Abstract
During ontogeny, the establishment of the hematopoietic system takes place in several phases, separated both in time and location. The process is initiated extra-embryonically in the yolk sac (YS) and concludes in the main arteries of the embryo with the formation of hematopoietic stem cells (HSC). Initially, it was thought that HSC-independent hematopoietic YS cells were transient, and only required to bridge the gap to HSC activity. However, in recent years it has become clear that these cells also contribute to embryonic organogenesis, including the emergence of HSCs. Furthermore, some of these early HSC-independent YS cells persist into adulthood as distinct hematopoietic populations. These previously unrecognized abilities of embryonic HSC-independent hematopoietic cells constitute a new field of interest. Here, we aim to provide a succinct overview of the current knowledge regarding the contribution of YS-derived hematopoietic cells to the development of the embryo and the adult hematopoietic system.
Keywords: yolk sac, lineage tracing, embryonic hematopoiesis, organogenesis, macrophage, hematopoietic waves, HSC-independent hematopoiesis
Introduction
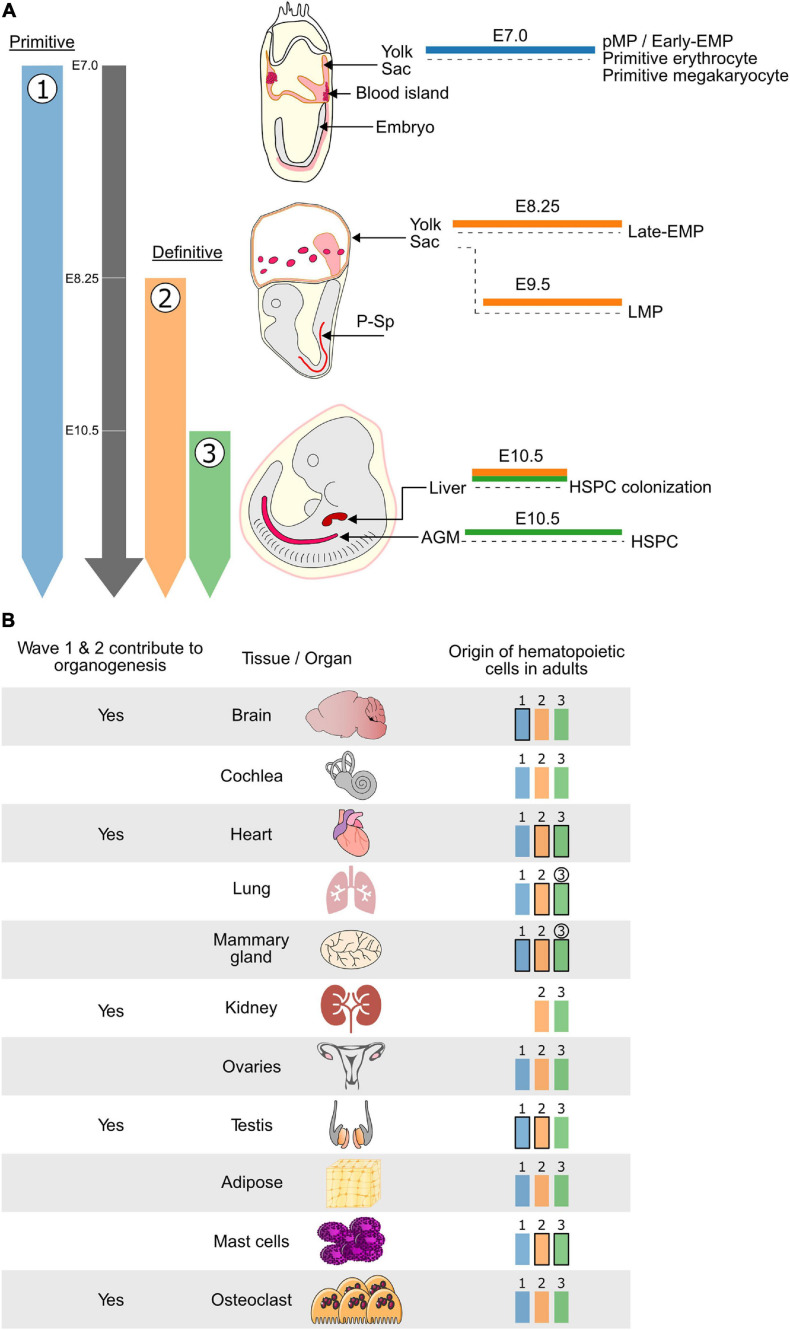
In mammals, the hematopoietic system is established during embryogenesis in three consecutive overlapping waves (Dzierzak and Bigas, 2018). In mice, the first wave, also termed primitive hematopoiesis, is initiated around embryonic day (E)7 and produces unipotent precursors that give rise to either primitive-erythrocytes, -megakaryocytes, or -macrophages (Palis et al., 1999; Tober et al., 2007). The primitive macrophage precursors have also been named primitive myeloid precursors (pMP). Furthermore, it has also been reported that the first wave may give rise to c-Myb independent erythroid–myeloid progenitors (early EMP) (Hoeffel et al., 2015). However, so far, macrophage (microglia) but not erythrocyte potential has been experimental confirmed for these so-called early EMP (Wittamer and Bertrand, 2020). Henceforth, we will refer to wave 1 myeloid cells as pMP/early EMP. The second wave of hematopoiesis marks the onset of definitive (erythroid) hematopoiesis and sees the emergence of both EMPs around E8.25 (late-EMPs) (McGrath et al., 2015; Palis, 2016), and lymphoid–myeloid progenitors (LMPs) (Adolfsson et al., 2005; Boiers et al., 2013) around E9.5. Around E10.5 the third wave generates both hematopoietic stem and progenitor cells (HSPC) (Figure 1A). The hematopoietic stem cells (HSCs) subsequently play a central role in maintaining the hematopoietic system for the lifetime of the organism (Medvinsky et al., 1993; Muller et al., 1994). Hematopoietic cells of all waves are generated from the mesoderm, which is known to give rise to both endothelial and hematopoietic lineages (Davidson and Zon, 2000; Dzierzak and Bigas, 2018). For the definitive hematopoietic waves (waves 2 and 3), it is now well established that hematopoiesis occurs via an endothelial-to-hematopoietic transition (EHT) from a specialized endothelial subpopulation known as hemogenic endothelium (HE) (Jaffredo et al., 1998; Zovein et al., 2008; Chen et al., 2009; Eilken et al., 2009; Lancrin et al., 2009; Boisset et al., 2010; Lacaud and Kouskoff, 2017; Garcia-Alegria et al., 2018; Ottersbach, 2019). The cellular origin of the first wave of primitive hematopoiesis is still disputed. It is unclear whether primitive hematopoiesis emerges directly from mesoderm, a hemangioblast, a HE, or another type of precursor (Amaya, 2013; Myers and Krieg, 2013). However, several recent studies suggest that primitive hematopoiesis (wave 1) is generated through a HE(-like) intermediate that has been termed hemogenic angioblast (Lancrin et al., 2009; Stefanska et al., 2017; Garcia-Alegria et al., 2018). Despite this potential common cellular origin, not all waves originate from the same anatomical site. The first two waves arise extra embryonically in the yolk sac (YS). In contrast, the third wave mainly takes place in the dorsal aorta within the aorta-gonad-mesonephros (AGM) region of the embryo, where HSC arise within so-called intra-aortic hematopoietic clusters (IAHC) (Boisset et al., 2010; Dzierzak and Bigas, 2018; Ottersbach, 2019). The HSC mature and amplify in the fetal liver (FL) before taking up residence in the bone marrow (BM).
FIGURE 1.
The hematopoietic system is established in developmental waves that differentially contribute to embryonic organogenesis and the adult hematopoietic system. (A) Schematic representing the timing and embryonic sites of hematopoietic activity during mouse development. The three waves of hematopoiesis are represented in Blue (wave 1: primitive erythrocytes, megakaryocytes, macrophages and Early EMP), Orange (wave 2: Late EMP and LMP) and Green (wave 3: HSPC). Wave 1 is known as the primitive wave while waves 2 and 3 constitute the definitive waves of hematopoiesis. Waves 1 and 2 which do not generate HSCs originate in the Yolk Sac while wave 3, which generates the HSPCs, is initiated in the AGM region of the embryo proper. Cells from both waves 2 and 3 can colonize the fetal liver where they can mature and expand before moving to their final destination. (B) (Left) HSC-independent macrophages which originate from the first two hematopoietic waves have been shown to play important roles during embryonic development of several tissues and organs. (Right) In addition to the wave 3 (Green) HSC-derived hematopoietic cells, the HSC-independent hematopoietic cells generated during wave 1 (Blue) and 2 (Orange) persist, to varying degrees, in adult tissues and organs. Waves that contribute most of the hematopoietic cells are depicted in bold and waves for which the contribution increases overtime are circled. Blue: hematopoietic wave 1 (primitive), Orange: hematopoietic wave 2 (Late-EMP/LMP), Green: hematopoietic wave 3 (HSC). E, embryonic day; Mφ, macrophage; HSC, hematopoietic stem cell; EMP, erythroid–myeloid progenitor; LMP, lymphoid–myeloid progenitor; HSPC, hematopoietic stem and progenitor cell; P-Sp, para-aortic splanchnopleura; AGM, aorta-gonad-mesonephros.
Initially, the YS waves of hematopoiesis were thought to provide an essential but transient blood supply for the embryo destined to be replaced by HSC-derived cells. However, there is accumulating evidence that HSC-independent blood cells can make significant contributions to the adult hematopoietic system. Furthermore, it is also becoming evident that in the embryo, the HSC-independent hematopoietic waves can play essential roles beyond erythrocyte-mediated oxygen exchange and early immune surveillance. These roles have been mainly assigned to myeloid-derived populations that have been shown to play crucial roles in embryonic organogenesis.
Here we provide a concise overview of the recent findings obtained in mouse models regarding the contribution of HSC-independent hematopoiesis to embryonic organogenesis and the adult hematopoietic system.
Resolving the Role and Origin of Hematopoietic Populations
The overlapping and transient nature of the different hematopoietic waves makes it challenging to determine their individual contributions towards organogenesis and the adult hematopoietic system. Transgenic mouse knockout (KO) and fate mapping models have been instrumental in shaping our current understanding of the contribution of the different hematopoietic waves towards the hematopoietic system (Table 1). KO models (via gene deletion/mutation or lineage-specific activation of diphtheria toxin) provide functional information for specific populations, while lineage tracing models (typically using fluorescent proteins) highlight the contribution of specific populations. However, the perfect model to delineate the hematopoietic waves does not exist, and it is important to consider the target cell type, wave specificity and labeling efficiency of the models used when interpreting results.
TABLE 1.
