Abstract
In vivo genome editing tools, such as those based on CRISPR, have been increasingly utilized in both basic and translational neuroscience research. There are currently nine in vivo non-CNS genome editing therapies in clinical trials, and the pre-clinical pipeline of major biotechnology companies demonstrate that this number will continue to grow. Several biotechnology companies commercializing in vivo genome editing and modification technologies are developing therapies for CNS disorders with accompanying large partnering deals. In this review, the authors discuss the current genome editing and modification therapy pipeline and those in development to treat CNS disorders. The authors also discuss the technical and commercial limitations to translation of these same therapies and potential avenues to overcome these hurdles.
Keywords: genome editing, neurological disease, CRISPR (clustered regularly interspaced short palindromic repeat)/Cas9 (CRISPR associated protein 9)-mediated genome editing, biotech companies, translational pipeline
Introduction
Genome Editing and Modification in the CNS
The possibility to introduce any desired modification in specific sites of the genome of cells, genome editing, is a longstanding ambition in biotechnology and molecular medicine and is now making precision medicine a real possibility for the treatment of genetic diseases.
A big step forward in the generation of new genome editing tools was the observation that the introduction of a double-strand-break (DSB) in the desired genomic site can strongly enhance the integration of a desired donor DNA sequence (Rouet et al., 1994). The discovery of zinc finger proteins (ZFP) dramatically changed the genome editing scenario as they are eukaryotic zinc ion-regulated small protein motifs able to bind DNA in a sequence-specific manner (Klug and Rhodes, 1987; Kim et al., 1996). When fused to transcriptional activator or repressors (ZFP-TFs), they modulate the expression of endogenous genes (Rebar et al., 2002). The next advance, the transcription activator-like effector (TALE) proteins from Xanthomonas bacteria, specifically recognize one single base instead of three bases (Boch et al., 2009; Moscou and Bogdanove, 2009), and can work as programmable nuclease, called TALEN (Li et al., 2011; Miller et al., 2011; Zhang et al., 2011). However, the cloning and protein engineering work for ZFNs and TALENs is complex. It requires two different effectors to cut each DNA strand as FokI works as a dimer and only laboratories with extensive expertise in molecular biology could take advantage of those techniques, thus not broadly adopted by the scientific community.
Conversely, the latest CRISPR tools are much simpler and more flexible to use and require minimal molecular skills to exploit them successfully in multiple genome editing strategies (Anzalone et al., 2020). The main simplification is that DNA target specificity is ensured by short nucleic acid sequences (short guide RNA, sgRNA) rather than protein modules and their cloning is thus faster and cheaper. Beside the classic Cas9 which induce genomic DSBs favoring gene inactivation or gene correction, the nickase Cas9 is the basic platform for the base editor tools that make direct C to T or A to G conversion at the target site (Komor et al., 2016; Nishida et al., 2016; Gaudelli et al., 2017). In addition, nuclease defective Cas9 (dCas9) can become a scaffold to which different effectors can be attached to deliver specific protein functions to genomic sites, such as transcriptional activators (CRISPRa), inhibitors (CRISPRi), epigenetic factors and histone modifiers (Shi et al., 2004; Mali et al., 2013; Perez-Pinera et al., 2013; Qi et al., 2013; Chavez et al., 2015; Hilton et al., 2015; Konermann et al., 2015; Thakore et al., 2015; Amabile et al., 2016; Liu et al., 2016; McDonald et al., 2016; Morita et al., 2016; Vojta et al., 2016; Xu et al., 2016; Xiong et al., 2017; Matharu and Ahituv, 2020). The more recent fusion of the dCas9 to a modified reverse transcriptase makes possible to rewrite new genetic information into a specified DNA site; in this case the prime editing exploits a guide RNA (prime editing guide RNA, pegRNA) that provides specificity and encodes the edit to be introduced at the same time (Anzalone et al., 2019).
Advances in genome editing strategies encouraged researchers to exploit those tools for preclinical studies even in the CNS, that has always represented a major challenge. The main reason is that neurons are post-mitotic cells and HDR is mainly restricted to cycling cells, specifically in S and G2, when homologous recombination between sister chromatids normally occurs (Lin et al., 2014). However, homology-independent targeted integration (HITI) and other similar systems have been recently described as improved NHEJ-based homology-independent strategy for targeted transgene integration, still based on CRISPR/Cas9, but also efficient in post-mitotic cells (Suzuki et al., 2016).
