Abstract
Apolipoprotein E (ApoE) is of great interest due to its role as a cholesterol/lipid transporter in the central nervous system (CNS) and as the most influential genetic risk factor for Alzheimer’s Disease (AD). Work over the last four decades has given us important insights into the structure of ApoE and how this might impact the neuropathology and pathogenesis of AD. In this review, we highlight the history and progress in the structural and molecular understanding of ApoE and discuss how these studies on ApoE have illuminated the physiology of ApoE, receptor binding, and interaction with amyloid-β (Aβ). We also identify future areas of study needed to advance our understanding of how ApoE influences neurodegeneration.
Keywords: Apolipoprotein E, ApoE, Amyloid-β, Aβ, Alzheimer’s Disease, AD, low density lipoprotein receptor, LDLR
In Brief
In this review, Chen, Strickland, and colleagues discuss the historical progression in understanding the structural and molecular properties of ApoE and describe further studies needed. These findings are used to illuminate some of the physiological and pathological consequences of ApoE.
Introduction
Apolipoprotein E (ApoE) was first identified in the 1970’s as one of the protein components of plasma very low-density lipoprotein (VLDL) (Shore et al., 1973; Utermann et al., 1975). It was found to play a critical role in plasma cholesterol metabolism and was soon realized to be a major risk factor in atherosclerosis and hypercholesterolemia (Mahley, 1988; Mahley et al., 2009). Early characterization of ApoE revealed six protein polymorphisms, separable by two-dimensional gel electrophoresis (Davignon et al., 1988; Rall et al., 1983). Three of these polymorphisms in humans are due to the presence of three ApoE isoforms (ApoE2, ApoE3, and ApoE4) characterized by a single amino acid substitution in ApoE2 and ApoE4, compared to the most prevalent isoform, ApoE3. The 3 other polymorphisms present in ApoE result from variable sialylation at Thr194 (Wernette-Hammond et al., 1989). Despite the impact on disease risk, ApoE isoforms differ from each other in its 299 amino acids at only two sites: ApoE4 is characterized by Arg112 and Arg158, ApoE3 by Cys112 and Arg158, and ApoE2 by Cys112 and Cys158 (Fig. 1A) (Weisgraber, 1994). Soon after the initial discovery of ApoE, the primary receptor for ApoE - low density lipoprotein receptor (LDLR) - was discovered in 1976 (Goldstein and Brown, 1976). Subsequent studies showed that ApoE was one of the primary ligands for LDLR, in addition to Apolipoprotein B (Innerarity and Mahley, 1978). Through its interaction with LDLR, ApoE plays a critical role in cellular uptake of ApoE-containing lipoproteins (Weisgraber, 1994).
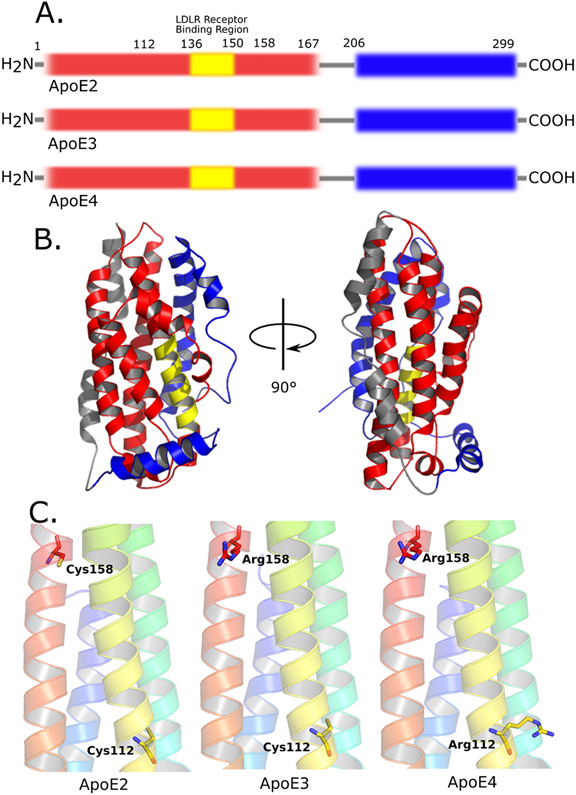
Figure 1. Illustration of domains and single residue substitutions of ApoE isoforms.