Mouse model for studying the contribution of different hematopoietic waves to hematopoietic system.
| Knockout models | Function | Affected wave | Major impact on hematopoietic waves | References | ||
| Csf1r (Dai et al., 2002) | Early/Late-EMP differentiation | 1, 2 | Lack of early EMP and reduced late-EMP contribution to TRM | Ginhoux et al., 2010; Hoeffel et al., 2012 | ||
| Csf1 (null mutation) (Marks and Lane, 1976) | Early EMP differentiation | 1 | Reduced early EMP contribution to TRM | Cecchini et al., 1994; Kondo and Duncan, 2009; Easley-Neal et al., 2019 | ||
| Myb (Mucenski et al., 1991) | Late-EMP and HSC differentiation | 2, 3 | Lack of late-EMP and HSC contribution to TRM | Schulz et al., 2012; Hoeffel et al., 2015 | ||
| Nur77 (Lee et al., 1995) | BM monocytes differentiation | Reduced BM monocytes/macrophages contribution to TRM | Hanna et al., 2011 | |||
| Plvap (Rantakari et al., 2015) | FL macrophage migration | Reduced FL monocytes/macrophages contribution to TRM | Rantakari et al., 2016 | |||
| Ccr2 (Hanna et al., 2011) | BM monocytes/macrophages migration | Reduced BM monocytes/macrophages contribution to TRM | Boring et al., 1997 | |||
| KitL (Ding et al., 2012) | Late-EMP and HSC maintenance | 2, 3 | Reduced late-EMP and HSC contribution to TRM | Ajami et al., 2007; Azzoni et al., 2018 | ||
| Cx3cr1 (Jung et al., 2000) | Mediates monocyte retention in the BM | None | Imai et al., 1997; Jacquelin et al., 2013 | |||
| Pu.1 (Scott et al., 1994; McKercher et al., 1996) | YS myeloid differentiation and HSC maintenance | 1, 2, 3 | Lack of EMP and HSC contribution to TRM | Scott et al., 1994; Olson et al., 1995; McKercher et al., 1996; Kim et al., 2004; Iwasaki et al., 2005; Kierdorf et al., 2013 | ||
| Runx1 (Okuda et al., 1996; Wang et al., 1996a; North et al., 1999) | Master regulator of hematopoiesis, expressed from HE onwards | 1, 2, 3 | Lack of EMP and HSC | Okuda et al., 1996; Wang et al., 1996a; North et al., 1999 | ||
| Cbfβ (Sasaki et al., 1996; Wang et al., 1996b; Niki et al., 1997) | Essential RUNX1 co-factor | 1, 2, 3 | Lack of EMP and HSC | Sasaki et al., 1996; Wang et al., 1996b; Niki et al., 1997 | ||
| Cbfβ (Tie2-Cbfβ) (Miller et al., 2002) | Essential RUNX1 co-factor | 1, 2, 3 | Lack of HSC | Chen et al., 2011 | ||
| Cbfβ (Ly6a-Cbfβ) (Chen et al., 2011) | Lack of EMP | Chen et al., 2011 | ||||
| Constitutive fate mapping models | Relevance |
Labeling efficiency (assay time point) |
References | |||
| LMP (Wave 2) | pMP/Early EMP (Wave 1) | Late-EMP (Wave 2) | HSC (Wave 3) | |||
| S100a4 (Bhowmick et al., 2004) (Transgene) | Active in FL monocytes and not in FL macrophages | Not done | ∼20% (5wo) | 64.5 ± 6.7% (5wo) | Not done | Hoeffel et al., 2015 |
| Not done | ∼20% (Adult) | ∼60% (Adult) | ∼100% (Adult) | Hashimoto et al., 2013 | ||
| Flt3 (Benz et al., 2008) (KI) | Active in HSC progeny | Not done | Not done | Not done | ∼80% (NB) | Hoeffel et al., 2015 |
| Not done | Not done | ∼20% (Adult) | ∼80% (Adult) | Hashimoto et al., 2013 | ||
| Not done | <2% (P8 onwards) | 10–30% (P8 onwards) | ∼80% (P8 onwards) | Gomez Perdiguero et al., 2015 | ||
| Not done | Not done | 10–20% (4wo) | ∼80% (4wo) | Schulz et al., 2012 | ||
| Ms4a3 (Liu et al., 2019) (KI) | Active in GMP | Not done | None (NB onwards) | Trace (NB onwards) | BM GMP: 68.7 ± 1.58% (Adult) | Liu et al., 2019 |
| Tnfrs11a (Maeda et al., 2012) (KI) | Essential for osteoclast development. Tracks YS progenitors. | Not done | ∼80% (E14.5) | ∼90% (6wo) | ∼10% (E14.5 onwards) | Mass et al., 2016 |
| Tnfrs11a (Percin et al., 2018) (KI) | Not done | Not done | ∼90% (Adult) | None (Adult) | Percin et al., 2018 | |
| Rag1 (McCormack et al., 2003) (KI) | Lymphoid-specific activity | ∼100% (E14.5) | Trace (E14.5) | <5% (E14.5) | Not done | Boiers et al., 2013 |
| Inducible fate mapping models | Relevance | Pulse timepoint |
Labeling efficiency (assay time point) |
References | ||
| pMP/Early EMP (Wave 1) | Late-EMP (Wave 2) | HSC (Wave 3) | ||||
| Csf1r (Qian et al., 2011) (Transgene) | Mainly a myeloid marker | E8.5 | 63.2 ± 5.6% (E13.5) | Trace (E13.5) | Not done | Hoeffel et al., 2015 |
| ∼60% (E11.5) | Marked (E12.5) | Not done | Gomez Perdiguero et al., 2015 | |||
| Cx3cr1 (Yona et al., 2013) (KI) | Mainly a myeloid marker | E9.0 | ∼40% (E16.0); ∼30% (NB onwards) | Trace (6wo onwards) | Trace (6wo) | Hagemeyer et al., 2016 |
| Runx1 (Samokhvalov et al., 2007) (KI; driven by P2/Runx1b promoter) | Master regulator of hematopoiesis, expressed from HE onwards | E7.25-E7.5 | ∼30% (E10.5) | <5% (8wo) | <3% (8wo) | Ginhoux et al., 2010 |
| Not done | Not done | none (9–12mo) | Samokhvalov et al., 2007 | |||
| ∼30% (E10.5); ∼20% (E13.5) | Not done | Not done | Hoeffel et al., 2012 | |||
| E7.5 | 22.2 ± 0.9% (E13.5) | < 5% (E13.5); <10% (E16.5) | Trace (E13.5) | Hoeffel et al., 2015 | ||
| Not done | ∼10% (8wo) | ∼10% (8wo) | Ginhoux et al., 2010 | |||
| Not done | Not done | 1–10% (9–12mo) | Samokhvalov et al., 2007 | |||
| Not done | ∼12.5% (8wo) | ∼7.5% (8wo) | Hoeffel et al., 2012 | |||
| E8.5 | ∼15% (E13.5); <5% (E16.5) | ∼25% (E13.5); ∼30% (E16.5) | 1–3% (E11.5); <5% (E13.5) | Hoeffel et al., 2015 | ||
| Not done | ∼30% (8wo) | ∼30% (8wo) | Ginhoux et al., 2010 | |||
| Not done | Not done | 1–50% (9–12mo) | Samokhvalov et al., 2007 | |||
| Not done | ∼30% (8wo) | ∼30% (8wo) | Hoeffel et al., 2012 | |||
| E9.5 | <5% (E13.5) | ∼15% (E13.5) | ∼30% (E13.5) | Hoeffel et al., 2015 | ||
| Not done | ∼20% (8wo) | ∼30% (8wo) | Ginhoux et al., 2010 | |||
| Not done | Not done | 50–100% (9–12mo) | Samokhvalov et al., 2007 | |||
| Not done | ∼25% (8wo) | ∼30% (8wo) | Hoeffel et al., 2012 | |||
| Tie2 (Gomez Perdiguero et al., 2015) (KI) | Endothelial marker (including HE) also expressed in subset of HSC and myeloid cells | E7.5 | ∼60% (E12.5) | ∼40% (6–8wo) | ∼40% (E12.5) | Gomez Perdiguero et al., 2015 |
| E8.5 | ∼30% (E12.5) | ∼20% (6–8wo) | ∼75% (E12.5) | |||
| E9.5 | trace (E12.5) | ∼10% (6–8wo) | ∼80% (E12.5) | |||
| E10.5 | none (E12.5) | ∼5% (6–8wo) | ∼40% (E12.5) | |||
| Kit (Sheng et al., 2015) (KI) | Express in early HSPC and YS HE cells | E7.5 | ∼70% (E13.5) | Trace (E13.5) | Trace (6wo) | Sheng et al., 2015 |
| E8.5 | ∼70% (E13.5) | ∼40% (E13.5) | ∼60% (6wo) | |||
| E9.5 | ∼50% (6wo) | ∼50% (6wo) | ∼40% (6wo) | |||
| Cdh5 (Sorensen et al., 2009) (Transgene) | Endothelial marker (including HE) | E7.5 | ∼80% (E10.5); ∼90% (E14.5) | ∼80% (E10.5) | <10% (E14.5) | Gentek et al., 2018a |
| E10.5 | Trace (E14.5) | Not done | ∼80% (E14.5) | |||
EMP, erythroid–myeloid progenitors; HSC, hematopoietic stem cells; TRM, tissue-resident macrophages; FL, fetal liver; BM, bone marrow; pMP, primitive myeloid precursors; HSPC, hematopoietic stem and progenitors; YS, yolk sac; LMP, lymphomyeloid progenitors; GMP, granulocyte-macrophage progenitors; HE, hemogenic endothelium; KI, knock-in; NB, new born; wo, week old; mo, month old.
Knockout mouse models have demonstrated specific dependencies of (wave-specific) hematopoietic populations on distinct transcription factors and signaling pathways. Csf1r (Colony Stimulating Factor 1 Receptor, cytokine receptor) KO mainly disrupts the early EMP differentiation (wave 1) and to a lesser extent the late-EMP differentiation (wave 2) (Dai et al., 2002). Although these mice are viable, they display drastically reduced levels of microglia and YS macrophages (Ginhoux et al., 2010; Hoeffel et al., 2012). Csf1 (Marks and Lane, 1976) (Colony Stimulating Factor 1, cytokine) null mice display a similar but milder phenotype, with varying degrees of microglia and YS macrophages depletion, due to partial compensatory effects of the alternative CSF1R ligand IL-34 (Wiktor-Jedrzejczak et al., 1990; Cecchini et al., 1994; Kondo and Duncan, 2009; Greter et al., 2012; Wang et al., 2012; Easley-Neal et al., 2019). Myb (Mucenski et al., 1991) (MYB Proto-Oncogene, transcription factor) KO disrupts late-EMP and HSC differentiation (wave 2 and wave 3) and results in anemia and embryonic lethality around E15.5 (Schulz et al., 2012; Hoeffel et al., 2015). A similar phenotype is observed in KitL (KIT Ligand, cytokine) KO mice which die perinatally (Ding et al., 2012). Nur77 (Lee et al., 1995) (Nuclear Receptor Subfamily 4 Group A Member 1, nuclear receptor) KO is viable but lacks circulating monocytes due to disrupted BM HSC differentiation (Hanna et al., 2011). Disrupting cell migration has also emerged as a useful strategy. Plvap (Rantakari et al., 2015) (Plasmalemma Vesicle Associated Protein, membrane protein) and CCR2 (Boring et al., 1997) (C-C Motif Chemokine Receptor 2, chemokine receptor) KOs are viable but respectively show impaired FL and BM monocyte migration (Rantakari et al., 2016). Cx3cr1 (Jung et al., 2000) (C-X3-C Motif Chemokine Receptor 1, chemokine receptor) KO is viable but shows impairment of leukocyte migration (Imai et al., 1997; Jacquelin et al., 2013) regardless of their wave of origin. There are several other models which disrupt all waves of hematopoietic development. Pu.1 (Scott et al., 1994; McKercher et al., 1996) (Spi-1 Proto-Oncogene, transcription factor) KO mice have defective YS myelopoiesis and HSC maintenance and die shortly after birth (Olson et al., 1995; Kim et al., 2004; Kierdorf et al., 2013). Deletion of Runx1 (Okuda et al., 1996; Wang et al., 1996a; North et al., 1999) (RUNX Family Transcription Factor 1, transcription factor) or its essential co-factor Cbfβ (Sasaki et al., 1996; Wang et al., 1996b; Niki et al., 1997) (Core-Binding Factor Subunit Beta, transcription factor) is embryonically lethal and results in the complete absence of hematopoiesis apart from primitive erythroid cells. These KOs are useful when coupled with targeted approaches. For example, the Cbfβ KO model can be used to deplete EMP or HSC by combination with respectively Ly6a-Cbfβ or Tie2-Cbfβ rescue alleles (Chen et al., 2011).
