Abstract
Hepatic metabolism catalyzed by the cytochrome P450 (CYP) superfamily affects liver toxicity associated with exposures to natural compounds and xenobiotic agents. Previously we generated a battery of HepG2-derived stable cell lines that individually express 14 CYPs (1A1, 1A2, 1B1, 2A6, 2B6, 2C8, 2C9, 2C18, 2C19, 2D6, 2E1, 3A4, 3A5, and 3A7). In this study, we comprehensively characterized each cell line for its CYP expression and enzyme activity. Specifically, we measured the mRNA expression, protein expression, and metabolite formation. Using CYP3A4, 2D6, and 2C9-overexpressing cells as representatives, we examined the stability of these cells in long-term cultures for up to 10 passages. The results showed that CYPs can be stably overexpressed for up to 10 cell culture passages without losing their activities. The robustness of responses to stimuli among the cells at different passages was also investigated in CYP3A4-overexpressing cells and the response to amiodarone and dronedarone showed no difference between the cells at the passage 2 and 10. Moreover, the mRNA expression level of most CYPs was higher in CYP-overexpressing HepG2 cells than that in HepaRG cells and primary human hepatocytes. This study confirmed the stability of CYP-overexpressing HepG2 cell lines and provided useful information for a broader use of these cells in pharmacologic and toxicologic research.
Keywords: CYP, CYP-expressing HepG2 cells, stability, HepaRG, primary human hepatocytes
Introduction
Organic compounds absorbed from the gut are subject to complex metabolic transformations occurring in the liver. The metabolic products of these reactions may exhibit enhanced or reduced toxicity compared to the parent compounds. As reviewed by Guengerich et al, the human cytochrome P450 (CYP) family of enzymes catalyzes an array of chemical reactions, such as alkyl and aryl hydroxylations, N- and S-oxygenations and dealkylations, epoxidations, dehalogenations, desulfurations, ring expansions and closures, aldehyde scissions, dehydrations, desaturations, ester cleavages, cis/trans isomerizations, one-electron oxidations, and coupling reactions on a wide spectrum of organic compounds that are significant to health.1 Notable structurally diverse categories of compounds that are of environmental concern for humans and that are subject to CYP-catalyzed reactions include organophosphorus pesticides, such as methyl parathion, isocarbophos, and chlorpyrifos,2–4 polycyclic aromatic hydrocarbons,5 perfluoroalkyl substances,6 and polychlorinated biphenyls.7–9 CYP-mediated metabolism is also involved in the mechanisms of action for naturally-occurring hepatotoxic and hepatocarcinogenic agents that may contaminate foods or other items consumed by humans, including mycotoxins like aflatoxin B1,10,11 marine shellfish toxins like okadaic acid,12 and phytotoxins like the pyrrolizidine alkaloids produced by a variety of plant species.13 Furthermore, CYP-mediated liver toxicity is of significant concern for synthetic therapeutic agents14 and for herbal remedies.15 Thus, understanding the role of CYP metabolism in human liver injury is critically important for both environmental toxicology and pharmacology.
Drug-induced liver toxicity is one of the major reasons for drug termination during development and for black box warnings as well as withdrawals of the approved drugs.16–18 Over 50% of marketed drugs had at least one reported case of liver injury, according to a data analysis of LiverTox (http://livertox.nlm.nih.gov), a website documenting the hepatotoxicity of drugs.14 Drug toxicity is attributed to many factors, one of which is that drug-induced liver toxicity in humans is not accurately predicted in animal models. For example, more than 50% drug associated liver toxicity observed in human was not observed in experimental animals that are commonly used for drug safety testing. The poor concordance is partially due to the differences between humans and animals in hepatic drug metabolism.19
Liver toxicity testing using human hepatic cells is being used more frequently to comply with the 3R’s principle of “Reduction, Refinement, and Replacement” for animal research. While primary human hepatocytes are generally considered the “gold standard” for assessing hepatic metabolism and toxicity of xenobiotics,20,21 their usefulness for routine applications, large-scale toxicity testing, and chronic toxicity studies is limited due to the decreased metabolic activity over time, reduced life spans, limited availability, and inter-donor variability.22,23 Immortalized hepatic cell lines provide a valuable alternative owing to their characteristics such as the stable phenotypes, unlimited life span, ready availability, easy manipulation, and cost efficiency. Such characteristics enable hepatic cell lines to be useful in vitro tools for preclinical screening of drug candidates.24 Of a variety of hepatic cell lines, the well-characterized non-tumorigenic HepG2 cell line is one of the most frequently used in liver toxicity research and for elucidating mechanisms of toxicity.25–37 However, reduced expression of drug metabolizing enzymes is a major drawback of HepG2 cells, hampering their use in metabolism-related liver toxicity studies.23,38–40 Genetic modification of HepG2 cells incorporating one or more drug-metabolizing enzymes can overcome the low biotrans-formation capacity of HepG2 cells and thus improve their utility for metabolism and toxicity studies. As such, a number of hepatic cell lines with improved metabolic capacity have been generated recently.41
Using a lentiviral gene expression system, we have generated an array of HepG2-derived cell lines that each stably expressed one of fourteen CYPs including CYP1A1, 1A2, 1B1, 2A6, 2B6, 2C8, 2C9, 2C18, 2C19, 2D6, 2E1, 3A4, 3A5, and 3A7. These HepG2 derivatives showed improved functionality of CYP as using luminogenic substrates of CYPs to assess enzyme activity.42 In the subsequent studies, we demonstrated the suitability of these cells for identifying the specific CYPs accounting for the metabolism and cytotoxicity of drugs and compounds of environmental concern.43–45 Because a broad application of these cells in pharmacologic and toxicologic research is anticipated, further characterization is needed to ensure the validation of these CYP-overexpressing HepG2 cells as a screening tool for metabolism-related drug hepatotoxicity.
Toward this goal, in this study we first characterized CYP function of these cells at three levels: mRNA (using real-time PCR), protein (using Western blot), and enzyme activity (using mass spectrometry analysis for CYP-specific metabolite detection). Second, we evaluated the stability for long-term cell culture of HepG2 cells expressing CYP3A4, 2D6, and 2C9. For this purpose, we compared the consistency of gene and protein expression, and enzymatic activity of CYPs among the cells with different passage numbers. Third, using amiodarone and dronedarone as model drugs whose toxicities are metabolism-related, we investigated the robustness of CYP3A4-overexpressing cells at different passages in response to stimuli. Finally, as a reference we compared CYP gene expressions among CYP-overexpressing HepG2 cells, HepaRG cells, and primary human hepatocytes.
Materials and methods
Chemicals and reagents
Dimethylsulfoxide (DMSO), Williams’ Medium E, phenacetin, 7-ethoxy-4-(trifluoromethyl)coumarin (7-EFC), coumarin, bupropion, taxol paclitaxel, diclofenac, omeprazole, dextromethorphan, chlorzoxazone, and midazolam were from Sigma-Aldrich (St. Louis, MO). Fetal bovine serum (FBS) was purchased from Atlanta Biologicals (Lawrenceville, GA). Antibiotic-antimycotic was from Life Technologies (Grand Island, NY). For Western blot analysis, primary antibodies were purchased from Abcam (Cambridge, MA) or Santa Cruz Biotechnology (Santa Cruz, CA). For real-time PCR, expression assays were purchased from Thermo Fisher Scientific (Waltham, MA). Further descriptive information for antibodies and real-time PCR expression assays is presented in Supplemental Table 1.
Cell culture
The HepG2 cell line was from the American Type Culture Collection (ATCC; Manassas, VA). CYP-overexpressing cell lines were previously engineered in our laboratory.42 HepG2 cells and CYP-overexpressing HepG2 cell lines were cultured in Williams’ Medium E complete media containing 10% FBS and antibiotics (100 units/mL penicillin, 100 μg/mL streptomycin, and 0.25 μg/mL amphotericin B). Unless otherwise specified, cells were seeded at a density of 2.5 × 105 cells/ml in a volume of 100 μl per well in 96-well plates, or 3 × 105 in a volume of 2 ml in 12-well plates, or 5 × 106 in a volume of 15 ml in 100 mm tissue culture dishes.
Undifferentiated HepaRG cells were purchased from Biopredic International (Saint Grégoire, France) and cultured according to the manufacturer’s protocol. Briefly, the cells were plated at a density of 1.3 × 104 cells/cm2 and cultured in Williams’ Medium E supplemented with 2 mM L-glutamine (Sigma-Aldrich) and growth additives (Lonza, Walkersville, MD, USA) for 14 days. The cells were then differentiated by adding a differentiation supplement (Lonza) to the medium for an additional 14 days prior to RNA isolation. Culture medium was refreshed every 2–3 days.
