Abstract
A peptidoglycan (PG) cell wall is an essential component of nearly all bacteria, providing protection against turgor pressure. Metabolism of this PG meshwork must be spatially and temporally regulated in order to support growth and division. Despite being an active area of research for decades, we have only recently identified the primary PG synthesis complexes that function during elongation (RodA-PBP2) and division (FtsW-FtsI), and we are still uncovering the importance of the other seemingly redundant cell wall enzymes. In this review, we highlight the discovery of the monofunctional glycosyltransferases, RodA and FtsW, and describe how these findings have prompted a reevaluation of the auxiliary role of the bifunctional class A penicillin binding proteins (aPBPs) as well as the L,D-transpeptidases (LDTs). Specifically, recent work indicates that the aPBPs and LDTs function independently of the primary morphogenetic complexes to support growth, provide protection from stresses, mediate morphogenesis, and/or allow adaptation to different growth conditions. These paradigm-shifting studies have reframed our understanding of bacterial cell wall metabolism, which will only become more refined as emerging technology allows us to tackle the remaining questions surrounding PG biosynthesis.
Introduction:
The cell wall is a critical structural component of almost all bacteria, and regulating cell wall metabolism is central to growth and division. Made of peptidoglycan (PG), the cell wall is a rigid structure that provides protection from lysis as the cell experiences high intracellular turgor pressure. PG is both necessary and sufficient to maintain bacterial cell shape, and spatially regulated PG synthesis and remodeling underlies morphogenesis. PG metabolism is mediated by a host of enzymes, many of which are direct targets of highly effective antibiotics. Despite being studied for over half a century, we continue to discover new essential PG enzymes and uncover the unique roles of others that were assumed to be functionally redundant.
PG comprises alternating N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) sugars polymerized into glycan strands that are covalently linked to each other through short peptide bridges to create a large mesh (Figure 1). PG synthesis and remodeling are mediated by cell wall enzymes and are especially important during two essential periods of dramatic cell shape change—growth and division. Two major biosynthetic functions are required to create the PG mesh: transglycosylation to polymerize the glycan strands and transpeptidation to crosslink them together (Figure 1). In the best-studied transpeptidase reaction, D,D-transpeptidases (TPases) catalyze a 4-3 linkage between peptide stems, where the numbers correspond to the position of the crosslinked amino acids in the peptide stem [1]. Alternatively, L,D-TPases (LDTs) can form 3-3 linkages [1]. D,D-TPase activity is carried out by two classes of penicillin binding proteins (PBPs) with distinct roles. The aPBPs are bifunctional, containing a TPase domain and a glycosyltransferase 51 (GT51) domain, whereas the monofunctional bPBPs have only transpeptidase activity [2]. In addition to the glycosyltransferase activity of the aPBPs, some species also produce monofunctional GT51 domain-containing proteins [2]. Until recently, GT51 domain-containing proteins were the only known GTases for PG synthesis. However, transformative studies over the past five years revealed that the Shape, Elongation, Division, and Sporulation (SEDS) family proteins RodA and FtsW are the primary glycosyltransferases (GTases) associated with elongation and division, respectively.
Figure 1: The basics of peptidoglycan (PG) synthesis.
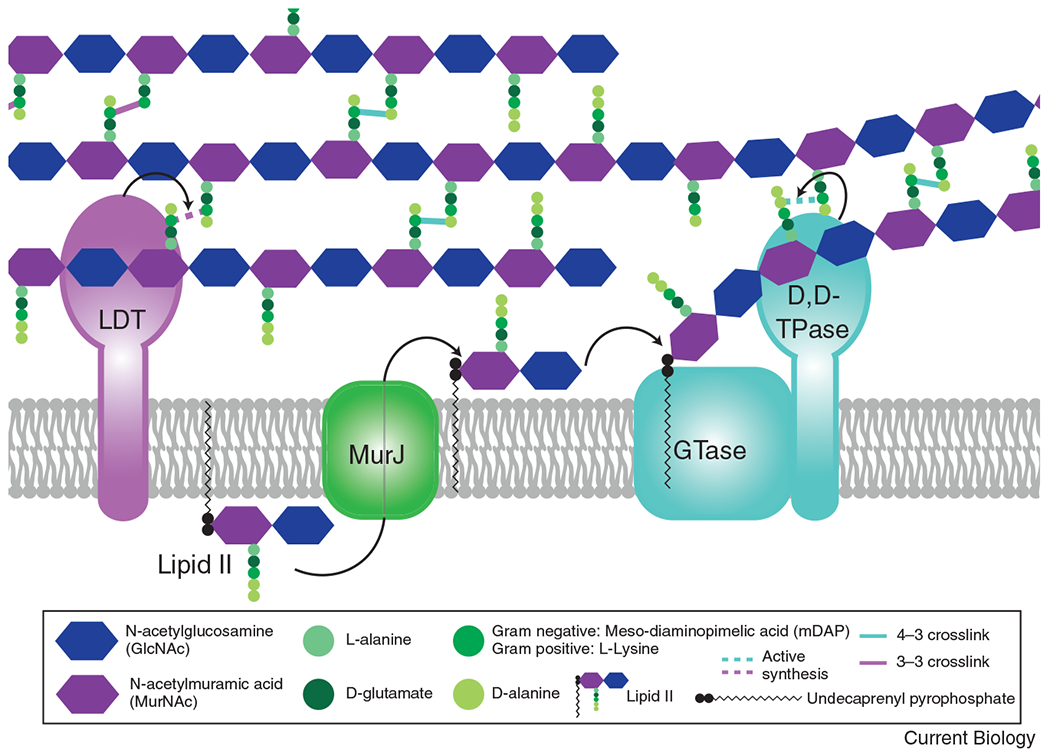
The PG precursor is synthesized in the cytoplasm to create the lipid-linked disaccharide-pentapeptide moiety, lipid II. Lipid II is then flipped across the cytoplasmic membrane by MurJ. Once in the periplasm, a glycosyltransferase (GTase) polymerizes the lipid II moiety onto the growing glycan strand, through the action of either a SEDS protein (RodA or FtsW), a bifunctional class A penicillin binding protein (aPBP), or a GT51 containing enzyme. Once polymerized, the peptide stem can then be crosslinked to a neighboring strand through the action of a transpeptidase (TPase). A monofunctional bPBP (PBP2 or FtsI) or a bifunctional aPBP acts as a D,D-TPase, creating a 4-3 linkage, shown in teal. For simplicity, only the SEDS/bPBP synthases are shown to depict transglycosylation and transpeptidation steps. The peptide stem can alternatively be crosslinked in a 3-3 linkage (shown in purple) through the action of an L,D-transpeptidase (LDT).