Preclinical Studies Using Genome Editing to Correct Neurological Diseases
ZFN and TALEN- based therapies have already been used in preclinical studies for several pathologies (Li et al., 2020). However, the technical limitations described above make these technologies challenging to be brought forward for treating CNS pathologies.
CRISPR-based genome editing to rescue neurological diseases has been recently tested in animal and in vitro human models. Several neurodevelopmental and neurodegenerative diseases have been tackled including Epilepsy, Autism Spectrum Disorder (ASD), Frontotemporal Dementia (FTD), Alzheimer’s, Huntington’s and Parkinson’s diseases (Yang et al., 2017; Kantor et al., 2018; Krishnan et al., 2020; Stepanichev, 2020; Turner et al., 2020; Vermilyea et al., 2020; Zhou et al., 2020). These approaches are based on either genome editing, silencing, or regulation, and they have been employed to overcome the limitations of classical gene therapy approaches.
Gene silencing and deletion of pathogenic repeats have been tested in animal and human models of Angelman Syndrome (AS), Fragile X syndrome (FXS), FTD and Alzheimer’s (Park et al., 2015, 2019; Xie et al., 2016; Gyorgy et al., 2018; Krishnan et al., 2020; Wolter et al., 2020). Although the results of these studies are promising showing rescue of the pathologies in vitro and in vivo, there are still preclinical tests to be performed in order to translate these approaches to the clinic. Some examples are the downstream effects of silencing a gene in a fully developed mature brain (Shitik et al., 2020), or the potential side effects of AAV integration in the DSBs (Wolter et al., 2020). Single hit mediated gene silencing of a pathogenic allele, as well as deletion of aberrant repeats, could have less impact on the immunological system. The disadvantages to these approaches are two-fold: the potential CRISPR-mediated off-target effects resulting in permanent changes to the genome and the delivery of these tools to patients. There is a massive ongoing effort to find better bioinformatic tools to predict off-target effects and in developing new delivery strategies to widely target CNS (Cota-Coronado et al., 2019).
CRISPRa, for example, has been already tested in in vivo animal models of neurodevelopmental and acquired epilepsies, and obesity (Matharu et al., 2019; Colasante et al., 2020a,b; Yamagata et al., 2020). These studies showed, for the first time, a long-lasting effect of endogenous gene upregulation either rescuing haploinsufficiency or modifying neuronal properties to treat pathological symptoms. Although there is great potential for effectively treating several CNS pathologies with CRISPRa, some hurdles for using this technology in humans still has to be addressed. These include the potential immunological response of the brain to long-term expression of dCAS9 (Crudele and Chamberlain, 2018) and a more efficient delivery (e.g., using smaller dCAS9).
On the other hand, the possibility of using genome editing to correct the pathological mutations is still an attractive prerogative of the CRISPR systems. Although the post-mitotic neuronal genome is difficult to modify, some recent techniques (Suzuki et al., 2016; Nishiyama et al., 2017; Yeh et al., 2019) allow gene modification in mature brain cells. In these studies, a successful insertion of new DNA in the genome of neurons has been shown to mildly rescue pathological conditions. Indeed, the main limitation is the low efficiency of the modifications that need to be addressed and improved before moving toward the clinic.
Furthermore, CRISPR base editors and CRISPR prime editing hold the potential to further improve the treatments for neurological diseases (Gaudelli et al., 2017; Anzalone et al., 2019; Duarte and Deglon, 2020). They are still behind in the preclinical pipeline due to the difficulties in the delivery of these constructs and the validation of the off-target effects. However, their ability to correct single mutations (Base editor) or longer DNA sequences (Prime) with high efficiency, without indels, is promising for future translational treatments. Recently, it has been shown that CRISPR base editing can be successfully employed in vivo to treat Amyotrophic Lateral Sclerosis (ALS) (Lim et al., 2020), splitting the base editors with an intein-mediated trans-splicing system, but the efficiency is still low.
Overall, all these different CRISPR-based technologies have been tested either in animal or in vitro human models, revealing an unprecedented potential for translation. The next steps are the refinement of the tools, in terms of delivery, efficiency and off-target effects in order to enable the development of an extensive commercial pipeline.
Discussion
The Current Therapeutic Pipeline to in Human Genome Editing
Despite the achievements in preclinical studies, therapeutic use of genome editing in the CNS is still in its infancy. Even though there are nine active clinical trials using in vivo genome editing1 (Hirakawa et al., 2020) none of them are to treat a CNS indication. Yet the potential of these technologies to treat CNS disorders is of great interest to pharmaceutical companies as seen from their pre-clinical pipelines (Table 1).
