Abstract
The systematic bioanalytical characterization of the protein product of the DMD gene, which is defective in the pediatric disorder Duchenne muscular dystrophy, led to the discovery of the membrane cytoskeletal protein dystrophin. Its full-length muscle isoform Dp427-M is tightly linked to a sarcolemma-associated complex consisting of dystroglycans, sarcoglyans, sarcospan, dystrobrevins and syntrophins. Besides these core members of the dystrophin–glycoprotein complex, the wider dystrophin-associated network includes key proteins belonging to the intracellular cytoskeleton and microtubular assembly, the basal lamina and extracellular matrix, various plasma membrane proteins and cytosolic components. Here, we review the central role of the dystrophin complex as a master node in muscle fibers that integrates cytoskeletal organization and cellular signaling at the muscle periphery, as well as providing sarcolemmal stabilization and contractile force transmission to the extracellular region. The combination of optimized tissue extraction, subcellular fractionation, advanced protein co-purification strategies, immunoprecipitation, liquid chromatography and two-dimensional gel electrophoresis with modern mass spectrometry-based proteomics has confirmed the composition of the core dystrophin complex at the sarcolemma membrane. Importantly, these biochemical and mass spectrometric surveys have identified additional members of the wider dystrophin network including biglycan, cavin, synemin, desmoglein, tubulin, plakoglobin, cytokeratin and a variety of signaling proteins and ion channels.
Keywords: cytoskeleton, dystrophin, dystrophin–glycoprotein complex, dystroglycan, sarcolemma
1. Introduction
Following the identification of one of the largest genes in the human genome, the DMD gene [1], and the initial characterization of its full-length protein product [2,3], it became clear that dystrophin exists in a variety of isoforms ranging in molecular mass from approximately 71 to 427 kDa [4]. Dystrophins are widely distributed in the body, including voluntary striated muscles, the heart, the brain and various other organs systems [5]. Since the large Dp427-M isoform was shown to be tightly associated with several glycoproteins in skeletal muscle fibers [6,7,8,9,10,11], it was concluded that dystrophin probably functions as a molecular anchoring system in the subsarcolemmal cytoskeleton [12,13]. Comprehensive subcellular fractionation studies in combination with both immunoblotting and mass spectrometry have established that dystrophin is highly enriched in the sarcolemma fraction from skeletal muscle [14,15]. Cell biological studies using immunofluorescence microscopy and immunoelectron microscopy agree with these findings and have localized dystrophin to the cytoplasmic site of the plasmalemma in skeletal muscle fibers [16,17]. Subsequent biochemical and cell biological studies confirmed that the dystrophin–glycoprotein complex provides a stabilizing linkage between the intracellular actin cytoskeleton and laminin-211 in the basal lamina of the extracellular matrix [11,18,19]. Modern mass spectrometry-based proteomic approaches have confirmed the composition of the core complex that associates with dystrophin and dystroglycans at the sarcolemma membrane [14,20,21,22,23,24].
The almost complete loss of dystrophin isoform Dp427-M and concomitant reduction in dystrophin-associated proteins triggers the complex pathophysiology of Duchenne muscular dystrophy [7,25], an X-linked and multifaceted disorder of early childhood that primarily affects striated muscles [26], causing respiratory dysfunction, late-onset cardiomyopathy and scoliosis. Functional disturbances of the nervous system [27] and abnormal energy metabolism are also distinctive features of dystrophinopathy [28,29]. The systematic usage of genetic animal models with dystrophic symptoms was instrumental for the elucidation of the body-wide effects of dystrophin deficiency [30,31,32,33]. Skeletal muscle abnormalities are characterized by a complex molecular and cellular pathogenesis, encompassing complex changes in contractile fibers and their supporting connective tissue and neuronal cells [34]. Cycles of degeneration and regeneration, impaired innervation patterns, progressive necrosis, fatty tissue replacement, reactive myofibrosis and sterile inflammation were established as secondary consequences of a primary defect in the dystrophin gene [35,36,37].
In addition to the involvement of sarcolemmal abnormalities in highly progressive Duchenne muscular dystrophy and more benign forms of Becker muscular dystrophy, changes in the dystrophin complex are also involved in sarcoglycanopathies and alpha-dystroglycanopathies. Sarcoglycanopathies represent subtypes of limb–girdle muscular dystrophy that are caused by primary abnormalities in the dystrophin-associated sarcoglycans and alpha-dystroglycanopathies are associated with the abnormal glycosylation of alpha-dystroglycan [38]. Caveolinopathies are another group of disorders that affect the plasma membrane via alterations in caveolae invaginations. Primary abnormalities in caveolin-3 are associated with certain forms of muscular dystrophy and the altered expression of this protein is postulated to also play a pathophysiological role in dystrophinopathy [39]. Importantly, new findings on the underlying mechanisms of dystrophic alterations are crucial for the identification of robust and disease-specific biomarker molecules [40,41,42,43] and the development of novel diagnostic approaches [44], as well as the design of innovative therapies to restore the dystrophin complex and counteract secondary aspects involved in progressive muscle wasting [45,46,47,48,49].
In this review, an overview of the core members of the dystrophin–glycoprotein complex in normal skeletal muscle is provided, including dystroglycans, sarcoglyans, sarcospan, dystrobrevins and syntrophins. This article then focuses on recent biochemical, proteomic and cell biological investigations aimed at the systematic identification of indirectly associated proteins belonging to the wider dystrophin network. This includes crucial members of the extracellular matrix such as biglycan, the surface desmoglein complex, and members of the cytoskeletal network including vimentin, tubulin, desmin and cytokeratin, as well as the cavin–caveolin complex, plakoglobin, ion channels and various signaling proteins. These newer findings have modified the initial view of dystrophin being a purely structural component that functions as a molecular anchor and shock absorber in the membrane cytoskeleton. The Dp427-M isoform of skeletal muscle acts probably as a crucial integrator at the fiber periphery and is, in conjunction with the integrin–laminin axis, majorly involved in lateral force transmission. In addition, the dystrophin complex has been implicated to provide a master node for cytoskeletal organization and cellular signaling events, which are characterized by the linkage of dystrophin to ion channels, the insulin signaling pathway, nitric oxide-based regulatory processes, kinase signaling pathways and excitation–contraction coupling [36].
2. The Core Dystrophin Complex in Skeletal Muscle
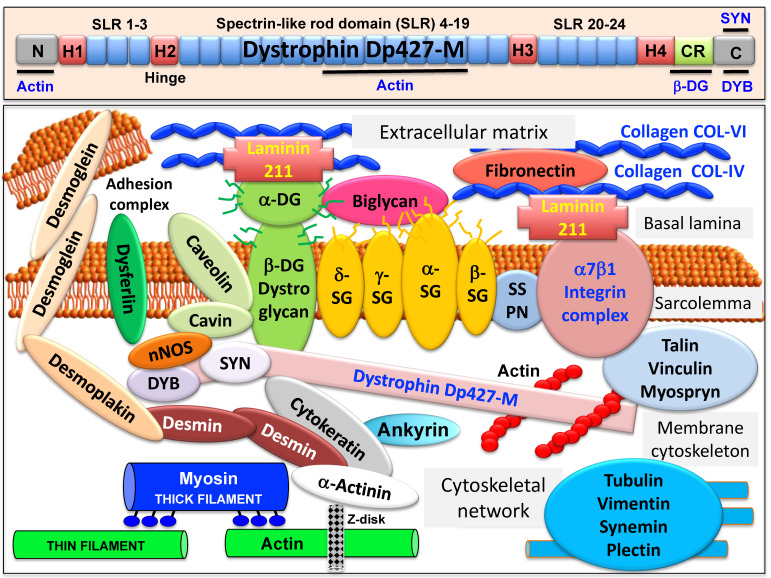
The full-length dystrophin isoform Dp427-M belongs to the class of giant muscle proteins [50] and consists of several distinct molecular domains as illustrated in the upper panel of Figure 1. This includes amino-terminal and central actin-binding domains, proline-rich hinge regions, spectrin-like rod domains and crucial carboxy-terminal binding sites for interactions with plasmalemmal and cytosolic components [51,52,53,54]. Dystrophin closely interacts with the integral proteins beta-dystroglycan, alpha/beta/gamma/delta-sarcoglyan and sarcospan of the sarcolemma, the extracellular receptor alpha-dystroglycan and laminin-211, the cytosolic components alpha/beta-dystrobrevin and alpha/beta-syntrophin, and the cortical actin cytoskeleton [9,10,11,12,13], as shown in the lower panel of Figure 1.
Figure 1.
Overview of the domain structure of dystrophin and the diverse interactions of the dystrophin–glycoprotein complex in skeletal muscle tissues. The upper panel shows a diagrammatic presentation of the main molecular domains of dystrophin isoform Dp427-M, including actin-binding sites at the N-terminus and central rod domain, proline-rich hinge regions (H1 to H4), spectrin-like rod (SLR) domains 1–3, 4–19 and 20–24, a cysteine-rich domain with binding sites for integral beta-dystroglycan (DG), the cysteine-rich domain (CR) and the C-terminus with binding sites for dystrobrevin (DYB) and syntrophin (SYN). The lower panel shows a model of the spatial configuration of the dystrophin complexome in skeletal muscle fibers. Shown is the dystrophin core complex consisting of the dystrophin isoform Dp427-M, dystroglycans (DG), sarcoglycans (SG), sarcospan (SSPN), syntrophins (SYN) and dystrobrevins (DYB), as well as the wider dystrophin-associated network that forms associations with the extracellular matrix, the sarcolemma, the cytoskeleton and the sarcomere.
Sedimentation analysis of the isolated dystrophin complex suggests a monomeric structure with an apparent molecular mass of 1.2 MDa [55]. In Duchenne muscular dystrophy, alterations in the expression of members of the dystrophin network are closely related to key pathophysiological features in dystrophin-deficient muscles, including degeneration-regeneration cycles, progressive fiber degeneration, fibrosis and chronic inflammation. Detailed reviews have been published on the composition of the core dystrophin complex [19,56,57,58], as well as the role of dystrophin and its associated glycoprotein complex in the multisystemic complications of dystrophinopathy and pathophysiological crosstalk throughout the body [34,35,36,37,59]. Therefore, this article does not attempt to recapitulate these biochemical and pathobiochemical issues in detail, but refers instead to specific aspects that are crucial for our general understanding of the wider functional role of the dystrophin complexome in normal skeletal muscle tissue.
2.1. The Dystrophin Node in Skeletal Muscle
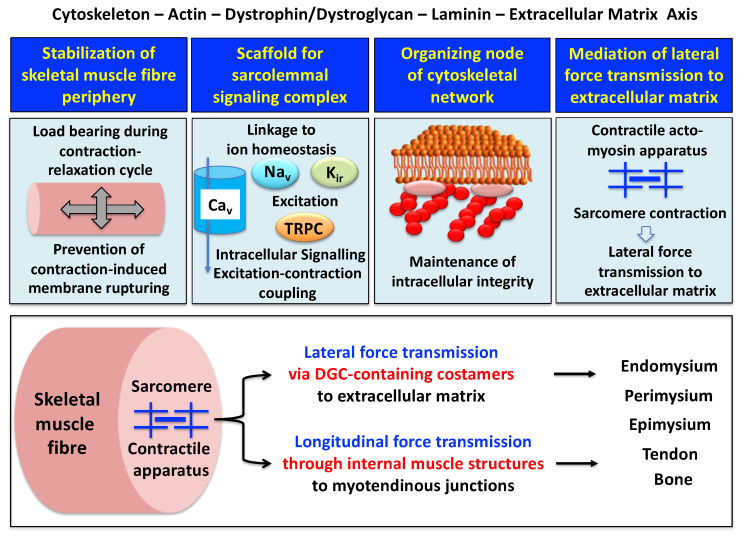
A model of the spatial configuration of the core dystrophin complex and its association with the extracellular matrix on the one hand and the intracellular cytoskeletal network of contractile fibers on the other hand is presented in the lower panel of above Figure 1. Specific aspects of dystrophin interaction patterns are discussed in detail in below sections. The cell biological concept that the dystrophin–glycoprotein complex occupies a central position at the fiber periphery is summarized in Figure 2. The diagram shows that the dystrophin-associated surface complex forms an organizing node that is majorly involved in (i) the provision of sarcolemmal membrane integrity via a stabilizing linkage between the intracellular actin cytoskeleton and the extracellular matrix protein laminin [11,13], (ii) the establishment of a molecular scaffold and anchoring system for ion channels and enzymes to mediate cellular signaling processes [60,61] (iii) the organization of actin filament attachment and its associated cytoskeletal network [62,63], and (iv) the mediation of lateral force transmission from sarcomeric contraction to the endomysium and its connected layers of the extracellular matrix [64,65].