This review will focus on recent advances in our understanding of the physiological functions and regulation of the major PG biosynthetic enzymes. The field currently posits that protein complexes containing a GTase of the SEDS family and a bPBP TPase are primarily responsible for cell wall synthesis during growth and division. These are supported by secondary PG enzymes, including aPBPs and LDTs, that promote PG enlargement, maintenance, and modification. The functions and importance of aPBPs and LDTs vary across bacterial species and growth conditions [3–6]. Through the essential activities of these PG enzymes and their regulators, a bacterial cell is able to coordinate the tight spatio-temporal control of PG synthesis necessary for survival.
SEDS family proteins function as glycosyltransferases
PG synthesis begins with roughly a dozen cytoplasmic enzymes that catalyze the synthesis of lipid II, the cell wall precursor molecule [1]. Lipid II, consisting of a lipid-linked disaccharide pentapeptide, is flipped across the cytoplasmic membrane by MurJ, where it becomes available for incorporation into the nascent cell wall [1] (Figure 1). The first step of incorporation links lipid II to a glycan strand via a GTase. Until recently, aPBPs and other GT51 domain-containing proteins like MtgA in Escherichia coli were thought to be the primary mediators of this function [1]. Surprisingly, however, some bacteria, notably Bacillus subtilis, are viable even when all known GT51-domain containing GTases are deleted [3,4,7]. This observation suggested the existence of novel enzymes with GTase activity and ultimately led to the paradigm-shifting discovery of the GTase activity of the SEDS family proteins, RodA and FtsW.
Multiple groups sought to identify the unknown essential cell wall factor(s) that functions as the primary GTase during elongation and/or division [3,4]. When looking for potential candidates, the SEDS family proteins RodA and FtsW stood out due to the conspicuous fact that each is a member of an essential morphogenetic complex—RodA being associated with the elongation machinery (elongasome/Rod complex) and FtsW with the division machinery (divisome). This was initially studied in a B. subtilis mutant lacking all four aPBPs (∆4), which, though viable, has an increased propensity for lysis and decreased cell width [3,4,7]. Consistent with a PG synthetic role for RodA, overproduction of RodA in the ∆4 mutant restored cells to wild type morphology and growth, while depletion of RodA in the ∆4 background was lethal [3,4]. Additionally, in E. coli, on deletion or inhibition of all aPBPs, elongasome-dependent PG synthesis still occurred, implicating RodA as the GTase [6]. In vitro assays of purified RodA in the presence of lipid II directly demonstrated its GTase activity and implicated two essential residues (W105 and D280) that are necessary for PG synthesis in vivo and in vitro [3].
Like RodA, the division-associated SEDS protein FtsW was recently demonstrated to be a GTase [5]. Previously, FtsW was proposed to function as a lipid II flippase, but in vitro assays established FtsW as the division-associated SEDS family GTase [5]. The in vitro GTase activity of FtsW requires its partner TPase, in this case FtsI [5]. However, FtsW’s GTase activity is not reliant on the TPase activity of FtsI, as either a catalytically dead variant or simply the transmembrane helix of FtsI is sufficient to stimulate PG polymerization in vitro [5]. These findings solidify the GTase activity of the SEDS protein FtsW and highlight the question of enzymatic regulation, which will be discussed in the following section. Overall, discovering the GTase activity of the SEDS proteins was a seminal advance in our understanding of how bacterial PG is synthesized during growth and division. As the essential functions of RodA and/or FtsW are required in nearly all bacteria, they present exciting new targets for antibiotic development; further work to fully characterize the catalytic mechanism and regulation of this group of enzymes is therefore crucial [4].