TABLE 1.
Companies with in vivo genome editing and regulation assets at preclinical stage [search on November 10, 2020].
| Company | Genome editing system | Approach | Affected Tissue/Organ/Therapeutic Area | Indication | Delivery | Target or Gene Delivered |
| Sangamo Therapeutics/Biogen | ZFP-TF | Gene downregulation | CNS | Tauopathies | AAV | Tau |
| Sangamo Therapeutics/Biogen | ZFP-TF | Gene downregulation | CNS | Synucleinopathies (Inc., Parkinson’s Disease) | AAV | Alpha-synuclein |
| Sangamo Therapeutics/Biogen | ZFP-TF | Gene downregulation | PNS and/or CNS | Neurological (Inc., a neuromuscular indication) | AAV | Unknown |
| Sangamo Therapeutics/Pfizer | ZFP-TF | Gene downregulation | CNS | ALS/FTD | AAV | Mutant C9ORF72 |
| Sangamo Therapeutics/Takeda | ZFP-TF | Gene downregulation | CNS | Huntington’s Disease | AAV | Mutant HTT |
| Sangamo Therapeutics | ZFP-TF | Gene downregulation | CNS | Prion | AAV | Unknown |
| Sangamo Therapeutics/Novartis | ZFP-TF | Gene downregulation | CNS | Neurodevelopmental Disorders (Inc., Autism Spectrum Disorder) | AAV | Unknown |
| Editas Medicine/Asklepios Biopharmaceutical | CRISPR/Cas9 | Unknown | PNS and/or CNS | Neurological | AAV | Unknown |
| Beam Therapeutics | CRISPR/dCas (base editor) | Correction or Silencing | CNS | Unknown | AAV | Unknown |
Public pre-clinical pipelines of biopharmaceutical companies using in vivo genome editing to treat CNS disorders. Queried the pipelines of genome editing companies and used search engines to find companies with publicly available information on its pipelines. Companies using genome editing and regulation technologies that do not publicize their pipelines are not shown.
Among the several biotech companies involved in genome editing and regulation, Sangamo Therapeutics (Sangamo), Editas Medicine and Beam Therapeutics are the only ones that have publicly stated their pipelines on in vivo genome editing therapies for the CNS. Interestingly, Beam Therapeutics, which uses CRISPR/Cas9-based base editing, has an undisclosed CNS project.
Sangamo and Biogen are co-developing up to another ten therapeutic candidates targeting a neurological indication using ZFP-TF, with one of the assets targeting a neuromuscular indication, whereas Editas Medicine and Asklepios BioPharmaceutical (AskBio) are developing a therapy utilizing AAV-CRISPR-Cas9. AskBio was acquired by Bayer in October 2020, positioning this large pharmaceutical company in the gene therapy and genome editing space2. Sangamo has disclosed that its pipeline includes therapies for tauopathies, synucleinopathies, Huntington’s disease, neurodevelopmental disorders, prion disease and ALS/FTD3.
On the other hand, other genome editing companies such as CRISPR Therapeutics, Intellia Therapeutics and Precision Biosciences have not entered the CNS space or have not yet disclosed their candidates.
Although there is great potential of prime editing it is too early for this technology to be added to commercial pipelines. Indeed, there are currently no publicized therapy assets using prime editing. To be noted, Beam Therapeutics licensed the IP for prime editing from Prime Medicine4.
Why are there still only few in vivo genome editing therapeutic programmes for the CNS? This is due to technical and commercial limitations. Biotechnology companies seek the indications with the largest patient population that are not adequately treated by current therapies. In this equation, companies also compute the risk of failure at a technical level. Delivering in vivo genome editing therapies to the CNS is technically harder than to other organ systems, which increases the risk of failure. In addition, CNS indications often have a more complex etiology than oncology or monogenic disorders in other organs. This can incentivize companies to invest in therapies that can target indications that have better defined genotype-phenotype relationships, such as oncology or monogenic disorders in the retina or liver.
The potential of off-target effects also plays an important role in the risk-aversion to the investment in CNS in vivo genome editing therapeutics. A permanent off-target change to the DNA could lead to material consequences for the patient. It is possible that biotechnology companies are waiting for increased specificity of CRISPR and other tools before targeting the CNS. In fact, seven out of nine disclosed in vivo genome editing therapies treating CNS indications (Table 1) are using tools acting on transcriptional regulation which leads to transient changes in neuronal gene expression, rather than genome modifications.
