of the erythromycin resistance gene ermE of S. erythraea, ermEp1 and its derivatives (Bibb et al., 1985); the phage I19 originated promoter SF14p (Labes et al., 1997); and the engineered kasOp∗ promoter (Takano et al., 2005; Wang et al., 2013). Inducible promoters commonly used to activate gene expression include: the thiostrepton-inducible promoter PtipA (Holmes et al., 1993), the synthetic resorcinol-inducible and cumate-inducible promoters (Horbal et al., 2014), and the synthetic tetracycline-inducible promoter tcp830 (Rodríguez-García et al., 2005).
Since the strategy of knocking in promoters with multiple operon structure by homologous double-crossover recombination is often time-consuming and laborious, (Zhang et al., 2017) reported an effective CRISPR-Cas9 knock-in strategy in Streptomyces, and this one-step strategy was applied to activate multiple silent BGCs in five Streptomyces species. Similarly, Tong et al. (2015) also adopted the CRISPR-Cas9 system (deemed CRISPRi) to control the expression of target genes in Actinomycetes. The combination of the CRISPR system and promoter engineering approaches makes the experimental operation and procedure relatively simple and efficient.
At present, progress in activating silent BGCs in Streptomyces through comprehensive multi-promoter insertion is limited. Constructing promoters with a wide range of transcription initiation activities, transcription strength and robustness would promote effective activation of silent BGCs, and gene expression balance needs to be taken into considerations as well. In short, promoter-based gene expression activation methods still need improvement.
Transcriptional Regulation Engineering
The biosynthesis of NPs in Streptomyces is regulated by precise regulatory systems, in which transcription factors (TFs) regulate the initiation level of transcription by binding to DNAs. In the era of synthetic biology, coordination of TFs regulations sometimes can activate silent BGCs, such as overexpression of positive regulatory genes or inactivation of negative regulatory genes in Streptomyces. For example, bldA of S. coelicolor can activate the expression of the antibiotics actinorhodin, undecylprodigiosin, and methylenomycin BGCs (Cuthbertson and Nodwell, 2013; Bhukya and Anand, 2017). Guo et al. (2018) used gene deletion, complementation, and overexpression to determine the MarR family transcriptional regulator (MFR) SAV4189 as an activator of avermectin biosynthesis in S. avermitilis. In addition to pathway-specific regulatory factors, global regulatory factors can also activate silent BGCs. For example, through genome sequencing analysis, gene knockout, and transcriptional analysis, the global regulator AdpA was found to be able to activate nikkomycin biosynthesis, and repress the biosynthesis of oviedomycin at the same time (Xu et al., 2017). Recently, Wang B. et al. (2019) reported a transcription factor decoy strategy for targeted activation of large silent polyketide synthase and non-ribosomal peptide synthetase, and discovered a novel oxazole family compound. Li et al. (2020) developed a base editing system that combines CRISPR-Cas9 with site-specific recombination to achieve successful genome editing in Streptomyces by programmed mutation of target genes, thereby achieving product biosynthesis (such as hygromycin B). Owing to their simplicity and ease of use, these strategies can be scaled up readily for the discovery of natural products in Streptomyces.
Ribosome Engineering
Ribosome engineering is an approach to discover microbes with certain spontaneous mutations in their ribosome or RNA polymerase, through screening antibiotic-resistant mutants on Petri dishes (Zhu et al., 2019). It is suitable for gene activation and strain improvement, resulting in the identification of novel secondary metabolites, as well as the enhancement of enzyme production and tolerance to toxic chemicals (Ochi, 2017).
The rpoB gene (encoding the RNA polymerase β-subunit) can activate silent BGCs in various Streptomyces by rifampicin resistance mutations (up to 70 times at the transcription level). Analysis of the metabolite profile showed that rpoB mutants produced many metabolites undetectable in wild-type strains (Tanaka et al., 2013). Li Z.Y. et al. (2019) used site-directed mutagenesis to generate ten mutants with point mutations in the highly conserved region of rpsL (encoding the ribosomal protein S12) or rpoB. Among them, L10/RpoB (H437Y) activated anthrachamycin biosynthesis in S. chattanoogensis L10 (CGMCC 2644). Zhang et al. designed a TTO (Transcription–Translation in One) method using a plug-and-play plasmid system to directly overexpress exogenous rpsL (encoding ribosomal protein S12) and rpoB (encoding RNA polymerase β subunit) genes containing beneficial mutations. This method overcomes the false positive problem in the traditional ribosome engineering method and was successfully applied to activate the silent BGCs in three Streptomyces strains, thus discovering two aromatic polyketide antibiotics (Zhang Q. et al., 2020). Moreover, the ppGpp can interact with RNA polymerase and affect the production of antibiotics (Artsimovitch et al., 2004). It is suggested that RNA polymerases carrying specific rif mutations in the β-subunit can functionally mimic modification induced by binding of ppGpp (Xu et al., 2002). So, some studies showed that rif mutations could alter the gene expression patterns of ppGpp. Thong et al. (2016) screened mutants resistant to rifampicin and found an unknown metabolite.