Figure 2.
Outline of the main functions of the dystrophin node and its associated protein complex as integrators of fiber stability, cellular signaling. cytoskeletal organization and lateral force transmission. The upper panels summarize the main functions of the trans-sarcolemmal axis formed by the intracellular actin cytoskeleton, the dystrophin–dystroglycan complex, the basal lamina component laminin and the extracellular matrix. The lower panel illustrates the physiological concept of force transmission in skeletal muscles, which can be divided into a laterally and a longitudinally acting system. In conjunction with other costameric proteins, the dystrophin–glycoprotein complex (DGC) is majorly involved in lateral force transmission to the extracellular matrix.
2.2. The Sarcolemmal Dystrophin Complex and Lateral Force Transmission
The peripheral structure of skeletal muscle fibers functions as an essential physical barrier with its protective basal lamina. The underlying sarcolemma membrane provides the physiological structure for the efficient exchange of ions, metabolites and signaling molecules within the contractile system [66]. The plasmalemma is connected to the terminal cisternae region of the sarcoplasmic reticulum at the triad junctions via its invaginations, the transverse tubules. This intricate membrane assembly and its associated Ca2+-handling apparatus is involved in the fine regulation of excitation–contraction coupling, muscle relaxation and ion homeostasis, and encounters enormous physical strain during contraction–relaxation cycles [67]. The dystrophin-associated complex is implicated to act as a biomolecular shock absorber by linking the basal lamina to the actin cytoskeleton and thus preventing rupturing of this muscle membrane system [68,69,70].
At specialized costamere regions, which play both a mechanical and a signaling role, the dystrophin complex forms in conjunction with the integrin–vinculin–talin axis a link to the contractile sarcomere units [63,71]. This bridging structure is postulated to provide an indirect means of lateral force transmission to the collagen-rich muscle exterior, in addition to longitudinal forces that are transmitted directly from the contractile apparatus through the cytosol to the myotendinous junction [72,73,74]. In skeletal muscle fibers, the characteristic longitudinal pattern of A bands and I bands reflect the organization of myosin-containing thick filaments and actin-containing thin filaments with their contractile sarcomeric units, which are positioned between Z discs. Following the energy-dependent crossbridge coupling between myosin heads and actin filaments, thin filaments slide past thick filaments. The force generated by this sarcomeric shortening event is partially transmitted by a lateral direct force between Z-disk structures and the M-line regions of neighboring myofibrils. Costamere structures at the fiber periphery play a central role as sensors of the relative mechanical load and support force transduction across the muscle plasma membrane. Contractile force is then further transmitted to the complex layers of the extracellular matrix, consisting of endomysium, perimysium and epimysium, towards the tendon and bone structure [65,72,73]. The second type of force transmission mechanism works by longitudinal means through internal muscle structures embedded in the cytosol. Both lateral and longitudinal coupling mechanisms act in parallel and ultimately transmit the force generated by the actomyosin apparatus in the sarcomere to the tendon and anchoring structures such as bone, as diagrammatically summarized in the lower panel of Figure 2. The dystrophin-associated dystroglycan subcomplex was shown to play a critical role in the sarcomeric cytoskeleton by limiting contraction-induced injury to skeletal muscle fibers [70].
The elucidation of the multifaceted functions of the dystrophin–glycoprotein complex in maintaining membrane stability during excitation–contraction–relaxation cycles, assisting lateral force transmission through costameres and providing a scaffold for anchoring surface receptors and maintaining cellular signaling mechanisms was carried out by multidisciplinary approaches. This included molecular genetics, biochemical purification strategies, structural/biophysical analysis, mass spectrometric proteomics analysis, bioinformatics, chemical crosslinking, cell biological characterization and comparative biomedical studies. In below sections, information on the findings of these bioanalyses is provided in relation to the individual members of the dystrophin–glycoprotein complex and its wider network of skeletal muscle proteins.
2.3. Muscle Dystrophin Dp427-M and Its Associated Glycoprotein Complex
The large muscle isoform of dystrophin is a rod-shaped protein [3] with considerable homology to the actinin superfamily of actin crosslinking components, which also includes utrophin and spectrin [75]. Both, dystrophin isoform Dp427-M of the sarcolemma and its autosomal homologue, utrophin isoform Up395-M of the neuromuscular junction, exhibit typical biochemical properties of cytoskeletal components, such as insolubility in non-ionic detergent and efficient extraction by alkaline treatment [76,77]. Compared to the main components of the contractile actomyosin apparatus and its regulatory sarcomeric elements, dystrophin represents a relatively minor component of the skeletal muscle fiber proteome. However, dystrophin isoform Dp427-M constitutes a considerable fraction of the subsarcolemmal cytoskeleton in contractile tissue [76]. This makes full-length dystrophin an important structural and functional component of the sarcolemmal lattice and costamere structures [71]. Besides being present in contractile fibers, dystrophin isoforms also exist in many non-muscle cells [78]. The various dystrophins are encoded by the 79 exon-spanning DMD gene, whereby seven different promoters drive the tissue-specific expression of the full-length isoforms Dp427-B in brain, Dp427-M in muscle and Dp427-P in Purkinje cells [4], as well as the shorter isoforms Dp260-R in retina [79], Dp140-B/K in brain and kidney [80], Dp116-S in Schwann cells [81] and Dp71-G in the brain [82] and a variety of other tissues including the spleen [83]. The promoter for Dp71 also produces the shortest known dystrophin isoform named Dp45, which is located in the central nervous system [84]. Of note, the central nervous system displays one of the greatest varieties of dystrophin isoforms, which are involved in synaptic modulation, neuronal excitability and signal integration. Brain Dp427-B is present in neurons of the cerebral cortex and in cerebellar Purkinje cells, Dp140-B is highly expressed during brain development and Dp71-G is located in both neurons and glia cells in the dentate gyrus [82]. Cognitive impairments and emotional disturbances in Duchenne patients are probably linked to altered dystrophin expression in the central nervous system and this is reflected by structural brain abnormalities [85]. The formation of dystrophin complexes and their involvement in dystrophinopathy-associated brain defects has been reviewed by Waite et al. [86].
The composition of the dystrophin–glycoprotein complex has been extensively investigated using a combination of digitonin-based solubilization, wheat germ agglutinin lectin chromatography, ion exchange chromatography and density gradient ultracentrifugation [7,14,55,87], as well as various chemical crosslinking and immunoprecipitation approaches [8,21,22,23,88,89]. Differential detergent extraction procedures [90], two-dimensional gel electrophoresis [10] or alkaline dissociation [87] can be used to isolate individual dystrophin subcomplexes or separate the dystrophin-associated glycoprotein complex from homogeneous dystrophin molecules. Based on these analyses, the core members of the dystrophin-associated complex can be divided into (i) cytosolic components alpha/beta-dystrobrevin [91,92] and alpha/beta-syntrophin [93,94] that interact with the cysteine-rich domain of dystrophin; (ii) integral glycoproteins, including the alpha/beta/gamma/delta-sarcoglyan subcomplex [95,96,97], the highly hydrophobic protein sarcospan [98,99,100] and the main carboxy-terminal dystrophin-binding partner beta-dystroglycan [101]; (iii) laminin-211 [102] and its extracellular receptor alpha-dystroglycan [103], which is a proteolytic cleavage product of the pre-dystroglycan molecule [104]; and (iv) the intracellular actin cytoskeleton that links to an amino-terminal and a rod domain site of full-length dystrophin [51,55,105].
As reviewed by Tarakci and Berger [97], the sarcoglycan subcomplex is initially assembled by the formation of a core between beta-sarcoglycan and delta-sarcoglycan, which subsequently recruits the other two sarcoglycans. Through interactions with sarcospan and additional dystrophin-associated proteins, the sarcoglycan complex secures the formation and mechanical maintenance of the sarcolemmal dystrophin complex. Besides its integrating role in membrane stabilization, the sarcoglycan subcomplex can be chemically modified during fiber contraction, which provides the transduction of information on relative contractile force into cellular signaling [97]. Interestingly, both components of the dystroglycan subcomplex are products of the same gene, DAG1, which encodes a pre-pro-protein version of alpha/beta-dystroglycan that includes a signaling peptide and both subunits [101,106]. The precursor protein is extensively modified by N- and O-glycosylation and undergoes proteolytic processing that generates the integral glycoprotein beta-dystroglycan and the extracellular laminin-binding receptor alpha-dystroglycan [103]. Thus, the two dystroglycans form the backbone of the trans-sarcolemmal linkage between the basal lamina component laminin-211 and the dystrophin-associated actin cytoskeleton in the subsarcolemmal region of skeletal muscle [11,13]. The phosphorylation of beta-dystroglycan, especially intracellular tyrosine residues [107], is a crucial step during interactions with signaling proteins [108]. The phosphorylation of the cysteine-rich region in the carboxy-terminal domain of dystrophin also plays a key role in strengthening the interaction with beta-dystroglycan [109]. Thus, post-translational modifications are important modulators of dynamic associations within the dystrophin–dystroglycan axis.
3. Proteomic and Biochemical Characterization of the Dystrophin Network in Skeletal Muscle
Over the last two decades, biochemical analyses and mass spectrometry-based proteomics were instrumental to confirm the composition of the dystrophin–glycoprotein complex. Based on these findings, proteomic approaches were extremely helpful for the subsequent identification of novel binding proteins that belong to the wider dystrophin complexome.
3.1. Proteomics of the Dystrophin Complex from Skeletal Muscle
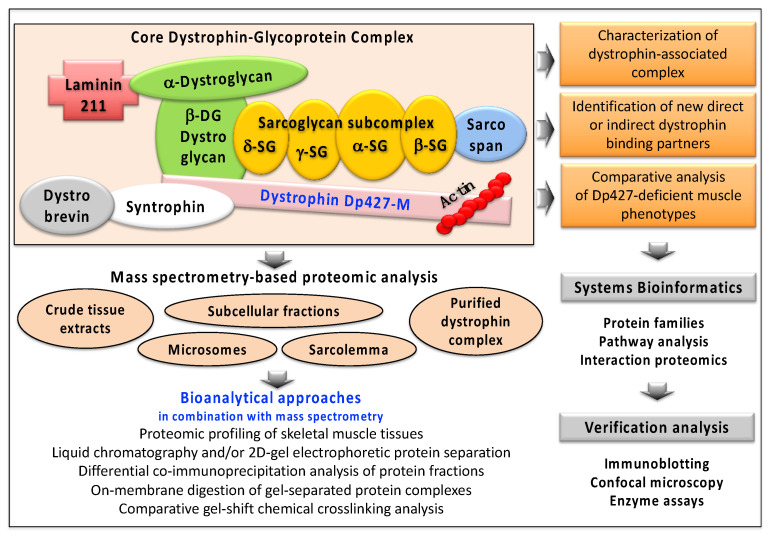
Following the discovery and initial biochemical and cell biological characterization of dystrophin, a variety of large-scale screening analyses were carried out to identify proteins that exhibit a close linkage to this membrane cytoskeletal component. Figure 3 illustrates how proteomic surveys and the usage of advanced mass spectrometry has helped to refine this search for novel dystrophin-binding partners and related proteins.
Figure 3.
Outline of the biochemical, proteomic and bioinformatic strategy to identify and characterize novel binding proteins of the dystrophin complexome. Mass spectrometry-based proteomic studies have used various starting materials, such as crude tissue extracts, subcellular fractions or purified protein complexes. Listed are the various bioanalytical techniques that are routinely used for the efficient separation of dystrophin-associated protein populations and their subsequent mass spectrometric and bioinformatic evaluation. To verify proteomic findings, often immuno-blotting, biochemical assays and cell biological methods are used.
Biochemical and proteomic screening studies have corroborated the direct binding partners of dystrophin, i.e., the integral glycoprotein beta-dystroglycan of the sarcolemma and the cytosolic components dystrobrevin, syntrophin and cortical actin [20,21,22,23,24,110,111]. The other main components of the core dystrophin complex were also identified by proteomics, including the sarcoglycans of the plasma membrane, the highly hydrophobic protein sarcospan and laminin-211 of the basal lamina [21,23]. In addition, comprehensive mass spectrometric studies identified further interaction patterns based on indirect associations with dystrophin isoform Dp427-M [21,22,23,24,89]. These new components of the wider dystrophin network include the extracellular matrix components collagen, fibronectin and biglycan, the plasmalemma proteins integrin, cavin and caveolin, and the cytoskeletal proteins cytokeratin, synemin, actinin, desmin, plectin, desmoglein, desmoplakin and tubulin.