Regulation of the SEDS-bPBP cell wall synthetic complexes
Unlike the SEDS family GTases, the essential monofunctional bPBPs required for elongation and division are well-characterized TPases. These TPase enzymes are PBP2 for elongation and FtsI (PBP3) for division in E. coli. Since the bPBPs are known to interact directly with the SEDS family proteins [5,8–10] and crosslinking is required for PG mesh formation after strand polymerization, it is logical to hypothesize that their activities and regulation are linked. Disrupting the balance between GTase and TPase activities can be lethal, as during treatment with β-lactam antibiotics, suggesting that the SEDS-bPBP pairs require a high level of coordination [11].
Regulation of SEDS-bPBP activity was first explored in E. coli, where RodA-PBP2 were already known to be spatially regulated by the components of the elongasome. The elongasome consists of the actin homolog MreB, regulatory proteins MreC, MreD, and RodZ, and the PG synthase complex RodA-PBP2. These proteins form a dynamic complex that moves around the cell’s circumference, driven by cell wall synthesis [12] (Figure 2). A PG synthesis activation pathway for the elongasome was first suggested when a hyperactive variant of PBP2 was found to suppress mutations in or loss of the elongasome proteins MreC, MreD, or RodZ, and to stimulate the in vitro and in vivo cell wall synthesis activity of RodA-PBP2 [13]. Further work implicated MreC and MreD in regulating RodA-PBP2 activity. MreD, which directly interacts with PBP2, is proposed to hold the PG synthases in an inactive state [14]. Crystal structures of the MreC-PBP2 complex from Helicobacter pylori revealed an MreC-dependent conformational change of PBP2 [15]. Collectively, these data led to a model wherein MreC binding to PBP2 displaces MreD and activates the RodA-PBP2 PG synthases [13,14]. Additionally, a recent structure of RodA in complex with its cognate bPBP established the role of PBP2’s pedestal domain as an allosteric activator of RodA, providing molecular resolution of the coordinated activity of the SEDS-bPBP pairs [8]. Two lines of evidence also suggest positive feedback from RodA-PBP2 to assembly of the elongasome in E. coli: (1) activating mutations in PBP2 increase the number of moving MreB foci, while decreasing MreB filament length [13] and (2) single molecule tracking indicated that PBP2 binding to a substrate (independent of its catalytic activity) initiates elongasome recruitment to sites of new PG synthesis [16].
Figure 2: Dynamics and functions of distinct PG synthetic enzymes.
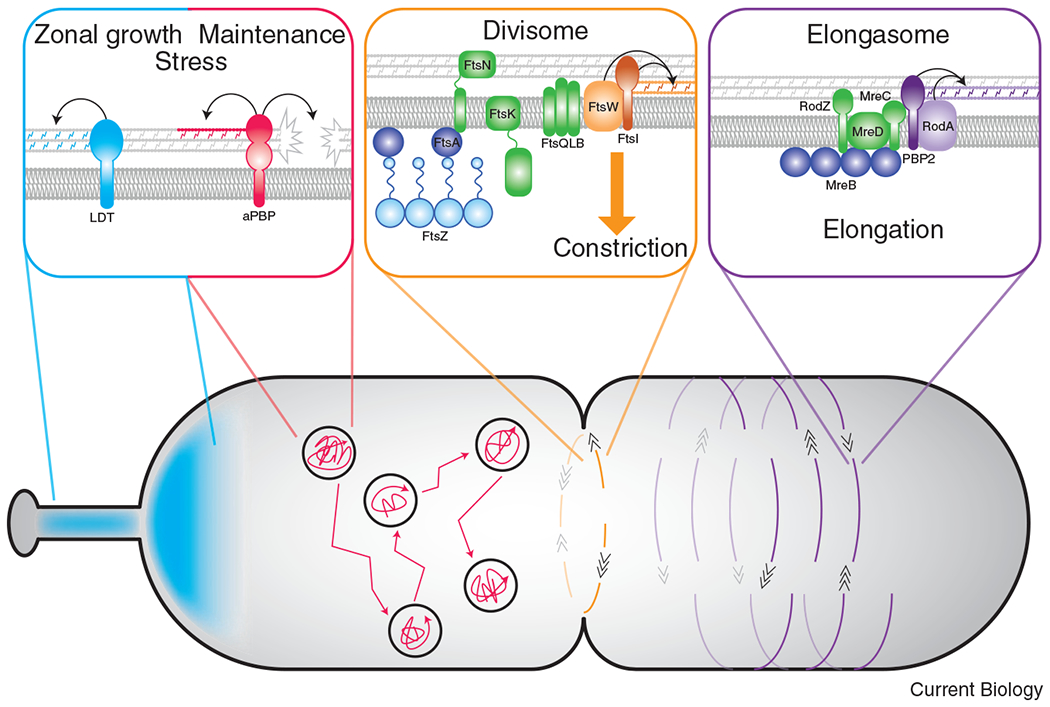
SEDS-bPBP pairs are associated with morphogenetic complexes to direct spatially-regulated PG synthesis, while movement of the aPBPs and LDTs are independent of the division/elongation machinery. Left panel: The importance of LDT activity varies among species. In some organisms, LDT-mediated crosslinking is important in specific regions in a cell, such as during stalk formation or unipolar growth. LDT activity also appears important for resistance to antibiotic treatment. Movement of the aPBPs is diffusive followed by periods of confinement. The bifunctional aPBPs likely act as repair machinery necessary for efficient growth and/or PG synthesis during stress conditions. Middle panel: In almost all bacteria with a cell wall, PG synthesis associated with the divisome is carried out by the FtsW-FtsI enzyme pair. The movement of FtsW-FtsI is bidirectional around the short axis of the division plane, directed by treadmilling of FtsZ filaments. The conserved activation pathway for FtsW-FtsI likely requires the divisome proteins FtsA, FtsK, FtsN, and FtsQLB. Right panel: In many rod-shaped organisms, RodA-PBP2 synthesize PG for elongation and are regulated by elongasome proteins, notably MreC. The elongation machinery moves bidirectionally about the circumference of the short axis of the cell but, unlike the divisome, movement is directed by PG synthesis rather than MreB dynamics.
