Abstract
The neurological damage of Alzheimer’s disease (AD) is thought to be irreversible upon onset of dementia-like symptoms, as it takes years to decades for occult pathologic changes to become symptomatic. It is thus necessary to identify individuals at risk for the development of the disease before symptoms manifest in order to provide early intervention. Surrogate markers are critical for early disease detection, stratification of patients in clinical trials, prediction of disease progression, evaluation of response to treatment, and also insight into pathomechanisms. Here, we review the evidence for a number of microRNAs that may serve as biomarkers with possible mechanistic insights into the AD pathophysiologic processes, years before the clinical manifestation of the disease.
Keywords: plasma microRNAome, MCI, epigenetic dysregulation
1. Introduction
Neurodegenerative diseases are chronic illnesses that cause progressive and protracted neurological decline, with late onset, sporadic Alzheimer’s disease (AD) being the most common. AD results in devastating cognitive, emotional, physical, social, and financial consequences for the patients and their families alike. Impaired cognitive function is a hallmark of AD and often one of the first symptoms. However, changes in cognitive function develop slowly over time, and patients are diagnosed at an advanced stage of cellular and molecular pathology [1,2]. The mechanisms that initiate AD and enable its progression are not well understood but result in the unrelenting spread of neuronal damage, including loss of synapses, metabolic and structural abnormalities, inflammation, and ultimately cell death, believed to be strongly related to the combination of pathohistological hallmarks, neuritic plaques and neurofibrillary tangles, and their building blocks, amyloid-beta and tau, respectively [3]. Extracellular amyloid-beta (Aβ40–42) peptide deposits in the form of neuritic plaques are the sine qua non for AD diagnosis on brain autopsy; however, the degree of amyloidosis does not correlate with the severity of AD symptoms [4]. Intracellular hyperphosphorylated tau accumulations give rise to the second AD hallmark, neurofibrillary tangles, but the biochemical basis for aberrant tau phosphorylation in AD is poorly understood [5]. Nevertheless, cerebrospinal fluid (CSF) levels of Aβ42, t-tau (total tau), and p-tau (tau phosphorylated at threonine 181) are considered core biomarkers to support AD diagnosis [6,7,8]. Two major problems are associated with current, conventional AD biomarkers.
First, as recognized by the National Institute of Aging and Alzheimer’s Association (NIA-AA), the current biomarkers are relatively difficult to access because the mechanisms that isolate and protect the central nervous system from injury also act as barriers to diagnosis [9]. To that end, recently reported plasma biomarker p-tau217 represents a step forward and a promise as p-tau217 plasma levels distinguished AD from other neurodegenerative diseases in multiple international cohorts [10]. However, it is unknown how early in the trajectory of AD p-tau217 plasma levels start to increase, which represents the remaining gap of knowledge about prognostic AD biomarkers that could identify the population at risk and the population in which therapeutic interventions may be deployed to halt molecular events predating cognitive decline. Second, cortical measurements of amyloid-beta and tau depositions via magnetic resonance imaging (MRI) and positron emission tomography (PET) [9] are expensive and thus not accessible on the scale needed for a public health challenge such as AD [8,11]. Last but not the least, the NIA-AA research framework aims to define AD biologically and thus seeks to expand AD biomarkers according to the scientific progress in the field of AD pathogenesis [9].
2. Small Non-Coding RNAs Regulate Brain Health
The failure to diagnose AD at an early stage of molecular pathology is considered to be the major reason why treatments have failed in clinical trials [12]. The hippocampus is among the first brain structures to show AD-associated pathology—neuritic plaques, neurofibrillary tangles, and synaptic and neuronal loss [3,13,14]. Deregulated hippocampal gene expression has been observed during aging and neurodegeneration in animal models and in human patients [15,16,17,18,19,20]. Small non-coding RNAs (ncRNAs) regulate gene expression in response to genome–environment interactions. In order to be applicable in routine screenings in primary care settings, AD biomarkers must be minimally invasive and inexpensive while able to identify individuals in need of further, more invasive, and more expensive diagnostic procedures [21]. Circulating small ncRNAs are expected to serve as such biomarkers [22,23,24,25,26,27,28,29,30,31,32]. The purpose of this review is to highlight evidence that evaluating circulating levels of a set of small ncRNAs might serve as AD prognostic and diagnostic biomarkers for consideration by the NIAA-AA research framework [9]. We deem this consideration to be particularly timely because RNA therapeutics are emerging as a promising approach in central nervous system (CNS) diseases [33,34,35,36].
The vast majority (~95%) of circulating small ncRNAs are microRNAs (miRNAs, miRs), ~19–22 nucleotides long, evolutionary conserved, and most extensively studied members of the ncRNA family [37]. They are transcribed by RNA polymerase II or RNA polymerase III into primary miRNA (Pri-miRNA) and further digested by microprocessor complexes to a Pre-miRNA, which is a hairpin loop-like structure roughly 70 nucleotides in length. This structure is then transported out of the nucleus into the cytoplasm via an exportin five transporter and then is processed by Dicer and TAR-RNA binding proteins (TRBP) to a miRNA duplex structure. Helicase further separates the duplex structure into two single strands, a guide strand and a passenger strand. The passenger strand is degraded, while the guide strand goes on to complex with Ago2 proteins to form the mature miRNA and serves as a regulator of post-transcription gene expression via pairing with messenger RNA (mRNA) [37]. Accumulated evidence suggests that miRNAs are secreted in extracellular space in vesicle-free form or in a microvesicle-encapsulated form bound to Ago2 proteins that provide resistance to nucleases, conveying stability to miRNAs in biofluids [38]. Consistently, miRNAs are extremely stable in cell-free environments and resistant to thaw–freeze cycles [23].