At present, in addition to the conventional modification of ribosomes through mutagenesis, other ribosomal regulatory elements have also been engineered. Siu and Chen (2019) proposed a new class of riboregulators called toehold-gated gRNA (thgRNA) by integrating toehold riboswitches into sgRNA scaffolds and demonstrated their programmability for multiplexed regulation in E. coli with minimal cross-talks. In the future, this approach could also be tested in Streptomyces for gene expression regulation.
Promoter engineering can activate a single gene expression in BGCs, and it can also activate the full-length BGCs to produce the corresponding NPs. This method can be further developed for high-throughput activation of silent BGCs. Knockout of negative regulatory genes is one method to explore new NPs. However, in Streptomyces, the traditional gene knockout strategy is often completed by plasmid-mediated homologous recombination, which is usually time-consuming and laborious. Due to the differences in the source, structure and functions of BGCs, more attempts and innovations are needed to unlock the transcriptional regulation of BGCs.
Discussion
At present, in addition to the methods mentioned in this review, the silent BGCs can also be activated by changing the culture conditions. Bode et al. (2002) defined it as one strain many compounds (OSMAC), that is, by adjusting the culture parameters of Streptomyces, such as medium composition, culture temperature, pH, aeration, and container type, to induce the expression of silent BGCs. Later, on the basis of OSMAC, other strategies were derived, such as the addition of low-concentration antibiotics, signal molecules and histone deacetylase inhibitors and other inducers (Seyedsayamdost, 2014), as well as co-cultivation strategies. In 2019, a review discussed the use of microbial culture techniques to expand the range of NPs available in the laboratory in recent years, mainly including methods such as adding physical scaffolds, adding small molecule elicitors, and co-cultivating with another microorganism (Tomm et al., 2019). Although these methods are relatively economical and simple, they are particularly suitable for Streptomyces species with incomplete genome information or genetic isolation defects.
Due to the high investment and low return rate of silent BGCs activation, the discovery of new NPs has entered a bottleneck. Through combining bioinformatics analysis with multi-omics data to explore the genomic data, insights to regulate and activate BGCs could be elicited. These methods can not only act alone to produce NPs, but can also be combined with each other. There is still an urgent requirement to develop better methods to activate silent BGCs. For example, structured data can be used to further elucidate the detailed mechanism, automation can help improving high-throughput capabilities, and AI can be employed to assist experiment design. Perhaps combining in situ activation with simulation analysis, heterologous expression and other strategies, more precise transcription activation could be achieved for silent BGCs exploration.
Author Contributions
ZL, YZ, and CH: writing—original draft. YL: writing—review and editing and project administration. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by the Key-Area Research and Development Program of Guangdong Province (2020B0303070002), the National Natural Science Foundation of China (Grant No. 32071426), the National Key R&D Program of China (2018YFA0903300), and the Natural Science Foundation of Tianjin Province (19JCYBJC24200).
References
- Ahmed Y., Rebets Y., Estevez M. R., Zapp J., Myronovskyi M., Luzhetskyy A. (2020). Engineering of Streptomyces lividans for heterologous expression of secondary metabolite gene clusters. Microb. Cell Fact. 19:5. 10.1186/s12934-020-1277-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti F., Leng D. J., Wilkening I., Song L., Tosin M., Corre C. (2019). Triggering the expression of a silent gene cluster from genetically intractable bacteria results in scleric acid discovery. Chem. Sci. 10 453–463. 10.1039/c8sc03814g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artsimovitch I., Patlan V., Sekine S., Vassylyeva M. N., Hosaka T., Ochi K., et al. (2004). Structural basis for transcription regulation by alarmone ppGpp. Cell 117 299–310. 10.1016/s0092-8674(04)00401-5 [DOI] [PubMed] [Google Scholar]