Constitutive lineage tracing models rely on lineage-specific promoter activity to drive Cre recombinase expression, which in turn irreversibly activates or deletes a target gene (Hoess and Abremski, 1984; Sauer and Henderson, 1988). Such models have been established to trace long term lineage contribution of HSC- and YS-derived hematopoietic cells. Flt3-Cre, Ms4a3-Cre, S100a4-Cre predominantly track HSC progeny, albeit with several restrictions. Flt3-Cre (Schulz et al., 2012; Hashimoto et al., 2013; Gomez Perdiguero et al., 2015; Hoeffel et al., 2015) and S100a4-Cre (Hashimoto et al., 2013; Hoeffel et al., 2015) mark the majority of HSC-derived cells (>80%). However, both also mark some YS-derived hematopoietic cells (Table 1). In contrast, Ms4a3-Cre does not mark any YS cells from the first or second wave and only marks HSC-derived GMPs (∼70%) (Liu et al., 2019). The Tnfrs11a-Cre model is currently best suited to track YS hematopoiesis with little (Maeda et al., 2012; Mass et al., 2016) or no (Percin et al., 2018) HSC labeling. However, this model cannot distinguish the two YS waves of hematopoiesis. Currently, the only option to track LMP progeny is the Rag1-Cre fate mapping model (Boiers et al., 2013) which marks all FL B and T cells alongside a small number of myeloid cells.
Inducible tracing models [tamoxifen-inducible Cre-mediated recombination (Metzger et al., 1995; Feil et al., 1997)] add an extra layer of specificity that can overcome certain limitations of the constitutive models. This approach allows not only for reporter activation or gene deletion in specific cell types but also during a defined developmental time window. The latter has allowed for the specific marking of the first hematopoietic wave in the YS (pMP/early EMP) using multiple models (Table 1). In this context, a caveat of the Csf1r-Mer-iCre-Mer and Cx3cr1-CreER based systems is that they only label myeloid progeny (Gomez Perdiguero et al., 2015; Hoeffel et al., 2015; Hagemeyer et al., 2016). In contrast, Tie2-Mer-iCre-Mer, Kit-Mer-Cre-Mer, Runx1-Mer-Cre-Mer, and Cdh5-CreERT2 provide less restricted marking. Distinguishing progeny from late-EMP (wave 2) and HSC (wave 3) is still challenging, as illustrated in Table 1 (Samokhvalov et al., 2007; Ginhoux et al., 2010; Hoeffel et al., 2012, 2015; Gentek et al., 2018a).
HSC-Independent Hematopoietic Cells Contribute to the Adult Hematopoietic System
In the adult hematopoietic system, several hematopoietic populations have been shown to consist of cells with an HSC-independent embryonic origin (Figure 1B). This has been best studied for tissue-resident macrophages (TRM), which were traditionally thought to be continuously replenished by BM HSC-derived monocytes. However, this view was challenged by the discovery of radiation-resistant and self-repopulating Langerhans cells (Merad et al., 2002), microglia (Ajami et al., 2007; Ginhoux et al., 2010), and alveolar macrophages (Guilliams et al., 2013; Hashimoto et al., 2013; Jakubzick et al., 2013) in BM transplantation and parabiosis studies. Subsequent lineage tracing studies have convincingly demonstrated that microglia are the progeny of myeloid cells produced during the first wave of hematopoiesis. Csf1r-Mer-iCre-Mer, Cx3cr1-CreER, Runx1-Mer-Cre-Mer, and Kit-Mer-Cre-Mer lineage tracing models all support the pMP/early EMP origin of microglia (Gomez Perdiguero et al., 2015; Hoeffel et al., 2015; Sheng et al., 2015; Hagemeyer et al., 2016). This has been further strengthened by the analysis of KitL and c-Myb KO models in which the number of late-EMP and HSC (waves 2 and 3) is drastically decreased, while the microglia population remains unaffected (Ding et al., 2012; Schulz et al., 2012; Azzoni et al., 2018).
It is now widely accepted that TRM populations do not have a unified common origin. Some tissues retain and maintain YS-derived cells while in others they are replaced or co-exist with BM-HSC-derived cells (for review Ginhoux and Guilliams, 2016; Mass, 2018; Wittamer and Bertrand, 2020). Below, we highlight recent findings concerning the persistence of YS-derived embryonic hematopoietic cells in adults.
HSC-Independent Origin of Adult TRM
Alveolar macrophages (AM) and interstitial macrophages (IM) are two major subsets of lung TRM (Lehnert et al., 1985; Liegeois et al., 2018). Around E10.5, YS pMPs/early EMPs (Tan and Krasnow, 2016) seed in the primordial lung buds. Parabiosis, adoptive transfer, and lineage tracing experiments have shown that a subset of pMP/early EMP-derived IM (Hoeffel et al., 2012, 2015; Guilliams et al., 2013; Gomez Perdiguero et al., 2015; Tan and Krasnow, 2016; Liu et al., 2019) and AM (van de Laar et al., 2016; Li et al., 2020) persist into adulthood. The functional significance of these sub-populations, if any, is currently unknown. However, it has been shown that FL monocyte-derived TRM possess enhanced mitochondrial respiratory and glycolytic capacity (Li et al., 2020) versus their HSC-independent counterparts.
Adipose tissue macrophages (ATM), are the most abundant immune cells in adipose tissues and play a prominent role in regulating inflammation and insulin sensitivity (Russo and Lumeng, 2018). ATM, which can proliferate and self-renew, are partially derived from embryonic HSC-independent cells (E9.0 pulsed Cx3Cr1-CreER) (Hassnain Waqas et al., 2017; Waqas et al., 2017). Further studies using the Ms4a3-Cre fate-mapping model, which traces BM-HSC monocyte-derived cells (Jaitin et al., 2019; Liu et al., 2019), demonstrated that half of the white adipose tissue ATM is Ms4a3 positive under normal physiological conditions. Interestingly, a high-fat diet increases the proportion of HSC-derived monocytes within ATM population (Jaitin et al., 2019).
The Ms4a3-Cre fate-mapping model has also revealed that adult renal macrophages (RM) are of mixed origin (Schulz et al., 2012; Epelman et al., 2014; Gomez Perdiguero et al., 2015; Hagemeyer et al., 2016; Mass et al., 2016; Liu et al., 2019; Munro et al., 2019). Furthermore, YS-derived RM (E9.5 pulsed Cx3cr1-CreER) are more proliferative than their HSC-derived counterpart and their contribution to the kidney TRM population expands with age (Ide et al., 2020).
Osteoclasts are multinucleated myeloid cells that resorb bone tissue and are critical for the development, repair, and remodeling of the skeleton (Udagawa et al., 1990; Takahashi et al., 1994). The HSC-independent origin of osteoclast was first suggested by ex vivo co-culture experiments (Thesingh, 1986) and has been recently confirmed in vivo using Csf1r-Mer-iCre-Mer and Cx3cr1-CreER models (Jacome-Galarza et al., 2019; Yahara et al., 2020). Early/late EMP-derived osteoclasts not only persist but also expand in the adult BM (Yahara et al., 2020). Furthermore, a subpopulation of early/late EMP-derived osteoclasts was found to reside in the spleen. Interestingly, this spleen population can fuse with BM-derived monocytes creating long-lived osteoclast syncytia with a mixed origin (Jacome-Galarza et al., 2019; Yahara et al., 2020).
Based on CD206 and MHCII expression, three testis subpopulations of TRM can be distinguished. These populations possess different levels of phagocytic capacity (Lokka et al., 2020). Tracing (E8.5 pulsed Csf1r-Mer-iCre-Mer, E13.5 pulsed Cx3cr1-CreER) and KO (Ccr2, Nur77, Plvap) studies have shown that all three waves of hematopoiesis contribute to adult testis TRM. Strikingly, antibody-based macrophage depletion experiments have demonstrated that adult BM-derived cells play no part in testis TRM maintenance (Lokka et al., 2020).
Finally characterization of Csf1 KOs, the Csf1r-EGFP constitutive tracing model, and E8.5 pulse-labeled Csf1r-Mer-iCre-Mer mice point to an HSC-independent origin for part of the TRM in adult cochlea (Kishimoto et al., 2019), ovaries (Jokela et al., 2020), and mammary glands (Gouon-Evans et al., 2000; Jappinen et al., 2019; Stewart et al., 2019).
Beyond Tissue Resident-Macrophages: Mast- and Lymphoid-Cells
Mast cells (MC) can be classified into two groups. Connective tissue MC (CTMC) populate the skin, tongue, trachea, esophagus, adipose tissues, and peritoneal- and pleural cavities while mucosal MC (MMC) are found in the gut and respiratory mucosa. The exclusive BM HSC-derived (van Furth and Cohn, 1968) origin of MC was first challenged by transplantation assays that showed BM only minimally contributes to MC repopulation in MC-depleted hosts (Kitamura et al., 1977, 1978). Subsequent fate-mapping studies using Csf1r-Mer-iCre-Mer and Runx1-Mer-Cre-Mer suggested that the majority of MMC are derived from HSC, whereas CTMC are largely derived from the HSC-independent EMP (Li Z. et al., 2018). Interestingly, EMP-derived and HSC-derived MC have distinct transcriptional profiles suggesting distinct biological functions (Gentek et al., 2018a; Li Z. et al., 2018). If the CTMCs derived from HSC-independent cells can persist in significant numbers in the adult is unclear. Csf1r-Mer-iCre-Mer and Runx1-Mer-Cre-Mer fate-mapping studies suggest that they can, while Cdh5-CreERT2 based lineage tracing suggests a mostly fetal HSC origin of adult CTMC (Gentek et al., 2018a). These contradictory findings highlight that data from fate-mapping models should be interpreted with caution and that currently, no model can definitively distinguish the progeny of late-EMP from fetal HSC.
