Abstract
With the approval and distribution of demonstrably safe COVID-19 vaccines bearing exceptionally high efficacy profiles, it may be tempting to envision a return to “normal” in the coming months. However, if there is one lesson to be learned from the ongoing pandemic, it is that, in a world of evolving zoonotic viruses, we must be better prepared for the next deadly outbreak. While the acute nature of the COVID-19 pandemic demanded a highly specific approach, it is advisable to consider the breadth of seemingly endless possibilities in our approach to managing the next inevitable occurrence of an outbreak. Though there is little chance of discovering a “magic pill” to combat all future pathogens, the highly conserved nature of non-surface viral proteins exposes an “Achilles’ heel” in the structural genome of viral pathogens. Herein, we consider the potential of targeting such proteins to develop broad-spectrum therapeutics for the future. To illustrate this point, we outline the therapeutic potential of targeting the nonstructural protein 16 methyltransferase, which is conserved across most coronaviruses.
Keywords: COVID-19, methyltransferase, coronavirus, mRNA capping
1. Introduction
The novel virus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) replicates in the host cytoplasm, underlying its dependence upon an autonomous mRNA capping mechanism (cap-1/m7GpppNm) [1]. This capping mechanism is achieved by the highly conserved nonstructural protein (NSP) 16, which is a 2′-O-methyltransferase (2′-O-MTase). The catalytic activity of this enzyme is essential both for replication in the host and for evasion of the host’s innate immune response, thereby allowing replication [1,2,3,4]. There is a high level of NSP16 structural conservation across most coronaviruses, including SARS-CoV (Figure 1) and Middle East respiratory syndrome coronavirus (MERS-CoV) [1,2,3,4]. Since all of these viruses have posed significant health risks in recent years, the structural conservation makes NSP16 an attractive therapeutic target for both the current pandemic as well as future outbreaks by yet-to-be discovered coronavirus variants.
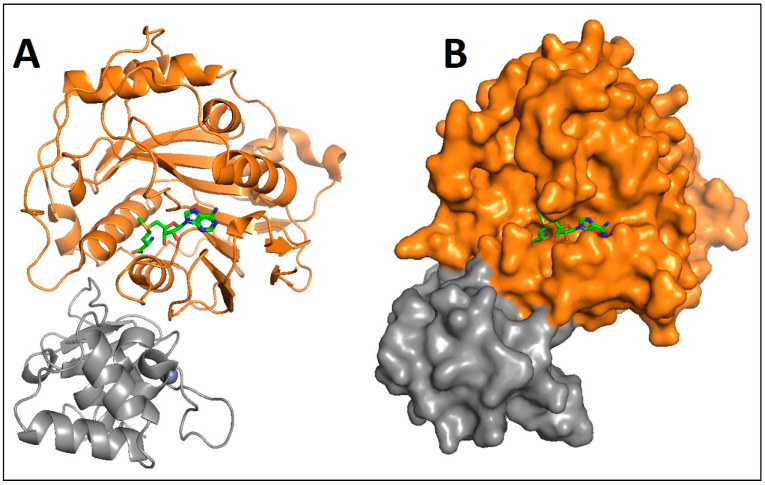
Figure 1.
Ribbon (A) and Surface (B) representation of NSP16 (orange) in complex with NSP10 (grey). S-adenosylhomocysteine (green) is shown at the active site of NSP16. The crystal structure for the NSP16/10 complex was downloaded from the RCSB protein data bank (pdb ID 3R24) [5]. Figures were captured using MacPyMol.
2. Discussion and Conclusions
The Middle East respiratory syndrome coronavirus (MERS-CoV) NSP16 is an S-adenosyl-L-methionine (SAM)-dependent 2-O-methyltransferase (2-O-MTase) that methylates the ribose 2-OH of the first transcribed nucleotide (N1) of viral RNA cap structures. This 2-O-MTase activity is regulated by NSP10. The 2-O methylation prevents virus detection by innate cell immunity mechanisms and viral translation inhibition by the interferon-stimulated IFIT-1 protein. The mechanism for binding the RNA-substrate, SAM, capping the RNA substrate and releasing SAH is illustrated in Aouadi et al. 2017 [4]. The NSP16/NSP10 2-O-MTase activity is sensitive to known MTase inhibitors including sinefungin [2,4]. Thus, there is great potential in the identification of both active site and distal site inhibitors of NSP16 as a means of preventing coronaviruses from propagating.