When used in an optimized way, the chemical crosslinking technique can be utilized for the stabilization of fragile protein complexes that would otherwise disintegrate during elaborate subcellular fractionation and extensive biochemical isolation procedures. In protein biochemistry, chemical crosslinking is defined as the intra- and/or inter-molecular stabilization of protein molecules via covalent bonding. Since the chemical crosslinking technique is capable of determining molecular changes in macromolecular oligomerization, it is an ideal bioanalytical tool for studying dynamic protein interaction patterns. The select joining of two or more protein species is achieved by the incubation of protein mixtures with a large variety of crosslinking agents that differ in their molecular spacer arm length, their solubility and chemical reactivity. Frequently used crosslinkers contain reactive groups that are suitable for optimum interactions with functional groups in protein molecules such as primary amines. For example, the crosslinker used in below Figure 4 is a homo-bifunctional, water-soluble, non-cleavable and amine-reactive agent that is highly useful for joining proteins in complex assemblies under physiological conditions. Following the addition of crosslinking agents, protein samples are usually incubated for 30 min at 25 °C for 30 min and then the reaction quenched by the addition of ammonium acetate. A convenient way to analyze the occurrence of crosslinked protein complexes is the determination of an altered electrophoretic mobility in one- or two-dimensional gels [89]. A crucial issue with this method is the potential occurrence of random crosslinking events. However, this can be avoided by employing a low ratio of crosslinking agent to protein and using proper conditions in relation to length of incubation time, efficient quenching of the crosslinking reaction, temperature, buffering and pH-value [112]. Chemical crosslinking has been established as an excellent method in proteomics for studying sensitive protein–protein interactions [113]. Since a large collection of crosslinking molecules exist that greatly vary in their specific spacer arm length, experiments with different agents can be designed to determine spatial constraints within proteins. If a chemical crosslinker with an extremely short spacer arm length is employed, this approach can even stabilize protein interactions within biological membranes that exist in extremely close proximity to another [114].
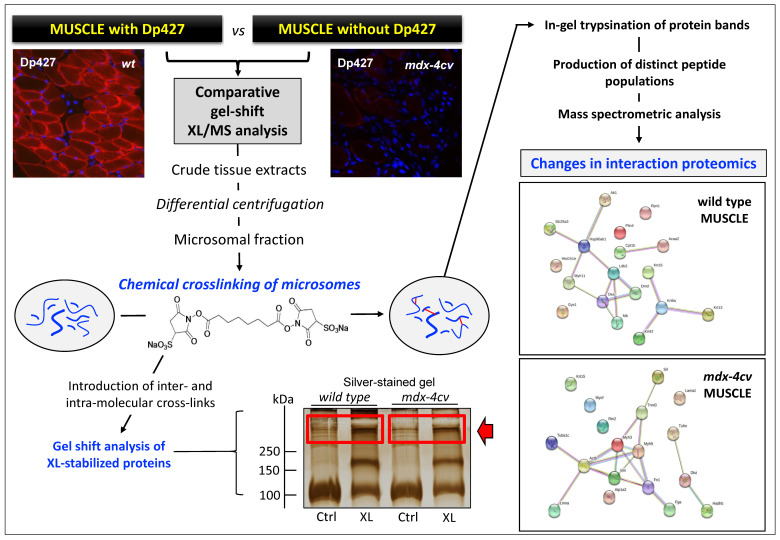
Figure 4.
Gel shift-based chemical crosslinking mass spectrometry (XL-MS) analysis of skeletal muscle specimens. Shown is the flow chart of a typical interaction proteomic and bioinformatic analysis using a combination of chemical crosslinking with the 11.4 Å agent BS3 (bis [sulfosuccinimidyl] suberate), gel electrophoretic separation of crosslinked protein complexes, mass spectrometric analysis for the identification of proteins with a crosslinker-induced shift in electrophoretic mobility and the bioinformatic evaluation of potential protein–protein interactions. As previously described in detail [112], the various experimental steps involved in this particular type of gel-shift XL-MS analysis include (i) the preparation of crude skeletal muscle tissue extracts by homogenization, (ii) the enrichment of the microsomal membrane fraction using differential centrifugation, (iii) the joining of proteins by incubation with a homo-bifunctional, water-soluble, non-cleavable and amine-reactive crosslinking agent, (iv) the quenching of the crosslinking reaction, (v) extraction of crosslinked proteins and gel electrophoretic separation, (vi) excision of crosslinked protein bands of high molecular mass, (vii) in-gel proteolysis of proteins using trypsin, (viii) separation of peptides by liquid chromatography, (ix) mass spectrometric identification of proteins, and (x) systems bioinformatic analysis of altered protein–protein interaction patterns. The position of the analyzed high-molecular-mass protein bands, using comparative mass spectrometry, are highlighted by a red box and are marked by an arrow. Protein interaction patterns were visualized with the bioinformatic STRING program [121]. The immunofluorescence microscopical images show transverse section of wild-type versus dystrophic mdx-4cv mouse gastrocnemius muscle. The sarcolemmal localization of dystrophin was stained in red with an antibody to isoform Dp427-M and nuclei were counterstained in blue with the fluorescent stain DAPI (4′,6-diamidino-2-phenylindole) that labels DNA molecules [111].
One of the earliest biochemical studies on the identification of dystrophin-associated proteins used chemical crosslinking [8] and these findings were confirmed by combining protein crosslinking with immunoblotting [88]. The combination of chemical crosslinking and mass spectrometric analysis established that a subpopulation of the sarcolemmal protein cavin-1 exists in a surface complex with beta-dystroglycan in skeletal muscle [89], which has previously been demonstrated in cardiac fibers [21]. Cavins are adapter proteins and mediate in conjunction with the structural caveolin proteins the formation and organization of small invaginations of the muscle plasma membrane, the bulb-like membrane pits called caveolae [115]. These structures are involved in lipid storage, endocytosis, cellular signaling and mechano-protection [116]. Freeze-fracture electron microscopy has established that the number and structure of caveolae are changed in dystrophinopathy, suggesting an important role of abnormal cavins in muscular dystrophy [39,117]. Of note, the crucial repair protein myoferlin [118] is also closely associated with caveolin and intrinsically involved in the maintenance and structural integrity of the sarcolemmal membrane, together with the Ca2+-dependent repair protein dysferlin [119].
Figure 4 outlines how a chemical crosslinking mass spectrometry (XL-MS) approach can be employed to study novel protein–protein interaction patterns, which are indirectly linked to dystrophin expression levels [112]. A variety of highly useful bioinformatic programs are available for the visualization of potential protein–protein interactions [120], as shown in the below images, which has been carried out with the STRING program [121]. In recent years, a variety of advances have occurred in XL-MS methodology, such as the development of MS-cleavable crosslinkers and software programs (XlinkX; MeroX; StavroX) that enable the identification of peptides directly from mass spectrometric data, and thus do not rely on gel electrophoresis or immunoblotting [113].
Co-sedimentation analysis of the dystrophin complex in combination with mass spectrometry identified a potential interaction with desmoglein [24]. The dystrophin–glycoprotein complex was enriched by established methods using an advanced subcellular membrane fractionation protocol [15] and detergent solubilization [6], followed by ion exchange and lectin affinity chromatography [7] and a final density gradient ultracentrifugation step [87]. The dystrophin complex was then separated by gradient gel electrophoresis and protein bands digested by on-membrane trypsination [20]. The co-sedimented proteins in the enriched dystrophin fraction were identified by peptide mass spectrometry. Besides the confirmatory identification of dystrophin and its tightly associated glycoproteins by a large number of unique peptide sequences, cytokeratin and the two cytolinker proteins desmoglein and desmoplakin [122] were shown to be present in the dystrophin fraction [24]. This finding agrees with the cell biological concept of the dystrophin complexome being involved in providing a structural lattice for linking individual components of the cytoskeletal network in skeletal muscle fibers. This includes dystrophin interactions with cytokeratin [123,124], synemin [125], plectin [126,127,128], as well as actinin, desmin and tubulin [129]. Intermediate filaments that contain the cytokeratin isoform K19 were shown to directly interact with the actin-binding region of Dp427-M [124].
Dystrophin was established to be directly involved in the organization and stabilization of costameric microtubules [129]. These diverse interactions of dystrophin with intermediate filaments and microtubules suggest that the Dp427-M isoform functions as a cytolinker molecule that maintains close interactions between the sarcolemmal dystroglycan complex and the intracellular cytoskeleton in contractile fibers. In relation to other cytosolic proteins, a close linkage of the dystrophin complex was shown with plakoglobin, which in turn binds to the insulin receptor [130], and the neuronal isoform nNOS of nitric oxide synthase [131]. Both proposed protein interactions provide key hubs for cellular signaling mechanisms within contractile fibers, such as organization of caveolae and the regulation of skeletal muscle size. Dystrophin-associated nitric oxide synthase, the enzyme responsible for the production of the key signaling molecule nitric oxide, was shown to play a regulatory role in costameres [132] and is involved in muscle fatigue, muscular atrophy and certain forms of muscular dystrophy [36,133,134]. In skeletal muscle tissues, nitric oxide levels are low under resting conditions and the production rate of this signaling molecule is greatly enhanced during repetitive muscle contractions. Crucial regulatory functions of nitric oxide in skeletal muscle include auto-regulation of blood flow, metabolic and bioenergetic integration at the level of glucose homeostasis and mitochondrial respiration, modulation of excitation–contraction coupling and contractile force generation, as well as myocyte differentiation [135,136]. The influence of nitric oxide on skeletal muscle function involves changes in redox-sensitive protein species, the activation of the second messenger molecule cyclic guanosine monophosphate and interactions with reactive oxygen species [135].
The localization and continued functioning of nNOS has a crucial regulatory impact on arteriolar blood flow within skeletal muscles. This is clearly shown by the fact that functional ischemia occurs in connection with reduced nitric oxide levels. Narrowing of blood vessels results in oxygen deficiency which in turn renders muscle fibers more susceptible to metabolic stress and cellular degeneration in muscular dystrophy [137,138,139]. Stabilization of nNOS enzyme at the sarcolemma depends on the structural and functional integrity of the sarcoglycan subcomplex. The importance of nNOS-associated signaling mechanisms is highlighted by the pathophysiological consequence of the lack of this enzyme in sarcoglycanopathy, which results in a severely dystrophic phenotype [133,134]. Disturbed allosteric interactions between phosphofructokinase and nNOS were shown to occur in dystrophin-deficient fibers [140], which may contribute to abnormal glycolytic activity patterns in muscular dystrophy [141,142]. The above listed studies allowed to draw the model of the spatial configuration of the dystrophin complex presented in the lower panel of above Figure 1.
A clear linkage to the extracellular matrix via interactions between the dystrophin/dystroglycan-containing sarcolemma and the basal lamina and its connections to layers of different types of collagen is supported by the localization of fibronectin and biglycan in close proximity to dystrophin and utrophin [24,143,144]. Both proteins are key extracellular matrix proteins and are majorly involved in the stabilization of damaged muscle fibers [24,145]. Biglycan appears to be critical for the expression levels of dystrobrevins, syntrophins and sarcoglycans [146,147], which represent dystrophin-associated proteins of high abundance that are routinely identified in crude muscle extracts and purified preparations of the dystrophin–glycoprotein complex [14,24,111,112,148]. In contrast, the highly hydrophobic protein sarcospan of 25 kDa [149] is more difficult to characterize by mass spectrometry. This component of the dystrophin complex is usually only recognized by a small number of unique peptides [19]. Interestingly, studies on subcomplex formation between sarcospan and sarcoglycans indicate a potential indirect linkage via these proteins between the dystrophin complex and the integrin complex of the sarcolemma [150,151].