In summary, overcoming some technical limitations that are specific for CNS, such as temporal and spatial control of tool expression, delivery and targetability (Wang et al., 2020); as well as accuracy and efficacy (Zhang et al., 2015) could increase the interest of biotechnology companies toward in vivo genome editing for CNS disorders, and therefore also increase investments and number of therapies in the clinic.
Partnerships
Biopharmaceutical companies developing in vivo genome editing therapies and advanced therapeutics are partnering with other biotechnology companies in order to make progress on some of those key limitations. For example, the partnership with AskBio will enable Editas Medicine to leverage its knowledge and IP on capsid development and its AAV delivery system in order to overcome the aforementioned bottlenecks of in vivo genome editing in the CNS5. In the transient gene therapy space, Roche and Spark Therapeutics partnered with Dyno Therapeutics in order to use Dyno Therapeutics’ CapsidMapTM platform to develop optimized AAV vectors for gene therapies targeting CNS and liver6. Those novel AAVs will have optimized tissue targeting and “immune-evading” properties.
Some CNS indications, however, have already an attractive commercial proposition. In fact, there are indications such as Huntington’s and ALS, for which there is a large therapeutic unmet need and the etiology is clear and are therefore suitable indications to be treated with in vivo genome editing. For this reason, large biopharmaceutical companies have partnered with genome editing companies to treat CNS disorders (Table 2).
TABLE 2.
Licensing deals from co-developed in vivo genome editing and regulation CNS assets (excluding AskBio/Editas) https://investor.sangamo.com/news-releases/news-release-details/sangamo-announces-global-collaboration-novartis-develop-genomic.
| Licensee | Licensor | Phase | Indication | Upfront ($m) | Milestone Payments (Up to $m) | Year |
| Pfizer | Sangamo Therapeutics | Pre-clinical | ALS/FTLD | 12 | 150 | 2018 |
| Takeda Pharmaceutical Company | Sangamo Therapeutics | Pre-clinical | Huntington’s | Unknown | Unknown | 2019 |
| Biogen | Sangamo Therapeutics | Pre-clinical | Tauopathies, Synucleinopathies (Inc., Parkinson’s disease), a neuromuscular target and up to nine other undisclosed neurological indication | 350 | 2,370 | 2020 |
| Novartis | Sangamo Therapeutics | Pre-clinical | Neurodevelopmental Disorders (Inc., Autism Spectrum Disorder) | 75 | 720 | 2020 |
Sangamo has positioned itself as the leader in in vivo genome editing for CNS disorders with its ZFP-TF technology. With four large collaborations with Pfizer, Takeda Pharmaceutical Company (Takeda), Biogen and Novartis (Table 2), it has managed, at least publicly, to become the biopharmaceutical company with the largest amount of genome editing therapeutic assets for CNS indications.
All disclosed CNS in vivo genome editing therapeutics are in early stages, but their potential is reflected in the large partnering and licensing deals (Table 2).
Sangamo signed two collaboration agreements with Pfizer and Takeda for the development of therapies for ALS/FTLD and Huntington’s, respectively. Under the collaboration with Pfizer, Sangamo will receive a $12m upfront payment from Pfizer7. In this agreement, Sangamo will be responsible for developing ZFP-TF candidates and Pfizer responsible for research, development, manufacturing and commercialization for the ZFP-TF program. Sangamo is eligible to receive development and commercial milestones of up to $150m, as well as tiered royalties on net sales.
More recently, Sangamo announced a global collaboration with Biogen to develop gene regulation therapies for tauopathies including Alzheimer’s disease, for synucleinopathies including Parkinson’s disease, a third undisclosed neuromuscular disease target, and up to nine additional undisclosed neurological disease targets. Sangamo will use its ZFP-TF platform to develop these assets. Biogen paid $350m upfront with up to $2.37b in development, regulatory, and commercial milestone payments8. In July 2020, Sangamo and Novartis announced a global collaboration to develop and commercialize gene regulation therapies to address three neurodevelopmental diseases, including autism spectrum disorder. The target genes are undisclosed. Novartis will pay $75m to Sangamo as an upfront license fee payment with a potential $720m in other development and commercial milestone payments. The agreement also stipulates that Sangamo is eligible to receive a high single-digit to sub-teen double-digit royalties on net commercial sales arising from the collaboration9.