Although aging represents the largest risk for sporadic, late onset AD, sociological, medical, and nutritional factors are thought to play a role [39,40]. Some of these epigenetic factors become influential rather early in life and under the umbrella term of “early life stress (ELS)” affect the onset of dementia as shown in monozygotic twin studies [41,42]. ELS rodent models suggest specific pathways through which genomic and epigenomic interaction affects neuroinflammation, cerebral lipid metabolism, brain insulin resistance, and myelin disintegration, in a manner that may ultimately lead to AD-associated histopathology [39].
A recent review of 191 miRNAs across 35 human studies measuring miRNA levels in blood, serum or plasma identified 30 miRNAs, replicated in more than one study, as dysregulated miRNAs in psychoses [43]. These 30 miRNAs are known to regulate pathways governing brain development and plasticity [43]. Consistently, depletion of the enzyme needed for miRNA biogenesis, Dicer, results is neurodevelopmental abnormalities including reduced progenitor numbers, abnormal neuronal differentiation, and thinner cortical wall in the mouse brain [44]. Further, miRNAs expressed in the human brain [45] regulate neuronal differentiation and synaptic plasticity [46,47]. Recent studies point toward a widespread involvement of ncRNA families in local translation within neuronal cells, including less abundant small RNAs (PIWI-interacting RNAs (piRNAs), endogenous small interfering RNAs (endo-siRNAs)) and long ncRNAs (circular RNAs (circRNAs), long intergenic ncRNAs (lincRNAs)). The emerging picture is an intricate crosstalk between different ncRNA families, mRNAs, and RNA-binding proteins (RBPs) orchestrating local dendritic proteome in an activity-dependent manner. The alterations in the activity of dendritic miRNAs, miR-132, miR-134, and miR-138 at the synapse have been linked with the pathogenesis of psychoses and neurodegenerative diseases [48], as the role of miRNAs in learning and memory in health and in AD has been extensively documented [33,49,50,51,52,53]. Consistently, miRNAs implicated in cognitive dysfunction associated with neurodegeneration were identified in CSF and postmortem brains of AD patients and also in mouse models of AD [33,54].
3. miRNAs as Diagnostic and Prognostic AD Biomarker Candidates
The majority of studies focused on epigenetic dysregulation as the source of biomarkers of preclinical AD have focused on the use of peripheral blood. Peripheral blood mononuclear cells (PBMCs) are one of the major cellular components to blood and are one the major sources of miRNAs [23]. They are of particular interest due to their innate ability to respond to internal and external stimuli and store information via epigenetic mechanism including DNA methylation, histone acetylation, and small ncRNA, including miRNA [23]. As potential biomarkers for prodromal and presymptomatic stages of AD, miRNAs, thanks to novel methods for their detection, represent a cost-efficient approach to screening persons for AD risk to identify individuals in need for further evaluation such as CSF analysis and imaging studies measuring AT(N) markers.
Kenny et al. (2019) evaluated diagnostic and prognostic potential of plasma miRNA profiling by seeking to differentiate between aged controls and patients with MCI and AD and also to predict cognitive decline in a five-year longitudinal study [55]. The discovery cohort included 31 control subjects and 30 patients with minimal cognitive impairment (MCI) enrolled in single-center community-based study with neurological, neuropsychological, and genetic information available. The longitudinal cohort included 18 subjects with progressions from cognitively healthy to dementia or stable MCI with longitudinal samples collected at 1–2-year intervals over a 5-year time period. miR-206 emerged as a biomarker for AD and for predicting the conversion from MCI to AD [55]. Altered levels of miR-206 in MCI and AD may be related to its target, brain-derived neurotrophic factor (BDNF) [56]. BDNF protein and mRNA levels have been reported to decrease in individuals with MCI and AD, resulting in synaptic loss and cell death, central to AD pathology [57,58,59,60].
Two recent studies, through diverse approaches, assessed performance of CSF miRNAs as biomarkers for cognitive decline. Using next-generation sequencing to study small ncRNAome in CSF exosomes, Jain and colleagues (2019) identified a combined signature consisting of three miRNAs and three piRNAs suitable to detect AD with an area under the curve (AUC) of 0.83 in a replication cohort and, importantly, predicted the conversion of MCI patients to AD with an AUC of 0.86 for the piRNA signature [54]. When combined with pTau and Aβ 42/40 ratio, the small ncRNA signature yielded an AUC of 0.98 [54]. Sandau and colleagues (2020) measured the expression of 17 miRNAs, implicated in AD based on available literature, in CSF samples from AD, MCI, and cognitively intact controls with the objective to test the ability of these implicated miRs to identify prodromal AD in MCI [61]. Five miRNAs (miR-142-3p, miR-146a-5p, miR-146b-5p, miR-193a-5p, miR-365a-3p) showed a linear trend of decreasing median expression across the ordered diagnoses (control to MCI to AD) and, importantly, when combined with Aβ42:T-tau levels, predicted MCI with increased classification performance [61]. The five CSF miRNAs suggested to predict AD in subjects with MCI [61] did not overlap with three CSF exosomal miRNAs (miR-34c, miR-30a, and miR-27a) within the small ncRNA signature to detect and predict conversion to AD [54], but common to both sets were the predicted targets-various cellular processes maintaining neuronal homeostasis, including response to pathologic stress [62].