Terminally differentiated, cryopreserved HepaRG cells were obtained from ThermoFisher and cultured according to the manufacturer’s protocol with minor modifications. Briefly, HepaRG cells were thawed and resus-pended in Williams’ Medium E supplemented with 2 mM GlutaMAX and the Thaw, Plate, & General Purpose Medium Supplement (ThermoFisher). Resuspended HepaRG cells were seeded in 60 mm tissue culture dishes (4.5 × 106/dish) and cultured for 24 hours. Cells were then cultured in Williams’ Medium E supplemented with 2 mM GlutaMAX and the Maintenance/Metabolism Medium Supplement (ThermoFisher) for 7 days prior to RNA isolation. Culture medium was refreshed every 2 days.
Cryopreserved primary human hepatocytes, pooled from 10 donors, were purchased from In Vitro ADMET Laboratories (Columbia, MD). Dishes used for primary human hepatocytes were pre-coated with PureCol® (Advanced BioMatrix, Carlsbad, CA) following the manufacturer’s protocol. Cells were thawed and seeded in 60 mm tissue culture dishes (4.5 × 106/dish) for 24 h prior to RNA isolation. Cells were maintained in Universal Primary Cell Plating Medium (UPCM™) provided by the supplier. All cells were maintained in a humidified incubator containing 5% CO2 at 37 °C.
CYP activity measurement by mass spectrometry
The activities of CYPs in transduced HepG2 cells were measured by quantifying the rate of formation of metabolites after the incubation with CYP-specific substrates. Each individual CYP cell line was seeded in 12-well plates for 24 h prior to the addition of its substrate (Supplemental Table 1) for 2 or 24 h and then the cell culture supernatant was harvested. The supernatants were diluted with four volumes of acetonitrile containing 100 ng/ml of the internal standard 4-hydroxy bupropion-D6. Samples were vortexed, centrifuged for 5 min at 13,000 × g, and the supernatants were loaded into sample vials for UPLC–MS analysis. Two μl of the supernatants were injected onto a Waters ACQUITY UPLC System coupled with a Waters Acquity QDa mass detector. The metabolites were eluted on an ACQUITY UPLC HSS T3 column (2.1 mm × 50 mm, 1.8 μm) at 40°C with mobile phases of LC–MS grade water (A) and acetonitrile (B), both containing 0.1% formic acid, at a flow rate of 0.5 ml/min. Elution started with 0% solvent B followed by a linear gradient of 0–100% solvent B in 1.2 min, returning to 0% B in 0.1 min, and maintained for 1.2 min to re-equilibrate the column. The LC eluent was introduced to mass detector with an electrospray ion source using single ion recording mode. The monitored (M + H)+ ions were m/z 152 for acetaminophen (the metabolite of phenacetin by CYP1A1 and 1A2), m/z 256 for hydroxybupropion (the metabolite of bupropion by CYP2B6), m/z 871 for 6α-hydroxypaclitaxel (the metabolite of taxol paclitaxel by CYP2C8), m/z 312 for 4-hydroxydiclofenac (the metabolite of diclofenac by CYP2C9), m/z 362 for 5-hydroxyomeprazole (the metabolite of omeprazole by CYP2C18 and 2C19), m/z 258 for dextrorphan (the metabolite of dextromethorphan by CYP2D6), and m/z 342 for 1-hydroxymidazolam (the metabolite of midazolam by CYP3A4, 3A5, and 3A7). Three (M-H)− ions were also monitored at m/z 229 for 7-hydroxy-4-(trifluoromethyl) coumarin (the metabolite of 7-ethoxy-4-(trifluoromethyl) coumarin by CYP1B1), m/z 161 for 7-hydroxycoumarin (the metabolite of coumarin by CYP2A6), and m/z 184 for 6-hydroxychlorzoxa-zone (the metabolite of chlorzoxazone by CYP2E1). All metabolites were identified and quantified in Waters Empower 3.0 (Milford, MA).
Lactate dehydrogenase assay
The cytotoxicity of amiodarone and dronedarone was assessed using a lactate dehydrogenase (LDH) assay as described previously.46
Western blot analysis
Cells were cultured in 100 mm tissue culture dishes and subcultured by trypsinization (trypsin–EDTA solution) at 95% confluency every two or three days. Whole-cell lysates from 4 × 106 cells at different passages were prepared using RIPA buffer containing Halt Protease Inhibitor Cocktail (ThermoFisher Scientific). The concentrations of the protein samples were determined using a Bio-Rad Protein Assay (Bio-Rad Laboratories, Hercules, CA). Standard Western blots were performed. Depending on the proteins of interest, antibodies were selected against CYP1A1, 1A2, 1B1, 2A6, 2B6, 2C8, 2C9, 2C18, 2C19, 2D6, 2E1, 3A4, 3A5, 3A7, and Myc, followed by an incubation with a secondary antibody conjugated with horseradish peroxidase (HRP) (Santa Cruz Biotechnology). GAPDH was used as the internal control. The protein signals were visualized with a FluorChem E System and quantified with AlphaView software (ProteinSimple, San Jose, CA).
RNA isolation and quantitative real-time PCR assay
Total RNA was isolated using an RNeasy mini kit (Qiagen, Germantown, MD). The purity and quality of RNA were examined using a NanoDrop 8000 (ThermoFisher Scientific). cDNAs were generated by reverse transcription of 2 μg total RNA using a high capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA). Quantitative real-time PCR (qPCR) for CYP1A1, 1A2, 1B1, 2A6, 2B6, 2C8, 2C9, 2C18, 2C19, 2D6, 2E1, 3A4, 3A5, 3A7, and GAPDH was performed to evaluate relative gene expression. qPCR reactions were performed using FastStart Universal Probe Master (Rox) (Millipore Sigma) with a Bio-Rad CFX96™ Real-Time PCR Detection System (Bio-Rad Laboratories) under universal cycling conditions (10 min at 95 °C; 15 s at 95 °C, 1 min at 60 °C, 40 cycles). Threshold cycle (Ct) was used to determine the relative expression levels of target genes. Ct values above 35 were deemed as non-detectable.47 Data normalization and analysis were conducted as described previously.48
Statistical analyses
Data are presented as the mean ± standard deviation (SD) of at least three independent experiments. Analyses were performed using GraphPad Prism 5 (GraphPad Software, San Diego, CA). Statistical significance was determined by one-way analysis of variance (ANOVA) followed by the Dunnett’s tests for pairwise-comparisons or two-way ANOVA followed by the Bonferroni post-test. The difference was considered statistically significant when p was less than 0.05.
Results
Characterization of the overexpression of CYPs in HepG2-derived cells
We first examined the gene expression of CYPs using real-time PCR. The calculation of relative gene expression was adopted from our previous study.23 The approximate abundance of each CYP is expressed as “Expression Value” using GAPDH as reference (Table 1). Expression Value implies the relative mRNA expression abundance of a CYP gene, arbitrarily assuming an expression level of the housekeeping gene GAPDH being 10,000 copies. The Expression Value of each CYP was defined using the equation: E = 2−(Ct of test gene-Ct of GAPDH) × 10,000. In parental HepG2 cells and the HepG2 cells transduced with empty vector control, the expression of CYP1A2, 1B1, 2C8, 2C9, 2C19, 2D6, and 3A4 was not detected (Ct > 35), while CYP1A1, 2A6, 2B6, 2C18, 2E1, 3A5, and 3A7 expressed at very low level (<50). As shown in Table 1, the expression level of any given CYP was not significantly different between the empty vector transduced-HepG2 control cells and parental HepG2 cells, suggesting that the expression of CYPs in HepG2 cells was not affected by the lentiviral transduction. As expected, the expression of CYP mRNA was dramatically increased in each of the corresponding CYP-overexpressing cell lines (Table 1).
Table 1.