- Bai C., Zhang Y., Zhao X., Hu Y., Xiang S., Miao J., et al. (2015). Exploiting a precise design of universal synthetic modular regulatory elements to unlock the microbial natural products in Streptomyces. Proc. Natl. Acad. Sci. U.S.A. 112 12181–12186. 10.1073/pnas.1511027112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltz R. H. (2010). Streptomyces and Saccharopolyspora hosts for heterologous expression of secondary metabolite gene clusters. J. Ind. Microbiol. Biotechnol. 37 759–772. 10.1007/s10295-010-0730-9 [DOI] [PubMed] [Google Scholar]
- Baltz R. H. (2016). Genetic manipulation of secondary metabolite biosynthesis for improved production in Streptomyces and other Actinomycetes. J. Ind. Microbiol. Biotechnol. 43 343–370. 10.1007/s10295-015-1682-x [DOI] [PubMed] [Google Scholar]
- Barreales E. G., Vicente C. M., De Pedro A., Santos-Aberturas J., Aparicio J. F. (2018). Promoter engineering reveals the importance of heptameric direct repeats for DNA binding by Streptomyces antibiotic regulatory protein-large ATP-binding regulator of the LuxR family (SARP-LAL) regulators in Streptomyces natalensis. Appl. Environ. Microbiol. 84:e00246-18. 10.1128/AEM.00246-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basitta P., Westrich L., Rösch M., Kulik A., Gust B., Apel A. K. (2017). AGOS: a Plug-and-play method for the assembly of artificial gene operons into functional biosynthetic gene clusters. ACS Synth. Biol. 6 817–825. 10.1021/acssynbio.6b00319 [DOI] [PubMed] [Google Scholar]
- Bauman K. D., Li J., Murata K., Mantovani S. M., Dahesh S., Nizet V., et al. (2019). Refactoring the cryptic streptophenazine biosynthetic gene cluster unites Phenazine, Polyketide, and Nonribosomal peptide biochemistry. Cell Chem. Biol. 26 724–736.e727. 10.1016/j.chembiol.2019.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhukya H., Anand R. (2017). TetR regulators: a structural and functional perspective. J. Indian Inst. Sci. 97 245–259. 10.1007/s41745-017-0025-5 [DOI] [Google Scholar]
- Bibb M. J., Janssen G. R., Ward J. M. (1985). Cloning and analysis of the promoter region of the erythromycin resistance gene (ermE) of Streptomyces erythraeus. Gene 38 215–226. 10.1016/0378-1119(85)90220-3 [DOI] [PubMed] [Google Scholar]
- Blodgett J. A. V., Zhang J. K., Metcalf W. W. (2005). Molecular cloning, sequence analysis, and heterologous expression of the Phosphinothricin Tripeptide biosynthetic gene cluster from Streptomyces viridochromogenes DSM 40736. Antimicrob. Agents Chemother. 49 230–240. 10.1128/aac.49.1.230-240.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode H. B., Bethe B., Höfs R., Zeeck A. (2002). Big effects from small changes: possible ways to explore nature’s chemical diversity. Chembiochem 3 619–627. [DOI] [PubMed] [Google Scholar]
- Bonet B., Teufel R., Crusemann M., Ziemert N., Moore B. S. (2015). Direct capture and heterologous expression of Salinispora natural product genes for the biosynthesis of enterocin. J. Nat. Prod. 78 539–542. 10.1021/np500664q [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodina I., Siebring J., Zhang J., Smith C. P., Van Keulen G., Dijkhuizen L., et al. (2008). Antibiotic overproduction inStreptomyces coelicolorA3(2) mediated by phosphofructokinase deletion. J. Biol. Chem. 283 25186–25199. 10.1074/jbc.m803105200 [DOI] [PubMed] [Google Scholar]
- Bu Q. T., Yu P., Wang J., Li Z. Y., Chen X. A., Mao X. M., et al. (2019). Rational construction of genome-reduced and high-efficient industrial Streptomyces chassis based on multiple comparative genomic approaches. Microb. Cell Fact 18:16. 10.1186/s12934-019-1055-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger B., Maffettone P. M., Gusev V. V., Aitchison C. M., Bai Y., Wang X., et al. (2020). A mobile robotic chemist. Nature 583 237–241. 10.1038/s41586-020-2442-2 [DOI] [PubMed] [Google Scholar]
- Casini A., Macdonald J. T., Jonghe J. D., Christodoulou G., Freemont P. S., Baldwin G. S., et al. (2014). One-pot DNA construction for synthetic biology: the Modular overlap-directed assembly with linkers (MODAL) strategy. Nucleic Acids Res. 42:e7. 10.1093/nar/gkt915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuthbertson L., Nodwell J. R. (2013). The TetR family of regulators. Microbiol. Mol. Biol. Rev. 77 440–475. 10.1128/MMBR.00018-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L., Liu J., Wang C., Liu H., Wen J. (2017). Enhancement of rapamycin production by metabolic engineering in Streptomyces hygroscopicus based on genome-scale metabolic model. J. Ind. Microbiol. Biotechnol. 44 259–270. 10.1007/s10295-016-1880-1 [DOI] [PubMed] [Google Scholar]