Here, we review the evidence from multiple studies in support of the notion that a few miRNAs may serve as prognostic and diagnostic AD biomarkers with a mechanistic dimension. This group of candidate AD biomarker miRNAs highlights dysregulation in cellular functions underlying pathophysiologic processes likely years before the manifestation of AD symptoms.
3.1. miR-29
β-site amyloid precursor protein cleaving enzyme 1 (BACE1) is a proteolytic enzyme that cleaves full-length amyloid precursor proteins (APP), contributing to the accumulation of neuritic plaques containing β-amyloid peptide Aβ40–42 in the neuropil, the AD hallmark. Hebert et al. (2008) reported high BACE 1 expression in postmortem brains of AD patients as well as a decreased expression of several miRNAs with candidate binding sites within 3′ UTRs of BACE1 [51]. They went on to demonstrate that BACE1 is a miRNA target gene and that the levels of expression of BACE1 and miR-29 a/b-1 do correlate in AD brains. The analysis of CSF samples of AD patients and non-demented controls by Muller et al. (2016) confirmed the association between the level of miR-29a expression and AD diagnosis [63]. miR-29a-3p levels have been reported as decreased in CSF of AD patients in other studies as well, although not always consistently [61]. To demonstrate the mechanistic relationship, Hebert et al. performed a series of in vitro functional essays proving that miR-29a/b-1 indeed can modulate BACE1 activity and thus β-amyloid production [51]. Excitingly, Pereira and colleagues (2016) developed technology to successfully deliver the recombinant pre-miR-29b into the cytoplasm in vitro, eliciting decreased expression of BACE1 and Aβ42 [64]. Together, these results suggest the pathophysiological, disease-mechanism-based connection between a level of a miRNA expression in clinically accessible biospecimens and the levels of an enzyme responsible for the accumulation of a histological hallmark of the disease. Equally exciting are the promising inroads into technology needed for successful delivery of miRNA-based therapeutics [64].
3.2. miR-34
A major risk factor for AD is aging, a process with many regulating genes including SIRT genes that encode sirtuins, proteins that govern cellular functions negatively impacted by aging including metabolism and response to oxidative stress and to DNA damage. Owczarz and colleagues (2017) showed that the PBMCs of long-lived individuals contained a low expression of SIRT1 mRNA coupled with high expression of miR-34a [65]. Data mining techniques to identify miRNA that both have a large number of targets and are differentially expressed in brains of AD patients identified increased expression of miR-34a in the temporal cortex of AD patients to correlate with the severity of AD pathology and also reported increased expression of miR-34a in 3xTg-AD mouse model [66]. Prior to those studies, Zovoilis and colleagues (2011) found miR-34c to be upregulated in the hippocampus of mouse models for amyloid deposition and age-associated memory decline and also in brain tissue from AD patients [33,67]. This group went further to show that the inhibition of miR-34c function ameliorated memory impairment and gene expression in a mouse model for amyloid deposition [33]. miR-34c expression is thought to grow in response to pathological stress [62] consistent with the evidence that stress-induced gene deregulation promotes cognitive decline [68]. Remarkably, miR-34c also belonged to a 6-member small ncRNA signature obtained from CSF exosomes suitable to detect AD and predict the conversion of MCI patients to AD [54]. The mechanistic dimension of miR-34 expression level as an AD biomarker is further strengthened by the evidence for the role of miR-34 in aging and neurodegeneration in Drosophila [69] and Caenorhabditis elegans [70], highlighting the evolutionary conserved regulation of fundamental cellular functions by miRNAs.
3.3. miR-146
In addition to miR34a, miR146a was in the 5-member group of miRNAs studied in the context of AD based on the evidence from data mining from various miRNA target gene databases predicting their role in cellular functions thought to be compromised in AD [66]. Furthermore, in the analysis of CSF samples from AD, MCI, and cognitively normal controls, miR-146a-5p was among five CSF miRNA with levels decreasing as normal controls developed MCI and MCI progressed to AD [61]. miR-146a-5p was also among four miRNAs that were upregulated in mouse models of AD (APPtg and TAUtg) and also in the brains of AD patients [71]. Interestingly, induction of miR-146a via gastrointestinal Gram-negative bacteria is thought to promote pathogenic stimulation of innate–immune and neuroinflammatory pathways relevant for inflammation-mediated pathologic consequences of amyloidosis in AD brain [72].
Additional miRNAs reported to be differentially expressed in postmortem brain tissue, CSF, PBMC, plasma, or serum of AD patients are presented in Table 1.
Table 1.