Patenting and Licensing
The commercialization route for biologics and advanced therapeutics, including genome editing therapeutics, is different from that of small molecules. Small molecule developers usually do not require a license for a critical technology (such as genome editing tools) in order to commercialize a therapy. In the case of advanced therapeutics, such as the use of CRISPR, any academic or commercial institution would require a license to key IP in order to have “freedom-to-operate” and to commercialize its CRISPR-based therapeutic. This is a major barrier to entry since developing a de novo genome editing tool in order to avoid expensive CRISPR licenses requires years of fundamental research (Brinegar et al., 2017).
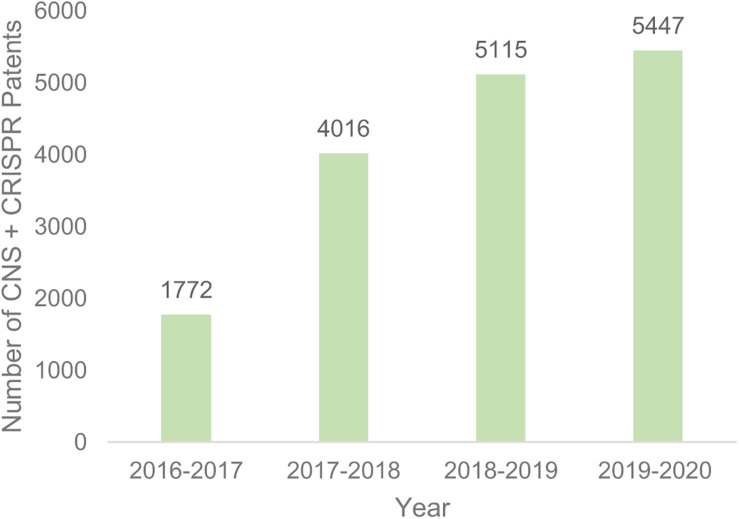
However, there are still 52,603 CRISPR patents filed globally (Google patents search 10/11/2020), of which 5,447 mention the CNS. The total number of patent filings that mention both CRISPR and CNS have been increasing since 2016 (Figure 1), demonstrating both the academic and institutional interest in the use of genome editing in the CNS.
FIGURE 1.
Number of patents filing that mention both CRISPR and CNS. Google patents search November 10 2020.
For each CRISPR patent filed, there can be multiple licenses. For example, the Broad Institute licensed its key patents, non-exclusively, to The Monsanto Company (part of Bayer) for use in agriculture (StatNews, 201610), but licensed it exclusively to Editas Medicine for human therapeutic use (Editas Medicine, 201411).
Conclusion and Future Perspectives
Although, as aforementioned, CRISPR/Cas9 tools can be designed and implemented much more easily than ZFPs, most of the preclinical studies that companies are running are based on ZFPs. This might be partially due to the more recent advent of CRISPR and the associated off-target effects, which have to be further tested. We now have several genome editing tools in our hands to really change the course of neurological disease treatment. Preclinical studies are promising and there are extensive efforts in the scientific community to find approaches to overcome the current barriers to developing a first in human genome editing therapeutic for CNS diseases. We envision that the next 5–10 years will be fundamental to understand whether we can completely eradicate some severe intractable neurological diseases using genome editing. The road to clinic is still full of hurdles but the speed of development in the field is one of the fastest ever seen in science.
Author Contributions
PL, GC, and GL conceived the review and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
PL was employed by Hummingbird Ventures. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. GL was supported by Epilepsy Research UK (F1707) and MRC New Investigator Award (MR/S011005/1). GC was supported by Associazione Gruppo Famiglie Dravet Grant 2019, Telethon GGP19249, CARIPLO Foundation (2016-0532), and the Italian Ministry of Health (GR-2016-02363972).