Comparison of CYP gene expressions in HepG2, HepG2-empty vector, CYP-overexpressing HepG2 cells and primary human hepatocytes.
| Expression Value* | Expression Value in CYP-overexpressing cells vs human hepatocytes (fold change level**) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| HepG2 | HepG2-empty vector | CYP-overexpressing HepG2 | Human Hepatocytes | |||||||
| CYP Gene | Average | S.D. | Average | S.D. | Cell lines | Average | S.D. | Average | S.D. | |
| CYP1A1 | 2.10 | 0.62 | 3.12 | 0.20 | HepG2-CYP1A1 | 9230.86 | 570.06 | 3.15 | 0.37 | +++ |
| CYP1A2 | N.D. | N.D. | HepG2-CYP1A2 | 6085.21 | 316.73 | 28.38 | 1.63 | +++ | ||
| CYP1B1 | N.D. | N.D. | HepG2-CYP1B1 | 7260.78 | 301.46 | 28.91 | 1.42 | +++ | ||
| CYP2A6 | 0.34 | 0.07 | 0.36 | 0.17 | HepG2-CYP2A6 | 14958.27 | 1494.98 | 1330.04 | 80.42 | ++ |
| CYP2B6 | 34.56 | 0.76 | 32.29 | 0.80 | HepG2-CYP2B6 | 6310.56 | 909.17 | 869.33 | 71.43 | + |
| CYP2C8 | N.D. | N.D. | HepG2-CYP2C8 | 3624.38 | 546.40 | 1122.95 | 44.64 | + | ||
| CYP2C9 | N.D. | N.D. | HepG2-CYP2C9 | 3848.98 | 196.82 | 1284.64 | 114.59 | + | ||
| CYP2C18 | 0.04 | 0.00 | 0.04 | 0.00 | HepG2-CYP2C18 | 7289.49 | 420.42 | 341.21 | 30.01 | ++ |
| CYP2C19 | N.D. | N.D. | HepG2-CYP2C19 | 4568.73 | 295.48 | 35.96 | 0.71 | +++ | ||
| CYP2D6 | N.D. | N.D. | HepG2-CYP2D6 | 525.41 | 83.72 | 13.50 | 3.36 | ++ | ||
| CYP2E1 | 0.04 | 0.01 | 0.04 | 0.00 | HepG2-CYP2E1 | 2005.09 | 132.22 | 5223.96 | 348.51 | − |
| CYP3A4 | N.D. | N.D. | HepG2-CYP3A4 | 1849.10 | 237.69 | 838.40 | 7.13 | + | ||
| CYP3A5 | 1.57 | 0.15 | 1.99 | 0.08 | HepG2-CYP3A5 | 10034.07 | 853.00 | 872.15 | 83.79 | ++ |
| CYP3A7 | 1.38 | 0.15 | 1.57 | 0.04 | HepG2-CYP3A7 | 6466.39 | 211.30 | 11.60 | 1.48 | +++ |
N.D., not detected.
Expression Value is calculated based on the assumption that the expression level of the housekeeping gene GAPDH is 10,000 copies (10, 000 is arbitrary). E = 2−ΔCt * 10,000; ΔCt = Ct of test gene-Ct of GAPDH.
Fold change level was defined as +++, fold change > 100;++, fold change = 10–100; +, fold change = 1–10; −, fold change = 0–1.
We next examined the CYP protein expression of each cell line using Western blotting (Figure 1). Both parental HepG2 cells and empty vector control cells showed negligible protein expression of all CYPs. Each CYP protein along with Myc-fusion protein was strongly expressed in its corresponding CYP-overexpressing cell line.
Figure 1.
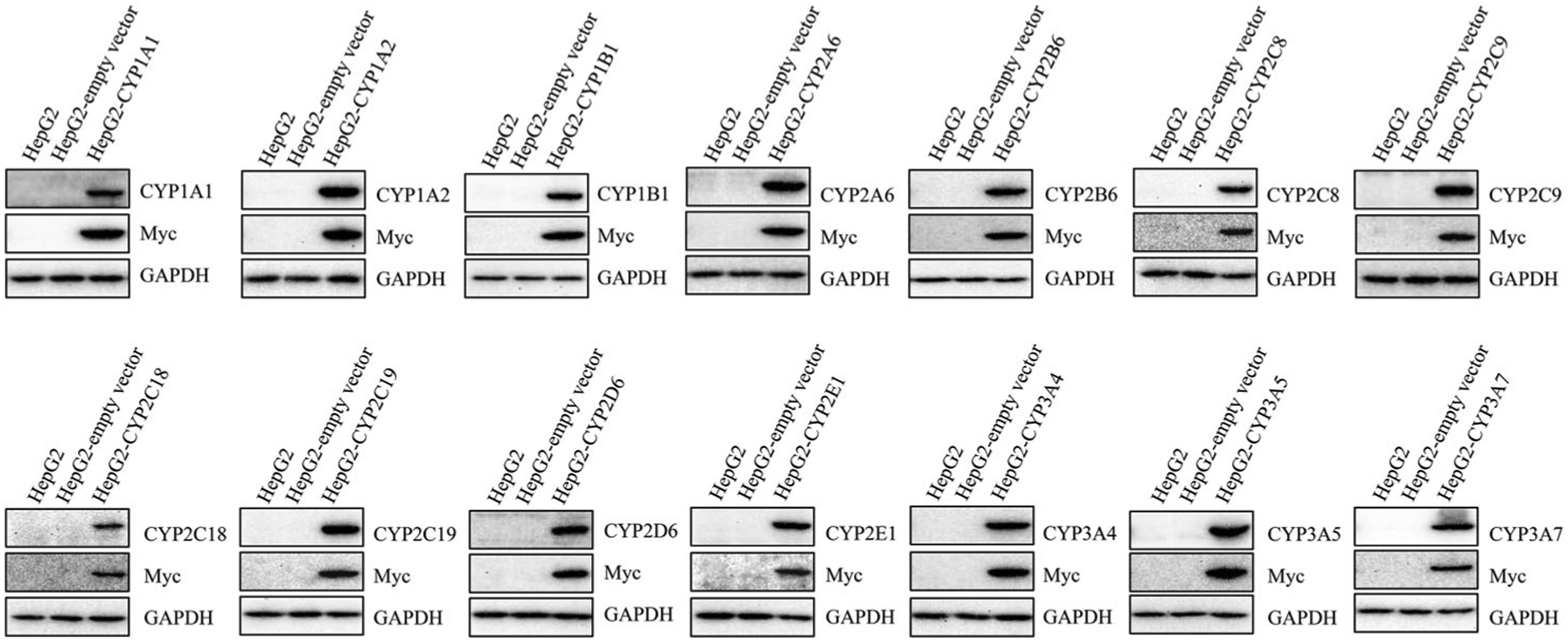
The expression of CYP proteins in stably transduced HepG2 cells. The protein levels of CYP isoforms in HepG2, HepG2-empty vector, and HepG2 CYP-overexpressing cells were detected by Western blotting. Myc was a fusion protein tag and GAPDH was used as a loading control.
To confirm further the metabolic functioning of the CYPs in the stably transduced HepG2 cells, we measured the enzymatic activity of each CYP using its corresponding specific substrate. Figure 2 depicts the representative UPLC-MS chromatograms of CYP-specific metabolites formation for the assessment of enzymatic activity. The substrate and metabolite of each CYP are listed in Supplementary Table 1. While the empty vector control cells had very little or no detectable activity for the enzymes, all CYP-overexpressing HepG2 cells displayed the enzymatic activities as demonstrated by the conversion of the substrates to their corresponding metabolites. Taken together, these results from different approaches ensured the functional human CYPs were transduced into HepG2 cell lines.
Figure 2.