Additional miRNAs reported to be differentially expressed in postmortem brain tissue, cerebrospinal fluid (CSF), peripheral blood mononuclear cells (PBMCs), plasma, or serum of Alzheimer’s disease (AD) patients.
| miRNA | Subjects | Material | Results | Reference |
|---|---|---|---|---|
| miR-125b | 10 AD patients, 5 controls | frontal cortex | increased in AD patients, correlating with increased tau phosphorylation | Banzhaf-Strathmann, Benito et al., 2014 [34] |
| miR-132-3p | hippocampi from 41 AD patients and 23 controls; prefrontal cortices from 49 individuals at Braak stage I-VI and 7 controls | hippocampus, prefrontal cortex |
decreased in AD | Lau, Bossers et al., 2013 [73] |
| miR-107 | 19 individuals, Braak stage 0-VI | temporal cortex (Brodmann area 21/22) | decreasing with increased Braak stage | Nelson and Wang, 2010 [74] |
| miR-212 miR-132 | brains from 11 AD, 7 “high pathology controls”, 9 controls; plasma from 16 AD patients, 16 AD-MCI subjects, and 31 controls | brains and neurally derived plasma exosomes | decreased in AD brains and neurally-derived plasma exosomes | Cha, Mengel et al., 2019 [75] |
| 12 miR signature | 48 AD patients, 22 controls | PBMC | separated AD patients from controls with 93.3% accuracy | Leidinger and Backes et al., 2013 [26] |
| miR-146a | 30 AD patients, 28 controls | Plasma | no correlation with AD diagnosis, but correlation with age and lower MMSE scores | Maffioletti, Milanesi et al., 2020 [76] |
| miR-501-3p | 27 AD patients, 18 controls | Serum | decreased in AD compared to age-matched controls; lower expression correlated with lower MMSE scores | Hara, Kikuchi et al., 2017 [30] |
| miR-125b | 22 AD patients, 18 subjects with non-inflammatory, and 8 subjects with inflammatory neurological conditions; 10 frontotemporal dementia patients | Serum | decreased in AD compared to non-inflammatory neurological conditions | Galimberti and Villa et al., 2014 [27] |
4. Conclusions
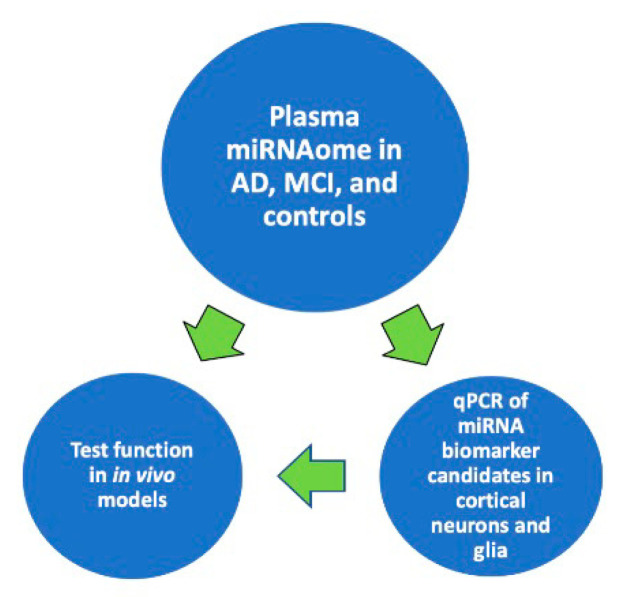
The accumulated evidence suggests that harnessing the power of ongoing large epidemiologic studies could provide robust classifier enabling the correlation of cognitive status in densely phenotyped individuals with plasma miRNAome to create and expand the tools for predicting and diagnosing AD. As shown in Diagram 1, it would then be possible to define the relationship between the expression of a circulating miRNA biomarker and the expression of that biomarker in the brain cells central to AD pathophysiology. Finally, it would be feasible to understand the functional consequence of a human disease miRNA biomarker in models of AD to enable mechanistic approaches toward the design of novel preventive and therapeutic interventions (Figure 1).
Figure 1.
Proposed approach toward discovering and evaluating circulating miRNA biomarker candidates for prognosing and diagnosing AD-associated cognitive decline.
Author Contributions
Conceptualization, C.K. and I.D.; writing—draft preparation, C.K.; writing—review and editing, I.D., A.F., J.K.B.; fund acquisition J.K.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Institutes of Health grants R21 AG059195 and RF1 AG057768-01A1.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mangialasche F., Solomon A., Winblad B., Mecocci P., Kivipelto M. Alzheimer’s disease: Clinical trials and drug development. Lancet Neurol. 2010;9:702–716. doi: 10.1016/S1474-4422(10)70119-8. [DOI] [PubMed] [Google Scholar]
- 2.Bateman R.J., Xiong C., Benzinger T.L., Fagan A.M., Goate A., Fox N.C., Marcus D.S., Cairns N.J., Xie X., Blazey T.M., et al. Clinical and biomarker changes in dominantly inherited Alzheimer’s disease. N. Engl. J. Med. 2012;367:795–804. doi: 10.1056/NEJMoa1202753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ittner L.M., Gotz J. Amyloid-beta and tau--a toxic pas de deux in Alzheimer’s disease. Nat. Rev. Neurosci. 2011;12:65–72. doi: 10.1038/nrn2967. [DOI] [PubMed] [Google Scholar]
- 4.Nelson P.T., Alafuzoff I., Bigio E.H., Bouras C., Braak H., Cairns N.J., Castellani R.J., Crain B.J., Davies P., Del Tredici K., et al. Correlation of Alzheimer disease neuropathologic changes with cognitive status: A review of the literature. J. Neuropathol. Exp. Neurol. 2012;71:362–381. doi: 10.1097/NEN.0b013e31825018f7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oba T., Saito T., Asada A., Shimizu S., Iijima K.M., Ando K. Microtubule Affinity Regulating Kinase 4 with an Alzheimer’s disease-related mutation promotes tau accumulation and exacerbates neurodegeneration. J. Biol. Chem. 2020;295:17138–17147. doi: 10.1074/jbc.RA120.014420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McKhann G.M., Knopman D.S., Chertkow H., Hyman B.T., Jack C.R., Jr., Kawas C.H., Klunk W.E., Koroshetz W.J., Manly J.J., Mayeux R., et al. The diagnosis of dementia due to Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:263–269. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dubois B., Feldman H.H., Jacova C., Hampel H., Molinuevo J.L., Blennow K., DeKosky S.T., Gauthier S., Selkoe D., Bateman R., et al. Advancing research diagnostic criteria for Alzheimer’s disease: The IWG-2 criteria. Lancet Neurol. 2014;13:614–629. doi: 10.1016/S1474-4422(14)70090-0. [DOI] [PubMed] [Google Scholar]