3.2. The Dystrophin Complex as a Cellular Signaling Node in Skeletal Muscle
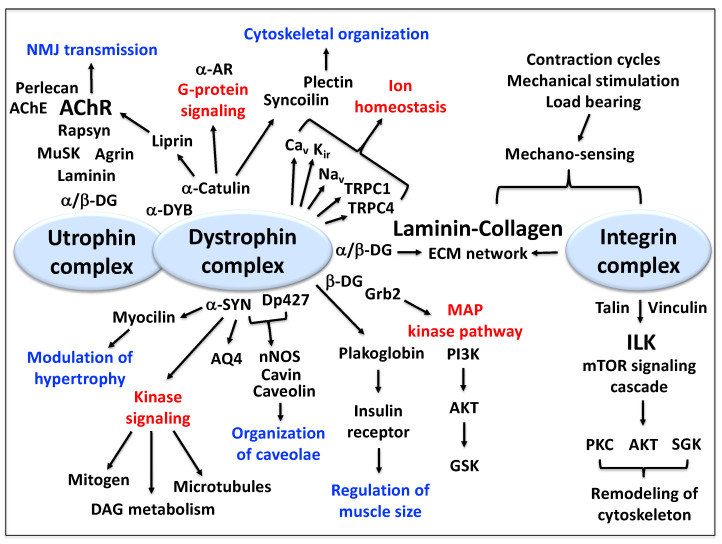
Besides providing the above-described stabilizing linkage between the basal lamina and the membrane cytoskeleton and thereby functioning as a molecular shock absorber, the dystrophin complex also acts as a critical hub for cellular signaling at the muscle plasma membrane [61]. The dystrophin complexome has been implicated to be involved in the modulation of hypertrophy, major kinase signaling cascades, the organization of caveolae structures, the regulation of skeletal muscle size, the mitogen-activated protein kinase pathway, the regulation of ion homeostasis, cytoskeletal organization, G-protein signaling and neuromuscular transmission in conjunction with its autosomal homologue utrophin, as well as mechano-sensing and cytoskeletal remodeling in association with the laminin-collagen bridge and the sarcolemmal integrin complex [36,61,152].
Following the characterization of dystrophin–dystroglycan interactions [153], it was shown that adaptor proteins of signal transduction and cell communication, such as the growth factor receptor-bound protein Grb2, are also associated with the dystrophin complex [154]. This finding linked dystrophin with cellular signaling processes and was followed by a large number of detailed studies into the physiological and cell biological role of the dystrophin complex. The signaling and regulatory function of the dystrophin complexome in skeletal muscle was shown to involve key physiological players of the muscle plasmalemma, such as channels for potassium, sodium, calcium and water [60]. This includes especially interactions between alpha-syntrophin and inward rectifier K+-channels (Kir2.1 at the neuromuscular junction) [155], Na+-channels (Nav1.4 and Nav1.5) [93], non-specific channels of the transient receptor potential cation channel family (TRPC1 and TRPC4) [156], and the aquaporin water channel (AQ4) [157,158]. Imaging by confocal microscopy techniques suggests furthermore co-localization of dystrophin and the voltage-sensing dihydropyridine receptor L-type Ca2+-channel (Cav1.1) [159]. The dystrophin complex appears to be intrinsically involved in the provision of a structural scaffold for the positioning of important ion channels and transporters.
Kinase-coupled pathways were demonstrated to be coupled to syntrophins and dystroglycans. This includes potential interactions between the non-receptor tyrosine kinase Src and beta-dystroglycan [160,161], as well as binding of various syntrophins to diacylglycerol kinase-zeta [162,163], microtubule-associated serine/threonine kinase MAST205 [164], and a specific mitogen-activated protein kinase named stress-activated protein kinase SAPK3 [165]. The proposed indirect linkage between the dystrophin complex and integrin [166] would suggest that the dystrophin complexome is also partially involved in mechano-sensing and the overall regulation of cytoskeletal remodeling via the phosphatidylinositol 3-kinase-related kinase mTOR (mechanistic/mammalian target of rapamycin) signaling cascade [167]. Other enzymes and regulatory factors involved in dystrophin-linked signaling mechanisms were identified as the ankyrin repeat-rich membrane spanning protein ARMS at the neuromuscular junction [168] and the nNOS isoform of nitric oxide synthase, which is involved in Ca2+/calmodulin dependent NO synthesis [36,140]. Myocilin, a modulator of muscle hypertrophy was shown to be linked to alpha-syntrophin [169]. Interestingly, alpha-dystrobrevin was demonstrated to interact with the cytoskeletal linker protein alpha-catulin, which acts as a scaffold structure for G-protein signaling pathways, [170,171,172], and is also linked to liprin and the guanidine nucleotide exchange factor Arhgef5 at the neuromuscular junction [173,174].
Although this article focuses on the dystrophin isoform Dp427-M and its associated components, it is important to briefly mention the crucial function of its autosomal homologue, the dystrophin-related protein utrophin of 395 kDa [175] and the overlapping localization of both membrane cytoskeletal proteins at the neuromuscular junction [176]. In analogy to dystrophin, utrophin also exists in a variety of isoforms, including Up71, G-utrophin, Up140, B-utrophin and A-utrophin. The domain structure of full-length utrophin is very similar to Dp427, especially at the carboxy-terminus, and is characterized by a major actin-binding site at the amino-terminus, a central spectrin-like rod domain and a carboxy-terminal interaction site with beta-dystroglycan [177]. At the motor endplate region, a very high density of utrophin isoform Up395-M exists at the post-synaptic membrane besides dystrophin [176,178]. Utrophin forms a tight complex with proteins involved in the regulation and maintenance of neurotransmission, such as agrin, the muscle-specific kinase MuSK, perlecan, acetylcholinesterase and the nicotinic acetylcholine receptor [179,180]. Figure 5 provides an overview of the complexity of the dystrophin-associated signaling processes and their potential interconnectivities.
Figure 5.
Overview of the function of the dystrophin complexome as a cellular signaling hub at the sarcolemma of skeletal muscle fibers. Shown is the close neighborhood relationship between the dystrophin core complex and the integrin complex within the plasma membrane, as well as the presence of the dystrophin and utrophin complex at the neuromuscular junction. Key pathways and functional results that are potentially linked to dystrophin are marked in red and blue, respectively. Abbreviations used: AChE, acetylcholinesterase; AChR, acetylcholine receptor; AKT, serine/threonine-specific protein kinase B; AQ4, aquaporin water channel; AR, adrenergic receptor; Cav, voltage-sensing L-type Ca2+-channel; DAG, diacylglycerol; DG, dystroglycan; Dp427, dystrophin protein of 427 kDa; DYB, dystrobrevin; ECM, extracellular matrix; Grb2, growth factor receptor-bound protein 2; GSK, glycogen synthase kinase; ILK, integrin-linked kinase; Kir, inward rectifier K+-channel; MAP, mitogen-activated protein; mTOR, mammalian/mechanistic target of rapamycin; MuSK, muscle-specific kinase; Nav, Na+-channel; NMJ, neuromuscular junction; nNOS, neuronal isoform of nitric oxide synthase; PI3K, phosphoinositide 3-kinase; PKC, protein kinase C; SGK, serine/threonine-protein kinase; SYN, syntrophin; TRPC, transient receptor potential cation channel.
4. Conclusions
Since the discovery of dystrophin over 30 years ago, a large body of scientific evidence has been gathered that strongly supports the idea that the full-length dystrophin isoform Dp427-M forms the core of a supramolecular protein assembly at the sarcolemma. This dystrophin complexome was shown to be intrinsically involved in linking the extracellular matrix component laminin-211 to the actin membrane cytoskeleton. The main members of the dystrophin-associated complex were identified as sarcolemmal proteins (beta-dystroglycan, alpha-sarcoglycan, beta-sarcoglycan, gamma-sarcoglycan, delta-sarcoglycan and sarcospan), extracellular proteins (alpha-dystroglycan and laminin-211) and cytosolic proteins (alpha-syntrophin, beta-syntrophin, alpha-dystrobrevin, beta-dystrobrevin and cortical actin). Closely interacting proteins with this dystrophin core complex are biglycan and collagens via their coupling to laminin in the extracellular matrix, and various cytoskeletal proteins through the intracellular network of intermediate filaments and microtubules with the actin membrane cytoskeleton. In addition, a variety of enzymes, ion channels and regulatory factors were found to exist in close proximity to dystrophin. These findings favor the systems biological concept of an integrative function of the dystrophin node in skeletal muscle tissues [181]. Dystrophin and its closely associated protein components appear to play a key role in cytoskeletal organization, the maintenance of fiber stability during cellular stresses caused by repeated excitation–contraction–relaxation cycles, the transmission of lateral force throughout the contracting muscle fiber system, and the provision of a structural basis for complex cellular signaling mechanisms.
Acknowledgments
We would like to thank our many national and international colleagues and collaborators for supporting our ongoing proteomics research into the identification and characterization of dystrophin and its associated protein complexes in health and disease.
Author Contributions
The first version of this article was written by P.D. and K.O., and then reviewed and edited by S.G., S.M., M.Z., H.S. and D.S. All authors have read and agreed to the published version of the manuscript.
Funding
Research was supported by the Kathleen Lonsdale Institute for Human Health Research at Maynooth University.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Koenig M., Hoffman E.P., Bertelson C.J., Monaco A.P., Feener C., Kunkel L.M. Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell. 1987;50:509–517. doi: 10.1016/0092-8674(87)90504-6. [DOI] [PubMed] [Google Scholar]
- 2.Hoffman E.P., Brown R.H., Jr., Kunkel L.M. Dystrophin: The protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 3.Koenig M., Monaco A.P., Kunkel L.M. The complete sequence of dystrophin predicts a rod-shaped cytoskeletal protein. Cell. 1988;53:219–228. doi: 10.1016/0092-8674(88)90383-2. [DOI] [PubMed] [Google Scholar]
- 4.Muntoni F., Torelli S., Ferlini A. Dystrophin and mutations: One gene, several proteins, multiple phenotypes. Lancet Neurol. 2003;2:731–740. doi: 10.1016/S1474-4422(03)00585-4. [DOI] [PubMed] [Google Scholar]
- 5.Petkova M.V., Morales-Gonzales S., Relizani K., Gill E., Seifert F., Radke J., Stenzel W., Garcia L., Amthor H., Schuelke M. Characterization of a Dmd (EGFP) reporter mouse as a tool to investigate dystrophin expression. Skelet. Muscle. 2016;6:25. doi: 10.1186/s13395-016-0095-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell K.P., Kahl S.D. Association of dystrophin and an integral membrane glycoprotein. Nature. 1989;338:259–262. doi: 10.1038/338259a0. [DOI] [PubMed] [Google Scholar]
- 7.Ervasti J.M., Ohlendieck K., Kahl S.D., Gaver M.G., Campbell K.P. Deficiency of a glycoprotein component of the dystrophin complex in dystrophic muscle. Nature. 1990;345:315–319. doi: 10.1038/345315a0. [DOI] [PubMed] [Google Scholar]
- 8.Yoshida M., Ozawa E. Glycoprotein complex anchoring dystrophin to sarcolemma. J. Biochem. 1990;108:748–752. doi: 10.1093/oxfordjournals.jbchem.a123276. [DOI] [PubMed] [Google Scholar]
- 9.Ervasti J.M., Campbell K.P. Membrane organization of the dystrophin-glycoprotein complex. Cell. 1991;66:1121–1131. doi: 10.1016/0092-8674(91)90035-W. [DOI] [PubMed] [Google Scholar]
- 10.Yamamoto H., Hagiwara Y., Mizuno Y., Yoshida M., Ozawa E. Heterogeneity of dystrophin-associated proteins. J. Biochem. 1993;114:132–139. doi: 10.1093/oxfordjournals.jbchem.a124128. [DOI] [PubMed] [Google Scholar]