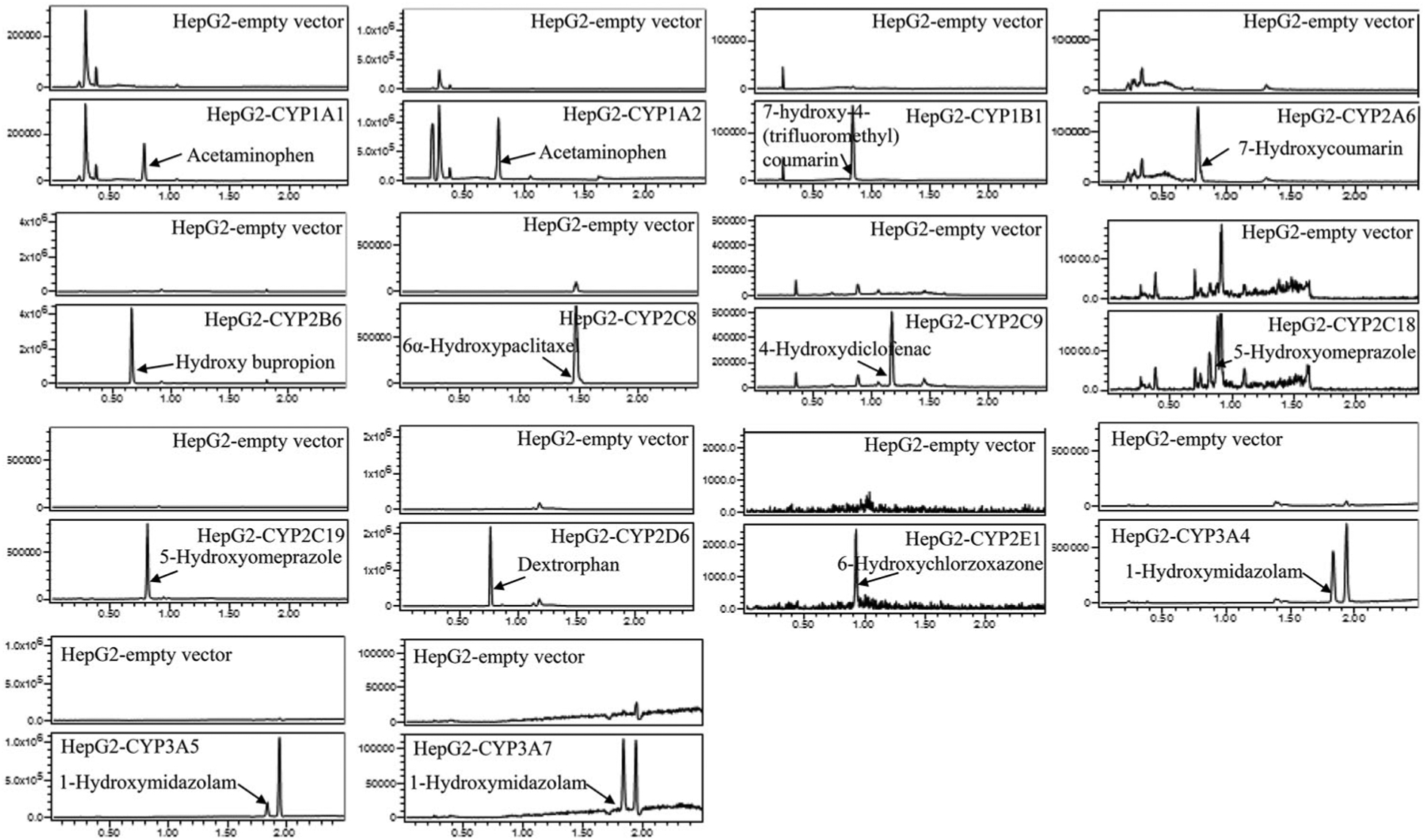
LC-MS chromatograms showing the CYP-specific metabolites formed by the HepG2 cells transduced with various CYPs.
Stability of the expression of CYPs in HepG2-derived cells
It has been reported that long-term culture impacts the characteristics of cells over time, such as the expression of proteins and responses to stimuli.49–53 Because CYP3A4, 2D6, and 2C9 together account for a large proportion of CYP family-dependent metabolism of xenobiotics in humans,54 we selected CYP3A4-, 2D6-, and 2C9-overexpressing cells as representatives to study the effects of cell passage number on cell morphology, CYP expression, and enzyme function. These CYP-expressing HepG2 cells were subcultured up to passage 10, and the mRNA and protein expression and enzyme activity of the cells from the passage 4, 6, 8, and 10 were compared with those of the cells at passage 2. No obvious morphological changes were observed using phase contrast microscopy regardless the passage number (data not shown). There was no significant decrease in the expression of gene (Figure 3A), protein (Figure 3B) and enzyme activity (Figure 3C) among the cells compared. The formation of enzyme-specific metabolites was not reduced among the cells with different passage numbers, even with the highest passage number of 10.
Figure 3.
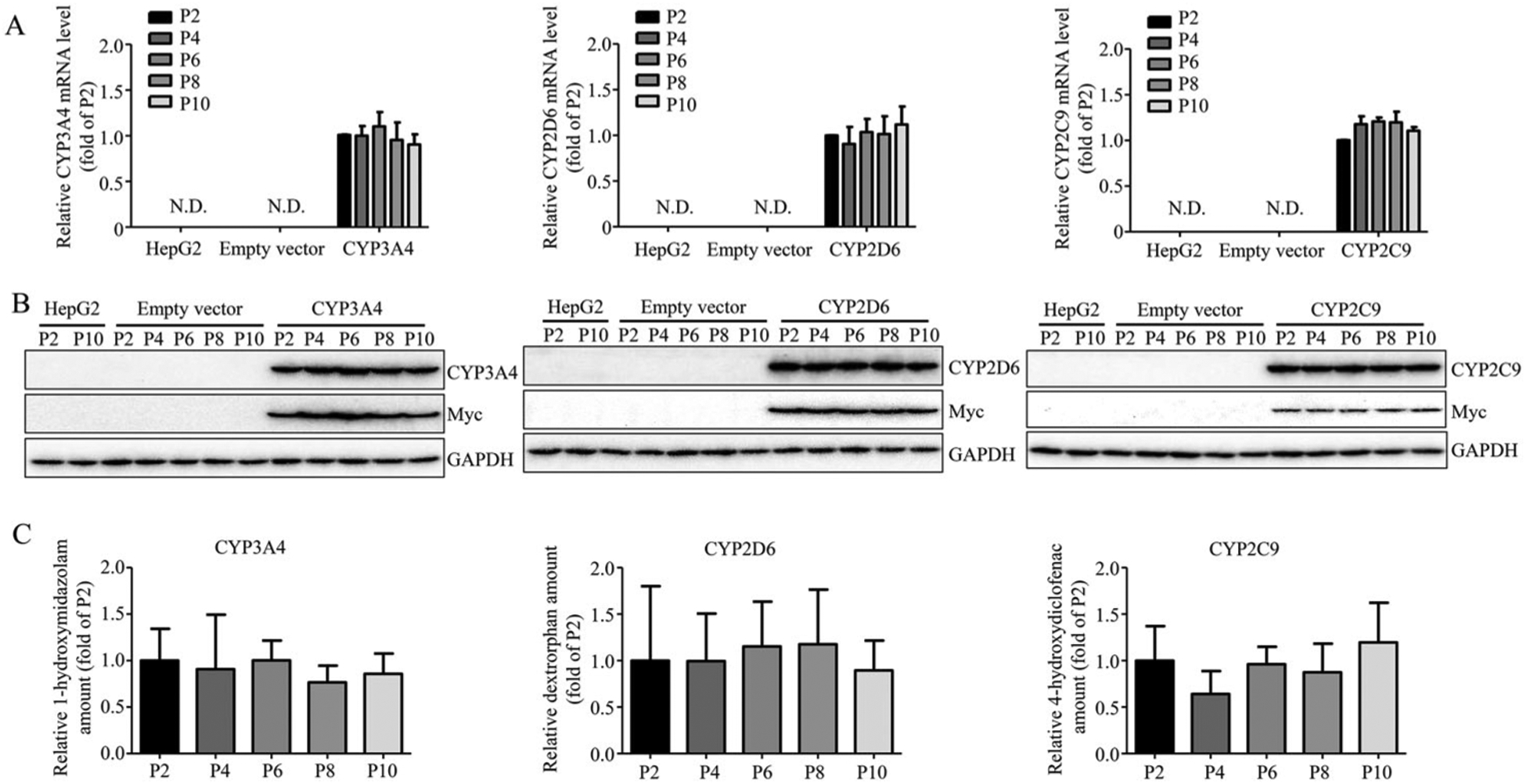
The stability of overexpressed CYP3A4, 2D6, and 2C9. The overexpression of CYP3A4, 2D6, and 2C9 at mRNA level (A), protein level (B), and enzymatic activity level (C) in HepG2-CYP3A4, HepG2-CYP2D6, HepG2-CYP2C9 cell lines was measured at indicated passage number by real-time PCR, Western blotting, and LC-MS. The results shown are mean ± S.D. from three independent experiments. Statistical analysis was conducted using one-way analysis of variance (ANOVA) followed by the Dunnett’s tests for pairwise-comparisons.
Collectively, our data demonstrated that the expression and function of CYPs were consistent among the cells at different passages encompassing 2–10. Next, we investigated whether the cell passage number could alter the cellular responses to stimuli by studying the responses to two drugs, amiodarone and dronedarone. Both amiodarone and dronedarone are used to treat cardiac arrhythmias and CYP3A4 is involved in the metabolism of both drugs. Earlier, we determined that the activation of CYP3A4 significantly increased the cytotoxicity of amiodarone, whereas CYP3A4 played the opposite role and counteracted the cytotoxicity of dronedarone.44,46 Here, we compared the CYP3A4 mediated toxicity between the cells at passage 2 and 10. Consistent with our previous studies, the cytotoxicity of amiodarone significantly increased in CYP3A4-overexpressing cells when compared to empty vector control cells. Conversely, the cytotoxicity of dronedarone significantly decreased in CYP3A4-overexpressing cells (Figure 4). These observations were consistent for cells at passage 2 and 10, demonstrating that the metabolic response to the insults was not affected by cell passaging.