Finally, YS-derived lymphoid cells have also been found to persist into adulthood. Early B and T-cells [B1a (Yoshimoto et al., 2011; Kobayashi et al., 2014) and γδ T (Boiers et al., 2013; Gentek et al., 2018b) cells], a primary source of innate immunity in early embryo development (Yoshimoto et al., 2012), persist into adulthood and remain functionally distinct from their HSC-derived counterparts. The ontogeny and contribution of these YS-derived lymphocytes has been reviewed previously (Yamane, 2018; Ghosn et al., 2019). Finally, the existence of EMP-derived NK cells, possessing a potent degranulation response, has been reported recently (Dege et al., 2020). This is particularly striking as NK cells are considered to be of lymphoid origin. However, it is unclear whether these EMP-derived NK cells are part of the myeloid lineage or if these findings have revealed lymphoid potential in EMP. Similarly, it is not clear to what extent EMP-derived NK cells persist into adulthood (Sun et al., 2011; Wu et al., 2014; Corat et al., 2017; Schlums et al., 2017).
HSC-Independent Macrophages Participate in Embryonic Organogenesis
The role of macrophages in tissue remodeling is an exciting field of ongoing research (Hoeffel and Ginhoux, 2018; Wittamer and Bertrand, 2020). The discovery of adult YS-derived TRM populations with an M2-like non-inflammatory phenotype, associated with wound healing and tissue repair, hints at potential roles in embryonic organogenesis (Takahashi et al., 1998; Rae et al., 2007; Pucci et al., 2009; Fantin et al., 2010; DeFalco et al., 2014; Italiani and Boraschi, 2014; Munro et al., 2019; Shigeta et al., 2019). However, identifying unique and specific roles of HSC-independent cells is complicated by the fact that embryonic organ development spans across all hematopoietic waves. Below we highlight the instances where specific roles for HSC-independent macrophages have been identified (Figure 1B).
HSC-Independent Embryonic Macrophages Guide Vascular Network Organization in Developing Organs
Vascular networks are established by tip- and stalk- endothelial cells. Endothelial tip-cells, guided by vascular endothelial growth factor (VEGF) gradients, drive the direction of the vessel while the endothelial stalk-cells follow and establish the vessel lumen (Herbert and Stainier, 2011). During embryonic organogenesis, macrophages have been shown to play an essential role in organizing endothelial networks. A role for HSC-independent macrophages in blood vessel anastomosis was first described in detail in the mouse embryonic hindbrain where it is entirely dependent on pMP/early EMP-derived macrophages (Fantin et al., 2010; Rymo et al., 2011). These macrophages invade the brain in a CSF1-dependent manner. Subsequently, upon brain vascularization, the macrophages closely associate with tip-endothelial cells. Macrophage depletion in the brain [Pu.1 (Scott et al., 1994; McKercher et al., 1996) and Csf1 KO (Wiktor-Jedrzejczak et al., 1990; Cecchini et al., 1994)], but not specific depletion of FL-derived macrophages (Lysm-Cre-mediated diphtheria toxin) (Clausen et al., 1999), significantly reduces the number of vessel intersections and thereby limits the overall complexity of the brain vascular network (Fantin et al., 2010; Rymo et al., 2011).
Hematopoietic stem cells-independent macrophages also play a role in kidney and testis vascular network formation. In mouse embryos, the mesonephros (a temporary kidney structure) and the gonads (which will give rise to the testis in males) are established near the extending nephric ducts around E9. Further extension of the nephric duct results in the generation of uretic buds and the metanephros (precursor to the adult kidneys) around E10-E11.5 (Takasato and Little, 2015). Proliferating primitive pMP/early EMP-derived macrophages (E7.5 pulsed Csf1r-Mer-iCre-Mer) are first detected in the gonadal region around E10.5 (DeFalco et al., 2014). By E11.5-E13.5, these macrophages closely associate with and engulf EC of the mesonephros vascular plexus and testis vasculature. Depletion of fetal macrophages (Cx3cr1-Cre-mediated diphtheria toxin) results in an enlarged mesonephros vascular plexus, reduced migration of EC into the testis, and impaired development of the coelomic artery. Blood vessels start entering the metanephros between E11.5-E12, and YS-derived macrophages are consistently found perivascular at developing vascular fronts (Rae et al., 2007; Hoeffel et al., 2015; Munro et al., 2019). Analysis of macrophage depleted E12.5 kidney explants (anti-CSF1R depletion) showed increased numbers of unconnected endothelial structures and a reduction in vascular network size, consistent with a role for macrophages in vessel anastomosis.
The developing heart harbors macrophages derived from both the HSC-independent and HSC-dependent hematopoietic waves (Ccr2-GFP, Cx3cr1-GFP) (Leid et al., 2016). HSC-independent macrophages (E7.5 pulsed Rosa26-tdCsf1r-MerCre) appear in the heart around E12.5 and predominantly populate the myocardium where they accumulate near perfused coronary vessels. Genetic depletion of macrophages (Csf1op/op) results in retarded primitive coronary plexus development. However, specific depletion of HSC-derived macrophages (Ccr2 KO) does not affect primitive coronary plexus development, indicating that HSC-independent macrophages are responsible for the modulation of the myocardial vascular network.
Finally, recent RNA-seq of a human Hofbauer cells (Zulu et al., 2019), a fetus-derived macrophage population found in the placenta, suggests that they may play a role in angiogenesis and remodeling (Thomas et al., 2021). Although Hofbauer cells have also been identified in mice (Takahashi et al., 1991), their role has not yet been investigated in vivo.
It is tempting to postulate a generalized role for HSC-independent macrophages in the establishment of vascular networks during embryonic organogenesis. In support of this, HSC-independent macrophages have a similar role in organizing vascular networks in zebrafish, independently of specific organs (Fantin et al., 2010).
HSC-Independent Macrophages Directly Support Organogenesis
Hematopoietic stem cells-independent macrophages have also been directly implicated in embryonic organ development. The central nervous system is arguably one of the best-studied systems in this context, with YS-derived microglia playing a role in multiple perinatal brain developmental events (Li and Barres, 2018; Low and Ginhoux, 2018). Around E14.5, microglia accumulate near developing axonal tracts and their genetic (Pu.1 KO)/antibody-based depletion (anti-CSF1R) or their inappropriate activation (E13.5 lipopolysaccharide maternal immune activation), affects the development of interneuronal connections and dopaminergic axon outgrowth (Squarzoni et al., 2014). Similarly, in the peripheral nervous system, macrophages/microglia are found in close contact with developing sensory neurons in dorsal root ganglia from E11. Genetic (Pu.1 KO) or antibody-based depletion (anti-CSF1R) of these macrophages alters the developmental trajectory of the sensory neurons (Angelim et al., 2018).
During gonad development, macrophages associate with and engulf mislocated germ (E10.5–E11.5) and Sertoli (E12.5) cells, and their absence (Cx3cr1-Cre/diphtheria toxin) results in irregularly branched and shortened testis cords (DeFalco et al., 2014). Furthermore, depletion of macrophages during embryogenesis (Csf1op/op, anti-CSF1R depletion) but not postnatally (Ccr2 KO, anti-CSF1 depletion at birth) results in impaired spermatogenesis after birth (Lokka et al., 2020). Similar observations have been made in kidney development where the clearance of rostral nephrogenic cells and uretic bud formation are delayed in the absence of YS-macrophages (Cx3cr1-Cre/diphtheria toxin) (Munro et al., 2019). These results suggest that embryonic macrophages are participating in gonad/testis and kidney development.
Yolk sac-derived osteoclasts are essential for normal skeletal development in the embryo and their absence (Csf1r KO model and Csf1-Cre-mediated Tnfrsf11a KO) disrupts tooth eruption, skull formation, long bone formation, and their timely hematopoietic colonization (Yoshida et al., 1990; Dougall et al., 1999; Dai et al., 2002; Jacome-Galarza et al., 2019). This phenotype is not observed when HSC-derived macrophages are deleted (Flt3-Cre-mediated and Csf1r KO) (Jacome-Galarza et al., 2019).
Finally, heart development also depends on HSC-independent macrophages which, interestingly, originate locally from HE cells populating the endocardium (Nakano et al., 2013; Yzaguirre and Speck, 2016; Shigeta et al., 2019). An important phase in heart development is the establishment and remodeling of the heart valves which starts around E9.5 and concludes after birth. Specific depletion of endocardial macrophages (Nfatc-Cre-mediated Csf1r KO) demonstrated that they are essential for heart valve development and that macrophages of other sources cannot compensate for their loss (Shigeta et al., 2019).
HSC-Independent Macrophages Support HSC Formation
Arguably the most striking function of HSC-independent macrophages is that they can affect HSC ontogeny. This has been studied in detail in zebrafish. HSCs generated in the dorsal aorta of zebrafish enter the circulation via the postcardinal vein (PCV) (Bertrand et al., 2010; Kissa and Herbomel, 2010; Lam et al., 2010). This requires newly formed HSC to traverse the mesenchyme separating the two vessels. Primitive macrophages accumulate in this subaortic mesenchyme and, via metalloproteinases mediated extracellular matrix degradation, create tracks for the HSC to enter the subaortic mesenchyme. These primitive macrophages then join the PCV from where they migrate to the zebrafish FL equivalent, known as the caudal hematopoietic tissue (CHT; Travnickova et al., 2015). Once the HSC reaches the CHT, a specific set of primitive VCAM+ macrophages (usher macrophages) interact with and “capture” passing HSPC and guide them into the CHT (Li D. et al., 2018).
In mice, HSC-independent macrophages also play an important role in HSC ontogeny. At E10.5, HSC-independent macrophages are found in close association with EC and IAHC in the AGM, where they possibly participate in moving KIT+ IAHC cells towards the aortic lumen (Mariani et al., 2019). The CXCL3 chemokine (expressed amongst others by HE and IAHC) is important for the macrophage accumulation in the AGM (Mariani et al., 2019). The deletion of its receptor, Cxcr3, results in increased numbers of macrophages in the YS and reduced numbers in the AGM. Reduction of the number of macrophages in the AGM, either by genetic (Cxcr3 KO) or chemical (clodronate and CSF1R inhibitor BLZ945) depletion, negatively affects HSC generation in the AGM (Mariani et al., 2019). Furthermore, both direct and indirect (transwell) co-culture experiments of AGM-derived aortic endothelial cells (including HE) with aortic macrophages result in an increase of the hematopoietic colony-forming capacity of the endothelial cells (Mariani et al., 2019). These results indicate an essential role for macrophage secreted factors in AGM EHT. RNA-seq of the aortic macrophages revealed that despite having an immune phenotype associated with anti-inflammatory or M2 type phenotype, they have a distinct pro-inflammatory transcriptome (Mariani et al., 2019). Currently, it is unclear if these macrophages promote emergence of all, or only subsets, of HSC.
Concluding Remarks
In the last decade, it has become clear that HSC-independent hematopoietic cells have previously unanticipated roles in both embryos and adults. They have been found to participate in organogenesis and persist in adults as distinct hematopoietic populations. There are however still many open questions about their exact role, origin, and contributions. In this context, the development of more precise and efficient genetic tracing models would be beneficial. Specifically, models that can efficiently differentiate wave 2 (late-EMP and LMP) from wave 3 (HSC) are needed.