- 8.Lee J.C., Kim S.J., Hong S., Kim Y. Diagnosis of Alzheimer’s disease utilizing amyloid and tau as fluid biomarkers. Exp. Mol. Med. 2019;51:1–10. doi: 10.1038/s12276-019-0250-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jack C.R., Jr., Bennett D.A., Blennow K., Carrillo M.C., Dunn B., Haeberlein S.B., Holtzman D.M., Jagust W., Jessen F., Karlawish J., et al. NIA-AA Research Framework: Toward a biological definition of Alzheimer’s disease. Alzheimers Dement. 2018;14:535–562. doi: 10.1016/j.jalz.2018.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palmqvist S., Janelidze S., Quiroz Y.T., Zetterberg H., Lopera F., Stomrud E., Su Y., Chen Y., Serrano G.E., Leuzy A., et al. Discriminative Accuracy of Plasma Phospho-tau217 for Alzheimer Disease vs Other Neurodegenerative Disorders. JAMA. 2020;324:772–781. doi: 10.1001/jama.2020.12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verberk I.M.W., Slot R.E., Verfaillie S.C.J., Heijst H., Prins N.D., van Berckel B.N.M., Scheltens P., Teunissen C.E., van der Flier W.M. Plasma Amyloid as Prescreener for the Earliest Alzheimer Pathological Changes. Ann. Neurol. 2018;84:648–658. doi: 10.1002/ana.25334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schneider L.S., Mangialasche F., Andreasen N., Feldman H., Giacobini E., Jones R., Mantua V., Mecocci P., Pani L., Winblad B., et al. Clinical trials and late-stage drug development for Alzheimer’s disease: An appraisal from 1984 to 2014. J. Intern. Med. 2014;275:251–283. doi: 10.1111/joim.12191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mesulam M.M. Neuroplasticity failure in Alzheimer’s disease: Bridging the gap between plaques and tangles. Neuron. 1999;24:521–529. doi: 10.1016/S0896-6273(00)81109-5. [DOI] [PubMed] [Google Scholar]
- 14.Haass C., Selkoe D.J. Soluble protein oligomers in neurodegeneration: Lessons from the Alzheimer’s amyloid beta-peptide. Nat. Rev. Mol. Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 15.Lu T., Pan Y., Kao S.Y., Li C., Kohane I., Chan J., Yankner B.A. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- 16.Berchtold N.C., Cribbs D.H., Coleman P.D., Rogers J., Head E., Kim R., Beach T., Miller C., Troncoso J., Trojanowski J.Q., et al. Gene expression changes in the course of normal brain aging are sexually dimorphic. Proc. Natl. Acad. Sci. USA. 2008;105:15605–15610. doi: 10.1073/pnas.0806883105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Selwood S.P., Parvathy S., Cordell B., Ryan H.S., Oshidari F., Vincent V., Yesavage J., Lazzeroni L.C., Murphy G.M., Jr. Gene expression profile of the PDAPP mouse model for Alzheimer’s disease with and without Apolipoprotein E. Neurobiol. Aging. 2009;30:574–590. doi: 10.1016/j.neurobiolaging.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 18.Cao K., Chen-Plotkin A.S., Plotkin J.B., Wang L.S. Age-correlated gene expression in normal and neurodegenerative human brain tissues. PLoS ONE. 2010;5:e13098. doi: 10.1371/journal.pone.0013098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peleg S., Sananbenesi F., Zovoilis A., Burkhardt S., Bahari-Javan S., Agis-Balboa R.C., Cota P., Wittnam J.L., Gogol-Doering A., Opitz L., et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328:753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- 20.Ray M., Zhang W. Analysis of Alzheimer’s disease severity across brain regions by topological analysis of gene co-expression networks. BMC Syst. Biol. 2010;4:136. doi: 10.1186/1752-0509-4-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Molinuevo J.L., Ayton S., Batrla R., Bednar M.M., Bittner T., Cummings J., Fagan A.M., Hampel H., Mielke M.M., Mikulskis A., et al. Current state of Alzheimer’s fluid biomarkers. Acta Neuropathol. 2018;136:821–853. doi: 10.1007/s00401-018-1932-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheinerman K.S., Tsivinsky V.G., Umansky S.R. Analysis of organ-enriched microRNAs in plasma as an approach to development of Universal Screening Test: Feasibility study. J. Transl. Med. 2013;11:304. doi: 10.1186/1479-5876-11-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rao P., Benito E., Fischer A. MicroRNAs as biomarkers for CNS disease. Front. Mol. Neurosci. 2013;6:39. doi: 10.3389/fnmol.2013.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen R.J., Kelly G., Sengupta A., Heydendael W., Nicholas B., Beltrami S., Luz S., Peixoto L., Abel T., Bhatnagar S. MicroRNAs as biomarkers of resilience or vulnerability to stress. Neuroscience. 2015;305:36–48. doi: 10.1016/j.neuroscience.2015.07.045. [DOI] [PubMed] [Google Scholar]