Though it has not yet been directly shown, a similar mechanism of regulation for the divisome complex seems likely. Like the elongasome, the divisome is a multi-protein complex made up of a cytoskeletal protein (the tubulin homolog FtsZ), regulators (including FtsA, FtsK, the FtsQ-FtsL-FtsB complex, and FtsN), and a SEDS-bPBP pair (FtsW-FtsI). Also like the elongasome, the divisome moves dynamically, but its movement is driven by treadmilling of FtsZ filaments rather than by PG synthesis in E. coli and B. subtilis [12] (Figure 2). A large body of genetic evidence suggests that the divisome requires activation to drive constriction. The current model in E. coli proposes that FtsA acts through FtsN to initiate constriction [17]. Genetic evidence implicates other division proteins in this activation pathway, as suppressors of an inactive FtsN variant were found in the division proteins FtsB and FtsL [18,19].
The constriction activation pathway in E. coli is presumed to culminate in PG synthesis by FtsW and FtsI. The most direct evidence in support of this model comes from hyperactive mutants of FtsW and FtsI (called FtsW-FtsI*) that were identified in Caulobacter crescentus. The FtsW-FtsI* mutant enzymes are proposed to be biased towards an active state, as FtsW-FtsI*-producing cells constrict faster and are shorter than wild type [20–22]. These hyperactive division mutants appear to function comparably to their elongasome counterparts, as one of the mutations in FtsW (A246T) is directly analogous to an E. coli RodA(A234T) variant with increased PG synthesis activity in vivo [13]. In Caulobacter, the FtsW-FtsI* mutant is able to render the normally essential division protein FzlA nonessential, suggesting that FzlA participates in the activation of FtsW-FtsI [23]. Hyperactivity of FtsW-FtsI* both impacts the rate of constriction and increases sensitivity to cell wall antibiotics that primarily target PBP2 or FtsI, reflecting the importance of FtsW-FtsI regulation in building a robust cell wall [20,21,23]. The evolving model for divisome regulation requires biochemical exploration and confirmation, but it highlights the complex regulation of the PG synthetic machinery required to maintain cell envelope integrity during growth and division. Moreover, as divisome regulation is explored in more bacteria, we are likely to uncover variations on the themes described here, as some of the characterized components of the E. coli or C. crescentus activation pathways are not broadly conserved.
Bifunctional PBPs fulfill a PG repair role and are required under stress conditions
With the discovery of the SEDS proteins as the primary GTases for growth and division, the physiological function of the aPBPs must now be re-examined. Emerging research is revealing that the aPBPs fulfill a unique maintenance role during growth and stress conditions. Recent evidence establishes a division of labor where the SEDS-bPBP complexes build a PG foundation at the site of elongation or division, while the aPBPs can expand and repair the PG mesh for growth and support as needed [6] (Figure 2). In E. coli, the two major aPBPs - PBP1a and PBP1b - are individually dispensable, yet it is clear that they collectively fulfill an essential role, as loss of both is lethal [24]. PBP1a and PBP1b are also regulated, requiring interaction with their respective activators, LpoA and LpoB [25,26]. In E. coli, the aPBPs were thought to be coordinated with the primary PG synthetic machinery since there is in vitro and in vivo evidence of an interaction between the aPBPs and bPBPs [27,28]. The aPBPs also have a direct interaction with divisome proteins that stimulate the aPBP’s GTase activity. In E. coli, the divisome proteins ZipA and FtsN increase the GTase activity in vitro of PBP1a and PBP1b, respectively [29]. However, in vivo single molecule studies revealed that the movements of the aPBPs are not linked to the cytoskeletally-associated SEDS-bPBP pairs, implying spatial separation of aPBP activity from the dynamic morphogenetic complexes [6,30,31] (Figure 2).