www.clinicaltrials.gov 2020 Search 13 November 2020
References
- Amabile A., Migliara A., Capasso P., Biffi M., Cittaro D., Naldini L., et al. (2016). Inheritable silencing of endogenous genes by hit-and-run targeted epigenetic editing. Cell 167 219–232.e214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzalone A. V., Koblan L. W., Liu D. R. (2020). Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 38 824–844. 10.1038/s41587-020-0561-9 [DOI] [PubMed] [Google Scholar]
- Anzalone A. V., Randolph P. B., Davis J. R., Sousa A. A., Koblan L. W., Levy J. M., et al. (2019). Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576 149–157. 10.1038/s41586-019-1711-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch J., Scholze H., Schornack S., Landgraf A., Hahn S., Kay S., et al. (2009). Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326 1509–1512. 10.1126/science.1178811 [DOI] [PubMed] [Google Scholar]
- Brinegar K., Yetisen A. K., Choi S., Vallillo E., Ruiz-Esparza G. U., Prabhakar A. M., et al. (2017). The commercialization of genome-editing technologies. Crit. Rev. Biotechnol. 37 924–932. [DOI] [PubMed] [Google Scholar]
- Chavez A., Scheiman J., Vora S., Pruitt B. W., Tuttle M., Iyer E. P. R., et al. (2015). Highly efficient Cas9-mediated transcriptional programming. Nat. Methods 12 326–328. 10.1038/nmeth.3312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasante G., Lignani G., Brusco S., Di Berardino C., Carpenter J., Giannelli S., et al. (2020a). dCas9-Based Scn1a gene activation restores inhibitory interneuron excitability and attenuates seizures in dravet syndrome mice. Mol. Ther. 28 235–253. 10.1016/j.ymthe.2019.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasante G., Qiu Y., Massimino L., Di Berardino C., Cornford J. H., Snowball A., et al. (2020b). In vivo CRISPRa decreases seizures and rescues cognitive deficits in a rodent model of epilepsy. Brain 143 891–905. 10.1093/brain/awaa045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cota-Coronado A., Diaz-Martinez N. F., Padilla-Camberos E., Diaz-Martinez N. E. (2019). Editing the central nervous system through CRISPR/Cas9 systems. Front. Mol. Neurosci. 12:110. 10.3389/fnmol.2019.00110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crudele J. M., Chamberlain J. S. (2018). Cas9 immunity creates challenges for CRISPR gene editing therapies. Nat. Commun. 9:3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte F., Deglon N. (2020). Genome editing for CNS disorders. Front. Neurosci. 14:579062. 10.3389/fnins.2020.579062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudelli N. M., Komor A. C., Rees H. A., Packer M. S., Badran A. H., Bryson D. I., et al. (2017). Programmable base editing of A∗T to G∗C in genomic DNA without DNA cleavage. Nature 551 464–471. 10.1038/nature24644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyorgy B., Loov C., Zaborowski M. P., Takeda S., Kleinstiver B. P., Commins C., et al. (2018). CRISPR/Cas9 mediated disruption of the swedish APP allele as a therapeutic approach for early-onset Alzheimer’s disease. Mol. Ther. Nucleic Acids 11 429–440. 10.1016/j.omtn.2018.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton I. B., D’ippolito A. M., Vockley C. M., Thakore P. I., Crawford G. E., Reddy T. E., et al. (2015). Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 33 510–517. 10.1038/nbt.3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa M. P., Krishnakumar R., Timlin J. A., Carney J. P., Butler K. S. (2020). Gene editing and CRISPR in the clinic: current and future perspectives. Biosci. Rep. 40:BSR20200127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantor B., Tagliafierro L., Gu J., Zamora M. E., Ilich E., Grenier C., et al. (2018). Downregulation of SNCA expression by targeted editing of DNA methylation: a potential strategy for precision therapy in PD. Mol. Ther. 26 2638–2649. 10.1016/j.ymthe.2018.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. G., Cha J., Chandrasegaran S. (1996). Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. U.S.A. 93 1156–1160. 10.1073/pnas.93.3.1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug A., Rhodes D. (1987). Zinc fingers: a novel protein fold for nucleic acid recognition. Cold Spring Harb. Symp. Quant. Biol. 52 473–482. 10.1101/sqb.1987.052.01.054 [DOI] [PubMed] [Google Scholar]