Figure 4.
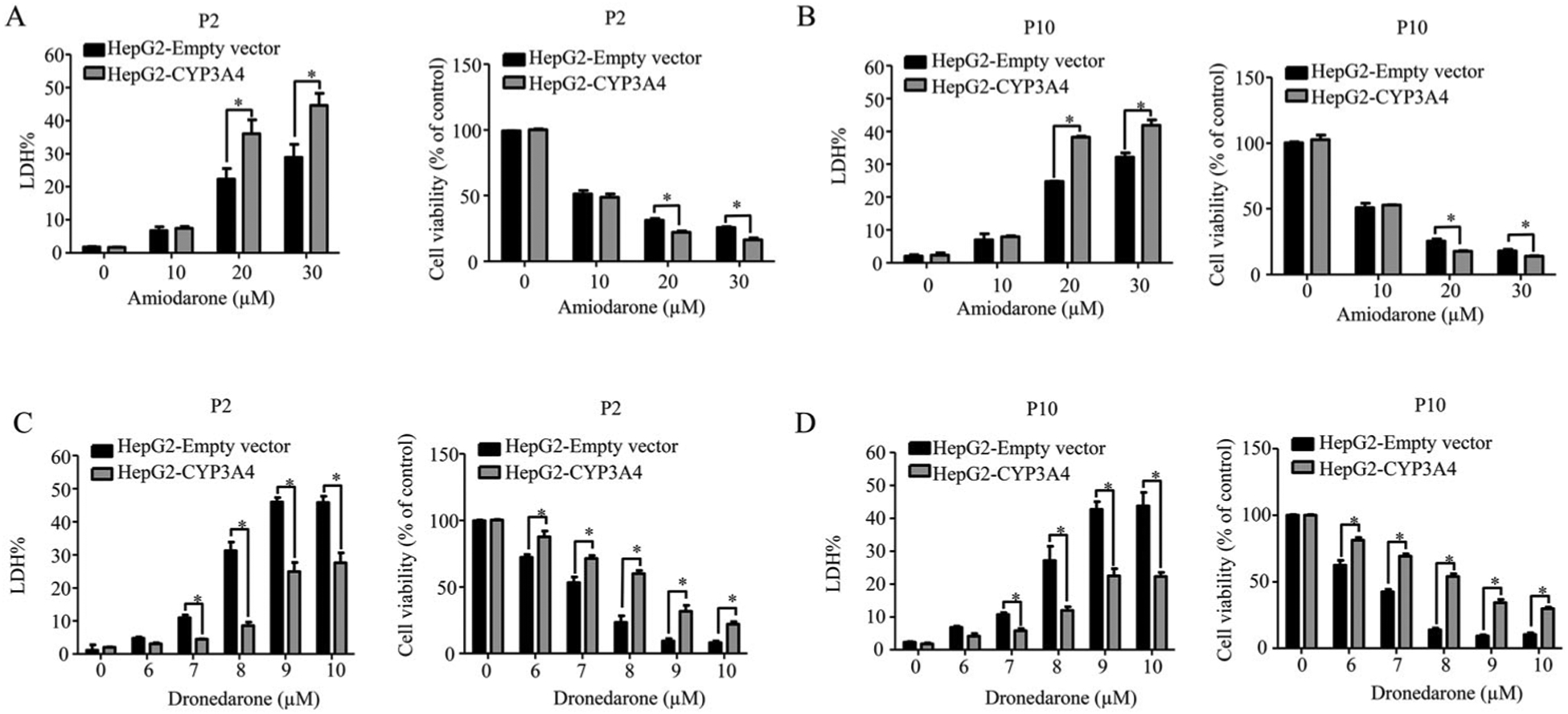
The robustness of CYP3A4-overexpressing cells with different passages in response to the toxicity of amiodarone and dronedarone. Empty vector-transduced or CYP3A4-overexpressing HepG2 cells at passage number 2 and 10 were treated with the indicated concentrations of amiodarone (A and B) or dronedarone (C and D) for 24 h. Cytotoxicity was measured using LDH assays. The results shown are mean ± S.D. from three independent experiments.*, p < 0.05 compared with that of empty vector control.
Comparison of CYP gene expression in CYP-overexpressing HepG2 cells, primary human hepatocytes, and HepaRG cells
Primary human hepatocytes and HepaRG cells have been used for drug metabolism and hepatotoxicity studies because of their metabolic capability. To provide a reference for researchers who intend to choose a cell type for certain purposes based on their experimental designs, CYP-overexpressing HepG2 cells, primary human hepatocytes, and terminally differentiated HepaRG cells were compared.
Primary human hepatocytes pooled from 10 donors (demographic data is presented in Supplemental Table 2) showed abundant gene expression of CYP2A6, 2B6, 2C8, 2C9, 2C18, 2E1, 3A4, and 3A5, but low expression of CYP1A1, 1A2, 1B1, 2C19, 2D6, and 3A7 (Table 1). With the exception of CYP2E1, the gene expression of all the other CYPs was higher in our CYP-overexpressing HepG2 cells than in primary human hepatocytes.
CYP gene expression in HepaRG cells obtained from two different vendors (Biopredic International and ThermoFisher) was profiled. Although the expression of the CYPs showed some variation (notably CYP2C8) between HepaRG cells from the two different sources, the expression of most CYPs was comparable (Table 2). The expression of CYP3A4 was high in the HepaRG cells, the mRNA levels of CYP1A2, 1B1, 2C18, 2C19, and 3A7 were low (< 100), and CYP2D6 was undetectable. Except for CYP3A4, the expression level of the CYPs in our CYP-overexpressing HepG2 cells was higher compared to HepaRG cells (Table 2).
Table 2.
Comparison of CYP gene expressions in CYP-overexpressing HepG2 cells and HepaRG cells.
| Expression Value* | Expression Value in #1 HepaRG vs #2 HepaRG cells (fold change level**) | Expression Value in CYP-overexpressing HepG2 vs #1 HepaRG cells (fold change level**) | Expression Value in CYP-overexpressing HepG2 vs #2 HepaRG cells (fold change level**) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CYP-overexpressing HepG2 | HepaRG cells (#1, Biopredic Int’l) | HepaRG cells (#2, ThermoFisher) | ||||||||
| CYP Gene | Cell lines | Average | S.D. | Average | S.D. | Average | S.D. | |||
| CYP1A1 | HepG2-CYP1A1 | 9230.86 | 570.06 | 117.64 | 6.85 | 166.22 | 72.00 | − | ++ | ++ |
| CYP1A2 | HepG2-CYP1A2 | 6085.21 | 316.73 | 21.49 | 1.33 | 6.77 | 0.51 | + | +++ | +++ |
| CYP1B1 | HepG2-CYP1B1 | 7260.78 | 301.46 | 31.08 | 0.47 | 38.83 | 7.04 | − | +++ | +++ |
| CYP2A6 | HepG2-CYP2A6 | 14958.27 | 1494.98 | 608.36 | 86.43 | 229.15 | 32.51 | + | ++ | ++ |
| CYP2B6 | HepG2-CYP2B6 | 6310.56 | 909.17 | 384.08 | 49.17 | 213.38 | 47.93 | + | ++ | ++ |
| CYP2C8 | HepG2-CYP2C8 | 3624.38 | 546.40 | 1124.47 | 49.17 | 106.53 | 25.47 | ++ | ++ | + |
| CYP2C9 | HepG2-CYP2C9 | 3848.98 | 196.82 | 2166.48 | 271.9 | 1743.30 | 607.69 | + | + | + |
| CYP2C18 | HepG2-CYP2C18 | 7289.49 | 420.42 | 64.10 | 21.23 | 38.68 | 5.46 | + | +++ | +++ |
| CYP2C19 | HepG2-CYP2C19 | 4568.73 | 295.48 | 80.18 | 7.60 | 16.75 | 11.03 | + | +++ | ++ |
| CYP2D6 | HepG2-CYP2D6 | 525.41 | 83.72 | N.D. | N.D. | N.A. | N.A. | N.A. | ||
| CYP2E1 | HepG2-CYP2E1 | 2005.09 | 132.22 | 626.62 | 31.11 | 314.79 | 53.41 | + | + | + |
| CYP3A4 | HepG2-CYP3A4 | 1849.10 | 237.69 | 11885.27 | 1109.14 | 3692.14 | 974.57 | + | − | − |
| CYP3A5 | HepG2-CYP3A5 | 10034.07 | 853.00 | 963.19 | 48.86 | 273.15 | 13.52 | + | ++ | ++ |
| CYP3A7 | HepG2-CYP3A7 | 6466.39 | 211.30 | 3.09 | 0.27 | 0.73 | 0.23 | + | +++ | +++ |
N.D., not detected. N.A., not available.