The process of developing a broad-spectrum coronavirus methyltransferase inhibitor can be further facilitated by screening commercially available molecules from existing databases for virtual screening and molecular docking [6,7,8]. An example of this followed a similar approach targeting a 16N methyltransferase with molecular docking techniques through which raltegravir and maraviroc emerged as potential candidates for the treatment of coronavirus infections [9]. Both of these antiviral drugs were originally approved in 2007 for HIV indications. Thus, the repurposing of drugs combined with targeting-based drug development is an efficient methodology of drug development for future viral infections.
The greater value in targeting such an enzyme is that it is highly conserved across a range of viruses. As a result, in addition to developing a therapeutic that will target the coronavirus associated with the current outbreak, one would also be developing a broad-spectrum therapeutic that will be useful against known coronaviruses, future coronaviruses, and perhaps other groups of viral pathogens altogether. This “broad-spectrum” approach, when more broadly applied across a range of viral threats, represents a more efficient and comprehensive means of proactively managing future pandemics.
Conflicts of Interest
The authors declare no conflict of interest. Brown is a guest editor for the Special Issue of Diseases, “Epigenetics and Disease”, and receives no remuneration for this work.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Dong S., Sun J., Mao Z., Wang L., Lu Y.-L., Li J. A guideline for homology modeling of the proteins from newly discovered betacoronavirus, 2019 novel coronavirus (2019-nCoV) J. Med. Virol. 2020:1–7. doi: 10.1002/jmv.25768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ke M., Chen Y., Wu A., Sun Y., Su C., Wu H., Jin X., Tao J., Wang Y., Ma X., et al. Short peptides derived from the interaction domain of SARS coronavirus nonstructural protein nsp10 can suppress the 2′-O-methyltransferase activity of nsp10/nsp16 complex. Virus Res. 2012;167:322–328. doi: 10.1016/j.virusres.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Decroly E., Debarnot C., Ferron F., Bouvet M., Coutard B., Imbert I., Gluais L., Papageorgiou N., Sharff A., Bricogne G., et al. Crystal structure and functional analysis of the SARS-Coronavirus RNA cap 2′-O-Methyltransferase nsp10/nsp16 complex. PLoS Pathog. 2011;7:e1002059. doi: 10.1371/journal.ppat.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aouadi W., Blanjoie A., Vasseur J.-J., Debart F., Canard B., Decroly E. Binding of the methyl donor S-Adenosyl-L-Methionine to Middle East respiratory syndrome Coronavirus 2′-O-Methyltransferase nsp16 promotes recruitment of the allosteric activator nsp10. J. Virol. 2017;91:e02217-16. doi: 10.1128/JVI.02217-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y., Su C., Ke M., Jin X., Xu L., Zhang Z., Wu A., Sun Y., Yang Z., Tien P., et al. Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2′-O-methylation by nsp16/nsp10 protein complex. PLoS Pathog. 2011;7:e1002294. doi: 10.1371/journal.ppat.1002294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sohag A.A.M., Hannan M.A., Rahman S., Hossain M., Hasan M., Khan M.K., Khatun A., Dash R., Uddin M.J. Revisiting potential druggable targets against SARS-CoV-2 and repurposing therapeutics under preclinical study and clinical trials: A comprehensive review. Drug Dev Res. 2020;81:919–941. doi: 10.1002/ddr.21709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chéron N., Yu C., Kolawole A., Shakhnovich E., Wobus C. Archives of Virology. Volume 160. Springer; Berlin, Germany: 2015. Repur-Posing of rutin for the inhibition of norovirus replication; pp. 2353–2358. [DOI] [PubMed] [Google Scholar]
- 8.Irwin J.J., Shoichet B.K. ZINC—A free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2015;45:177–182. doi: 10.1021/ci049714+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tazikeh-Lemeski E., Moradi S., Raoufi R., Shahlaei M., Janlou M.A.M., Zolghadri S. Targeting SARS-COV-2 non-structural protein 16: A virtual drug repurposing study. J. Biomol. Struct. Dyn. 2020;23:1–14. doi: 10.1080/07391102.2020.1779133. [DOI] [PMC free article] [PubMed] [Google Scholar]