Despite their apparent redundancy, there are morphological consequences of loss of aPBP activity. Loss of the aPBPs in E. coli or B. subtilis decreases cell width, reduces the rate of cell wall incorporation, and impacts overall cell wall integrity [6,7,27,31]. In E. coli, deletion of PBP1b results in reduced cell wall stiffness, suggesting an ability of aPBPs to find and repair cell wall defects [31]. Other work in B. subtilis discovered a balance between the two PG synthesis systems to achieve optimize cell size: the elongasome decreases cell width, and the aPBPs increase cell width [32]. This supportive role of the aPBPs has been corroborated in other species including Streptococcus pneumoniae, where the aPBPs modify and strengthen the newly synthesized PG made by the SEDS-bPBP complexes [33]. These findings imply a conserved role of the aPBPs in normal growth as maintenance enzymes that fill in the gaps in the cell wall as they arise (Figure 2).
Though the aPBPs do not appear to function as the primary PG synthases in normal growth, recent studies reveal a major responsibility of these enzymes during fluctuating environmental conditions. For instance, growth of E. coli in acidic or alkaline environments was found to be dependent on the presence of PBP1b and PBP1a, respectively [34]. This impact on viability likely does not involve a novel enzymatic activity - the global cell wall composition is unchanged in altered pH [35] - but rather reflects the ability of specific synthases to retain enzymatic activity and act as repair enzymes under conditions when others are inactive [34]. Interestingly, PBP1b also appears critical in the context of β-lactam sensitivity, as loss of PBP1b or its regulator LpoB results in hypersensitivity to cell wall-targeting antibiotics [36]. The aPBPs also appear to be important for adapting to osmotic changes and mechanical stresses, as well as for initiating de novo PG synthesis [37,38]. These findings are likely the beginning of a complete understanding of the PG enzymes and their distinct roles in cell wall homeostasis and repair.
LDT activity can protect against cell envelope stress
Among the cell wall enzymes, the less-well studied LDTs occupy a unique category within the TPases. Though the LDTs crosslink peptide stems, they are not members of the PBP family. Rather, they have minimal amino acid similarity and a distinct active site from the PBPs, explaining their insensitivity to inhibition by penicillin [39]. In E. coli, the predominant 4–3 crosslink is between a meso-diaminopimelic acid (mDAP) in the third position of a tetra-peptide to a terminal D-alanine (D-ala) in the fourth position of a neighboring tetra-peptide stem. However, roughly 3–10% of the crosslinks in E. coli consist of a 3–3 linkage, mDAP-mDAP, which is synthesized by the LDTs from already processed tetrapeptides [40] (Figure 1). Similar to the SEDS-bPBP pairs, PG remodeling by the LDTs functions in concert with a GTase, specifically an aPBP [41,42]. Though nonessential in E. coli, LDTs appear to be important under stress conditions: inhibiting the LDTs with copper ions results in cell envelope defects [43], increased LDT activity provides resistance to cell lysis during OM stress [41], and LDT-mediated crosslinking can confer resistance to broad spectrum β-lactam antibiotics in E. coli and Enterococcus faecium [42,44].
In contrast to E. coli, LDTs appear to play a major role in organisms that exhibit polar growth, notably Mycobacterium tuberculosis and Agrobacterium tumefaciens (Figure 2). LDT-generated 3-3 linkages comprise about 80% of the total crosslinks in M. tuberculosis and more than 50% of the crosslinks in A. tumefaciens [45,46]. In these organisms, the abundance of 3-3 crosslinks appears to be important for resistance to certain cell wall-targeting agents. For instance, loss of LDT activity in M. tuberculosis results in growth defects, loss of virulence, and increased sensitivity to the β-lactam antibiotics that target PBPs [47]. Similarly, the PG of A. tumefaciens is highly resistant to lysozyme-mediated degradation [48]. The distinct chemistry of the cell wall resulting from LDT activity likely underlies these protective effects.
LDTs are also important for generating spatially discrete areas within the cell wall of uniquely crosslinked PG and for formation of polar structures, as was found in C. crescentus [48,49]. C. crescentus forms a unipolar appendage called the stalk, which contains all elements of the cell envelope and requires PG synthesis for its formation (Figure 2). Interestingly, stalk morphology is unaffected by treatment with β-lactams and cell wall digesting enzymes like lysozyme, which can be attributed to increased LDT activity in the stalk [48,50]. Though the physiological significance of high LDT activity in the stalk is unknown, it highlights a spatially-regulated PG metabolic activity that provides protection against PG assaults. As with the aPBPs, we are only now learning the importance of these previously overlooked enzymes as we push the limits of stress and culture conditions and explore PG metabolism in diverse bacteria.
Conclusions and Perspectives:
Only a decade ago, our understanding of the bacterial cell wall synthesis machinery was profoundly different than it is today. Previously, the primary biosynthetic PG machinery was thought to consist of the monofunctional TPases and the GTase activity of aPBPs. The SEDS family proteins had an unknown essential function and we knew very little about the activation of cell wall synthesis during growth and division. The SEDS-bPBP complexes are now characterized as the primary morphogenetic PG enzymes, and we are beginning to understand their activity and regulation. In retrospect, it seems the SEDS GTases were hidden in plain sight - some bacteria lack aPBPs, but encode at least one bPBP and a SEDS protein [3] – making it somewhat surprising their activity was not discovered earlier. Despite the progress made in recent years, there are still many unknowns. For one, the activation pathway and regulatory machinery for SEDS-bPBP pairs is undoubtedly complex. In addition to identifying the activating factors, the link between these enzymes and the dynamic cytoskeletal complexes with which they associate is poorly understood. Through single molecule studies we are now able to observe the directed movement of the cytoskeletal components and associated enzymes, which will begin to reveal more about the distribution of morphogenetic proteins and corresponding PG synthesis [12]. A more in depth understanding of these highly conserved PG synthetic enzymes is also pivotal as they can be exploited as antimicrobial targets.