Expression Value is calculated based on the assumption that the expression level of the housekeeping gene GAPDH is 10,000 copies (10, 000 is arbitrary). E = 2−ΔCt * 10,000; ΔCt = Ct of test gene-Ct of GAPDH.
Fold change level was defined as +++, fold change > 100; ++, fold change = 10–100; +, fold change = 1–10; −, fold change = 0–1.
Discussion
To study CYP-dependent drug and chemical metabolism and to identify the role of a specific CYP in xenobiotic-induced toxicity, a number of HepG2-derived cell lines have been developed, transiently or stably expressing one or more CYPs.11,41,42,55–57 Among these cells, our laboratory established an array of cell lines encompassing 14 CYPs.42 The utility of our CYP-overexpressing HepG2 cell lines has been proved in the studies focusing on metabolism-associated toxicity induced by amiodarone,44 dronedarone,46 and sertraline.45 The usefulness of these cells for the research of drug-drug interactions has also been demonstrated.58,59 In the current study, our efforts focused on addressing the stability and response robustness of these cells to make them available for a broader use by the pharmacologic and toxicologic communities.
For a quick reference, we listed the information of materials used for real-time PCR (expression assays), Western blotting (antibodies), and mass spectrometry analysis (CYP substrates and metabolites) in Supplementary Table 1. It is worth mentioning that some commercially available primers/probes and antibodies we tested were not suitable for the detection methods used in this study, possibly due to the low specificity; thus, attention should be paid when selecting specific reagents.
The choice of a cell model strongly depends on the research goals. Primary human hepatocytes are the most widely accepted cell model for pharmacological and toxicological studies, taking metabolic capability into account. The HepaRG cell line has recently emerged as a surrogate for primary human hepatocytes. Hepatic cell lines have been used for years; however, they are not the best representative of human liver cells due to the lack of metabolizing enzymes. To overcome it, we incorporated CYP metabolic capability in HepG2 cells by transducing lentiviral cDNA expression vectors containing the individual CYP sequences.42 Unlike the parental HepG2 cells that exhibit limited drug metabolic capacity,23,60 our cell lines express 14 CYPs individually at the transcriptional, translational, and functional levels (Table 1). To understand better the suitability of CYP-overexpressing HepG2 cell lines and provide a reference profile, comparisons were made among these three types of cells. HepaRG cells did not fully recapitulate the enzyme profile of primary human hepatocytes (Table 2). In particular, CYP2D6 was not detected in HepaRG cells, although that was expected since the donor for HepaRG cells was a poor CYP2D6 metabolizer.61,62 CYP2D6 has an important role in drug metabolism because it is the primary enzyme to metabolize 15–25% of clinically used drugs, including antiarrhythmics, antipsychotics, β-blockers, and anti-cancer drugs.54 The lack of CYP2D6 in HepaRG cell line limits its use for certain pharmacological and toxicological studies involving CYP2D6 activity.55
The gene expression of CYP2E1 in primary human hepatocytes was higher than in HepaRG and CYP2E1-overexpressing HepG2 cells (Tables 1 and 2). CYP2E1 enzyme expression is inducible by many of its substrates (e.g. alcohol) with complex mechanisms including transcriptional regulation.63 In addition, CYP2E1 is inducible under diverse pathophysiological conditions, including diabetes, obesity, fasting, and alcoholic liver disease.64 The high expression level of CYP2E1 observed in the primary hepatocytes could be explained by the lifestyle and disease conditions of the donors (Supplemental Table 2). For example, half of the donors were obese (BMI ≥ 30) and 4 out of the 10 donors consumed alcohol frequently.
Except for the CYPs noted, the overall individual gene expression of CYP-overexpressing HepG2 cells was much higher than that of primary human hepatocytes and HepaRG cells. From the metabolic standpoint, our engineered cells represent an improvement from the parental HepG2 cells and can be considered as surrogate enzyme sources for exploring the role of specific CYPs that contribute to the metabolism and toxicity of drugs. However, we are aware that to make the in vitro toxicity studies more human-relevant, the physiological levels of metabolic enzymes should be incorporated. To achieve this goal, our laboratory is currently conducting the experiments to establish cell lines expressing CYPs at the levels comparable to primary human hepatocytes, by adjusting the multiplicity of infection (MOI) in the experiment of lentiviral plasmid transfection.
In summary, we have demonstrated the metabolic stability and response robustness of our CYP-overexpressing HepG2 cell lines. These cells can provide a practical in vitro approach for screening drug and chemical metabolism, for metabolism-associated drug toxicity investigation, and for drug-drug interaction studies.
Supplementary Material
Funding
XL and DL were supported by appointments to the Postgraduate Research Program at the National Center for Toxicological Research administered by the Oak Ridge Institute for Science Education through an interagency agreement between the U.S. Department of Energy and the U.S. FDA.
Footnotes
Publisher's Disclaimer: Disclaimer
Publisher's Disclaimer: This article is not an official guidance or policy statement of the U.S. FDA. No official support or endorsement by the U.S. FDA is intended or should be inferred.
References
- [1].Guengerich FP. Common and uncommon cytochrome P450 reactions related to metabolism and chemical toxicity. Chem Res Toxicol. 2001;14(6):611–650. doi: 10.1021/tx0002583. [DOI] [PubMed] [Google Scholar]
- [2].Ellison CA, Tian Y, Knaak JB, Kostyniak PJ, Olson JR. Human hepatic cytochrome P450-specific metabolism of the organophosphorus pesticides methyl parathion and diazinon. Drug Metab Dispos. 2012;40(1):1–5. doi: 10.1124/dmd.111.042572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Zhuang XM, Wei X, Tan Y, et al. Contribution of carboxylesterase and cytochrome P450 to the bioactivation and detoxification of isocarbophos and its enantiomers in human liver microsomes. Toxicol Sci. 2014;140(1):40–48. doi: 10.1093/toxsci/kfu067. [DOI] [PubMed] [Google Scholar]
- [4].D’Agostino J, Zhang H, Kenaan C, Hollenberg PF. Mechanism-based inactivation of human cytochrome P450 2B6 by chlorpyrifos. Chem Res Toxicol. 2015;28(7):1484–1495. doi: 10.1021/acs.chemrestox.5b00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Rendic S, Guengerich FP. Contributions of human enzymes in carcinogen metabolism. Chem Res Toxicol. 2012;25(7):1316–1383. doi: 10.1021/tx300132k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Benskin JP, Holt A, Martin JW. Isomer-specific biotransformation rates of a perfluorooctane sulfonate (PFOS)-precursor by cytochrome P450 isozymes and human liver microsomes. Environ Sci Technol. 2009;43(22):8566–8572. doi: 10.1021/es901915f. [DOI] [PubMed] [Google Scholar]
- [7].Uwimana E, Li X, Lehmler HJ. Human liver microsomes atropselectively metabolize 2,2’,3,4’,6-Pentachlorobiphenyl (PCB 91) to a 1,2-shift product as the major metabolite. Environ Sci Technol. 2018;52(10):6000–6008. doi: 10.1021/acs.est.8b00612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Nagayoshi H, Kakimoto K, Konishi Y, Kajimura K, Nakano T. Determination of the human cytochrome P450 monooxygenase catalyzing the enantioselective oxidation of 2,2’,3,5’,6-pentachlorobiphenyl (PCB 95) and 2,2’,3,4,4’,5’,6-heptachlorobiphenyl (PCB 183). Environ Sci Pollut Res Int. 2018;25(17):16420–16426. doi: 10.1007/s11356-017-0434-z. [DOI] [PubMed] [Google Scholar]