It is also essential to acquire more detailed knowledge of the different hematopoietic waves, both mechanistically and in terms of their exact sites of origin. Indeed, the observation that heart-specific HE can give rise to a specialized population of macrophages, raises the question whether other specialized hematopoietic cells are produced in a site or organ-specific way. Both in the AGM and the YS, multiple sites of hematopoietic emergence have been described (Muller et al., 1994; Medvinsky and Dzierzak, 1996; de Bruijn et al., 2000; Li et al., 2012; Frame et al., 2016; Kasaai et al., 2017). Closer investigation of these known sites as well as the identification of new sites could reveal the existence of new, functionally unique, hematopoietic populations. Furthermore, understanding if and how the distinct hematopoietic cells generated by the different waves interact to optimize blood production is equally fascinating. Altogether such knowledge could provide cues to develop better strategies for in vitro generation of HSCs and/or specific blood lineages from embryonic- and induced pluripotent stem cells (ES and IPSC). IPSC generated from different cellular sources may be inherently primed towards specific hematopoietic lineages. Additionally, it may be beneficial to incorporate mature hematopoietic cells into in vitro blood production protocols. Along these lines, it has been recently reported that macrophages can support the in vitro production of mature enucleated erythroid cells (Lopez-Yrigoyen et al., 2019).
Finally, findings in animal model systems are starting to be confirmed in humans. Macrophages have been found to accumulate in the human AGM at the time of HSC formation (Travnickova et al., 2015), and single-cell sequencing indicates that human microglia are also derived from HSC-independent hematopoietic waves (Bian et al., 2020). In conclusion, it is now well established that HSC-independent hematopoiesis is essential for embryonic organogenesis and its progeny can, and does, persist after birth. This has opened up a new and fascinating field of hematopoietic research.
Author Contributions
WHN and ML wrote the manuscript. MZHF produced the figure. GL revised and edited the manuscript. All authors approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Renaud Mevel for critical reading of the manuscript. Due to space limitations we were, regrettably, unable to cite all manuscripts that have contributed to the discussed research topic.
Footnotes
Funding. The work in our laboratory is supported by Cancer Research UK C5759/A20971 (GL) and Blood Cancer UK 19014 (GL).
References
- Adolfsson J., Mansson R., Buza-Vidas N., Hultquist A., Liuba K., Jensen C. T., et al. (2005). Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell 121 295–306. 10.1016/j.cell.2005.02.013 [DOI] [PubMed] [Google Scholar]
- Ajami B., Bennett J. L., Krieger C., Tetzlaff W., Rossi F. M. (2007). Local self-renewal can sustain CNS microglia maintenance and function throughout adult life. Nat. Neurosci. 10 1538–1543. 10.1038/nn2014 [DOI] [PubMed] [Google Scholar]
- Amaya E. (2013). The hemangioblast: a state of competence. Blood 122 3853–3854. 10.1182/blood-2013-10-533075 [DOI] [PubMed] [Google Scholar]
- Angelim M., Maia L., Mouffle C., Ginhoux F., Low D., Amancio-Dos-Santos A., et al. (2018). Embryonic macrophages and microglia ablation alter the development of dorsal root ganglion sensory neurons in mouse embryos. Glia 66 2470–2486. 10.1002/glia.23499 [DOI] [PubMed] [Google Scholar]
- Azzoni E., Frontera V., McGrath K. E., Harman J., Carrelha J., Nerlov C., et al. (2018). Kit ligand has a critical role in mouse yolk sac and aorta-gonad-mesonephros hematopoiesis. EMBO Rep. 19:, e45477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz C., Martins V. C., Radtke F., Bleul C. C. (2008). The stream of precursors that colonizes the thymus proceeds selectively through the early T lineage precursor stage of T cell development. J. Exp. Med. 205 1187–1199. 10.1084/jem.20072168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand J. Y., Chi N. C., Santoso B., Teng S., Stainier D. Y., Traver D. (2010). Haematopoietic stem cells derive directly from aortic endothelium during development. Nature 464 108–111. 10.1038/nature08738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhowmick N. A., Chytil A., Plieth D., Gorska A. E., Dumont N., Shappell S., et al. (2004). TGF-beta signaling in fibroblasts modulates the oncogenic potential of adjacent epithelia. Science 303 848–851. 10.1126/science.1090922 [DOI] [PubMed] [Google Scholar]
- Bian Z., Gong Y., Huang T., Lee C. Z. W., Bian L., Bai Z., et al. (2020). Deciphering human macrophage development at single-cell resolution. Nature 582 571–576. 10.1038/s41586-020-2316-7 [DOI] [PubMed] [Google Scholar]
- Boiers C., Carrelha J., Lutteropp M., Luc S., Green J. C., Azzoni E., et al. (2013). Lymphomyeloid contribution of an immune-restricted progenitor emerging prior to definitive hematopoietic stem cells. Cell Stem Cell 13 535–548. 10.1016/j.stem.2013.08.012 [DOI] [PubMed] [Google Scholar]
- Boisset J. C., van Cappellen W., Andrieu-Soler C., Galjart N., Dzierzak E., Robin C. (2010). In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature 464 116–120. 10.1038/nature08764 [DOI] [PubMed] [Google Scholar]
- Boring L., Gosling J., Chensue S. W., Kunkel S. L., Farese R. V., Jr., Broxmeyer H. E., et al. (1997). Impaired monocyte migration and reduced type 1 (Th1) cytokine responses in C-C chemokine receptor 2 knockout mice. J. Clin. Invest. 100 2552–2561. 10.1172/jci119798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchini M. G., Dominguez M. G., Mocci S., Wetterwald A., Felix R., Fleisch H., et al. (1994). Role of colony stimulating factor-1 in the establishment and regulation of tissue macrophages during postnatal development of the mouse. Development 120 1357–1372. [DOI] [PubMed] [Google Scholar]
- Chen M. J., Li Y., De Obaldia M. E., Yang Q., Yzaguirre A. D., Yamada-Inagawa T., et al. (2011). Erythroid/myeloid progenitors and hematopoietic stem cells originate from distinct populations of endothelial cells. Cell Stem Cell 9 541–552. 10.1016/j.stem.2011.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M. J., Yokomizo T., Zeigler B. M., Dzierzak E., Speck N. A. (2009). Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature 457 887–891. 10.1038/nature07619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen B. E., Burkhardt C., Reith W., Renkawitz R., Forster I. (1999). Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic. Res. 8 265–277. [DOI] [PubMed] [Google Scholar]
- Corat M. A., Schlums H., Wu C., Theorell J., Espinoza D. A., Sellers S. E., et al. (2017). Acquired somatic mutations in PNH reveal long-term maintenance of adaptive NK cells independent of HSPCs. Blood 129 1940–1946. 10.1182/blood-2016-08-734285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X. M., Ryan G. R., Hapel A. J., Dominguez M. G., Russell R. G., Kapp S., et al. (2002). Targeted disruption of the mouse colony-stimulating factor 1 receptor gene results in osteopetrosis, mononuclear phagocyte deficiency, increased primitive progenitor cell frequencies, and reproductive defects. Blood 99 111–120. 10.1182/blood.v99.1.111 [DOI] [PubMed] [Google Scholar]
- Davidson A. J., Zon L. I. (2000). Turning mesoderm into blood: the formation of hematopoietic stem cells during embryogenesis. Curr. Top Dev. Biol. 50 45–60. 10.1016/s0070-2153(00)50003-9 [DOI] [PubMed] [Google Scholar]
- de Bruijn M. F., Speck N. A., Peeters M. C., Dzierzak E. (2000). Definitive hematopoietic stem cells first develop within the major arterial regions of the mouse embryo. EMBO J. 19 2465–2474. 10.1093/emboj/19.11.2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFalco T., Bhattacharya I., Williams A. V., Sams D. M., Capel B. (2014). Yolk-sac-derived macrophages regulate fetal testis vascularization and morphogenesis. Proc. Natl. Acad. Sci. U. S. A. 111 E2384–E2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dege C., Fegan K. H., Creamer J. P., Berrien-Elliott M. M., Luff S. A., Kim D., et al. (2020). Potently cytotoxic natural killer cells initially emerge from Erythro-myeloid progenitors during mammalian development. Dev. Cell 53 229–239e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Saunders T. L., Enikolopov G., Morrison S. J. (2012). Endothelial and perivascular cells maintain haematopoietic stem cells. Nature 481 457–462. 10.1038/nature10783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougall W. C., Glaccum M., Charrier K., Rohrbach K., Brasel K., De Smedt T., et al. (1999). RANK is essential for osteoclast and lymph node development. Genes Dev. 13 2412–2424. 10.1101/gad.13.18.2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzierzak E., Bigas A. (2018). Blood development: hematopoietic stem cell dependence and independence. Cell Stem Cell 22 639–651. 10.1016/j.stem.2018.04.015 [DOI] [PubMed] [Google Scholar]
- Easley-Neal C., Foreman O., Sharma N., Zarrin A. A., Weimer R. M. (2019). CSF1R Ligands IL-34 and CSF1 are differentially required for microglia development and maintenance in white and Gray matter brain regions. Front. Immunol. 10:2199. 10.3389/fimmu.2019.02199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilken H. M., Nishikawa S., Schroeder T. (2009). Continuous single-cell imaging of blood generation from haemogenic endothelium. Nature 457 896–900. 10.1038/nature07760 [DOI] [PubMed] [Google Scholar]
- Epelman S., Lavine K. J., Beaudin A. E., Sojka D. K., Carrero J. A., Calderon B., et al. (2014). Embryonic and adult-derived resident cardiac macrophages are maintained through distinct mechanisms at steady state and during inflammation. Immunity 40 91–104. 10.1016/j.immuni.2013.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantin A., Vieira J. M., Gestri G., Denti L., Schwarz Q., Prykhozhij S., et al. (2010). Tissue macrophages act as cellular chaperones for vascular anastomosis downstream of VEGF-mediated endothelial tip cell induction. Blood 116 829–840. 10.1182/blood-2009-12-257832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R., Wagner J., Metzger D., Chambon P. (1997). Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem. Biophys. Res. Commun. 237 752–757. 10.1006/bbrc.1997.7124 [DOI] [PubMed] [Google Scholar]
- Frame J. M., Fegan K. H., Conway S. J., McGrath K. E., Palis J. (2016). Definitive hematopoiesis in the yolk sac emerges from Wnt-responsive hemogenic endothelium independently of circulation and arterial identity. Stem Cells 34 431–444. 10.1002/stem.2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Alegria E., Menegatti S., Fadlullah M. Z. H., Menendez P., Lacaud G., Kouskoff V. (2018). Early human hemogenic endothelium generates primitive and definitive hematopoiesis In Vitro. Stem Cell Rep.orts 11 1061–1074. 10.1016/j.stemcr.2018.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentek R., Ghigo C., Hoeffel G., Bulle M. J., Msallam R., Gautier G., et al. (2018a). Hemogenic endothelial fate mapping reveals dual developmental origin of mast cells. Immunity 48 1160–1171e5. [DOI] [PubMed] [Google Scholar]