- Du D., Wang L., Tian Y., Liu H., Tan H., Niu G. (2015). Genome engineering and direct cloning of antibiotic gene clusters via phage ϕBT1 integrase-mediated site-specific recombination in Streptomyces. Sci. Rep. 5:8740. 10.1038/srep08740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D. G., Young L., Chuang R. Y., Venter J. C., Hutchison C. A., III, Smith H. O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6 343–345. 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- Guo J., Zhang X., Lu X., Liu W., Chen Z., Li J., et al. (2018). SAV4189, a MarR-family regulator in Streptomyces avermitilis, activates Avermectin biosynthesis. Front. Microbiol. 9:1358. 10.3389/fmicb.2018.01358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackl S., Bechthold A. (2015). The Gene bldA, a regulator of morphological differentiation and antibiotic production in Streptomyces. Arch. Pharm. 348 455–462. 10.1002/ardp.201500073 [DOI] [PubMed] [Google Scholar]
- Holmes D. J., Caso J. L., Thompson C. J. (1993). Autogenous transcriptional activation of a thiostrepton-induced gene in Streptomyces lividans. EMBO J. 12 3183–3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horbal L., Fedorenko V., Luzhetskyy A. (2014). Novel and tightly regulated resorcinol and cumate-inducible expression systems for Streptomyces and other Actinobacteria. Appl. Microbiol. Biotechnol. 98 8641–8655. 10.1007/s00253-014-5918-x [DOI] [PubMed] [Google Scholar]
- Horbal L., Marques F., Nadmid S., Mendes M. V., Luzhetskyy A. (2018). Secondary metabolites overproduction through transcriptional gene cluster refactoring. Metab. Eng. 49 299–315. 10.1016/j.ymben.2018.09.010 [DOI] [PubMed] [Google Scholar]
- Hover B. M., Kim S.-H., Katz M., Charlop-Powers Z., Owen J. G., Ternei M. A., et al. (2018). Culture-independent discovery of the malacidins as calcium-dependent antibiotics with activity against multidrug-resistant Gram-positive pathogens. Nat. Microbiol. 3 415–422. 10.1038/s41564-018-0110-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji C. H., Kim J. P., Kang H. S. (2018). Library of synthetic Streptomyces regulatory sequences for use in promoter engineering of natural product biosynthetic gene clusters. ACS Synth. Biol. 7 1946–1955. 10.1021/acssynbio.8b00175 [DOI] [PubMed] [Google Scholar]
- Jiang M., Yin M., Wu S., Han X., Ji K., Wen M., et al. (2017). GdmRIII, a TetR family transcriptional regulator, controls Geldanamycin and Elaiophylin biosynthesis in Streptomyces autolyticus CGMCC0516. Sci. Rep. 7:4803. 10.1038/s41598-017-05073-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Zhao X., Gabrieli T., Lou C., Ebenstein Y., Zhu T. F. (2015). Cas9-Assisted Targeting of chromosome segments CATCH enables one-step targeted cloning of large gene clusters. Nat. Commun. 6:8101. 10.1038/ncomms9101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A. C., Gust B., Kulik A., Heide L., Buttner M. J., Bibb M. J. (2013). Phage P1-derived artificial chromosomes facilitate heterologous expression of the FK506 gene cluster. PLoS One 8:e69319. 10.1371/journal.pone.0069319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallifidas D., Jiang G., Ding Y., Luesch H. (2018). Rational engineering of Streptomyces albus J1074 for the overexpression of secondary metabolite gene clusters. Microb. Cell Fact 17:25. 10.1186/s12934-018-0874-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H. S., Charlop-Powers Z., Brady S. F. (2016). Multiplexed CRISPR/Cas9- and TAR-mediated promoter engineering of natural product biosynthetic gene clusters in yeast. ACS Synth. Biol. 5 1002–1010. 10.1021/acssynbio.6b00080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz M., Hover B. M., Brady S. F. (2016). Culture-independent discovery of natural products from soil metagenomes. J. Ind. Microbiol. Biotechnol. 43 129–141. [DOI] [PubMed] [Google Scholar]
- Kenshole E., Herisse M., Michael M., Pidot S. J. (2021). Natural product discovery through microbial genome mining. Curr. Opin. Chem. Biol. 60 47–54. 10.1016/j.cbpa.2020.07.010 [DOI] [PubMed] [Google Scholar]
- Kim H., Ji C. H., Je H. W., Kim J. P., Kang H. S. (2020). mpCRISTAR: multiple plasmid approach for CRISPR/Cas9 and TAR-mediated multiplexed refactoring of natural product biosynthetic gene clusters. ACS Synth. Biol. 9 175–180. 10.1021/acssynbio.9b00382 [DOI] [PubMed] [Google Scholar]