- 11.Ervasti J.M., Campbell K.P. A role for the dystrophin-glycoprotein complex as a transmembrane linker between laminin and actin. J. Cell Biol. 1993;122:809–823. doi: 10.1083/jcb.122.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campbell K.P. Three muscular dystrophies: Loss of cytoskeleton-extracellular matrix linkage. Cell. 1995;80:675–679. doi: 10.1016/0092-8674(95)90344-5. [DOI] [PubMed] [Google Scholar]
- 13.Ohlendieck K. Towards an understanding of the dystrophin-glycoprotein complex: Linkage between the extracellular matrix and the membrane cytoskeleton in muscle fibers. Eur. J. Cell Biol. 1996;69:1–10. [PubMed] [Google Scholar]
- 14.Murphy S., Ohlendieck K. Mass spectrometric identification of dystrophin, the protein product of the Duchenne muscular dystrophy gene, in distinct muscle surface membranes. Int. J. Mol. Med. 2017;40:1078–1088. doi: 10.3892/ijmm.2017.3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohlendieck K., Ervasti J.M., Snook J.B., Campbell K.P. Dystrophin-glycoprotein complex is highly enriched in isolated skeletal muscle sarcolemma. J. Cell Biol. 1991;112:135–148. doi: 10.1083/jcb.112.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bonilla E., Samitt C.E., Miranda A.F., Hays A.P., Salviati G., DiMauro S., Kunkel L.M., Hoffman E.P., Rowland L.P. Duchenne muscular dystrophy: Deficiency of dystrophin at the muscle cell surface. Cell. 1988;54:447–452. doi: 10.1016/0092-8674(88)90065-7. [DOI] [PubMed] [Google Scholar]
- 17.Watkins S.C., Hoffman E.P., Slayter H.S., Kunkel L.M. Immunoelectron microscopic localization of dystrophin in myofibres. Nature. 1988;333:863–866. doi: 10.1038/333863a0. [DOI] [PubMed] [Google Scholar]
- 18.Ervasti J.M., Sonnemann K.J. Biology of the striated muscle dystrophin-glycoprotein complex. Int. Rev. Cytol. 2008;265:191–225. doi: 10.1016/S0074-7696(07)65005-0. [DOI] [PubMed] [Google Scholar]
- 19.Murphy S., Ohlendieck K. The biochemical and mass spectrometric profiling of the dystrophin complexome from skeletal muscle. Comput. Struct. Biotechnol. J. 2015;14:20–27. doi: 10.1016/j.csbj.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewis C., Ohlendieck K. Mass spectrometric identification of dystrophin isoform Dp427 by on-membrane digestion of sarcolemma from skeletal muscle. Anal. Biochem. 2010;404:197–203. doi: 10.1016/j.ab.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 21.Yoon J.H., Johnson E., Xu R., Martin L.T., Martin P.T., Montanaro F. Comparative proteomic profiling of dystroglycan-associated proteins in wild type, mdx, and Galgt2 transgenic mouse skeletal muscle. J. Proteome Res. 2012;11:4413–4424. doi: 10.1021/pr300328r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson E.K., Li B., Yoon J.H., Flanigan K.M., Martin P.T., Ervasti J., Montanaro F. Identification of new dystroglycan complexes in skeletal muscle. PLoS ONE. 2013;8:e73224. doi: 10.1371/journal.pone.0073224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turk R., Hsiao J.J., Smits M.M., Ng B.H., Pospisil T.C., Jones K.S., Campbell K.P., Wright M.E. Molecular signatures of membrane protein complexes underlying muscular dystrophy. Mol. Cell. Proteom. 2016;15:2169–2185. doi: 10.1074/mcp.M116.059188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murphy S., Brinkmeier H., Krautwald M., Henry M., Meleady P., Ohlendieck K. Proteomic profiling of the dystrophin complex and membrane fraction from dystrophic mdx muscle reveals decreases in the cytolinker desmoglein and increases in the extracellular matrix stabilizers biglycan and fibronectin. J. Muscle Res. Cell. Motil. 2017;38:251–268. doi: 10.1007/s10974-017-9478-4. [DOI] [PubMed] [Google Scholar]
- 25.Ohlendieck K., Matsumura K., Ionasescu V.V., Towbin J.A., Bosch E.P., Weinstein S.L., Sernett S.W., Campbell K.P. Duchenne muscular dystrophy: Deficiency of dystrophin-associated proteins in the sarcolemma. Neurology. 1993;43:795–800. doi: 10.1212/WNL.43.4.795. [DOI] [PubMed] [Google Scholar]
- 26.Guiraud S., Aartsma-Rus A., Vieira N.M., Davies K.E., van Ommen G.J., Kunkel L.M. The pathogenesis and therapy of muscular dystrophies. Annu. Rev. Genom. Hum. Genet. 2015;16:281–308. doi: 10.1146/annurev-genom-090314-025003. [DOI] [PubMed] [Google Scholar]
- 27.Doorenweerd N. Combining genetics, neuropsychology and neuroimaging to improve understanding of brain involvement in Duchenne muscular dystrophy—A narrative review. Neuromuscul. Disord. 2020;30:437–442. doi: 10.1016/j.nmd.2020.05.001. [DOI] [PubMed] [Google Scholar]
- 28.Goemans N., Buyse G. Current treatment and management of dystrophinopathies. Curr. Treat. Options Neurol. 2014;16:287. doi: 10.1007/s11940-014-0287-4. [DOI] [PubMed] [Google Scholar]
- 29.Mercuri E., Bönnemann C.G., Muntoni F. Muscular dystrophies. Lancet. 2019;394:2025–2038. doi: 10.1016/S0140-6736(19)32910-1. [DOI] [PubMed] [Google Scholar]
- 30.Partridge T.A. The mdx mouse model as a surrogate for Duchenne muscular dystrophy. FEBS J. 2013;280:4177–4186. doi: 10.1111/febs.12267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McGreevy J.W., Hakim C.H., McIntosh M.A., Duan D. Animal models of Duchenne muscular dystrophy: From basic mechanisms to gene therapy. Dis. Model Mech. 2015;8:195–213. doi: 10.1242/dmm.018424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson K., Faelan C., Patterson-Kane J.C., Rudmann D.G., Moore S.A., Frank D., Charleston J., Tinsley J., Young G.D., Milici A.J. Duchenne and becker muscular dystrophies: A review of animal models, clinical end points, and biomarker quantification. Toxicol. Pathol. 2017;45:961–976. doi: 10.1177/0192623317734823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mázala D.A., Novak J.S., Hogarth M.W., Nearing M., Adusumalli P., Tully C.B., Habib N.F., Gordish-Dressman H., Chen Y.W., Jaiswal J.K., et al. TGF-β-driven muscle degeneration and failed regeneration underlie disease onset in a DMD mouse model. JCI Insight. 2020;5:e135703. doi: 10.1172/jci.insight.135703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shin J., Tajrishi M.M., Ogura Y., Kumar A. Wasting mechanisms in muscular dystrophy. Int. J. Biochem. Cell Biol. 2013;45:2266–2279. doi: 10.1016/j.biocel.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holland A., Murphy S., Dowling P., Ohlendieck K. Pathoproteomic profiling of the skeletal muscle matrisome in dystrophinopathy associated myofibrosis. Proteomics. 2016;16:345–366. doi: 10.1002/pmic.201500158. [DOI] [PubMed] [Google Scholar]
- 36.Allen D.G., Whitehead N.P., Froehner S.C. Absence of dystrophin disrupts skeletal muscle signaling: Roles of Ca2+, reactive oxygen species, and nitric oxide in the development of muscular dystrophy. Physiol. Rev. 2016;96:253–305. doi: 10.1152/physrev.00007.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tidball J.G., Welc S.S., Wehling-Henricks M. Immunobiology of inherited muscular dystrophies. Compr. Physiol. 2018;8:1313–1356. doi: 10.1002/cphy.c170052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liewluck T., Milone M. Untangling the complexity of limb-girdle muscular dystrophies. Muscle Nerve. 2018;58:167–177. doi: 10.1002/mus.26077. [DOI] [PubMed] [Google Scholar]
- 39.Pradhan B.S., Prószyński T.J. A role for caveolin-3 in the pathogenesis of muscular dystrophies. Int. J. Mol. Sci. 2020;21:8736. doi: 10.3390/ijms21228736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murphy S., Zweyer M., Mundegar R.R., Swandulla D., Ohlendieck K. Proteomic serum biomarkers for neuromuscular diseases. Expert Rev. Proteom. 2018;15:277–291. doi: 10.1080/14789450.2018.1429923. [DOI] [PubMed] [Google Scholar]
- 41.Dowling P., Murphy S., Zweyer M., Raucamp M., Swandulla D., Ohlendieck K. Emerging proteomic biomarkers of X-linked muscular dystrophy. Expert Rev. Mol. Diagn. 2019;19:739–755. doi: 10.1080/14737159.2019.1648214. [DOI] [PubMed] [Google Scholar]
- 42.Grounds M.D., Terrill J.R., Al-Mshhdani B.A., Duong M.N., Radley-Crabb H.G., Arthur P.G. Biomarkers for Duchenne muscular dystrophy: Myonecrosis, inflammation and oxidative stress. Dis. Model Mech. 2020;13:dmm043638. doi: 10.1242/dmm.043638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Al-Khalili Szigyarto C. Duchenne Muscular Dystrophy: Recent advances in protein biomarkers and the clinical application. Expert Rev. Proteom. 2020;17:365–375. doi: 10.1080/14789450.2020.1773806. [DOI] [PubMed] [Google Scholar]
- 44.Thompson R., Spendiff S., Roos A., Bourque P.R., Warman Chardon J., Kirschner J., Horvath R., Lochmüller H. Advances in the diagnosis of inherited neuromuscular diseases and implications for therapy development. Lancet Neurol. 2020;19:522–532. doi: 10.1016/S1474-4422(20)30028-4. [DOI] [PubMed] [Google Scholar]
- 45.Partridge T.A. Impending therapies for Duchenne muscular dystrophy. Curr. Opin. Neurol. 2011;24:415–422. doi: 10.1097/WCO.0b013e32834aa3f1. [DOI] [PubMed] [Google Scholar]
- 46.Spinazzola J.M., Kunkel L.M. Pharmacological therapeutics targeting the secondary defects and downstream pathology of Duchenne muscular dystrophy. Expert Opin. Orphan Drugs. 2016;4:1179–1194. doi: 10.1080/21678707.2016.1240613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Waldrop M.A., Flanigan K.M. Update in Duchenne and Becker muscular dystrophy. Curr. Opin. Neurol. 2019;32:722–727. doi: 10.1097/WCO.0000000000000739. [DOI] [PubMed] [Google Scholar]
- 48.Werneck L.C., Lorenzoni P.J., Ducci R.D., Fustes O.H., Kay C.S.K., Scola R.H. Duchenne muscular dystrophy: An historical treatment review. Arq. Neuropsiquiatr. 2019;77:579–589. doi: 10.1590/0004-282x20190088. [DOI] [PubMed] [Google Scholar]
- 49.Salmaninejad A., Jafari Abarghan Y., Bozorg Qomi S., Bayat H., Yousefi M., Azhdari S., Talebi S., Mojarrad M. Common therapeutic advances for Duchenne muscular dystrophy (DMD) Int. J. Neurosci. 2020 doi: 10.1080/00207454.2020.1740218. [DOI] [PubMed] [Google Scholar]
- 50.Murphy S., Dowling P., Zweyer M., Swandulla D., Ohlendieck K. Proteomic profiling of giant skeletal muscle proteins. Expert Rev. Proteom. 2019;16:241–256. doi: 10.1080/14789450.2019.1575205. [DOI] [PubMed] [Google Scholar]
- 51.Koenig M., Kunkel L.M. Detailed analysis of the repeat domain of dystrophin reveals four potential hinge segments that may confer flexibility. J. Biol. Chem. 1990;265:4560–4566. doi: 10.1016/S0021-9258(19)39599-7. [DOI] [PubMed] [Google Scholar]
- 52.Fealey M.E., Horn B., Coffman C., Miller R., Lin A.Y., Thompson A.R., Schramel J., Groth E., Hinderliter A., Cembran A., et al. Dynamics of dystrophin’s actin-binding domain. Biophys. J. 2018;115:445–454. doi: 10.1016/j.bpj.2018.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abdullah M., Hassan A., Rashid S., Naeem M. Tyrosine phosphorylation as a regulator of dystrophin and beta-dystroglycan interaction: A molecular insight. J. Mol. Graph Model. 2020;99:107623. doi: 10.1016/j.jmgm.2020.107623. [DOI] [PubMed] [Google Scholar]
- 54.Delalande O., Molza A.E., Dos Santos Morais R., Chéron A., Pollet É., Raguenes-Nicol C., Tascon C., Giudice E., Guilbaud M., Nicolas A., et al. Dystrophin’s central domain forms a complex filament that becomes disorganized by in-frame deletions. J. Biol. Chem. 2018;293:6637–6646. doi: 10.1074/jbc.M117.809798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rybakova I.N., Ervasti J.M. Dystrophin-glycoprotein complex is monomeric and stabilizes actin filaments in vitro through a lateral association. J. Biol. Chem. 1997;272:28771–28778. doi: 10.1074/jbc.272.45.28771. [DOI] [PubMed] [Google Scholar]