As the genomes of a growing number of bacteria have been sequenced, we can identify most of the cell wall family enzymes of either known or unknown physiological function. With this knowledge, we can continue to dissect the importance of apparently “redundant” PG enzymes, which are likely not redundant at all. The recent work on the aPBPs and LDTs has scratched the surface of identifying conditions when apparently nonessential PG enzymes become essential for viability. Future work will expand this paradigm to other cell wall enzymes, potential activators, and novel cell wall chemistries. One avenue that will inform this work is the study of PG biochemistry and regulation in a diversity of bacterial systems. Many of the discoveries discussed above were only possible as a result of studying different bacterial systems, each offering unique technical strengths and/or occupying distinct biological niches. This could immediately be applied to one outstanding question in this realm which is the apparent lack of aPBP activators, e.g. the LpoA/B proteins, outside of the γ-proteobacteria. It is unlikely that the aPBPs do not require activators and/or regulators in other species; rather we simply have not yet discovered them. Ultimately, we marvel at the rapid advancement of the field of PG biogenesis in recent years and anticipate an increasing rate of discovery as tool development continues to push the limits of what is discoverable.
Acknowledgements
We would like to thank members of the Goley lab including Christopher Mahone, Jordan Barrows, Erika Smith, and Wanda Figueroa-Cuilan for helpful discussions and feedback. We also want to acknowledge Ryan McQuillen for advice on figures. Work on cell wall metabolism in the Goley lab is supported by the National Institutes of Health through grants R01GM108640 and R35GM136221 (to EDG). AKD is supported in part by the National Institutes of Health under grant T32GM007445.
References:
- 1.Zhao H, Patel V, Helmann JD, and Dörr T (2017). Don’t let sleeping dogmas lie: new views of peptidoglycan synthesis and its regulation. Mol. Microbiol 106, 847–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Egan AJF, Biboy J, van’t Veer I, Breukink E, and Vollmer W (2015). Activities and regulation of peptidoglycan synthases. Philos. Trans. R. Soc. B Biol. Sci 370, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meeske AJ, Riley EP, Robins WP, Uehara T, Mekalanos JJ, Kahne D, Walker S, Kruse AC, Bernhardt TG, and Rudner DZ (2016). SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature 537, 634–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Emami K, Guyet A, Kawai Y, Devi J, Wu LJ, Allenby N, Daniel RA, and Errington J (2017). RodA as the missing glycosyltransferase in Bacillus subtilis and antibiotic discovery for the peptidoglycan polymerase pathway. Nat. Microbiol 2, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taguchi A, Welsh MA, Marmont LS, Lee W, Sjodt M, Kruse AC, Kahne D, Bernhardt TG, and Walker S (2019). FtsW is a peptidoglycan polymerase that is functional only in complex with its cognate penicillin-binding protein. Nat. Microbiol 4, 587–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho H, Wivagg CN, Kapoor M, Barry Z, Rohs PDA, Suh H, Marto JA, Garner EC, and Bernhardt TG (2016). Bacterial cell wall biogenesis is mediated by SEDS and PBP polymerase families functioning semi-autonomously. Nat. Microbiol 1, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McPherson DC, and Popham DL (2003). Peptidoglycan synthesis in the absence of class A penicillin-binding proteins in Bacillus subtilis. J. Bacteriol 185, 1423–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sjodt M, Rohs PDA, Gilman MSA, Erlandson SC, Zheng S, Green AG, Brock KP, Taguchi A, Kahne D, Walker S, et al. (2020). Structural coordination of polymerization and crosslinking by a SEDS–bPBP peptidoglycan synthase complex. Nat. Microbiol 5, 813–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fraipont C, Alexeeva S, Wolf B, Der Ploeg R, Schloesser M, Den Blaauwen T, and Nguyen-Distèche M (2011). The integral membrane FtsW protein and peptidoglycan synthase PBP3 form a subcomplex in Escherichia coli. Microbiology 157, 251–259. [DOI] [PubMed] [Google Scholar]
- 10.Fay A, Meyer P, and Dworkin J (2010). Interactions between late-acting proteins required for peptidoglycan synthesis during sporulation. J. Mol. Biol 399, 547–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho H, Uehara T, and Bernhardt TG (2014). Beta-lactam antibiotics induce a lethal malfunctioning of the bacterial cell wall synthesis machinery. Cell 159, 1300–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wagstaff J, and Löwe J (2018). Prokaryotic cytoskeletons: Protein filaments organizing small cells. Nat. Rev. Microbiol 16, 187–201. [DOI] [PubMed] [Google Scholar]