- Gentek R., Ghigo C., Hoeffel G., Jorquera A., Msallam R., Wienert S., et al. (2018b). Epidermal gammadelta T cells originate from yolk sac hematopoiesis and clonally self-renew in the adult. J. Exp. Med. 215 2994–3005. 10.1084/jem.20181206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosn E., Yoshimoto M., Nakauchi H., Weissman I. L., Herzenberg L. A. (2019). Hematopoietic stem cell-independent hematopoiesis and the origins of innate-like B lymphocytes. Development 146 dev170571. 10.1242/dev.170571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F., Greter M., Leboeuf M., Nandi S., See P., Gokhan S., et al. (2010). Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330 841–845. 10.1126/science.1194637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F., Guilliams M. (2016). Tissue-resident macrophage ontogeny and homeostasis. Immunity 44 439–449. 10.1016/j.immuni.2016.02.024 [DOI] [PubMed] [Google Scholar]
- Gomez Perdiguero E., Klapproth K., Schulz C., Busch K., Azzoni E., Crozet L., et al. (2015). Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature 518 547–551. 10.1038/nature13989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouon-Evans V., Rothenberg M. E., Pollard J. W. (2000). Postnatal mammary gland development requires macrophages and eosinophils. Development 127 2269–2282. [DOI] [PubMed] [Google Scholar]
- Greter M., Lelios I., Pelczar P., Hoeffel G., Price J., Leboeuf M., et al. (2012). Stroma-derived interleukin-34 controls the development and maintenance of langerhans cells and the maintenance of microglia. Immunity 37 1050–1060. 10.1016/j.immuni.2012.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilliams M., De Kleer I., Henri S., Post S., Vanhoutte L., De Prijck S., et al. (2013). Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J. Exp. Med. 210 1977–1992. 10.1084/jem.20131199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeyer N., Kierdorf K., Frenzel K., Xue J., Ringelhan M., Abdullah Z., et al. (2016). Transcriptome-based profiling of yolk sac-derived macrophages reveals a role for Irf8 in macrophage maturation. EMBO J. 35 1730–1744. 10.15252/embj.201693801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna R. N., Carlin L. M., Hubbeling H. G., Nackiewicz D., Green A. M., Punt J. A., et al. (2011). The transcription factor NR4A1 (Nur77) controls bone marrow differentiation and the survival of Ly6C- monocytes. Nat. Immunol. 12 778–785. 10.1038/ni.2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto D., Chow A., Noizat C., Teo P., Beasley M. B., Leboeuf M., et al. (2013). Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity 38 792–804. 10.1016/j.immuni.2013.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassnain Waqas S. F., Noble A., Hoang A. C., Ampem G., Popp M., Strauss S., et al. (2017). Adipose tissue macrophages develop from bone marrow-independent progenitors in Xenopus laevis and mouse. J. Leukoc. Biol. 102 845–855. 10.1189/jlb.1a0317-082rr [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert S. P., Stainier D. Y. (2011). Molecular control of endothelial cell behaviour during blood vessel morphogenesis. Nat. Rev. Mol. Cell Biol. 12 551–564. 10.1038/nrm3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffel G., Chen J., Lavin Y., Low D., Almeida F. F., See P., et al. (2015). C-Myb(+) erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages. Immunity 42 665–678. 10.1016/j.immuni.2015.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffel G., Ginhoux F. (2018). Fetal monocytes and the origins of tissue-resident macrophages. Cell Immunol. 330 5–15. 10.1016/j.cellimm.2018.01.001 [DOI] [PubMed] [Google Scholar]
- Hoeffel G., Wang Y., Greter M., See P., Teo P., Malleret B., et al. (2012). Adult Langerhans cells derive predominantly from embryonic fetal liver monocytes with a minor contribution of yolk sac-derived macrophages. J. Exp. Med. 209 1167–1181. 10.1084/jem.20120340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoess R. H., Abremski K. (1984). Interaction of the bacteriophage P1 recombinase Cre with the recombining site loxP. Proc. Natl. Acad. Sci. U. S. A. 81 1026–1029. 10.1073/pnas.81.4.1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide S., Yahara Y., Kobayashi Y., Strausser S. A., Ide K., Watwe A., et al. (2020). Yolk-sac-derived macrophages progressively expand in the mouse kidney with age. Elife 9:, e51756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai T., Hieshima K., Haskell C., Baba M., Nagira M., Nishimura M., et al. (1997). Identification and molecular characterization of fractalkine receptor CX3CR1, which mediates both leukocyte migration and adhesion. Cell 91 521–530. 10.1016/s0092-8674(00)80438-9 [DOI] [PubMed] [Google Scholar]
- Italiani P., Boraschi D. (2014). From monocytes to M1/M2 macrophages: phenotypical vs. functional differentiation. Front. Immunol. 5:514. 10.3389/fimmu.2014.00514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H., Somoza C., Shigematsu H., Duprez E. A., Iwasaki-Arai J., Mizuno S., et al. (2005). Distinctive and indispensable roles of PU.1 in maintenance of hematopoietic stem cells and their differentiation. Blood 106 1590–1600. 10.1182/blood-2005-03-0860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacome-Galarza C. E., Percin G. I., Muller J. T., Mass E., Lazarov T., Eitler J., et al. (2019). Developmental origin, functional maintenance and genetic rescue of osteoclasts. Nature 568 541–545. 10.1038/s41586-019-1105-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquelin S., Licata F., Dorgham K., Hermand P., Poupel L., Guyon E., et al. (2013). CX3CR1 reduces Ly6Chigh-monocyte motility within and release from the bone marrow after chemotherapy in mice. Blood 122 674–683. 10.1182/blood-2013-01-480749 [DOI] [PubMed] [Google Scholar]
- Jaffredo T., Gautier R., Eichmann A., Dieterlen-Lièvre F. (1998). Intraaortic hemopoietic cells are derived from endothelial cells during ontogeny. Development 22, 4574–4583. [DOI] [PubMed] [Google Scholar]
- Jaitin D. A., Adlung L., Thaiss C. A., Weiner A., Li B., Descamps H., et al. (2019). Lipid-associated macrophages control metabolic homeostasis in a Trem2-dependent manner. Cell 178 686–698e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubzick C., Gautier E. L., Gibbings S. L., Sojka D. K., Schlitzer A., Johnson T. E., et al. (2013). Minimal differentiation of classical monocytes as they survey steady-state tissues and transport antigen to lymph nodes. Immunity 39 599–610. 10.1016/j.immuni.2013.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jappinen N., Felix I., Lokka E., Tyystjarvi S., Pynttari A., Lahtela T., et al. (2019). Fetal-derived macrophages dominate in adult mammary glands. Nat. Commun. 10:, 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jokela H., Lokka E., Kiviranta M., Tyystjarvi S., Gerke H., Elima K., et al. (2020). Fetal-derived macrophages persist and sequentially maturate in ovaries after birth in mice. Eur. J. Immunol. 50 1500–1514. 10.1002/eji.202048531 [DOI] [PubMed] [Google Scholar]
- Jung S., Aliberti J., Graemmel P., Sunshine M. J., Kreutzberg G. W., Sher A., et al. (2000). Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol. Cell Biol. 20 4106–4114. 10.1128/mcb.20.11.4106-4114.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasaai B., Caolo V., Peacock H. M., Lehoux S., Gomez-Perdiguero E., Luttun A., et al. (2017). Erythro-myeloid progenitors can differentiate from endothelial cells and modulate embryonic vascular remodeling. Sci. Rep. 7:, 43817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierdorf K., Erny D., Goldmann T., Sander V., Schulz C., Perdiguero E. G., et al. (2013). Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat. Neurosci. 16 273–280. 10.1038/nn.3318 [DOI] [PubMed] [Google Scholar]
- Kim H. G., de Guzman C. G., Swindle C. S., Cotta C. V., Gartland L., Scott E. W., et al. (2004). The ETS family transcription factor PU.1 is necessary for the maintenance of fetal liver hematopoietic stem cells. Blood 104 3894–3900. 10.1182/blood-2002-08-2425 [DOI] [PubMed] [Google Scholar]
- Kishimoto I., Okano T., Nishimura K., Motohashi T., Omori K. (2019). Early development of resident macrophages in the mouse cochlea depends on yolk sac hematopoiesis. Front. Neurol. 10:1115. 10.3389/fneur.2019.01115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissa K., Herbomel P. (2010). Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature 464 112–115. 10.1038/nature08761 [DOI] [PubMed] [Google Scholar]
- Kitamura Y., Go S., Hatanaka K. (1978). Decrease of mast cells in W/Wv mice and their increase by bone marrow transplantation. Blood 52 447–452. 10.1182/blood.v52.2.447.bloodjournal522447 [DOI] [PubMed] [Google Scholar]
- Kitamura Y., Shimada M., Hatanaka K., Miyano Y. (1977). Development of mast cells from grafted bone marrow cells in irradiated mice. Nature 268 442–443. 10.1038/268442a0 [DOI] [PubMed] [Google Scholar]
- Kobayashi M., Shelley W. C., Seo W., Vemula S., Lin Y., Liu Y., et al. (2014). Functional B-1 progenitor cells are present in the hematopoietic stem cell-deficient embryo and depend on Cbfbeta for their development. Proc. Natl. Acad. Sci. U. S. A. 111 12151–12156. 10.1073/pnas.1407370111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y., Duncan I. D. (2009). Selective reduction in microglia density and function in the white matter of colony-stimulating factor-1-deficient mice. J. Neurosci. Res. 87 2686–2695. 10.1002/jnr.22096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacaud G., Kouskoff V. (2017). Hemangioblast, hemogenic endothelium, and primitive versus definitive hematopoiesis. Exp. Hematol. 49 19–24. 10.1016/j.exphem.2016.12.009 [DOI] [PubMed] [Google Scholar]
- Lam E. Y., Hall C. J., Crosier P. S., Crosier K. E., Flores M. V. (2010). Live imaging of Runx1 expression in the dorsal aorta tracks the emergence of blood progenitors from endothelial cells. Blood 116 909–914. 10.1182/blood-2010-01-264382 [DOI] [PubMed] [Google Scholar]
- Lancrin C., Sroczynska P., Stephenson C., Allen T., Kouskoff V., Lacaud G. (2009). The haemangioblast generates haematopoietic cells through a haemogenic endothelium stage. Nature 457 892–895. 10.1038/nature07679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. L., Wesselschmidt R. L., Linette G. P., Kanagawa O., Russell J. H., Milbrandt J. (1995). Unimpaired thymic and peripheral T cell death in mice lacking the nuclear receptor NGFI-B (Nur77). Science 269 532–535. 10.1126/science.7624775 [DOI] [PubMed] [Google Scholar]