- Kouprina N., Larionov V. (2016). Transformation-associated recombination (TAR) cloning for genomics studies and synthetic biology. Chromosoma 125 621–632. 10.1007/s00412-016-0588-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouprina N., Larionov V. (2019). TAR cloning: perspectives for functional genomics, biomedicine, and biotechnology. Mol. Ther. Methods Clin. Dev. 14 16–26. 10.1016/j.omtm.2019.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause J., Handayani I., Blin K., Kulik A., Mast Y. (2020). Disclosing the potential of the SARP-Type regulator PapR2 for the activation of antibiotic gene clusters in Streptomycetes. Front. Microbiol. 11:225. 10.3389/fmicb.2020.00225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labes G., Bibb M., Wohlleben W. (1997). Isolation and characterization of a strong promoter element from the Streptomyces ghanaensis phage I19 using the gentamicin resistance gene (aacC1) of Tn 1696 as reporter. Microbiology 143(Pt 5), 1503–1512. 10.1099/00221287-143-5-1503 [DOI] [PubMed] [Google Scholar]
- Lee N., Hwang S., Kim J., Cho S., Palsson B., Cho B.-K. (2020). Mini review: genome mining approaches for the identification of secondary metabolite biosynthetic gene clusters in Streptomyces. Comput. Struct. Biotechnol. J. 18 1548–1556. 10.1016/j.csbj.2020.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N., Hwang S., Lee Y., Cho S., Palsson B., Cho B. K. (2019). Synthetic biology tools for novel secondary metabolite discovery in Streptomyces. J. Microbiol. Biotechnol. 29 667–686. 10.4014/jmb.1904.04015 [DOI] [PubMed] [Google Scholar]
- Lewis K. (2020). The science of antibiotic discovery. Cell 181 29–45. 10.1016/j.cell.2020.02.056 [DOI] [PubMed] [Google Scholar]
- Li L., Liu X., Jiang W., Lu Y. (2019). Recent advances in synthetic biology approaches to optimize production of bioactive natural products in Actinobacteria. Front. Microbiol. 10:2467. 10.3389/fmicb.2019.02467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. Y., Bu Q. T., Wang J., Liu Y., Chen X. A., Mao X. M., et al. (2019). Activation of anthrachamycin biosynthesis in Streptomyces chattanoogensis L10 by site-directed mutagenesis of rpoB. J. Zhejiang. Univ. Sci. B 20 983–994. 10.1631/jzus.B1900344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Zhao Y., Ruan L., Yang S., Ge M., Jiang W., et al. (2015). A stepwise increase in pristinamycin II biosynthesis by Streptomyces pristinaespiralis through combinatorial metabolic engineering. Metab. Eng. 29 12–25. 10.1016/j.ymben.2015.02.001 [DOI] [PubMed] [Google Scholar]
- Li S., Liu Q., Zhong Z., Deng Z., Sun Y. (2020). Exploration of Hygromycin B biosynthesis utilizing CRISPR-Cas9-associated base editing. ACS Chem. Biol. 15 1417–1423. 10.1021/acschembio.0c00071 [DOI] [PubMed] [Google Scholar]
- Liu H., Jiang H., Haltli B., Kulowski K., Muszynska E., Feng X., et al. (2009). Rapid cloning and heterologous expression of the meridamycin biosynthetic gene cluster using a versatile Escherichia coli-Streptomyces artificial chromosome vector, pSBAC. J. Nat. Prod. 72 389–395. 10.1021/np8006149 [DOI] [PubMed] [Google Scholar]
- Liu L., Cheng Y., Lyu M., Zhao X., Wen Y., Li J., et al. (2019). AveI, an AtrA homolog of Streptomyces avermitilis, controls avermectin and oligomycin production, melanogenesis, and morphological differentiation. Appl. Microbiol. Biotechnol. 103 8459–8472. 10.1007/s00253-019-10062-3 [DOI] [PubMed] [Google Scholar]
- Loman N. J., Pallen M. J. (2015). Science China life sciences twenty years of bacterial genome sequencing. Nat. Rev. Microbiol. 13 787–794. 10.1038/nrmicro3565 [DOI] [PubMed] [Google Scholar]
- Luo Y., Enghiad B., Zhao H. (2016). New tools for reconstruction and heterologous expression of natural product biosynthetic gene clusters. Nat. Prod. Rep. 33 174–182. 10.1039/c5np00085h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Huang H., Liang J., Wang M., Lu L., Shao Z., et al. (2013). Activation and characterization of a cryptic polycyclic tetramate macrolactam biosynthetic gene cluster. Nat. Commun. 4:2894. 10.1038/ncomms3894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Zhang L., Barton K. W., Zhao H. (2015). Systematic Identification of a Panel of Strong Constitutive Promoters from Streptomyces albus. ACS Synth. Biol. 4 1001–1010. 10.1021/acssynbio.5b00016 [DOI] [PubMed] [Google Scholar]