- 25.Kumar S., Vijayan M., Bhatti J.S., Reddy P.H. MicroRNAs as Peripheral Biomarkers in Aging and Age-Related Diseases. Prog. Mol. Biol. Transl. Sci. 2017;146:47–94. doi: 10.1016/bs.pmbts.2016.12.013. [DOI] [PubMed] [Google Scholar]
- 26.Leidinger P., Backes C., Deutscher S., Schmitt K., Mueller S.C., Frese K., Haas J., Ruprecht K., Paul F., Stahler C., et al. A blood based 12-miRNA signature of Alzheimer disease patients. Genome Biol. 2013;14:R78. doi: 10.1186/gb-2013-14-7-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galimberti D., Villa C., Fenoglio C., Serpente M., Ghezzi L., Cioffi S.M., Arighi A., Fumagalli G., Scarpini E. Circulating miRNAs as potential biomarkers in Alzheimer’s disease. J. Alzheimers Dis. 2014;42:1261–1267. doi: 10.3233/JAD-140756. [DOI] [PubMed] [Google Scholar]
- 28.Cheng L., Doecke J.D., Sharples R.A., Villemagne V.L., Fowler C.J., Rembach A., Martins R.N., Rowe C.C., Macaulay S.L., Masters C.L., et al. Prognostic serum miRNA biomarkers associated with Alzheimer’s disease shows concordance with neuropsychological and neuroimaging assessment. Mol. Psychiatry. 2015;20:1188–1196. doi: 10.1038/mp.2014.127. [DOI] [PubMed] [Google Scholar]
- 29.Schwarz E.C., Backes C., Knorck A., Ludwig N., Leidinger P., Hoxha C., Schwar G., Grossmann T., Muller S.C., Hart M., et al. Deep characterization of blood cell miRNomes by NGS. Cell Mol. Life Sci. 2016;73:3169–3181. doi: 10.1007/s00018-016-2154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hara N., Kikuchi M., Miyashita A., Hatsuta H., Saito Y., Kasuga K., Murayama S., Ikeuchi T., Kuwano R. Serum microRNA miR-501-3p as a potential biomarker related to the progression of Alzheimer’s disease. Acta Neuropathol. Commun. 2017;5:10. doi: 10.1186/s40478-017-0414-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagaraj S., Laskowska-Kaszub K., Debski K.J., Wojsiat J., Dabrowski M., Gabryelewicz T., Kuznicki J., Wojda U. Profile of 6 microRNA in blood plasma distinguish early stage Alzheimer’s disease patients from non-demented subjects. Oncotarget. 2017;8:16122–16143. doi: 10.18632/oncotarget.15109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Basavaraju M., de Lencastre A. Alzheimer’s disease: Presence and role of microRNAs. Biomol. Concepts. 2016;7:241–252. doi: 10.1515/bmc-2016-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zovoilis A., Agbemenyah H.Y., Agis-Balboa R.C., Stilling R.M., Edbauer D., Rao P., Farinelli L., Delalle I., Schmitt A., Falkai P., et al. microRNA-34c is a novel target to treat dementias. EMBO J. 2011;30:4299–4308. doi: 10.1038/emboj.2011.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Banzhaf-Strathmann J., Benito E., May S., Arzberger T., Tahirovic S., Kretzschmar H., Fischer A., Edbauer D. MicroRNA-125b induces tau hyperphosphorylation and cognitive deficits in Alzheimer’s disease. EMBO J. 2014;33:1667–1680. doi: 10.15252/embj.201387576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salta E., De Strooper B. Noncoding RNAs in neurodegeneration. Nat. Rev. Neurosci. 2017;18:627–640. doi: 10.1038/nrn.2017.90. [DOI] [PubMed] [Google Scholar]
- 36.Hanna J., Hossain G.S., Kocerha J. The Potential for microRNA Therapeutics and Clinical Research. Front. Genet. 2019;10:478. doi: 10.3389/fgene.2019.00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 38.Kumar S., Reddy P.H. Are circulating microRNAs peripheral biomarkers for Alzheimer’s disease? Biochim. Biophys. Acta. 2016;1862:1617–1627. doi: 10.1016/j.bbadis.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lemche E. Early Life Stress and Epigenetics in Late-onset Alzheimer’s Dementia: A Systematic Review. Curr. Genom. 2018;19:522–602. doi: 10.2174/1389202919666171229145156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nativio R., Donahue G., Berson A., Lan Y., Amlie-Wolf A., Tuzer F., Toledo J.B., Gosai S.J., Gregory B.D., Torres C., et al. Dysregulation of the epigenetic landscape of normal aging in Alzheimer’s disease. Nat. Neurosci. 2018;21:497–505. doi: 10.1038/s41593-018-0101-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gatz M., Pedersen N.L., Berg S., Johansson B., Johansson K., Mortimer J.A., Posner S.F., Viitanen M., Winblad B., Ahlbom A. Heritability for Alzheimer’s Disease- The Study of Dementia in Swedish Twins. J. Gerontol. 1997;52:M117-25. doi: 10.1093/gerona/52A.2.M117. [DOI] [PubMed] [Google Scholar]