- 13.Rohs PDA, Buss J, Sim SI, Squyres GR, Srisuknimit V, Smith M, Cho H, Sjodt M, Kruse AC, Garner EC, et al. (2018). A central role for PBP2 in the activation of peptidoglycan polymerization by the bacterial cell elongation machinery. PLoS Genet. 14, 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu X, Biboy J, Vollmer W, and den Blaauwen T (2019). MreC and MreD balance the interaction between the elongasome proteins PBP2 and RodA. bioRxiv. 10.1101/769984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Contreras-Martel C, Martins A, Ecobichon C, Trindade DM, Matteï PJ, Hicham S, Hardouin P, Ghachi M. El, Boneca IG, and Dessen A (2017). Molecular architecture of the PBP2-MreC core bacterial cell wall synthesis complex. Nat. Commun 8, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Özbaykal G, Wollrab E, Simon F, Vigouroux A, Cordier B, Aristov A, Chaze T, Matondo M, and van Teeffelen S (2020). The transpeptidase PBP2 governs initial localization and activity of the major cell-wall synthesis machinery in E. coli. Elife 9, 1–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pichoff S, Du S, and Lutkenhaus J (2015). The bypass of ZipA by overexpression of FtsN requires a previously unknown conserved FtsN motif essential for FtsA-FtsN interaction supporting a model in which FtsA monomers recruit late cell division proteins to the Z ring. Mol. Microbiol 95, 971–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu B, Persons L, Lee L, and de Boer PAJ (2015). Roles for both FtsA and the FtsBLQ subcomplex in FtsN-stimulated cell constriction in Escherichia coli. Mol. Microbiol 95, 945–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsang MJ, and Bernhardt TG (2015). A role for the FtsQLB complex in cytokinetic ring activation revealed by an ftsL allele that accelerates division. Mol. Microbiol 95, 925–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lambert A, Vanhecke A, Archetti A, Holden S, Schaber F, Pincus Z, Laub MT, Goley E, and Manley S (2018). Constriction rate modulation can drive cell size control and homeostasis in C. crescentus. iScience 4, 180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Modell JW, Kambara TK, Perchuk BS, and Laub MT (2014). A DNA damage-Induced, SOS-independent checkpoint regulates cell division in Caulobacter crescentus. PLoS Biol. 12, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Modell JW, Hopkins AC, and Laub MT (2011). A DNA damage checkpoint in Caulobacter crescentus inhibits cell division through a direct interaction with FtsW. Genes Dev. 25, 1328–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lariviere PJ, Mahone CR, Santiago-Collazo G, Howell M, Daitch AK, Zeinert R, Chien P, Brown PJB, and Goley ED (2019). An Essential Regulator of Bacterial Division Links FtsZ to Cell Wall Synthase Activation. Curr. Biol 29, 1460–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yousif SY, Broome-Smith JK, and Spratt BG (1985). Lysis of Escherichia coli by β-lactam antibiotics: Deletion analysis of the role of penicillin-binding proteins 1A and 1B. J. Gen. Microbiol 131, 2839–2847. [DOI] [PubMed] [Google Scholar]
- 25.Typas A, Banzhaf M, Van Den Berg Van Saparoea B, Verheul J, Biboy J, Nichols RJ, Zietek M, Beilharz K, Kannenberg K, Von Rechenberg M, et al. (2010). Regulation of peptidoglycan synthesis by outer-membrane proteins. Cell 143, 1097–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paradis-Bleau C, Markovski M, Uehara T, Lupoli TJ, Walker S, Kahne DE, and Bernhardt TG (2010). Lipoprotein cofactors located in the outer membrane activate bacterial cell wall polymerases. Cell 143, 1110–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Banzhaf M, van den Berg van Saparoea B, Terrak M, Fraipont C, Egan A, Philippe J, Zapun A, Breukink E, Nguyen-Distèche M, den Blaauwen T, et al. (2012). Cooperativity of peptidoglycan synthases active in bacterial cell elongation. Mol. Microbiol. 85, 179–194. [DOI] [PubMed] [Google Scholar]
- 28.Bertsche U, Kast T, Wolf B, Fraipont C, Aarsman MEG, Kannenberg K, Von Rechenberg M, Nguyen-Distèche M, Den Blaauwen T, Höltje JV, et al. (2006). Interaction between two murein (peptidoglycan) synthases, PBP3 and PBP1B, in Escherichia coli. Mol. Microbiol 61, 675–690. [DOI] [PubMed] [Google Scholar]
- 29.Pazos M, Peters K, Casanova M, Palacios P, VanNieuwenhze M, Breukink E, Vicente M, and Vollmer W (2018). Z-ring membrane anchors associate with cell wall synthases to initiate bacterial cell division. Nat. Commun 9, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee TK, Meng K, Shi H, and Huang KC (2016). Single-molecule imaging reveals modulation of cell wall synthesis dynamics in live bacterial cells. Nat. Commun 7, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vigouroux A, Cordier B, Aristov A, Alvarez L, Özbaykal G, Chaze T, Oldewurtel ER, Matondo M, Cava F, Bikard D, et al. (2020). Class-A penicillin binding proteins do not contribute to cell shape but repair cell-wall defects. Elife 9, 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dion MF, Kapoor M, Sun Y, Wilson S, Ryan J, Vigouroux A, van Teeffelen S, Oldenbourg R, and Garner EC (2019). Bacillus subtilis cell