- Mao D., Okada B. K., Wu Y., Xu F., Seyedsayamdost M. R. (2018). Recent advances in activating silent biosynthetic gene clusters in bacteria. Curr. Opin. Microbiol. 45 156–163. 10.1016/j.mib.2018.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín L., Gutiérrez-Del-Río I., Yagüe P., Manteca Á, Villar C. J., Lombó F. (2017). De novo biosynthesis of Apigenin, Luteolin, and Eriodictyol in the Actinomycete Streptomyces albus and production improvement by feeding and spore conditioning. Front. Microbiol. 8:921. 10.3389/fmicb.2017.00921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myronovskyi M., Luzhetskyy A. (2016). Native and engineered promoters in natural product discovery. Nat. Prod. Rep. 33 1006–1019. 10.1039/c6np00002a [DOI] [PubMed] [Google Scholar]
- Myronovskyi M., Luzhetskyy A. (2019). Heterologous production of small molecules in the optimized Streptomyces hosts. Nat. Prod. Rep. 36 1281–1294. 10.1039/c9np00023b [DOI] [PubMed] [Google Scholar]
- Myronovskyi M., Rosenkranzer B., Nadmid S., Pujic P., Normand P., Luzhetskyy A. (2018). Generation of a cluster-free Streptomyces albus chassis strains for improved heterologous expression of secondary metabolite clusters. Metab. Eng. 49 316–324. 10.1016/j.ymben.2018.09.004 [DOI] [PubMed] [Google Scholar]
- Nah H. J., Pyeon H. R., Kang S. H., Choi S. S., Kim E. S. (2017). Cloning and heterologous expression of a large-sized natural product biosynthetic gene cluster in Streptomyces species. Front. Microbiol. 8:394. 10.3389/fmicb.2017.00394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochi K. (2017). “Chapter 9 - Cryptic pathways and implications for novel drug discovery,” in Microbial Resources, ed. Kurtböke I. (Cambridge, MA: Academic Press; ), 189–203. [Google Scholar]
- Onaka H. (2017). Novel antibiotic screening methods to awaken silent or cryptic secondary metabolic pathways in Actinomycetes. J. Antibiot. 70 865–870. 10.1038/ja.2017.51 [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. (1981). Yeast transformation: a model system for the study of recombination. Proc. Natl. Acad. Sci. U.S.A. 78 6354–6358. 10.1073/pnas.78.10.6354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyeon H. R., Nah H. J., Kang S. H., Choi S. S., Kim E. S. (2017). Heterologous expression of pikromycin biosynthetic gene cluster using Streptomyces artificial chromosome system. Microb. Cell Fact 16:96. 10.1186/s12934-017-0708-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H., Shi C., Zhao H. (2020). Computational tools for discovering and engineering natural product biosynthetic pathways. iScience 23 100795. 10.1016/j.isci.2019.100795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-García A., Combes P., Pérez-Redondo R., Smith M. C., Smith M. C. (2005). Natural and synthetic tetracycline-inducible promoters for use in the antibiotic-producing bacteria Streptomyces. Nucleic Acids Res. 33:e87. 10.1093/nar/gni086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutledge P. J., Challis G. L. (2015). Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Microbiol. 13 509–523. 10.1038/nrmicro3496 [DOI] [PubMed] [Google Scholar]
- Seyedsayamdost M. R. (2014). High-throughput platform for the discovery of elicitors of silent bacterial gene clusters. Proc. Natl. Acad. Sci. U.S.A. 111 7266–7271. 10.1073/pnas.1400019111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z., Rao G., Li C., Abil Z., Luo Y., Zhao H. (2013). Refactoring the silent spectinabilin gene cluster using a plug-and-play scaffold. ACS Synth. Biol. 2 662–669. 10.1021/sb400058n [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu K. H., Chen W. (2019). Riboregulated toehold-gated gRNA for programmable CRISPR-Cas9 function. Nat. Chem. Biol. 15 217–220. 10.1038/s41589-018-0186-1 [DOI] [PubMed] [Google Scholar]
- Song C., Luan J., Li R., Jiang C., Hou Y., Cui Q., et al. (2020). RedEx: a method for seamless DNA insertion and deletion in large multimodular polyketide synthase gene clusters. Nucleic Acids Res. 48:gkaa956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano E., Kinoshita H., Mersinias V., Bucca G., Hotchkiss G., Nihira T., et al. (2005). A bacterial hormone (the SCB1) directly controls the expression of a pathway-specific regulatory gene in the cryptic type I polyketide biosynthetic gene cluster of Streptomyces coelicolor. Mol. Microbiol. 56 465–479. 10.1111/j.1365-2958.2005.04543.x [DOI] [PubMed] [Google Scholar]