- 56.Gao Q.Q., McNally E.M. The dystrophin complex: Structure, function, and implications for therapy. Compr. Physiol. 2015;5:1223–1239. doi: 10.1002/cphy.c140048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gawor M., Prószyński T.J. The molecular cross talk of the dystrophin-glycoprotein complex. Ann. NY Acad. Sci. 2018;1412:62–72. doi: 10.1111/nyas.13500. [DOI] [PubMed] [Google Scholar]
- 58.Bhat H.F., Mir S.S., Dar K.B., Bhat Z.F., Shah R.A., Ganai N.A. ABC of multifaceted dystrophin glycoprotein complex (DGC) J. Cell. Physiol. 2018;233:5142–5159. doi: 10.1002/jcp.25982. [DOI] [PubMed] [Google Scholar]
- 59.Holland A., Carberry S., Ohlendieck K. Proteomics of the dystrophin-glycoprotein complex and dystrophinopathy. Curr. Protein Pept. Sci. 2013;14:680–697. doi: 10.2174/13892037113146660083. [DOI] [PubMed] [Google Scholar]
- 60.Leyva-Leyva M., Sandoval A., Felix R., González-Ramírez R. Biochemical and functional interplay between Ion channels and the components of the dystrophin-associated glycoprotein complex. J. Membr. Biol. 2018;251:535–550. doi: 10.1007/s00232-018-0036-9. [DOI] [PubMed] [Google Scholar]
- 61.Constantin B. Dystrophin complex functions as a scaffold for signalling proteins. Biochim. Biophys. Acta. 2014;1838:635–642. doi: 10.1016/j.bbamem.2013.08.023. [DOI] [PubMed] [Google Scholar]
- 62.Rybakova I.N., Amann K.J., Ervasti J.M. A new model for the interaction of dystrophin with F-actin. J. Cell Biol. 1996;135:661–672. doi: 10.1083/jcb.135.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rybakova I.N., Patel J.R., Ervasti J.M. The dystrophin complex forms a mechanically strong link between the sarcolemma and costameric actin. J. Cell Biol. 2000;150:1209–1214. doi: 10.1083/jcb.150.5.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Peter A.K., Cheng H., Ross R.S., Knowlton K.U., Chen J. The costamere bridges sarcomeres to the sarcolemma in striated muscle. Prog. Pediatr. Cardiol. 2011;31:83–88. doi: 10.1016/j.ppedcard.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ramaswamy K.S., Palmer M.L., van der Meulen J.H., Renoux A., Kostrominova T.Y., Michele D.E., Faulkner J.A. Lateral transmission of force is impaired in skeletal muscles of dystrophic mice and very old rats. J. Physiol. 2011;589:1195–1208. doi: 10.1113/jphysiol.2010.201921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Campbell K.P., Stull J.T. Skeletal muscle basement membrane-sarcolemma-cytoskeleton interaction minireview series. J. Biol. Chem. 2003;278:12599–12600. doi: 10.1074/jbc.R300005200. [DOI] [PubMed] [Google Scholar]
- 67.Shishmarev D. Excitation-contraction coupling in skeletal muscle: Recent progress and unanswered questions. Biophys. Rev. 2020;12:143–153. doi: 10.1007/s12551-020-00610-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Petrof B.J., Shrager J.B., Stedman H.H., Kelly A.M., Sweeney H.L. Dystrophin protects the sarcolemma from stresses developed during muscle contraction. Proc. Natl. Acad. Sci. USA. 1993;90:3710–3714. doi: 10.1073/pnas.90.8.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sarkis J., Vié V., Winder S.J., Renault A., Le Rumeur E., Hubert J.F. Resisting sarcolemmal rupture: Dystrophin repeats increase membrane-actin stiffness. FASEB J. 2013;27:359–367. doi: 10.1096/fj.12-208967. [DOI] [PubMed] [Google Scholar]
- 70.Rader E.P., Turk R., Willer T., Beltrán D., Inamori K., Peterson T.A., Engle J., Prouty S., Matsumura K., Saito F., et al. Role of dystroglycan in limiting contraction-induced injury to the sarcomeric cytoskeleton of mature skeletal muscle. Proc. Natl. Acad. Sci. USA. 2016;113:10992–10997. doi: 10.1073/pnas.1605265113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ervasti J.M. Costameres: The Achilles’ heel of Herculean muscle. J. Biol. Chem. 2003;278:13591–13594. doi: 10.1074/jbc.R200021200. [DOI] [PubMed] [Google Scholar]
- 72.Hughes D.C., Wallace M.A., Baar K. Effects of aging, exercise, and disease on force transfer in skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 2015;309:E1–E10. doi: 10.1152/ajpendo.00095.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jaka O., Casas-Fraile L., López de Munain A., Sáenz A. Costamere proteins and their involvement in myopathic processes. Expert Rev. Mol. Med. 2015;17:e12. doi: 10.1017/erm.2015.9. [DOI] [PubMed] [Google Scholar]
- 74.Purslow P.P. The Structure and Role of Intramuscular Connective Tissue in Muscle Function. Front. Physiol. 2020;11:495. doi: 10.3389/fphys.2020.00495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Murphy A.C., Young P.W. The actinin family of actin cross-linking proteins—A genetic perspective. Cell. Biosci. 2015;5:49. doi: 10.1186/s13578-015-0029-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ohlendieck K., Campbell K.P. Dystrophin constitutes 5% of membrane cytoskeleton in skeletal muscle. FEBS Lett. 1991;283:230–234. doi: 10.1016/0014-5793(91)80595-T. [DOI] [PubMed] [Google Scholar]
- 77.Ohlendieck K. Characterisation of the dystrophin-related protein utrophin in highly purified skeletal muscle sarcolemma vesicles. Biochim. Biophys. Acta. 1996;1283:215–222. doi: 10.1016/0005-2736(96)00102-2. [DOI] [PubMed] [Google Scholar]
- 78.Tokarz S.A., Duncan N.M., Rash S.M., Sadeghi A., Dewan A.K., Pillers D.A. Redefinition of dystrophin isoform distribution in mouse tissue by RT-PCR implies role in nonmuscle manifestations of duchenne muscular dystrophy. Mol. Genet. Metab. 1998;65:272–281. doi: 10.1006/mgme.1998.2763. [DOI] [PubMed] [Google Scholar]
- 79.Ueda H., Baba T., Ohno S. Current knowledge of dystrophin and dystrophin-associated proteins in the retina. Histol. Histopathol. 2000;15:753–760. doi: 10.14670/HH-15.753. [DOI] [PubMed] [Google Scholar]
- 80.Lidov H.G., Kunkel L.M. Dystrophin and Dp140 in the adult rodent kidney. Lab. Invest. 1998;78:1543–1551. [PubMed] [Google Scholar]
- 81.Mahyoob Rani A.Q., Maeta K., Kawaguchi T., Awano H., Nagai M., Nishio H., Matsuo M. Schwann cell-specific Dp116 is expressed in glioblastoma cells, revealing two novel DMD gene splicing patterns. Biochem. Biophys. Rep. 2019;20:100703. doi: 10.1016/j.bbrep.2019.100703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Naidoo M., Anthony K. Dystrophin Dp71 and the neuropathophysiology of duchenne muscular dystrophy. Mol. Neurobiol. 2020;57:1748–1767. doi: 10.1007/s12035-019-01845-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dowling P., Gargan S., Zweyer M., Henry M., Meleady P., Swandulla D., Ohlendieck K. Proteome-wide changes in the mdx-4cv spleen due to pathophysiological crosstalk with dystrophin-deficient skeletal muscle. iScience. 2020;23:101500. doi: 10.1016/j.isci.2020.101500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Romo-Yáñez J., Rodríguez-Martínez G., Aragón J., Siqueiros-Márquez L., Herrera-Salazar A., Velasco I., Montanez C. Characterization of the expression of dystrophins and dystrophin-associated proteins during embryonic neural stem/progenitor cell differentiation. Neurosci. Lett. 2020;736:135247. doi: 10.1016/j.neulet.2020.135247. [DOI] [PubMed] [Google Scholar]
- 85.Angelini C., Pinzan E. Advances in imaging of brain abnormalities in neuromuscular disease. Ther. Adv. Neurol. Disord. 2019;12:1756286419845567. doi: 10.1177/1756286419845567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Waite A., Brown S.C., Blake D.J. The dystrophin-glycoprotein complex in brain development and disease. Trends Neurosci. 2012;35:487–496. doi: 10.1016/j.tins.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 87.Ervasti J.M., Kahl S.D., Campbell K.P. Purification of dystrophin from skeletal muscle. J. Biol. Chem. 1991;266:9161–9165. doi: 10.1016/S0021-9258(18)31565-5. [DOI] [PubMed] [Google Scholar]
- 88.Finn D.M., Ohlendieck K. Oligomerization of beta-dystroglycan in rabbit diaphragm and brain as revealed by chemical crosslinking. Biochim. Biophys. Acta. 1998;1370:325–336. doi: 10.1016/S0005-2736(97)00283-6. [DOI] [PubMed] [Google Scholar]
- 89.Murphy S., Zweyer M., Mundegar R.R., Swandulla D., Ohlendieck K. Chemical crosslinking analysis of β-dystroglycan in dystrophin-deficient skeletal muscle. HRB Open Res. 2018;1:17. doi: 10.12688/hrbopenres.12846.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yoshida M., Suzuki A., Yamamoto H., Noguchi S., Mizuno Y., Ozawa E. Dissociation of the complex of dystrophin and its associated proteins into several unique groups by n-octyl beta-D-glucoside. Eur. J. Biochem. 1994;222:1055–1061. doi: 10.1111/j.1432-1033.1994.tb18958.x. [DOI] [PubMed] [Google Scholar]
- 91.Sadoulet-Puccio H.M., Feener C.A., Schaid D.J., Thibodeau S.N., Michels V.V., Kunkel L.M. The genomic organization of human dystrobrevin. Neurogenetics. 1997;1:37–42. doi: 10.1007/s100480050006. [DOI] [PubMed] [Google Scholar]
- 92.Nakamori M., Takahashi M.P. The role of alpha-dystrobrevin in striated muscle. Int. J. Mol. Sci. 2011;12:1660–1671. doi: 10.3390/ijms12031660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gee S.H., Gee R., Madhaven S.H., Levinson S.R., Caldwell J.H., Sealock R., Froehner S.C. Interaction of muscle and brain sodium channels with multiple members of the syntrophin family of dystrophin-associated proteins. J. Neurosci. 1998;18:128–137. doi: 10.1523/JNEUROSCI.18-01-00128.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bhat S.S., Ali R., Khanday F.A. Syntrophins entangled in cytoskeletal meshwork: Helping to hold it all together. Cell Prolif. 2019;52:e12562. doi: 10.1111/cpr.12562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Roberds S.L., Anderson R.D., Ibraghimov-Beskrovnaya O., Campbell K.P. Primary structure and muscle-specific expression of the 50-kDa dystrophin-associated glycoprotein (adhalin) J. Biol. Chem. 1993;268:23739–23742. doi: 10.1016/S0021-9258(20)80440-2. [DOI] [PubMed] [Google Scholar]
- 96.Anastasi G., Cutroneo G., Rizzo G., Favaloro A. Sarcoglycan subcomplex in normal and pathological human muscle fibers. Eur. J. Histochem. 2007;51:29–33. [PubMed] [Google Scholar]
- 97.Tarakci H., Berger J. The sarcoglycan complex in skeletal muscle. Front. Biosci. 2016;21:744–756. doi: 10.2741/4418. [DOI] [PubMed] [Google Scholar]
- 98.Crosbie R.H., Heighway J., Venzke D.P., Lee J.C., Campbell K.P. Sarcospan, the 25-kDa transmembrane component of the dystrophin-glycoprotein complex. J. Biol. Chem. 1997;272:31221. doi: 10.1074/jbc.272.50.31221. [DOI] [PubMed] [Google Scholar]
- 99.Crosbie R.H., Lim L.E., Moore S.A., Hirano M., Hays A.P., Maybaum S.W., Collin H., Dovico S.A., Stolle C.A., Fardeau M., et al. Molecular and genetic characterization of sarcospan: Insights into sarcoglycan-sarcospan interactions. Hum. Mol. Genet. 2000;9:2019–2027. doi: 10.1093/hmg/9.13.2019. [DOI] [PubMed] [Google Scholar]