- Komor A. C., Kim Y. B., Packer M. S., Zuris J. A., Liu D. R. (2016). Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533 420–424. 10.1038/nature17946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S., Brigham M. D., Trevino A. E., Joung J., Abudayyeh O. O., Barcena C., et al. (2015). Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517 583–588. 10.1038/nature14136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan G., Zhang Y., Gu Y., Kankel M. W., Gao F. B., Almeida S. (2020). CRISPR deletion of the C9ORF72 promoter in ALS/FTD patient motor neurons abolishes production of dipeptide repeat proteins and rescues neurodegeneration. Acta Neuropathol. 140 81–84. 10.1007/s00401-020-02154-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Yang Y., Hong W., Huang M., Wu M., Zhao X. (2020). Applications of genome editing technology in the targeted therapy of human diseases: mechanisms, advances and prospects. Signal Transduct. Target. Ther. 5:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Huang S., Zhao X., Wright D. A., Carpenter S., Spalding M. H., et al. (2011). Modularly assembled designer TAL effector nucleases for targeted gene knockout and gene replacement in eukaryotes. Nucleic Acids Res. 39 6315–6325. 10.1093/nar/gkr188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C. K. W., Gapinske M., Brooks A. K., Woods W. S., Powell J. E., Zeballos C. M., et al. (2020). Treatment of a mouse model of ALS by in vivo base editing. Mol. Ther. 28 1177–1189. 10.1016/j.ymthe.2020.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S., Staahl B. T., Alla R. K., Doudna J. A. (2014). Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife 3:e04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. S., Wu H., Ji X., Stelzer Y., Wu X., Czauderna S., et al. (2016). Editing DNA methylation in the mammalian genome. Cell 167 233–247.e217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Aach J., Stranges P. B., Esvelt K. M., Moosburner M., Kosuri S., et al. (2013). CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 31 833–838. 10.1038/nbt.2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matharu N., Ahituv N. (2020). Modulating gene regulation to treat genetic disorders. Nat. Rev. Drug Discov. 19 757–775. 10.1038/s41573-020-0083-7 [DOI] [PubMed] [Google Scholar]
- Matharu N., Rattanasopha S., Tamura S., Maliskova L., Wang Y., Bernard A., et al. (2019). CRISPR-mediated activation of a promoter or enhancer rescues obesity caused by haploinsufficiency. Science 363:eaau0629. 10.1126/science.aau0629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald J. I., Celik H., Rois L. E., Fishberger G., Fowler T., Rees R., et al. (2016). Reprogrammable CRISPR/Cas9-based system for inducing site-specific DNA methylation. Biol. Open 5 866–874. 10.1242/bio.019067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. C., Tan S., Qiao G., Barlow K. A., Wang J., Xia D. F., et al. (2011). A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 29 143–148. 10.1038/nbt.1755 [DOI] [PubMed] [Google Scholar]
- Morita S., Noguchi H., Horii T., Nakabayashi K., Kimura M., Okamura K., et al. (2016). Targeted DNA demethylation in vivo using dCas9-peptide repeat and scFv-TET1 catalytic domain fusions. Nat. Biotechnol. 34 1060–1065. 10.1038/nbt.3658 [DOI] [PubMed] [Google Scholar]
- Moscou M. J., Bogdanove A. J. (2009). A simple cipher governs DNA recognition by TAL effectors. Science 326:1501. 10.1126/science.1178817 [DOI] [PubMed] [Google Scholar]
- Nishida K., Arazoe T., Yachie N., Banno S., Kakimoto M., Tabata M., et al. (2016). Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353:aaf8729. 10.1126/science.aaf8729 [DOI] [PubMed] [Google Scholar]
- Nishiyama J., Mikuni T., Yasuda R. (2017). Virus-Mediated genome editing via homology-directed repair in mitotic and postmitotic cells in mammalian brain. Neuron 96 755–768.e755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C. Y., Halevy T., Lee D. R., Sung J. J., Lee J. S., Yanuka O., et al. (2015). Reversion of FMR1 methylation and silencing by editing the triplet repeats in fragile X iPSC-derived neurons. Cell Rep. 13 234–241. 10.1016/j.celrep.2015.08.084 [DOI] [PubMed] [Google Scholar]
- Park H., Oh J., Shim G., Cho B., Chang Y., Kim S., et al. (2019). In vivo neuronal gene editing via CRISPR-Cas9 amphiphilic nanocomplexes alleviates deficits in mouse models of Alzheimer’s disease. Nat. Neurosci. 22 524–528. 10.1038/s41593-019-0352-0 [DOI] [PubMed] [Google Scholar]
- Perez-Pinera P., Kocak D. D., Vockley C. M., Adler A. F., Kabadi A. M., Polstein L. R., et al. (2013). RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat. Methods 10 973–976. 10.1038/nmeth.2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L. S., Larson M. H., Gilbert L. A., Doudna J. A., Weissman J. S., Arkin A. P., et al. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152 1173–1183. 10.1016/j.cell.2013.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebar E. J., Huang Y., Hickey R., Nath A. K., Meoli D., Nath S., et al. (2002). Induction of angiogenesis in a mouse model using engineered transcription factors. Nat. Med. 8 1427–1432. 10.1038/nm1202-795 [DOI] [PubMed] [Google Scholar]