- Tan G. Y., Liu T. (2017). Rational synthetic pathway refactoring of natural products biosynthesis in Actinobacteria. Metab. Eng. 39 228–236. 10.1016/j.ymben.2016.12.006 [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Kasahara K., Hirose Y., Murakami K., Kugimiya R., Ochi K. (2013). Activation and products of the cryptic secondary metabolite biosynthetic gene clusters by rifampin resistance (rpoB) mutations in Actinomycetes. J. Bacteriol. 195 2959–2970. 10.1128/JB.00147-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao W., Chen L., Zhao C., Wu J., Yan D., Deng Z., et al. (2019). In Vitro packaging mediated one-step targeted cloning of natural product pathway. ACS Synth. Biol. 8 1991–1997. 10.1021/acssynbio.9b00248 [DOI] [PubMed] [Google Scholar]
- Tao W., Yang A., Deng Z., Sun Y. (2018). CRISPR/Cas9-based editing of Streptomyces for discovery, characterization, and production of natural products. Front. Microbiol. 9:1660. 10.3389/fmicb.2018.01660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thong W. L., Shin-Ya K., Nishiyama M., Kuzuyama T. (2016). Methylbenzene-containing Polyketides from a Streptomyces that spontaneously acquired rifampicin resistance: structural elucidation and biosynthesis. J. Nat. Prod. 79 857–864. 10.1021/acs.jnatprod.5b00922 [DOI] [PubMed] [Google Scholar]
- Thong W. L., Shin-Ya K., Nishiyama M., Kuzuyama T. (2018). Discovery of an antibacterial isoindolinone-containing tetracyclic polyketide by cryptic gene activation and characterization of its biosynthetic gene cluster. ACS Chem. Biol. 13 2615–2622. 10.1021/acschembio.8b00553 [DOI] [PubMed] [Google Scholar]
- Tomm H. A., Ucciferri L., Ross A. C. (2019). Advances in microbial culturing conditions to activate silent biosynthetic gene clusters for novel metabolite production. J. Ind. Microbiol. Biotechnol. 46 1381–1400. 10.1007/s10295-019-02198-y [DOI] [PubMed] [Google Scholar]
- Tong Y., Charusanti P., Zhang L., Weber T., Lee S. Y. (2015). CRISPR-Cas9 Based engineering of actinomycetal genomes. ACS Synth. Biol. 4 1020–1029. 10.1021/acssynbio.5b00038 [DOI] [PubMed] [Google Scholar]
- Tong Y., Weber T., Lee S. Y. (2019a). CRISPR/Cas-based genome engineering in natural product discovery. Nat. Prod. Rep. 36 1262–1280. 10.1039/C8NP00089A [DOI] [PubMed] [Google Scholar]
- Tong Y., Whitford C. M., Robertsen H. L., Blin K., Jørgensen T. S., Klitgaard A. K., et al. (2019b). Highly efficient DSB-free base editing for streptomycetes with CRISPR-BEST. Proc. Natl. Acad. Sci. U.S.A. 116 20366–20375. 10.1073/pnas.1913493116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Santen J. A., Kautsar S. A., Medema M. H., Linington R. G. (2020). Microbial natural product databases: moving forward in the multi-omics era. Nat. Prod. Rep. 10.1039/D0NP00053A [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh C. T., Fischbach M. A. (2010). Natural products version 2.0: connecting genes to molecules. J. Am. Chem. Soc. 132 2469–2493. 10.1021/ja909118a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Guo F., Dong S. H., Zhao H. (2019). Activation of silent biosynthetic gene clusters using transcription factor decoys. Nat. Chem. Biol. 15 111–114. 10.1038/s41589-018-0187-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Chen X. A., Li Y. Q., Mao X. M. (2019). Identification of a secondary metabolism-responsive promoter by proteomics for over-production of natamycin in Streptomyces. Arch. Microbiol. 201 1459–1464. 10.1007/s00203-019-01710-3 [DOI] [PubMed] [Google Scholar]
- Wang H., Li Z., Jia R., Yin J., Li A., Xia L., et al. (2018). ExoCET: exonuclease in vitro assembly combined with RecET recombination for highly efficient direct DNA cloning from complex genomes. Nucleic Acids Res. 46:e28. 10.1093/nar/gkx1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Li X., Wang J., Xiang S., Feng X., Yang K. (2013). An engineered strong promoter for Streptomycetes. Appl. Environ. Microbiol. 79 4484–4492. 10.1128/AEM.00985-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H. Y., Li X. F., Li Z. Q., Zhan X. Q., Mao X. M., Li Y. Q. (2020). The application of regulatory cascades in Streptomyces: yield enhancement and metabolite mining. Front. Microbiol. 11:406. 10.3389/fmicb.2020.00406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Song Z., Xu X., Ma Z., Bechthold A., Yu X. (2019). ToyA, a positive pathway-specific regulator for toyocamycin biosynthesis in Streptomyces diastatochromogenes 1628. Appl. Microbiol. Biotechnol. 103 7071–7084. 10.1007/s00253-019-09959-w [DOI] [PubMed] [Google Scholar]