- 42.Roman G.C., Mancera-Paez O., Bernal C. Epigenetic Factors in Late-Onset Alzheimer’s Disease: MTHFR and CTH Gene Polymorphisms, Metabolic Transsulfuration and Methylation Pathways, and B Vitamins. Int. J. Mol. Sci. 2019;20:319. doi: 10.3390/ijms20020319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gruzdev S.K., Yakovlev A.A., Druzhkova T.A., Guekht A.B., Gulyaeva N.V. The Missing Link: How Exosomes and miRNAs can Help in Bridging Psychiatry and Molecular Biology in the Context of Depression, Bipolar Disorder and Schizophrenia. Cell Mol. Neurobiol. 2019;39:729–750. doi: 10.1007/s10571-019-00684-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kawase-Koga Y., Otaegi G., Sun T. Different timings of Dicer deletion affect neurogenesis and gliogenesis in the developing mouse central nervous system. Dev. Dyn. 2009;238:2800–2812. doi: 10.1002/dvdy.22109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sempere L.F., Freemantle S., Pitha-Rowe I., Moss E., Dmitrovsky E., Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Presutti C., Rosati J., Vincenti S., Nasi S. Non coding RNA and brain. BMC Neurosci. 2006;7(Suppl. 1):S5. doi: 10.1186/1471-2202-7-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fiore R., Siegel G., Schratt G. MicroRNA function in neuronal development, plasticity and disease. Biochim. Biophys. Acta. 2008;1779:471–478. doi: 10.1016/j.bbagrm.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 48.Bicker S., Lackinger M., Weiss K., Schratt G. MicroRNA-132, -134, and -138: A microRNA troika rules in neuronal dendrites. Cell Mol. Life Sci. 2014;71:3987–4005. doi: 10.1007/s00018-014-1671-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin Q., Wei W., Coelho C.M., Li X., Baker-Andresen D., Dudley K., Ratnu V.S., Boskovic Z., Kobor M.S., Sun Y.E., et al. The brain-specific microRNA miR-128b regulates the formation of fear-extinction memory. Nat. Neurosci. 2011;14:1115–1117. doi: 10.1038/nn.2891. [DOI] [PubMed] [Google Scholar]
- 50.Gao J., Wang W.Y., Mao Y.W., Graff J., Guan J.S., Pan L., Mak G., Kim D., Su S.C., Tsai L.H. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 2010;466:1105–1109. doi: 10.1038/nature09271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hebert S.S., Horre K., Nicolai L., Papadopoulou A.S., Mandemakers W., Silahtaroglu A.N., Kauppinen S., Delacourte A., De Strooper B. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/beta-secretase expression. Proc. Natl. Acad. Sci. USA. 2008;105:6415–6420. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aksoy-Aksel A., Zampa F., Schratt G. MicroRNAs and synaptic plasticity--a mutual relationship. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014;369:20130515. doi: 10.1098/rstb.2013.0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cohen J.E., Lee P.R., Chen S., Li W., Fields R.D. MicroRNA regulation of homeostatic synaptic plasticity. Proc. Natl. Acad. Sci. USA. 2011;108:11650–11655. doi: 10.1073/pnas.1017576108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jain G., Stuendl A., Rao P., Berulava T., Pena Centeno T., Kaurani L., Burkhardt S., Delalle I., Kornhuber J., Hull M., et al. A combined miRNA-piRNA signature to detect Alzheimer’s disease. Transl. Psychiatry. 2019;9:250. doi: 10.1038/s41398-019-0579-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kenny A., McArdle H., Calero M., Rabano A., Madden S.F., Adamson K., Forster R., Spain E., Prehn J.H.M., Henshall D.C., et al. Elevated Plasma microRNA-206 Levels Predict Cognitive Decline and Progression to Dementia from Mild Cognitive Impairment. Biomolecules. 2019;9:734. doi: 10.3390/biom9110734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee S.T., Chu K., Jung K.H., Kim J.H., Huh J.Y., Yoon H., Park D.K., Lim J.Y., Kim J.M., Jeon D., et al. miR-206 regulates brain-derived neurotrophic factor in Alzheimer disease model. Ann. Neurol. 2012;72:269–277. doi: 10.1002/ana.23588. [DOI] [PubMed] [Google Scholar]
- 57.Panja D., Bramham C.R. BDNF mechanisms in late LTP formation: A synthesis and breakdown. Neuropharmacology. 2014;76 Pt C:664–676. doi: 10.1016/j.neuropharm.2013.06.024. [DOI] [PubMed] [Google Scholar]
- 58.Edelmann E., Lessmann V., Brigadski T. Pre- and postsynaptic twists in BDNF secretion and action in synaptic plasticity. Neuropharmacology. 2014;76 Pt C:610–627. doi: 10.1016/j.neuropharm.2013.05.043. [DOI] [PubMed] [Google Scholar]
- 59.Holsinger R.M., Schnarr J., Henry P., Castelo V.T., Fahnestock M. Quantitation of BDNF mRNA in human parietal cortex by competitive reverse transcription-polymerase chain reaction: Decreased levels in Alzheimer’s disease. Brain Res. Mol. Brain. Res. 2000;76:347–354. doi: 10.1016/S0169-328X(00)00023-1. [DOI] [PubMed] [Google Scholar]
- 60.Peng S., Wuu J., Mufson E.J., Fahnestock M. Precursor form of brain-derived neurotrophic factor and mature brain-derived neurotrophic factor are decreased in the pre-clinical stages of Alzheimer’s disease. J. Neurochem. 2005;93:1412–1421. doi: 10.1111/j.1471-4159.2005.03135.x. [DOI] [PubMed] [Google Scholar]