- 100.Marshall J.L., Crosbie-Watson R.H. Sarcospan: A small protein with large potential for Duchenne muscular dystrophy. Skelet. Muscle. 2013;3:1. doi: 10.1186/2044-5040-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ibraghimov-Beskrovnaya O., Ervasti J.M., Leveille C.J., Slaughter C.A., Sernett S.W., Campbell K.P. Primary structure of dystrophin-associated glycoproteins linking dystrophin to the extracellular matrix. Nature. 1992;355:696–702. doi: 10.1038/355696a0. [DOI] [PubMed] [Google Scholar]
- 102.Carmignac V., Durbeej M. Cell-matrix interactions in muscle disease. J. Pathol. 2012;226:200–218. doi: 10.1002/path.3020. [DOI] [PubMed] [Google Scholar]
- 103.Bozzi M., Morlacchi S., Bigotti M.G., Sciandra F., Brancaccio A. Functional diversity of dystroglycan. Matrix Biol. 2009;28:179–187. doi: 10.1016/j.matbio.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 104.Barresi R., Campbell K.P. Dystroglycan: From biosynthesis to pathogenesis of human disease. J. Cell Sci. 2006;119:199–207. doi: 10.1242/jcs.02814. [DOI] [PubMed] [Google Scholar]
- 105.Mias-Lucquin D., Dos Santos Morais R., Chéron A., Lagarrigue M., Winder S.J., Chenuel T., Pérez J., Appavou M.S., Martel A., Alviset G., et al. How the central domain of dystrophin acts to bridge F-actin to sarcolemmal lipids. J. Struct. Biol. 2020;209:107411. doi: 10.1016/j.jsb.2019.107411. [DOI] [PubMed] [Google Scholar]
- 106.Ibraghimov-Beskrovnaya O., Milatovich A., Ozcelik T., Yang B., Koepnick K., Francke U., Campbell K.P. Human dystroglycan: Skeletal muscle cDNA, genomic structure, origin of tissue specific isoforms and chromosomal localization. Hum. Mol. Genet. 1993;2:1651–1657. doi: 10.1093/hmg/2.10.1651. [DOI] [PubMed] [Google Scholar]
- 107.Miller G., Moore C.J., Terry R., La Riviere T., Mitchell A., Piggott R., Dear T.N., Wells D.J., Winder S.J. Preventing phosphorylation of dystroglycan ameliorates the dystrophic phenotype in mdx mouse. Hum. Mol. Genet. 2012;21:4508–4520. doi: 10.1093/hmg/dds293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Moore C.J., Winder S.J. The inside and out of dystroglycan post-translational modification. Neuromuscul. Disord. 2012;22:959–965. doi: 10.1016/j.nmd.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 109.Swiderski K., Shaffer S.A., Gallis B., Odom G.L., Arnett A.L., Scott Edgar J., Baum D.M., Chee A., Naim T., Gregorevic P., et al. Phosphorylation within the cysteine-rich region of dystrophin enhances its association with β-dystroglycan and identifies a potential novel therapeutic target for skeletal muscle wasting. Hum. Mol. Genet. 2014;23:6697–6711. doi: 10.1093/hmg/ddu388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Murphy S., Zweyer M., Raucamp M., Henry M., Meleady P., Swandulla D., Ohlendieck K. Proteomic profiling of the mouse diaphragm and refined mass spectrometric analysis of the dystrophic phenotype. J. Muscle Res. Cell. Motil. 2019;40:9–28. doi: 10.1007/s10974-019-09507-z. [DOI] [PubMed] [Google Scholar]
- 111.Murphy S., Zweyer M., Henry M., Meleady P., Mundegar R.R., Swandulla D., Ohlendieck K. Proteomic analysis of the sarcolemma-enriched fraction from dystrophic mdx-4cv skeletal muscle. J. Proteom. 2019;191:212–227. doi: 10.1016/j.jprot.2018.01.015. [DOI] [PubMed] [Google Scholar]
- 112.Murphy S., Zweyer M., Mundegar R.R., Swandulla D., Ohlendieck K. Comparative gel-based proteomic analysis of chemically crosslinked complexes in dystrophic skeletal muscle. Electrophoresis. 2018;39:1735–1744. doi: 10.1002/elps.201800028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Iacobucci C., Götze M., Sinz A. Cross-linking/mass spectrometry to get a closer view on protein interaction networks. Curr. Opin. Biotechnol. 2020;63:48–53. doi: 10.1016/j.copbio.2019.12.009. [DOI] [PubMed] [Google Scholar]
- 114.Belsom A., Rappsilber J. Anatomy of a crosslinker. Curr. Opin. Chem. Biol. 2020;60:39–46. doi: 10.1016/j.cbpa.2020.07.008. [DOI] [PubMed] [Google Scholar]
- 115.Hill M.M., Bastiani M., Luetterforst R., Kirkham M., Kirkham A., Nixon S.J., Walser P., Abankwa D., Oorschot V.M., Martin S., et al. PTRF-Cavin, a conserved cytoplasmic protein required for caveola formation and function. Cell. 2008;132:113–124. doi: 10.1016/j.cell.2007.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lo H.P., Hall T.E., Parton R.G. Mechanoprotection by skeletal muscle caveolae. Bioarchitecture. 2016;6:22–27. doi: 10.1080/19490992.2015.1131891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bonilla E., Fischbeck K., Schotland D.L. Freeze-fracture studies of muscle caveolae in human muscular dystrophy. Am. J. Pathol. 1981;104:167–173. [PMC free article] [PubMed] [Google Scholar]
- 118.Demonbreun A.R., Quattrocelli M., Barefield D.Y., Allen M.V., Swanson K.E., McNally E.M. An actin-dependent annexin complex mediates plasma membrane repair in muscle. J. Cell Biol. 2016;213:705–718. doi: 10.1083/jcb.201512022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Harsini F.M., Bui A.A., Rice A.M., Chebrolu S., Fuson K.L., Turtoi A., Bradberry M., Chapman E.R., Sutton R.B. Structural basis for the distinct membrane binding activity of the homologous C2A domains of myoferlin and dysferlin. J. Mol. Biol. 2019;431:2112–2126. doi: 10.1016/j.jmb.2019.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Seungjin N., Paek E. Computational methods in mass spectrometry-based structural proteomics for studying protein structure, dynamics, and interactions. Comput. Struct. Biotechnol. J. 2020;18:1391–1402. doi: 10.1016/j.csbj.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P., et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucl. Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Boyer J.G., Bernstein M.A., Boudreau-Larivière C. Plakins in striated muscle. Muscle Nerve. 2010;41:299–308. doi: 10.1002/mus.21472. [DOI] [PubMed] [Google Scholar]
- 123.Ursitti J.A., Lee P.C., Resneck W.G., McNally M.M., Bowman A.L., O’Neill A., Stone M.R., Bloch R.J. Cloning and characterization of cytokeratins 8 and 19 in adult rat striated muscle. Interaction with the dystrophin glycoprotein complex. J. Biol. Chem. 2004;279:41830–41838. doi: 10.1074/jbc.M400128200. [DOI] [PubMed] [Google Scholar]
- 124.Stone M.R., O’Neill A., Catino D., Bloch R.J. Specific interaction of the actin-binding domain of dystrophin with intermediate filaments containing keratin 19. Mol. Biol. Cell. 2005;16:4280–4293. doi: 10.1091/mbc.e05-02-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bhosle R.C., Michele D.E., Campbell K.P., Li Z., Robson R.M. Interactions of intermediate filament protein synemin with dystrophin and utrophin. Biochem. Biophys. Res. Commun. 2006;346:768–777. doi: 10.1016/j.bbrc.2006.05.192. [DOI] [PubMed] [Google Scholar]
- 126.Hijikata T., Murakami T., Ishikawa H., Yorifuji H. Plectin tethers desmin intermediate filaments onto subsarcolemmal dense plaques containing dystrophin and vinculin. Histochem. Cell Biol. 2003;119:109–123. doi: 10.1007/s00418-003-0496-5. [DOI] [PubMed] [Google Scholar]
- 127.Rezniczek G.A., Konieczny P., Nikolic B., Reipert S., Schneller D., Abrahamsberg C., Davies K.E., Winder S.J., Wiche G. Plectin 1f scaffolding at the sarcolemma of dystrophic (mdx) muscle fibers through multiple interactions with beta-dystroglycan. J. Cell Biol. 2007;176:965–977. doi: 10.1083/jcb.200604179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hijikata T., Nakamura A., Isokawa K., Imamura M., Yuasa K., Ishikawa R., Kohama K., Takeda S., Yorifuji H. Plectin 1 links intermediate filaments to costameric sarcolemma through beta-synemin, alpha-dystrobrevin and actin. J. Cell Sci. 2008;121:2062–2074. doi: 10.1242/jcs.021634. [DOI] [PubMed] [Google Scholar]
- 129.Prins K.W., Humston J.L., Mehta A., Tate V., Ralston E., Ervasti J.M. Dystrophin is a microtubule-associated protein. J. Cell Biol. 2009;186:363–369. doi: 10.1083/jcb.200905048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Eid Mutlak Y., Aweida D., Volodin A., Ayalon B., Dahan N., Parnis A., Cohen S. A signaling hub of insulin receptor, dystrophin glycoprotein complex and plakoglobin regulates muscle size. Nat. Commun. 2020;11:1381. doi: 10.1038/s41467-020-14895-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Brenman J.E., Chao D.S., Gee S.H., McGee A.W., Craven S.E., Santillano D.R., Wu Z., Huang F., Xia H., Peters M.F., et al. Interaction of nitric oxide synthase with the postsynaptic density protein PSD-95 and 1-syntrophin mediated by PDZ domains. Cell. 1996;84:757–767. doi: 10.1016/S0092-8674(00)81053-3. [DOI] [PubMed] [Google Scholar]
- 132.Gorza L., Sorge M., Seclì L., Brancaccio M. Master Regulators of Muscle Atrophy: Role of Costamere Components. Cells. 2021;10:61. doi: 10.3390/cells10010061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Fanin M., Tasca E., Nascimbeni A.C., Angelini C. Sarcolemmal neuronal nitric oxide synthase defect in limb-girdle muscular dystrophy: An adverse modulating factor in the disease course? J. Neuropathol. Exp. Neurol. 2009;68:383–390. doi: 10.1097/NEN.0b013e31819cd612. [DOI] [PubMed] [Google Scholar]
- 134.Angelini C., Tasca E., Nascimbeni A.C., Fanin M. Muscle fatigue, nNOS and muscle fiber atrophy in limb girdle muscular dystrophy. Acta Myol. 2014;33:119–126. [PMC free article] [PubMed] [Google Scholar]
- 135.Stamler J.S., Meissner G. Physiology of nitric oxide in skeletal muscle. Physiol. Rev. 2001;81:209–237. doi: 10.1152/physrev.2001.81.1.209. [DOI] [PubMed] [Google Scholar]
- 136.Gantner B.N., LaFond K.M., Bonini M.G. Nitric oxide in cellular adaptation and disease. Redox Biol. 2020;34:101550. doi: 10.1016/j.redox.2020.101550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sato K., Yokota T., Ichioka S., Shibata M., Takeda S. Vasodilation of intramuscular arterioles under shear stress in dystrophin-deficient skeletal muscle is impaired through decreased nNOS expression. Acta Myol. 2008;27:30–36. [PMC free article] [PubMed] [Google Scholar]
- 138.Tidball J.G., Wehling-Henricks M. Nitric oxide synthase deficiency and the pathophysiology of muscular dystrophy. J. Physiol. 2014;592:4627–4638. doi: 10.1113/jphysiol.2014.274878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Nichols B., Takeda S., Yokota T. Nonmechanical roles of dystrophin and associated proteins in exercise, neuromuscular junctions, and brains. Brain Sci. 2015;5:275–298. doi: 10.3390/brainsci5030275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wehling-Henricks M., Oltmann M., Rinaldi C., Myung K.H., Tidball J.G. Loss of positive allosteric interactions between neuronal nitric oxide synthase and phosphofructokinase contributes to defects in glycolysis and increased fatigability in muscular dystrophy. Hum. Mol. Genet. 2009;18:3439–3451. doi: 10.1093/hmg/ddp288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Di Mauro S., Angelini C., Catani C. Enzymes of the glycogen cycle and glycolysis in various human neuromuscular disorders. J. Neurol. Neurosurg. Psychiatry. 1967;30:411–415. doi: 10.1136/jnnp.30.5.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chinet A.E., Even P.C., Decrouy A. Dystrophin-dependent efficiency of metabolic pathways in mouse skeletal muscles. Experientia. 1994;50:602–605. doi: 10.1007/BF01921731. [DOI] [PubMed] [Google Scholar]