- Lehnert B. E., Valdez Y. E., Holland L. M. (1985). Pulmonary macrophages: alveolar and interstitial populations. Exp. Lung. Res. 9 177–190. 10.3109/01902148509057522 [DOI] [PubMed] [Google Scholar]
- Leid J., Carrelha J., Boukarabila H., Epelman S., Jacobsen S. E., Lavine K. J. (2016). Primitive embryonic macrophages are required for coronary development and maturation. Circ. Res. 118 1498–1511. 10.1161/circresaha.115.308270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Xue W., Li M., Dong M., Wang J., Wang X., et al. (2018). VCAM-1(+) macrophages guide the homing of HSPCs to a vascular niche. Nature 564 119–124. 10.1038/s41586-018-0709-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Okreglicka K. M., Pohlmeier L. M., Schneider C., Kopf M. (2020). Fetal monocytes possess increased metabolic capacity and replace primitive macrophages in tissue macrophage development. EMBO J. 39:, e103205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Barres B. A. (2018). Microglia and macrophages in brain homeostasis and disease. Nat. Rev. Immunol. 18 225–242. 10.1038/nri.2017.125 [DOI] [PubMed] [Google Scholar]
- Li Z., Lan Y., He W., Chen D., Wang J., Zhou F., et al. (2012). Mouse embryonic head as a site for hematopoietic stem cell development. Cell Stem Cell 11 663–675. 10.1016/j.stem.2012.07.004 [DOI] [PubMed] [Google Scholar]
- Li Z., Liu S., Xu J., Zhang X., Han D., Liu J., et al. (2018). Adult connective tissue-resident mast cells originate from late Erythro-myeloid progenitors. Immunity 49 640–653e5. [DOI] [PubMed] [Google Scholar]
- Liegeois M., Legrand C., Desmet C. J., Marichal T., Bureau F. (2018). The interstitial macrophage: a long-neglected piece in the puzzle of lung immunity. Cell Immunol. 330 91–96. 10.1016/j.cellimm.2018.02.001 [DOI] [PubMed] [Google Scholar]
- Liu Z., Gu Y., Chakarov S., Bleriot C., Kwok I., Chen X., et al. (2019). Fate mapping via Ms4a3-expression history traces monocyte-derived cells. Cell 178 1509–1525e19. [DOI] [PubMed] [Google Scholar]
- Lokka E., Lintukorpi L., Cisneros-Montalvo S., Makela J. A., Tyystjarvi S., Ojasalo V., et al. (2020). Generation, localization and functions of macrophages during the development of testis. Nat. Commun. 11:, 4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Yrigoyen M., Yang C. T., Fidanza A., Cassetta L., Taylor A. H., McCahill A., et al. (2019). Genetic programming of macrophages generates an in vitro model for the human erythroid island niche. Nat. Commun. 10:, 881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low D., Ginhoux F. (2018). Recent advances in the understanding of microglial development and homeostasis. Cell Immunol. 330 68–78. 10.1016/j.cellimm.2018.01.004 [DOI] [PubMed] [Google Scholar]
- Maeda K., Kobayashi Y., Udagawa N., Uehara S., Ishihara A., Mizoguchi T., et al. (2012). Wnt5a-Ror2 signaling between osteoblast-lineage cells and osteoclast precursors enhances osteoclastogenesis. Nat. Med. 18 405–412. 10.1038/nm.2653 [DOI] [PubMed] [Google Scholar]
- Mariani S. A., Li Z., Rice S., Krieg C., Fragkogianni S., Robinson M., et al. (2019). Pro-inflammatory aorta-associated macrophages are involved in embryonic development of hematopoietic stem cells. Immunity 50 1439–1452e5. 10.1016/j.immuni.2019.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks S. C., Jr., Lane P. W. (1976). Osteopetrosis, a new recessive skeletal mutation on chromosome 12 of the mouse. J. Hered 67 11–18. 10.1093/oxfordjournals.jhered.a108657 [DOI] [PubMed] [Google Scholar]
- Mass E. (2018). Delineating the origins, developmental programs and homeostatic functions of tissue-resident macrophages. Int. Immunol. 30 493–501. 10.1093/intimm/dxy044 [DOI] [PubMed] [Google Scholar]
- Mass E., Ballesteros I., Farlik M., Halbritter F., Gunther P., Crozet L., et al. (2016). Specification of tissue-resident macrophages during organogenesis. Science 353:, aaf4238. 10.1126/science.aaf4238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack M. P., Forster A., Drynan L., Pannell R., Rabbitts T. H. (2003). The LMO2 T-cell oncogene is activated via chromosomal translocations or retroviral insertion during gene therapy but has no mandatory role in normal T-cell development. Mol. Cell Biol. 23 9003–9013. 10.1128/MCB.23.24.9003-9013.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath K. E., Frame J. M., Fegan K. H., Bowen J. R., Conway S. J., Catherman S. C., et al. (2015). Distinct sources of hematopoietic progenitors emerge before HSCs and provide functional blood cells in the mammalian embryo. Cell Rep. 11 1892–1904. 10.1016/j.celrep.2015.05.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKercher S. R., Torbett B. E., Anderson K. L., Henkel G. W., Vestal D. J., Baribault H., et al. (1996). Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 15 5647–5658. 10.1002/j.1460-2075.1996.tb00949.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvinsky A., Dzierzak E. (1996). Definitive hematopoiesis is autonomously initiated by the AGM region. Cell 86 897–906. 10.1016/S0092-8674(00)80165-8 [DOI] [PubMed] [Google Scholar]
- Medvinsky A. L., Samoylina N. L., Muller A. M., Dzierzak E. A. (1993). An early pre-liver intraembryonic source of CFU-S in the developing mouse. Nature 364 64–67. 10.1038/364064a0 [DOI] [PubMed] [Google Scholar]
- Merad M., Manz M. G., Karsunky H., Wagers A., Peters W., Charo I., et al. (2002). Langerhans cells renew in the skin throughout life under steady-state conditions. Nat. Immunol. 3 1135–1141. 10.1038/ni852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger D., Clifford J., Chiba H., Chambon P. (1995). Conditional site-specific recombination in mammalian cells using a ligand-dependent chimeric Cre recombinase. Proc. Natl. Acad. Sci. U. S. A. 92 6991–6995. 10.1073/pnas.92.15.6991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J., Horner A., Stacy T., Lowrey C., Lian J. B., Stein G., et al. (2002). The core-binding factor beta subunit is required for bone formation and hematopoietic maturation. Nat. Genet. 32 645–649. 10.1038/ng1049 [DOI] [PubMed] [Google Scholar]
- Mucenski M. L., McLain K., Kier A. B., Swerdlow S. H., Schreiner C. M., Miller T. A., et al. (1991). A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell 65 677–689. 10.1016/0092-8674(91)90099-K [DOI] [PubMed] [Google Scholar]
- Muller A. M., Medvinsky A., Strouboulis J., Grosveld F., Dzierzak E. (1994). Development of hematopoietic stem cell activity in the mouse embryo. Immunity 1 291–301. 10.1016/1074-7613(94)90081-7 [DOI] [PubMed] [Google Scholar]
- Munro D. A. D., Wineberg Y., Tarnick J., Vink C. S., Li Z., Pridans C., et al. (2019). Macrophages restrict the nephrogenic field and promote endothelial connections during kidney development. Elife 8:, e43271. 10.7554/eLife.43271.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers C. T., Krieg P. A. (2013). BMP-mediated specification of the erythroid lineage suppresses endothelial development in blood island precursors. Blood 122 3929–3939. 10.1182/blood-2013-03-490045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano H., Liu X., Arshi A., Nakashima Y., van Handel B., Sasidharan R., et al. (2013). Haemogenic endocardium contributes to transient definitive haematopoiesis. Nat. Commun. 4:, 1564. 10.1038/ncomms2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki M., Okada H., Takano H., Kuno J., Tani K., Hibino H., et al. (1997). Hematopoiesis in the fetal liver is impaired by targeted mutagenesis of a gene encoding a non-DNA binding subunit of the transcription factor, polyomavirus enhancer binding protein 2/core binding factor. Proc. Natl. Acad. Sci. U. S. A. 94 5697–5702. 10.1073/pnas.94.11.5697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- North T., Gu T. L., Stacy T., Wang Q., Howard L., Binder M., et al. (1999). Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development 126 2563–2575. [DOI] [PubMed] [Google Scholar]
- Okuda T., van Deursen J., Hiebert S. W., Grosveld G., Downing J. R. (1996). AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84 321–330. 10.1016/S0092-8674(00)80986-1 [DOI] [PubMed] [Google Scholar]
- Olson M. C., Scott E. W., Hack A. A., Su G. H., Tenen D. G., Singh H., et al. (1995). PU. 1 is not essential for early myeloid gene expression but is required for terminal myeloid differentiation. Immunity 3 703–714. 10.1016/1074-7613(95)90060-8 [DOI] [PubMed] [Google Scholar]
- Ottersbach K. (2019). Endothelial-to-haematopoietic transition: an update on the process of making blood. Biochem. Soc. Trans. 47 591–601. 10.1042/BST20180320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palis J. (2016). Hematopoietic stem cell-independent hematopoiesis: emergence of erythroid, megakaryocyte, and myeloid potential in the mammalian embryo. FEBS Lett. 590 3965–3974. 10.1002/1873-3468.12459 [DOI] [PubMed] [Google Scholar]
- Palis J., Robertson S., Kennedy M., Wall C., Keller G. (1999). Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development 126 5073–5084. [DOI] [PubMed] [Google Scholar]
- Percin G. I., Eitler J., Kranz A., Fu J., Pollard J. W., Naumann R., et al. (2018). CSF1R regulates the dendritic cell pool size in adult mice via embryo-derived tissue-resident macrophages. Nat. Commun. 9:, 5279. 10.1038/s41467-018-07685-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucci F., Venneri M. A., Biziato D., Nonis A., Moi D., Sica A., et al. (2009). gene signature shared by tumor-infiltrating Tie2-expressing monocytes, blood “resident” monocytes, and embryonic macrophages suggests common functions and developmental relationships. Blood 114 901–914. 10.1182/blood-2009-01-200931 [DOI] [PubMed] [Google Scholar]
- Qian B. Z., Li J., Zhang H., Kitamura T., Zhang J., Campion L. R., et al. (2011). CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature 475 222–225. 10.1038/nature10138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae F., Woods K., Sasmono T., Campanale N., Taylor D., Ovchinnikov D. A., et al. (2007). Characterisation and trophic functions of murine embryonic macrophages based upon the use of a Csf1r-EGFP transgene reporter. Dev. Biol. 308 232–246. 10.1016/j.ydbio.2007.05.027 [DOI] [PubMed] [Google Scholar]
- Rantakari P., Auvinen K., Jappinen N., Kapraali M., Valtonen J., Karikoski M., et al. (2015). The endothelial protein PLVAP in lymphatics controls the entry of lymphocytes and antigens into lymph nodes. Nat. Immunol. 16 386–396. 10.1038/ni.3101 [DOI] [PubMed] [Google Scholar]
- Rantakari P., Jappinen N., Lokka E., Mokkala E., Gerke H., Peuhu E., et al. (2016). Fetal liver endothelium regulates the seeding of tissue-resident macrophages. Nature 538 392–396. 10.1038/nature19814 [DOI] [PubMed] [Google Scholar]