- Xu J., Tozawa Y., Lai C., Hayashi H., Ochi K. (2002). A rifampicin resistance mutation in the rpoB gene confers ppGpp-independent antibiotic production in Streptomyces coelicolor A3(2). Mol. Genet. Genom. 268 179–189. 10.1007/s00438-002-0730-1 [DOI] [PubMed] [Google Scholar]
- Xu J., Zhang J., Zhuo J., Li Y., Tian Y., Tan H. (2017). Activation and mechanism of a cryptic oviedomycin gene cluster via the disruption of a global regulatory gene, adpA, in Streptomyces ansochromogenes. J. Biol. Chem. 292 19708–19720. 10.1074/jbc.M117.809145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Wang Y. M., Zhao Z. L., Gao G. X., Huang S. X., Kang Q. J., et al. (2016). Functional Genome mining for metabolites encoded by large gene clusters through Heterologous expression of a whole-genome bacterial artificial chromosome library in Streptomyces spp. Appl. Environ. Microbiol. 82 5795–5805. 10.1128/aem.01383-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Wright G. D. (2019). Heterologous expression-facilitated natural products’ discovery in Actinomycetes. J. Ind. Microbiol. Biotechnol. 46 415–431. 10.1007/s10295-018-2097-2 [DOI] [PubMed] [Google Scholar]
- Zhang J., Zhang D., Zhu J., Liu H., Liang S., Luo Y. (2020). Efficient multiplex genome editing in Streptomyces via engineered CRISPR-Cas12a systems. Front. Bioeng. Biotechnol. 8:726. 10.3389/fbioe.2020.00726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Ren J.-W., Wang W., Zhai J. A., Yang J., Liu N., et al. (2020). A versatile transcription-translation in one approach for activation of cryptic biosynthetic gene clusters. ACS Chem. Biol. 15 2551–2557. 10.1021/acschembio.0c00581 [DOI] [PubMed] [Google Scholar]
- Zhang M. M., Wong F. T., Wang Y., Luo S., Lim Y. H., Heng E., et al. (2017). CRISPR-Cas9 strategy for activation of silent Streptomyces biosynthetic gene clusters. Nat. Chem. Biol. 13 607–609. 10.1038/nchembio.2341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Buchholz F., Muyrers J. P., Stewart A. F. (1998). A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 20 123–128. 10.1038/2417 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Muyrers J. P., Testa G., Stewart A. F. (2000). DNA cloning by homologous recombination in Escherichia coli. Nat. Biotechnol. 18 1314–1317. 10.1038/82449 [DOI] [PubMed] [Google Scholar]
- Zhao Q., Wang L., Luo Y. (2019). Recent advances in natural products exploitation in Streptomyces via synthetic biology. Eng. Life Sci. 19 452–462. 10.1002/elsc.201800137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Li G., Chen Y., Lu Y. (2020). Challenges and advances in genome editing technologies in Streptomyces. Biomolecules 10:734. 10.3390/biom10050734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. W., Tian J. Z., Zheng G. S., Chen J., Sun C. W., Yang Z. Y., et al. (2020). Multiplex genome editing using a dCas9-cytidine deaminase fusion in Streptomyces. Sci. China Life Sci. 63 1053–1062. 10.1007/s11427-019-1559-y [DOI] [PubMed] [Google Scholar]
- Zhou M., Jing X., Xie P., Chen W., Wang T., Xia H., et al. (2012). Sequential deletion of all the polyketide synthase and nonribosomal peptide synthetase biosynthetic gene clusters and a 900-kb subtelomeric sequence of the linear chromosome of Streptomyces coelicolor. FEMS Microbiol. Lett. 333 169–179. 10.1111/j.1574-6968.2012.02609.x [DOI] [PubMed] [Google Scholar]
- Zhou Y., Murphy A. C., Samborskyy M., Prediger P., Dias L. C., Leadlay P. F. (2015). Iterative mechanism of macrodiolide formation in the anticancer compound conglobatin. Chem. Biol. 22 745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Duan Y., Huang Y. (2019). The Application of ribosome engineering to natural product discovery and yield improvement in Streptomyces. Antibiotics 8:133. 10.3390/antibiotics8030133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Li H., Yu P., Guo Y., Luo S., Chen Z., et al. (2017). SlnR is a positive pathway-specific regulator for salinomycin biosynthesis in Streptomyces albus. Appl. Microbiol. Biotechnol. 101 1547–1557. 10.1007/s00253-016-7918-5 [DOI] [PubMed] [Google Scholar]
- Ziermann R., Betlach M. C. (1999). Recombinant polyketide synthesis in Streptomyces: engineering of improved host strains. Biotechniques 26 106–110. 10.2144/99261st05 [DOI] [PubMed] [Google Scholar]