- [9].Uwimana E, Cagle B, Yeung C, et al. Atropselective oxidation of 2,2’,3,3’,4,6’-hexachlorobiphenyl (PCB 132) to hydroxylated metabolites by human liver microsomes and its implications for PCB 132 neurotoxicity. Toxicol Sci. 2019;171(2):406–420. doi: 10.1093/toxsci/kfz150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kamdem LK, Meineke I, Godtel-Armbrust U, Brockmoller J, Wojnowski L. Dominant contribution of P450 3A4 to the hepatic carcinogenic activation of aflatoxin B1. Chem Res Toxicol. 2006;19(4):577–586. doi: 10.1021/tx050358e. [DOI] [PubMed] [Google Scholar]
- [11].Tolosa L, Donato MT, Perez-Cataldo G, Castell JV, Gomez-Lechon MJ. Upgrading cytochrome P450 activity in HepG2 cells co-transfected with adenoviral vectors for drug hepatotoxicity assessment. Toxicol in Vitro. 2012;26(8):1272–1277. doi: 10.1016/j.tiv.2011.11.008. [DOI] [PubMed] [Google Scholar]
- [12].Hashizume T, Yoshitomi S, Asahi S, Matsumura S, Chatani F, Oda H. In vitro micronucleus test in HepG2 transformants expressing a series of human cytochrome P450 isoforms with chemicals requiring metabolic activation. Mutat Res. 2009;677(1–2):1–7. doi: 10.1016/j.mrgentox.2009.03.009. [DOI] [PubMed] [Google Scholar]
- [13].Yang M, Ruan J, Fu PP, Lin G. Cytotoxicity of pyrrolizidine alkaloid in human hepatic parenchymal and sinusoidal endothelial cells: Firm evidence for the reactive metabolites mediated pyrrolizidine alkaloid-induced hepatotoxicity. Chem Biol Interact. 2016;243:119–126. doi: 10.1016/j.cbi.2015.09.011. [DOI] [PubMed] [Google Scholar]
- [14].Bjornsson ES. Hepatotoxicity by drugs: The most common implicated agents. Int J Mol Sci. 2016;17:224. doi: 10.3390/ijms17020224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Liu Z, He X, Wang L, Zhang Y, Hai Y, Gao R. Chinese herbal medicine hepatotoxicity: the evaluation and recognization based on large-scale evidence database. Curr Drug Metab. 2019;20(2):138–146. doi: 10.2174/1389200219666180813144114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Thakkar S, Li T, Liu Z, Wu L, Roberts R, Tong W. Drug-induced liver injury severity and toxicity (DILIst): binary classification of 1279 drugs by human hepatotoxicity. Drug Discov Today. 2020;25(1):201–208. doi: 10.1016/j.drudis.2019.09.022. [DOI] [PubMed] [Google Scholar]
- [17].Kaplowitz N Idiosyncratic drug hepatotoxicity. Nat Rev Drug Discov. 2005;4(6):489–499. doi: 10.1038/nrd1750. [DOI] [PubMed] [Google Scholar]
- [18].Lee WM. Drug-induced hepatotoxicity. N Engl J Med. 2003;349(5):474–485. doi: 10.1056/NEJMra021844. [DOI] [PubMed] [Google Scholar]
- [19].Baillie TA, Rettie AE. Role of biotransformation in drug-induced toxicity: influence of intra- and inter-species differences in drug metabolism. Drug Metab Pharmacokinet. 2011;26(1):15–29. doi: 10.2133/dmpk.dmpk-10-rv-089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Gomez-Lechon MJ, Donato MT, Castell JV, Jover R. Human hepatocytes as a tool for studying toxicity and drug metabolism. Curr Drug Metab. 2003;4(4):292–312. doi: 10.2174/1389200033489424. [DOI] [PubMed] [Google Scholar]
- [21].Hewitt NJ, Lechon MJ, Houston JB, et al. Primary hepatocytes: current understanding of the regulation of metabolic enzymes and transporter proteins, and pharmaceutical practice for the use of hepatocytes in metabolism, enzyme induction, transporter, clearance, and hepatotoxicity studies. Drug Metab Rev. 2007;39(1):159–234. doi: 10.1080/03602530601093489. [DOI] [PubMed] [Google Scholar]
- [22].den Braver-Sewradj SP, den Braver MW, Vermeulen NP, Commandeur JN, Richert L, Vos JC. Inter-donor variability of phase I/phase II metabolism of three reference drugs in cryopreserved primary human hepatocytes in suspension and monolayer. Toxicol in Vitro. 2016;33:71–79. doi: 10.1016/j.tiv.2016.02.013. [DOI] [PubMed] [Google Scholar]
- [23].Guo L, Dial S, Shi L, et al. Similarities and differences in the expression of drug-metabolizing enzymes between human hepatic cell lines and primary human hepatocytes. Drug Metab Dispos. 2011;39(3):528–538. doi: 10.1124/dmd.110.035873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ren Z, Chen S, Ning B, Guo L. Use of liver-derived cell lines for the study of drug-induced liver injury. In: Chen M, Will Y, eds. Book Chapter in Drug-Induced Liver Toxicity. New York, NY: Human Press; 2018:151–177. [Google Scholar]
- [25].Bova MP, Tam D, McMahon G, Mattson MN. Troglitazone induces a rapid drop of mitochondrial membrane potential in liver HepG2 cells. Toxicol Lett. 2005;155(1):41–50. doi: 10.1016/j.toxlet.2004.08.009. [DOI] [PubMed] [Google Scholar]
- [26].Chen S, Dobrovolsky VN, Liu F, et al. The role of autophagy in usnic Acid-induced toxicity in hepatic cells. Toxicol Sci. 2014;142(1):33–44. doi: 10.1093/toxsci/kfu154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chen S, Wan L, Couch L, et al. Mechanism study of goldenseal-associated DNA damage. Toxicol Lett. 2013;221(1):64–72. doi: 10.1016/j.toxlet.2013.05.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Chen S, Xuan J, Couch L, et al. Sertraline induces endoplasmic reticulum stress in hepatic cells. Toxicology. 2014;322:78–88. doi: 10.1016/j.tox.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Chen S, Xuan J, Wan L, et al. Sertraline, an antidepressant, induces apoptosis in hepatic cells through the mitogen-activated protein kinase pathway. Toxicol Sci. 2014;137(2):404–415. doi: 10.1093/toxsci/kft254. [DOI] [PubMed] [Google Scholar]
- [30].Dykens JA, Jamieson J, Marroquin L, Nadanaciva S, Billis PA, Will Y. Biguanide-induced mitochondrial dysfunction yields increased lactate production and cytotoxicity of aerobically-poised HepG2 cells and human hepatocytes in vitro. Toxicol Appl Pharmacol. 2008;233(2):203–210. doi: 10.1016/j.taap.2008.08.013. [DOI] [PubMed] [Google Scholar]
- [31].Felser A, Blum K, Lindinger PW, Bouitbir J, Krahenbuhl S. Mechanisms of hepato-cellular toxicity associated with dronedarone-a comparison to amiodarone. Toxicol Sci. 2013;131(2):480–490. doi: 10.1093/toxsci/kfs298. [DOI] [PubMed] [Google Scholar]
- [32].Greer ML, Barber J, Eakins J, Kenna JG. Cell based approaches for evaluation of drug-induced liver injury. Toxicology. 2010;268(3):125–131. doi: 10.1016/j.tox.2009.08.007. [DOI] [PubMed] [Google Scholar]
- [33].Guo L, Zhang L, Sun Y, et al. Differences in hepatotoxicity and gene expression profiles by anti-diabetic PPAR gamma agonists on rat primary hepatocytes and human HepG2 cells. Mol Divers. 2006;10(3):349–360. doi: 10.1007/s11030-006-9038-0. [DOI] [PubMed] [Google Scholar]
- [34].Juan-Garcia A, Manyes L, Ruiz MJ, Font G. Involvement of enniatins-induced cytotoxicity in human HepG2 cells. Toxicol Lett. 2013;218(2):166–173. doi: 10.1016/j.toxlet.2013.01.014. [DOI] [PubMed] [Google Scholar]
- [35].Nguyen KC, Willmore WG, Tayabali AF. Cadmium telluride quantum dots cause oxidative stress leading to extrinsic and intrinsic apoptosis in hepatocellular carcinoma HepG2 cells. Toxicology. 2013;306:114–123. doi: 10.1016/j.tox.2013.02.010. [DOI] [PubMed] [Google Scholar]
- [36].O’Brien PJ, Irwin W, Diaz D, et al. High concordance of drug-induced human hepatotoxicity with in vitro cytotoxicity measured in a novel cell-based model using high content screening. Arch Toxicol. 2006;80(9):580–604. doi: 10.1007/s00204-006-0091-3. [DOI] [PubMed] [Google Scholar]