- Russo L., Lumeng C. N. (2018). Properties and functions of adipose tissue macrophages in obesity. Immunology 155 407–417. 10.1111/imm.13002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymo S. F., Gerhardt H., Wolfhagen Sand F., Lang R., Uv A., Betsholtz C. (2011). A two-way communication between microglial cells and angiogenic sprouts regulates angiogenesis in aortic ring cultures. PLoS One 6:e15846. 10.1371/journal.pone.0015846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samokhvalov I. M., Samokhvalova N. I., Nishikawa S. (2007). Cell tracing shows the contribution of the yolk sac to adult haematopoiesis. Nature 446 1056–1061. 10.1038/nature05725 [DOI] [PubMed] [Google Scholar]
- Sasaki K., Yagi H., Bronson R. T., Tominaga K., Matsunashi T., Deguchi K., et al. (1996). Absence of fetal liver hematopoiesis in mice deficient in transcriptional coactivator core binding factor beta. Proc. Natl. Acad. Sci. U. S. A. 93 12359–12363. 10.1073/pnas.93.22.12359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer B., Henderson N. (1988). Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc. Natl. Acad. Sci. U. S. A. 85 5166–5170. 10.1073/pnas.85.14.5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlums H., Jung M., Han H., Theorell J., Bigley V., Chiang S. C., et al. (2017). Adaptive NK cells can persist in patients with GATA2 mutation depleted of stem and progenitor cells. Blood 129 1927–1939. 10.1182/blood-2016-08-734236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz C., Gomez Perdiguero E., Chorro L., Szabo-Rogers H., Cagnard N., Kierdorf K., et al. (2012). A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 336 86–90. 10.1126/science.1219179 [DOI] [PubMed] [Google Scholar]
- Scott E. W., Simon M. C., Anastasi J., Singh H. (1994). Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science 265 1573–1577. 10.1126/science.8079170 [DOI] [PubMed] [Google Scholar]
- Sheng J., Ruedl C., Karjalainen K. (2015). Most tissue-resident macrophages except microglia are derived from fetal hematopoietic stem cells. Immunity 43 382–393. 10.1016/j.immuni.2015.07.016 [DOI] [PubMed] [Google Scholar]
- Shigeta A., Huang V., Zuo J., Besada R., Nakashima Y., Lu Y., et al. (2019). Endocardially derived macrophages are essential for valvular remodeling. Dev. Cell 48 617–630e3. 10.1016/j.devcel.2019.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen I., Adams R. H., Gossler A. (2009). DLL1-mediated Notch activation regulates endothelial identity in mouse fetal arteries. Blood 113 5680–5688. 10.1182/blood-2008-08-174508 [DOI] [PubMed] [Google Scholar]
- Squarzoni P., Oller G., Hoeffel G., Pont-Lezica L., Rostaing P., Low D., et al. (2014). Microglia modulate wiring of the embryonic forebrain. Cell Rep. 8 1271–1279. 10.1016/j.celrep.2014.07.042 [DOI] [PubMed] [Google Scholar]
- Stefanska M., Batta K., Patel R., Florkowska M., Kouskoff V., Lacaud G. (2017). Primitive erythrocytes are generated from hemogenic endothelial cells. Sci. Rep. 7:, 6401. 10.1038/s41598-017-06627-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart T. A., Hughes K., Hume D. A., Davis F. M. (2019). Developmental stage-specific distribution of macrophages in mouse mammary gland. Front. Cell Dev. Biol. 7:250. 10.3389/fcell.2019.00250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J. C., Beilke J. N., Bezman N. A., Lanier L. L. (2011). Homeostatic proliferation generates long-lived natural killer cells that respond against viral infection. J. Exp. Med. 208 357–368. 10.1084/jem.20100479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Donovan M. J., Rogers R. A., Ezekowitz R. A. (1998). Distribution of murine mannose receptor expression from early embryogenesis through to adulthood. Cell Tissue Res. 292 311–323. 10.1007/s004410051062 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Naito M., Katabuchi H., Higashi K. (1991). Development, differentiation, and maturation of macrophages in the chorionic villi of mouse placenta with special reference to the origin of Hofbauer cells. J. Leukoc. Biol. 50 57–68. 10.1002/jlb.50.1.57 [DOI] [PubMed] [Google Scholar]
- Takahashi N., Udagawa N., Tanaka S., Murakami H., Owan I., Tamura T., et al. (1994). Postmitotic osteoclast precursors are mononuclear cells which express macrophage-associated phenotypes. Dev. Biol. 163 212–221. 10.1006/dbio.1994.1137 [DOI] [PubMed] [Google Scholar]
- Takasato M., Little M. H. (2015). The origin of the mammalian kidney: implications for recreating the kidney in vitro. Development 142 1937–1947. 10.1242/dev.104802 [DOI] [PubMed] [Google Scholar]
- Tan S. Y., Krasnow M. A. (2016). Developmental origin of lung macrophage diversity. Development 143 1318–1327. 10.1242/dev.129122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thesingh C. W. (1986). Formation sites and distribution of osteoclast progenitor cells during the ontogeny of the mouse. Dev. Biol. 117 127–134. 10.1016/0012-1606(86)90355-6 [DOI] [PubMed] [Google Scholar]
- Thomas J. R., Appios A., Zhao X., Dutkiewicz R., Donde M., Lee C. Y. C., et al. (2021). Phenotypic and functional characterization of first-trimester human placental macrophages, Hofbauer cells. J. Exp. Med. 218:, e20200891. 10.1084/jem.20200891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tober J., Koniski A., McGrath K. E., Vemishetti R., Emerson R., de Mesy-Bentley K. K., et al. (2007). The megakaryocyte lineage originates from hemangioblast precursors and is an integral component both of primitive and of definitive hematopoiesis. Blood 109 1433–1441. 10.1182/blood-2006-06-031898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travnickova J., Tran Chau V., Julien E., Mateos-Langerak J., Gonzalez C., Lelievre E., et al. (2015). Primitive macrophages control HSPC mobilization and definitive haematopoiesis. Nat. Commun. 6:, 6227. 10.1038/ncomms7227 [DOI] [PubMed] [Google Scholar]
- Udagawa N., Takahashi N., Akatsu T., Tanaka H., Sasaki T., Nishihara T., et al. (1990). Origin of osteoclasts: mature monocytes and macrophages are capable of differentiating into osteoclasts under a suitable microenvironment prepared by bone marrow-derived stromal cells. Proc. Natl. Acad. Sci. U. S. A. 87 7260–7264. 10.1073/pnas.87.18.7260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Laar L., Saelens W., De Prijck S., Martens L., Scott C. L., Van Isterdael G., et al. (2016). Yolk sac macrophages, fetal liver, and adult monocytes can colonize an empty niche and develop into functional tissue-resident macrophages. Immunity 44 755–768. 10.1016/j.immuni.2016.02.017 [DOI] [PubMed] [Google Scholar]
- van Furth R., Cohn Z. A. (1968). The origin and kinetics of mononuclear phagocytes. J. Exp. Med. 128 415–435. 10.1084/jem.128.3.415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Stacy T., Binder M., Marin-Padilla M., Sharpe A. H., Speck N. A. (1996a). Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc. Natl. Acad. Sci. U. S. A. 93 3444–3449. 10.1073/pnas.93.8.3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Stacy T., Miller J. D., Lewis A. F., Gu T. L., Huang X., et al. (1996b). The CBFbeta subunit is essential for CBFalpha2 (AML1) function in vivo. Cell 87 697–708. 10.1016/S0092-8674(00)81389-6 [DOI] [PubMed] [Google Scholar]
- Wang Y., Szretter K. J., Vermi W., Gilfillan S., Rossini C., Cella M., et al. (2012). IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia. Nat. Immunol. 13 753–760. 10.1038/ni.2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waqas S. F. H., Hoang A. C., Lin Y. T., Ampem G., Azegrouz H., Balogh L., et al. (2017). Neuropeptide FF increases M2 activation and self-renewal of adipose tissue macrophages. J. Clin. Invest. 127:, 3559. 10.1172/JCI95841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiktor-Jedrzejczak W., Bartocci A., Ferrante A. W., Jr., Ahmed-Ansari A., Sell K. W., Pollard J. W., et al. (1990). Total absence of colony-stimulating factor 1 in the macrophage-deficient osteopetrotic (op/op) mouse. Proc. Natl. Acad. Sci. U. S. A. 87 4828–4832. 10.1073/pnas.87.12.4828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittamer V., Bertrand J. Y. (2020). Yolk sac hematopoiesis: does it contribute to the adult hematopoietic system? Cell Mol. Life Sci. 77 4081–4091. 10.1007/s00018-020-03527-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Li B., Lu R., Koelle S. J., Yang Y., Jares A., et al. (2014). Clonal tracking of rhesus macaque hematopoiesis highlights a distinct lineage origin for natural killer cells. Cell Stem Cell 14 486–499. 10.1016/j.stem.2014.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahara Y., Barrientos T., Tang Y. J., Puviindran V., Nadesan P., Zhang H., et al. (2020). Erythromyeloid progenitors give rise to a population of osteoclasts that contribute to bone homeostasis and repair. Nat. Cell Biol. 22 49–59. 10.1038/s41556-019-0437-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane T. (2018). Mouse yolk sac hematopoiesis. Front. Cell Dev. Biol. 6:80. 10.3389/fcell.2018.00080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yona S., Kim K. W., Wolf Y., Mildner A., Varol D., Breker M., et al. (2013). Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 38 79–91. 10.1016/j.immuni.2012.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H., Hayashi S., Kunisada T., Ogawa M., Nishikawa S., Okamura H., et al. (1990). The murine mutation osteopetrosis is in the coding region of the macrophage colony stimulating factor gene. Nature 345 442–444. 10.1038/345442a0 [DOI] [PubMed] [Google Scholar]
- Yoshimoto M., Montecino-Rodriguez E., Ferkowicz M. J., Porayette P., Shelley W. C., Conway S. J., et al. (2011). Embryonic day 9 yolk sac and intra-embryonic hemogenic endothelium independently generate a B-1 and marginal zone progenitor lacking B-2 potential. Proc. Natl. Acad. Sci. U. S. A. 108 1468–1473. 10.1073/pnas.1015841108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto M., Porayette P., Glosson N. L., Conway S. J., Carlesso N., Cardoso A. A., et al. (2012). Autonomous murine T-cell progenitor production in the extra-embryonic yolk sac before HSC emergence. Blood 119 5706–5714. 10.1182/blood-2011-12-397489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yzaguirre A. D., Speck N. A. (2016). Insights into blood cell formation from hemogenic endothelium in lesser-known anatomic sites. Dev. Dyn. 245 1011–1028. 10.1002/dvdy.24430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zovein A. C., Hofmann J. J., Lynch M., French W. J., Turlo K. A., Yang Y., et al. (2008). Fate tracing reveals the endothelial origin of hematopoietic stem cells. Cell Stem Cell 3 625–636. 10.1016/j.stem.2008.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zulu M. Z., Martinez F. O., Gordon S., Gray C. M. (2019). The elusive role of placental macrophages: the Hofbauer cell. J. Innate. Immun. 11 447–456. 10.1159/000497416 [DOI] [PMC free article] [PubMed] [Google Scholar]