diameter is determined by the opposing actions of two distinct cell wall synthetic systems. Nat. Microbiol 4, 1294–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Straume D, Piechowiak KW, Olsen S, Stamsås GA, Berg KH, Kjos M, Heggenhougen MV, Alcorlo M, Hermoso JA, and Håvarstein LS (2020). Class A PBPs have a distinct and unique role in the construction of the pneumococcal cell wall. PNAS 117, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mueller EA, Egan AJ, Breukink E, Vollmer W, and Levin PA (2019). Plasticity of Escherichia coli cell wall metabolism promotes fitness and antibiotic resistance across environmental conditions. Elife 8, 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peters K, Kannan S, Rao VA, Biboy J, Vollmer D, Erickson SW, Lewis RJ, Young KD, and Vollmer W (2016). The redundancy of peptidoglycan carboxypeptidases ensures robust cell shape maintenance in Escherichia coli. MBio 7, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garcia Del Portillo F, and De Pedro MA (1990). Differential effect of mutational impairment of penicillin-binding proteins 1A and 1B on Escherichia coli strains harboring thermosensitive mutations in the cell division genes ftsA, ftsQ, ftsZ, and pbpB. J. Bacteriol 172, 5863–5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ranjit DK, Jorgenson MA, and Young KD (2017). PBP1B glycosyltransferase and transpeptidase activities play different essential roles during the de novo regeneration of rod morphology in Escherichia coli. J. Bacteriol 199, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Auer GK, Lee TK, Rajendram M, Cesar S, Miguel A, Huang KC, and Weibel DB (2016). Mechanical genomics identifies diverse modulators of bacterial cell stiffness. Cell Syst. 2, 402–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tolufashe GF, Sabe VT, Ibeji CU, Ntombel T, Govender T, Maguire GEM, Kruger HG, Lamichhane G, and Honarparvar B (2018). Structure and function of L,D- and D,D-transpeptidase family enzymes from Mycobacterium tuberculosis. Curr. Med. Chem 26, 1–17. [DOI] [PubMed] [Google Scholar]
- 40.Glauner B, Holtje JV, and Schwarz U (1988). The composition of the murein of Escherichia coli. J. Biol. Chem 263, 10088–10095. [PubMed] [Google Scholar]
- 41.Morè N, Martorana AM, Biboy J, Otten C, Winkle M, Serrano CKG, Montón Silva A, Atkinson L, Yau H, Breukink E, et al. (2019). Peptidoglycan remodeling enables Escherichia coli to survive severe outer membrane assembly defect. MBio 10, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hugonnet JE, Mengin-Lecreulx D, Monton A, den Blaauwen T, Carbonnelle E, Veckerlé C, Yves VB, van Nieuwenhze M, Bouchier C, Tu K, et al. (2016). Factors essential for L,D-transpeptidase-mediated peptidoglycan cross-linking and β-lactam resistance in Escherichia coli. Elife 5, 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peters K, Pazos M, Edoo Z, Hugonnet JE, Martorana AM, Polissi A, VanNieuwenhze MS, Arthur M, and Vollmer W (2018). Copper inhibits peptidoglycan LD-transpeptidases suppressing β-lactam resistance due to bypass of penicillin-binding proteins. Proc. Natl. Acad. Sci. U. S. A 115, 10786–10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mainardi JL, Legrand R, Arthur M, Schoot B, Van Heijenoort J, and Gutmann L (2000). Novel mechanism of β-lactam resistance due to bypass of DD- transpeptidation in Enterococcus faecium. J. Biol. Chem 275, 16490–16496. [DOI] [PubMed] [Google Scholar]
- 45.Lavollay M, Arthur M, Fourgeaud M, Dubost L, Marie A, Veziris N, Blanot D, Gutmann L, and Mainardi JL (2008). The peptidoglycan of stationary-phase Mycobacterium tuberculosis predominantly contains cross-links generated by L,D-transpeptidation. J. Bacteriol 190, 4360–4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cameron TA, Anderson-Furgeson J, Zupan JR, Zik JJ, and Zambryski PC (2014). Peptidoglycan synthesis machinery in Agrobacterium tumefaciens during unipolar growth and cell division. MBio. 5, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schoonmaker MK, Bishai WR, and Lamichhane G (2014). Nonclassical transpeptidases of Mycobacterium tuberculosis alter cell size, morphology, the cytosolic matrix, protein localization, virulence, and resistance to β-lactams. J. Bacteriol 196, 1394–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stankeviciute G, Miguel AV, Radkov A, Chou S, Huang KC, and Klein EA (2019). Differential modes of crosslinking establish spatially distinct regions of peptidoglycan in Caulobacter crescentus. Mol. Microbiol 111, 995–1008. [DOI] [PubMed] [Google Scholar]
- 49.Billini M, Biboy J, Kühn J, Vollmer W, and Thanbichler M (2019). A specialized MreB-dependent cell wall biosynthetic complex mediates the formation of stalk-specific peptidoglycan in Caulobacter crescentus. 15, 1–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmidt J, and Stanier RY (1966). The development of cellular stalks in bacteria. J. Cell Biol 28, 423–436. [DOI] [PMC free article] [PubMed] [Google Scholar]