- Rouet P., Smih F., Jasin M. (1994). Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol. Cell. Biol. 14 8096–8106. 10.1128/mcb.14.12.8096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Lan F., Matson C., Mulligan P., Whetstine J. R., Cole P. A., et al. (2004). Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119 941–953. [DOI] [PubMed] [Google Scholar]
- Shitik E. M., Velmiskina A. A., Dolskiy A. A., Yudkin D. V. (2020). Reactivation of FMR1 gene expression is a promising strategy for fragile X syndrome therapy. Gene Ther. 27 247–253. 10.1038/s41434-020-0141-0 [DOI] [PubMed] [Google Scholar]
- Stepanichev M. (2020). Gene editing and Alzheimer’s disease: is there light at the end of the tunnel? Front. Genome Ed. 2:4. 10.3389/fgeed.2020.00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Tsunekawa Y., Hernandez-Benitez R., Wu J., Zhu J., Kim E. J., et al. (2016). In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature 540 144–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakore P. I., D’ippolito A. M., Song L., Safi A., Shivakumar N. K., Kabadi A. M., et al. (2015). Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat. Methods 12 1143–1149. 10.1038/nmeth.3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner T. J., Zourray C., Schorge S., Lignani G. (2020). Recent advances in gene therapy for neurodevelopmental disorders with epilepsy. J. Neurochem. 10.1111/jnc.15168 [Online ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermilyea S. C., Babinski A., Tran N., To S., Guthrie S., Kluss J. H., et al. (2020). In vitro CRISPR/Cas9-directed gene editing to model LRRK2 G2019S Parkinson’s disease in common marmosets. Sci. Rep. 10:3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojta A., Dobrinic P., Tadic V., Bockor L., Korac P., Julg B., et al. (2016). Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res. 44 5615–5628. 10.1093/nar/gkw159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Zhang F., Gao G. (2020). CRISPR-Based therapeutic genome editing: strategies and in vivo delivery by AAV vectors. Cell 181 136–150. 10.1016/j.cell.2020.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolter J. M., Mao H., Fragola G., Simon J. M., Krantz J. L., Bazick H. O., et al. (2020). Cas9 gene therapy for Angelman syndrome traps Ube3a-ATS long non-coding RNA. Nature 587 281–284. 10.1038/s41586-020-2835-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie N., Gong H., Suhl J. A., Chopra P., Wang T., Warren S. T. (2016). Reactivation of FMR1 by CRISPR/Cas9-mediated deletion of the expanded CGG-Repeat of the fragile X chromosome. PLoS One 11:e0165499. 10.1371/journal.pone.0165499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong T., Meister G. E., Workman R. E., Kato N. C., Spellberg M. J., Turker F., et al. (2017). Targeted DNA methylation in human cells using engineered dCas9-methyltransferases. Sci. Rep. 7:6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Tao Y., Gao X., Zhang L., Li X., Zou W., et al. (2016). A CRISPR-based approach for targeted DNA demethylation. Cell Discov 2:16009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata T., Raveau M., Kobayashi K., Miyamoto H., Tatsukawa T., Ogiwara I., et al. (2020). CRISPR/dCas9-based Scn1a gene activation in inhibitory neurons ameliorates epileptic and behavioral phenotypes of Dravet syndrome model mice. Neurobiol. Dis. 141:104954. 10.1016/j.nbd.2020.104954 [DOI] [PubMed] [Google Scholar]
- Yang S., Chang R., Yang H., Zhao T., Hong Y., Kong H. E., et al. (2017). CRISPR/Cas9-mediated gene editing ameliorates neurotoxicity in mouse model of Huntington’s disease. J. Clin. Invest. 127 2719–2724. 10.1172/jci92087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh C. D., Richardson C. D., Corn J. E. (2019). Advances in genome editing through control of DNA repair pathways. Nat. Cell Biol. 21 1468–1478. 10.1038/s41556-019-0425-z [DOI] [PubMed] [Google Scholar]
- Zhang F., Cong L., Lodato S., Kosuri S., Church G. M., Arlotta P. (2011). Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat. Biotechnol. 29 149–153. 10.1038/nbt.1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. H., Tee L. Y., Wang X. G., Huang Q. S., Yang S. H. (2015). Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol. Ther. Nucleic Acids 4:e264. 10.1038/mtna.2015.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Su J., Hu X., Zhou C., Li H., Chen Z., et al. (2020). Glia-to-Neuron conversion by CRISPR-CasRx alleviates symptoms of neurological disease in mice. Cell 181:e516. [DOI] [PubMed] [Google Scholar]