- 61.Sandau U.S., Wiedrick J.T., Smith S.J., McFarland T.J., Lusardi T.A., Lind B., Harrington C.A., Lapidus J.A., Galasko D.R., Quinn J.F., et al. Performance of Validated MicroRNA Biomarkers for Alzheimer’s Disease in Mild Cognitive Impairment. J. Alzheimers Dis. 2020;78:245–263. doi: 10.3233/JAD-200396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Haramati S., Navon I., Issler O., Ezra-Nevo G., Gil S., Zwang R., Hornstein E., Chen A. MicroRNA as repressors of stress-induced anxiety: The case of amygdalar miR-34. J. Neurosci. 2011;31:14191–14203. doi: 10.1523/JNEUROSCI.1673-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Muller M., Jakel L., Bruinsma I.B., Claassen J.A., Kuiperij H.B., Verbeek M.M. MicroRNA-29a Is a Candidate Biomarker for Alzheimer’s Disease in Cell-Free Cerebrospinal Fluid. Mol. Neurobiol. 2016;53:2894–2899. doi: 10.1007/s12035-015-9156-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pereira P.A., Tomas J.F., Queiroz J.A., Figueiras A.R., Sousa F. Recombinant pre-miR-29b for Alzheimer s disease therapeutics. Sci. Rep. 2016;6:19946. doi: 10.1038/srep19946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Owczarz M., Budzinska M., Domaszewska-Szostek A., Borkowska J., Polosak J., Gewartowska M., Slusarczyk P., Puzianowska-Kuznicka M. miR-34a and miR-9 are overexpressed and SIRT genes are downregulated in peripheral blood mononuclear cells of aging humans. Exp. Biol. Med. 2017;242:1453–1461. doi: 10.1177/1535370217720884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sarkar S., Jun S., Rellick S., Quintana D.D., Cavendish J.Z., Simpkins J.W. Expression of microRNA-34a in Alzheimer’s disease brain targets genes linked to synaptic plasticity, energy metabolism, and resting state network activity. Brain Res. 2016;1646:139–151. doi: 10.1016/j.brainres.2016.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Muller M., Kuiperij H.B., Claassen J.A., Kusters B., Verbeek M.M. MicroRNAs in Alzheimer’s disease: Differential expression in hippocampus and cell-free cerebrospinal fluid. Neurobiol. Aging. 2014;35:152–158. doi: 10.1016/j.neurobiolaging.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 68.Agis-Balboa R.C., Pinheiro P.S., Rebola N., Kerimoglu C., Benito E., Gertig M., Bahari-Javan S., Jain G., Burkhardt S., Delalle I., et al. Formin 2 links neuropsychiatric phenotypes at young age to an increased risk for dementia. EMBO J. 2017;36:2815–2828. doi: 10.15252/embj.201796821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu N., Landreh M., Cao K., Abe M., Hendriks G.J., Kennerdell J.R., Zhu Y., Wang L.S., Bonini N.M. The microRNA miR-34 modulates ageing and neurodegeneration in Drosophila. Nature. 2012;482:519–523. doi: 10.1038/nature10810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang J., Chen D., He Y., Melendez A., Feng Z., Hong Q., Bai X., Li Q., Cai G., Wang J., et al. MiR-34 modulates Caenorhabditis elegans lifespan via repressing the autophagy gene atg9. Age. 2013;35:11–22. doi: 10.1007/s11357-011-9324-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sierksma A., Lu A., Salta E., Vanden Eynden E., Callaerts-Vegh Z., D’Hooge R., Blum D., Buee L., Fiers M., De Strooper B. Deregulation of neuronal miRNAs induced by amyloid-beta or TAU pathology. Mol. Neurodegener. 2018;13:54. doi: 10.1186/s13024-018-0285-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao Y., Lukiw W.J. Microbiome-Mediated Upregulation of MicroRNA-146a in Sporadic Alzheimer’s Disease. Front. Neurol. 2018;9:145. doi: 10.3389/fneur.2018.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lau P., Bossers K., Janky R., Salta E., Frigerio C.S., Barbash S., Rothman R., Sierksma A.S., Thathiah A., Greenberg D., et al. Alteration of the microRNA network during the progression of Alzheimer’s disease. EMBO Mol. Med. 2013;5:1613–1634. doi: 10.1002/emmm.201201974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nelson P.T., Wang W.X. MiR-107 is reduced in Alzheimer’s disease brain neocortex: Validation study. J. Alzheimers Dis. 2010;21:75–79. doi: 10.3233/JAD-2010-091603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cha D.J., Mengel D., Mustapic M., Liu W., Selkoe D.J., Kapogiannis D., Galasko D., Rissman R.A., Bennett D.A., Walsh D.M. miR-212 and miR-132 Are Downregulated in Neurally Derived Plasma Exosomes of Alzheimer’s Patients. Front. Neurosci. 2019;13:1208. doi: 10.3389/fnins.2019.01208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Maffioletti E., Milanesi E., Ansari A., Zanetti O., Galluzzi S., Geroldi C., Gennarelli M., Bocchio-Chiavetto L. miR-146a Plasma Levels Are Not Altered in Alzheimer’s Disease but Correlate With Age and Illness Severity. Front. Aging Neurosci. 2019;11:366. doi: 10.3389/fnagi.2019.00366. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.