- [37].Maiuri AR, Breier AB, Turkus JD, Ganey PE, Roth RA. Calcium contributes to the cytotoxic interaction between diclofenac and cytokines. Toxicol Sci. 2016;149(2):372–384. doi: 10.1093/toxsci/kfv249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Brandon EF, Raap CD, Meijerman I, Beijnen JH, Schellens JH. An update on in vitro test methods in human hepatic drug biotransformation research: pros and cons. Toxicol Appl Pharmacol. 2003;189(3):233–246. doi: 10.1016/S0041-008X(03)00128-5. [DOI] [PubMed] [Google Scholar]
- [39].Westerink WM, Schoonen WG. Cytochrome P450 enzyme levels in HepG2 cells and cryopreserved primary human hepatocytes and their induction in HepG2 cells. Toxicol in Vitro. 2007;21(8):1581–1591. doi: 10.1016/j.tiv.2007.05.014. [DOI] [PubMed] [Google Scholar]
- [40].Wilkening S, Stahl F, Bader A. Comparison of primary human hepatocytes and hepatoma cell line Hepg2 with regard to their biotransformation properties. Drug Metab Dispos. 2003;31(8):1035–1042. doi: 10.1124/dmd.31.8.1035. [DOI] [PubMed] [Google Scholar]
- [41].Gomez-Lechon MJ, Tolosa L, Donato MT. Upgrading HepG2 cells with adenoviral vectors that encode drug-metabolizing enzymes: application for drug hepatotoxicity testing. Expert Opin Drug Metab Toxicol. 2017;13(2):137–148. doi: 10.1080/17425255.2017.1238459. [DOI] [PubMed] [Google Scholar]
- [42].Xuan J, Chen S, Ning B, Tolleson WH, Guo L. Development of HepG2-derived cells expressing cytochrome P450s for assessing metabolism-associated drug-induced liver toxicity. Chem Biol Interact. 2016;255:63–73. doi: 10.1016/j.cbi.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Chen S, Wu Q, Ning B, Bryant M, Guo L. The role of hepatic cytochrome P450s in the cytotoxicity of dronedarone. Arch Toxicol. 2018;92(6):1969–1981. doi: 10.1007/s00204-018-2196-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wu Q, Ning B, Xuan J, Ren Z, Guo L, Bryant MS. The role of CYP 3A4 and 1A1 in amiodarone-induced hepatocellular toxicity. Toxicol Lett. 2016;253:55–62. doi: 10.1016/j.toxlet.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Chen S, Wu Q, Li X, et al. The role of hepatic cytochrome P450s in the cytotoxicity of sertraline. Arch Toxicol. 2020;94(7):2401–2411. doi: 10.1007/s00204-020-02753-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Chen S, Ren Z, Yu D, Ning B, Guo L. DNA damage-induced apoptosis and mitogen-activated protein kinase pathway contribute to the toxicity of dronedarone in hepatic cells. Environ Mol Mutagen. 2018;59(4):278–289. doi: 10.1002/em.22173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].McCall MN, McMurray HR, Land H, Almudevar A. On non-detects in qPCR data. Bioinformatics. 2014;30(16):2310–2316. doi: 10.1093/bioinformatics/btu239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Guo L, Li Q, Xia Q, Dial S, Chan PC, Fu P. Analysis of gene expression changes of drug metabolizing enzymes in the livers of F344 rats following oral treatment with kava extract. Food Chem Toxicol. 2009;47(2):433–442. doi: 10.1016/j.fct.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Esquenet M, Swinnen JV, Heyns W, Verhoeven G. LNCaP prostatic adenocarcinoma cells derived from low and high passage numbers display divergent responses not only to androgens but also to retinoids. J Steroid Biochem Mol Biol. 1997;62(5–6):391–399. doi: 10.1016/s0960-0760(97)00054-x. [DOI] [PubMed] [Google Scholar]
- [50].Chang-Liu CM, Woloschak GE. Effect of passage number on cellular response to DNA-damaging agents: cell survival and gene expression. Cancer Lett. 1997;113(1–2):77–86. doi: 10.1016/s0304-3835(97)04599-0. [DOI] [PubMed] [Google Scholar]
- [51].Yu H, Cook TJ, Sinko PJ. Evidence for diminished functional expression of intestinal transporters in Caco-2 cell monolayers at high passages. Pharm Res. 1997;14(6):757–762. 10.1023/a:1012150405949. [DOI] [PubMed] [Google Scholar]
- [52].Wenger SL, Senft JR, Sargent LM, Bamezai R, Bairwa N, Grant SG. Comparison of established cell lines at different passages by karyotype and comparative genomic hybridization. Biosci Rep. 2004;24(6):631–639. doi: 10.1007/s10540-005-2797-5. [DOI] [PubMed] [Google Scholar]
- [53].Sambuy Y, De Angelis I, Ranaldi G, Scarino ML, Stammati A, Zucco F. The Caco-2 cell line as a model of the intestinal barrier: influence of cell and culture-related factors on Caco-2 cell functional characteristics. Cell Biol Toxicol. 2005;21(1):1–26. doi: 10.1007/s10565-005-0085-6. [DOI] [PubMed] [Google Scholar]
- [54].Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 2013;138(1):103–141. doi: 10.1016/j.pharmthera.2012.12.007. [DOI] [PubMed] [Google Scholar]
- [55].Donato MT, Jover R, Gomez-Lechon MJ. Hepatic cell lines for drug hepatotoxicity testing: limitations and strategies to upgrade their metabolic competence by gene engineering. Curr Drug Metab. 2013;14(9):946–968. doi: 10.2174/1389200211314090002. [DOI] [PubMed] [Google Scholar]
- [56].Tolosa L, Jiménez N, Pérez G, Castell JV, Gómez-Lechón MJ, Donato MT. Customised in vitro model to detect human metabolism-dependent idiosyncratic drug-induced liver injury. Arch Toxicol. 2018;92(1):383–399. doi: 10.1007/s00204-017-2036-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Hashizume T, Yoshitomi S, Asahi S, et al. Advantages of human hepatocyte-derived transformants expressing a series of human cytochrome p450 isoforms for genotoxicity examination. Toxicol Sci. 2010;116(2):488–497. doi: 10.1093/toxsci/kfq154. [DOI] [PubMed] [Google Scholar]
- [58].Vagiannis D, Novotna E, Skarka A, et al. Ensartinib (X-396) effectively modulates pharmacokinetic resistance mediated by ABCB1 and ABCG2 drug efflux transporters and CYP3A4 biotransformation enzyme. Cancers (Basel). 2020;12(4):813. doi: 10.3390/cancers12040813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Hofman J, Sorf A, Vagiannis D, et al. Brivanib exhibits potential for pharmacokinetic drug-drug interactions and the modulation of multidrug resistance through the inhibition of human ABCG2 drug efflux transporter and CYP450 biotransformation enzymes. Mol Pharm. 2019;16(11):4436–4450. doi: 10.1021/acs.molpharmaceut.9b00361. [DOI] [PubMed] [Google Scholar]
- [60].Yoshitomi S, Ikemoto K, Takahashi J, Miki H, Namba M, Asahi S. Establishment of the transformants expressing human cytochrome P450 subtypes in HepG2, and their applications on drug metabolism and toxicology. Toxicol in Vitro. 2001;15(3):245–256. doi: 10.1016/S0887-2333(01)00011-X. [DOI] [PubMed] [Google Scholar]
- [61].Guillouzo A, Corlu A, Aninat C, Glaise D, Morel F, Guguen-Guillouzo C. The human hepatoma HepaRG cells: a highly differentiated model for studies of liver metabolism and toxicity of xenobiotics. Chem Biol Interact. 2007;168(1):66–73. doi: 10.1016/j.cbi.2006.12.003. [DOI] [PubMed] [Google Scholar]
- [62].Kanebratt KP, Andersson TB. Evaluation of HepaRG cells as an in vitro model for human drug metabolism studies. Drug Metab Dispos. 2008;36(7):1444–1452. doi: 10.1124/dmd.107.020016. [DOI] [PubMed] [Google Scholar]
- [63].Gonzalez FJ. The 2006 Bernard B. Brodie Award Lecture. Cyp2e1. Drug Metab Dispos. 2007;35(1):1–8. doi: 10.1124/dmd.106.012492. [DOI] [PubMed] [Google Scholar]
- [64].Caro AA, Cederbaum AI. Oxidative stress, toxicology, and pharmacology of CYP2E1. Annu Rev Pharmacol Toxicol. 2004;44:27–42. doi: 10.1146/annurev.pharmtox.44.101802.121704. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