- 143.Amenta A.R., Yilmaz A., Bogdanovich S., McKechnie B.A., Abedi M., Khurana T.S., Fallon J.R. Biglycan recruits utrophin to the sarcolemma and counters dystrophic pathology in mdx mice. Proc. Natl. Acad. Sci. USA. 2011;108:762–767. doi: 10.1073/pnas.1013067108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bowe M.A., Mendis D.B., Fallon J.R. The small leucine-rich repeat proteoglycan biglycan binds to alpha-dystroglycan and is upregulated in dystrophic muscle. J. Cell Biol. 2000;148:801–810. doi: 10.1083/jcb.148.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Nastase M.V., Young M.F., Schaefer L. Biglycan: A multivalent proteoglycan providing structure and signals. J. Histochem. Cytochem. 2012;60:963–975. doi: 10.1369/0022155412456380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mercado M.L., Amenta A.R., Hagiwara H., Rafii M.S., Lechner B.E., Owens R.T., McQuillan D.J., Froehner S.C., Fallon J.R. Biglycan regulates the expression and sarcolemmal localization of dystrobrevin, syntrophin, and nNOS. FASEB J. 2006;20:1724–1726. doi: 10.1096/fj.05-5124fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Rafii M.S., Hagiwara H., Mercado M.L., Seo N.S., Xu T., Dugan T., Owens R.T., Hook M., McQuillan D.J., Young M.F., et al. Biglycan binds to alpha- and gamma-sarcoglycan and regulates their expression during development. J. Cell Physiol. 2006;209:439–447. doi: 10.1002/jcp.20740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Murphy S., Henry M., Meleady P., Zweyer M., Mundegar R.R., Swandulla D., Ohlendieck K. Simultaneous pathoproteomic evaluation of the dystrophin-glycoprotein complex and secondary changes in the mdx-4cv mouse model of duchenne muscular dystrophy. Biology. 2015;4:397–423. doi: 10.3390/biology4020397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Shu C., Parfenova L., Mokhonova E., Collado J.R., Damoiseaux R., Campagna J., John V., Crosbie R.H. High-throughput screening identifies modulators of sarcospan that stabilize muscle cells and exhibit activity in the mouse model of Duchenne muscular dystrophy. Skelet. Muscle. 2020;10:26. doi: 10.1186/s13395-020-00244-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Miller G., Wang E.L., Nassar K.L., Peter A.K., Crosbie R.H. Structural and functional analysis of the sarcoglycan-sarcospan subcomplex. Exp. Cell Res. 2007;313:639–651. doi: 10.1016/j.yexcr.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Marshall J.L., Oh J., Chou E., Lee J.A., Holmberg J., Burkin D.J., Crosbie-Watson R.H. Sarcospan integration into laminin-binding adhesion complexes that ameliorate muscular dystrophy requires utrophin and alpha7 integrin. Hum. Mol. Genet. 2015;24:2011–2022. doi: 10.1093/hmg/ddu615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Le Rumeur E., Winder S.J., Hubert J.F. Dystrophin: More than just the sum of its parts. Biochim. Biophys. Acta. 2010;1804:1713–1722. doi: 10.1016/j.bbapap.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 153.Jung D., Yang B., Meyer J., Chamberlain J.S., Campbell K.P. Identification and characterization of the dystrophin anchoring site on beta-dystroglycan. J. Biol. Chem. 1995;270:27305–27310. doi: 10.1074/jbc.270.45.27305. [DOI] [PubMed] [Google Scholar]
- 154.Yang B., Jung D., Motto D., Meyer J., Koretzky G., Campbell K.P. SH3 domain-mediated interaction of dystroglycan and Grb2. J. Biol. Chem. 1995;270:11711–11714. doi: 10.1074/jbc.270.20.11711. [DOI] [PubMed] [Google Scholar]
- 155.Leonoudakis D., Conti L.R., Anderson S., Radeke C.M., McGuire L.M., Adams M.E., Froehner S.C., Yates J.R., 3rd, Vandenberg C.A. Protein trafficking and anchoring complexes revealed by proteomic analysis of inward rectifier potassium channel (Kir2.x)-associated proteins. J. Biol. Chem. 2004;279:22331–22346. doi: 10.1074/jbc.M400285200. [DOI] [PubMed] [Google Scholar]
- 156.Sabourin J., Lamiche C., Vandebrouck A., Magaud C., Rivet J., Cognard C., Bourmeyster N., Constantin B. Regulation of TRPC1 and TRPC4 cation channels requires an alpha1-syntrophin-dependent complex in skeletal mouse myotubes. J. Biol. Chem. 2009;284:36248–36261. doi: 10.1074/jbc.M109.012872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Adams M.E., Mueller H.A., Froehner S.C. In vivo requirement of the alpha-syntrophin PDZ domain for the sarcolemmal localization of nNOS and aquaporin-4. J. Cell Biol. 2001;155:113–122. doi: 10.1083/jcb.200106158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Neely J.D., Amiry-Moghaddam M., Ottersen O.P., Froehner S.C., Agre P., Adams M.E. Syntrophin-dependent expression and localization of Aquaporin-4 water channel protein. Proc. Natl. Acad. Sci. USA. 2001;98:14108–14113. doi: 10.1073/pnas.241508198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Friedrich O., von Wegner F., Chamberlain J.S., Fink R.H., Rohrbach P. L-type Ca2+ channel function is linked to dystrophin expression in mammalian muscle. PLoS ONE. 2008;3:e1762. doi: 10.1371/journal.pone.0001762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sotgia F., Lee H., Bedford M.T., Petrucci T., Sudol M., Lisanti M.P. Tyrosine phosphorylation of beta-dystroglycan at its WW domain binding motif, PPxY, recruits SH2 domain containing proteins. Biochemistry. 2001;40:14585–14592. doi: 10.1021/bi011247r. [DOI] [PubMed] [Google Scholar]
- 161.Thompson O., Kleino I., Crimaldi L., Gimona M., Saksela K., Winder S.J. Dystroglycan, Tks5 and Src mediated assembly of podosomes in myoblasts. PLoS ONE. 2008;3:e3638. doi: 10.1371/journal.pone.0003638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Abramovici H., Hogan A.B., Obagi C., Topham M.K., Gee S.H. Diacylglycerol kinase-zeta localization in skeletal muscle is regulated by phosphorylation and interaction with syntrophins. Mol. Biol. Cell. 2003;14:4499–4511. doi: 10.1091/mbc.e03-03-0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Abramovici H., Gee S.H. Morphological changes and spatial regulation of diacylglycerol kinase-zeta, syntrophins, and Rac1 during myoblast fusion. Cell Motil. Cytoskelet. 2007;64:549–567. doi: 10.1002/cm.20204. [DOI] [PubMed] [Google Scholar]
- 164.Lumeng C., Phelps S., Crawford G.E., Walden P.D., Barald K., Chamberlain J.S. Interactions between beta 2-syntrophin and a family of microtubule-associated serine/threonine kinases. Nat. Neurosci. 1999;2:611–617. doi: 10.1038/10165. [DOI] [PubMed] [Google Scholar]
- 165.Hasegawa M., Cuenda A., Spillantini M.G., Thomas G.M., Buée-Scherrer V., Cohen P., Goedert M. Stress-activated protein kinase-3 interacts with the PDZ domain of alpha1-syntrophin. A mechanism for specific substrate recognition. J. Biol. Chem. 1999;274:12626–12631. doi: 10.1074/jbc.274.18.12626. [DOI] [PubMed] [Google Scholar]
- 166.Boppart M.D., Mahmassani Z.S. Integrin signaling: Linking mechanical stimulation to skeletal muscle hypertrophy. Am. J. Physiol. Cell Physiol. 2019;317:C629–C641. doi: 10.1152/ajpcell.00009.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ilha J., do Espírito-Santo C.C., de Freitas G.R. mTOR Signaling Pathway and Protein Synthesis: From Training to Aging and Muscle Autophagy. Adv. Exp. Med. Biol. 2018;1088:139–151. doi: 10.1007/978-981-13-1435-3_7. [DOI] [PubMed] [Google Scholar]
- 168.Luo S., Chen Y., Lai K.O., Arévalo J.C., Froehner S.C., Adams M.E., Chao M.V., Ip N.Y. {alpha}-Syntrophin regulates ARMS localization at the neuromuscular junction and enhances EphA4 signaling in an ARMS-dependent manner. J. Cell Biol. 2005;169:813–824. doi: 10.1083/jcb.200412008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Joe M.K., Kee C., Tomarev S.I. Myocilin interacts with syntrophins and is member of dystrophin-associated protein complex. J. Biol. Chem. 2012;287:13216–13227. doi: 10.1074/jbc.M111.224063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Lyssand J.S., Whiting J.L., Lee K.S., Kastl R., Wacker J.L., Bruchas M.R., Miyatake M., Langeberg L.K., Chavkin C., Scott J.D., et al. Alpha-dystrobrevin-1 recruits alpha-catulin to the alpha1D-adrenergic receptor/dystrophin-associated protein complex signalosome. Proc. Natl. Acad. Sci. USA. 2010;107:21854–21859. doi: 10.1073/pnas.1010819107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Oh H.J., Abraham L.S., van Hengel J., Stove C., Proszynski T.J., Gevaert K., DiMario J.X., Sanes J.R., van Roy F., Kim H. Interaction of α-catulin with dystrobrevin contributes to integrity of dystrophin complex in muscle. J. Biol. Chem. 2012;287:21717–21728. doi: 10.1074/jbc.M112.369496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Gingras J., Gawor M., Bernadzki K.M., Grady R.M., Hallock P., Glass D.J., Sanes J.R., Proszynski T.J. Α-Dystrobrevin-1 recruits Grb2 and α-catulin to organize neurotransmitter receptors at the neuromuscular junction. J. Cell Sci. 2016;129:898–911. doi: 10.1242/jcs.181180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Bernadzki K.M., Gawor M., Pęziński M., Mazurek P., Niewiadomski P., Rędowicz M.J., Prószyński T.J. Liprin-α-1 is a novel component of the murine neuromuscular junction and is involved in the organization of the postsynaptic machinery. Sci. Rep. 2017;7:9116. doi: 10.1038/s41598-017-09590-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Bernadzki K.M., Daszczuk P., Rojek K.O., Pęziński M., Gawor M., Pradhan B.S., de Cicco T., Bijata M., Bijata K., Włodarczyk J., et al. Arhgef5 binds α-dystrobrevin 1 and regulates neuromuscular junction integrity. Front. Mol. Neurosci. 2020;13:104. doi: 10.3389/fnmol.2020.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Karpati G., Carpenter S., Morris G.E., Davies K.E., Guerin C., Holland P. Localization and quantitation of the chromosome 6-encoded dystrophin-related protein in normal and pathological human muscle. J. Neuropathol. Exp. Neurol. 1993;52:119–128. doi: 10.1097/00005072-199303000-00004. [DOI] [PubMed] [Google Scholar]
- 176.Ohlendieck K., Ervasti J.M., Matsumura K., Kahl S.D., Leveille C.J., Campbell K.P. Dystrophin-related protein is localized to neuromuscular junctions of adult skeletal muscle. Neuron. 1991;7:499–508. doi: 10.1016/0896-6273(91)90301-F. [DOI] [PubMed] [Google Scholar]
- 177.Blake D.J., Tinsley J.M., Davies K.E. Utrophin: A structural and functional comparison to dystrophin. Brain Pathol. 1996;6:37–47. doi: 10.1111/j.1750-3639.1996.tb00781.x. [DOI] [PubMed] [Google Scholar]
- 178.Matsumura K., Ervasti J.M., Ohlendieck K., Kahl S.D., Campbell K.P. Association of dystrophin-related protein with dystrophin-associated proteins in mdx mouse muscle. Nature. 1992;360:588–591. doi: 10.1038/360588a0. [DOI] [PubMed] [Google Scholar]
- 179.Belhasan D.C., Akaaboune M. The role of the dystrophin glycoprotein complex on the neuromuscular system. Neurosci. Lett. 2020;722:134833. doi: 10.1016/j.neulet.2020.134833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ng S.Y., Ljubicic V. Recent insights into neuromuscular junction biology in Duchenne muscular dystrophy: Impacts, challenges, and opportunities. EBioMedicine. 2020;61:103032. doi: 10.1016/j.ebiom.2020.103032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Smith L.R., Meyer G., Lieber R.L. Systems analysis of biological networks in skeletal muscle function. Wiley Interdiscip. Rev. Syst. Biol. Med. 2013;5:55–71. doi: 10.1002/wsbm.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]