A) Figure shows early domain categorizations of ApoE with thrombin cleavage. Red: the N-terminal domain of ApoE (1-167); yellow: the LDLR receptor binding domain (136-150); grey: the hinge region (167-206) where cleavage site is located; blue: the C-terminal domain (206-299). Also, the amino acids that distinguish the different ApoE isoforms are shown at position 112 and 158.
B) The full-length 3D structure of ApoE3 by NMR (PDB:2l7b). Colors are coded to illustrate the different domains of ApoE mentioned in figure 1A.
C) Positions of single residue substitution among ApoE isoforms shown in 3D structure. ApoE2: Cys112 & Cys 158; ApoE3: Cys112 & Arg158; ApoE4: Arg112 & Arg 158. Figures are generated from PDB files from left to right: 1le2, 1ple, 1le4.
In addition to the critical role that ApoE plays in peripheral cholesterol metabolism, ApoE was found to be the primary apolipoprotein lipid and cholesterol transporter in the central nervous system (CNS) (Boyles et al., 1985; Mahley, 1988). ApoE is produced by almost all cell types in the brain: astrocytes, which secrete the vast majority of ApoE under physiological conditions; microglia, under pathological conditions, and neurons, under certain injury conditions (Blue et al., 1983; Boyles et al., 1985; DeMattos et al., 1998; Newman et al., 1985; Pitas et al., 1987; Weisgraber, 1994; Williams et al., 1985). Characterization of ApoE secreted from astrocytes revealed that ApoE is secreted in lipoprotein particles similar in size to high density lipoproteins (HDL) (Fagan et al., 1999; Pitas et al., 1987). In 1993, APOE genotype was found to strongly modulate risk for Alzheimer’s Disease (AD) (Corder et al., 1993; Strittmatter et al., 1993a, 1993b). A single copy of the ε4 allele increases AD risk by about 3-fold, with two copies of the ε4 allele increasing disease risk about 12-fold relative to individuals who have 2 copies of the ε3 allele. The ε2 allele decreases AD risk by about 0.6 (Corder et al., 1993; Holtzman and Herz, 2012). ApoE was also identified as a core component in cerebral and vascular amyloid plaques (Näslund et al., 1995; Wisniewski and Frangione, 1992). Early studies with amyloid-β (Aβ) depositing mice (PDAPP or Tg2576 mice) mice found that in the absence of murine ApoE, Aβ still deposited in the brain, but did not form or formed very little β-pleated sheet structures in the brain parenchyma or in cerebral amyloid angiopathy (CAA) (Bales et al., 1997; Holtzman et al., 2000). Interestingly, expression of human ApoE3 and ApoE4 in PDAPP mice in the absence of murine ApoE delayed the onset of Aβ deposition relative to mouse ApoE or no ApoE, suggesting that human ApoE may play role in Aβ clearance as well as in Aβ fibril formation (Holtzman et al., 1999). Later studies in PDAPP mice expressing human ApoE2, ApoE3, and ApoE4 revealed that the amount of Aβ pathology is dependent on ApoE isoform: ApoE4 leads to the most Aβ pathology and ApoE2 the least with ApoE3 being intermediate between the two other isoforms (Bales et al., 2009; Fagan et al., 2002; Holtzman et al., 2000). Similar findings with ApoE isoform-dependent effects on Aβ were seen in different APP/PS1 transgenic mice (Kim et al., 2011; Kuszczyk et al., 2013; Youmans et al., 2012). Apart from ApoE-Aβ interaction, it was reported that ApoE isoform modulates extracellular, soluble, monomeric Aβ levels (Castellano et al., 2011) and that the levels of ApoE receptors, such as LDLR or LRP1, also modulate Aβ pathology (Kanekiyo et al., 2013; Kim et al., 2009; Verghese et al., 2013). In addition to the importance of ApoE in Aβ pathology, ApoE has been shown to dramatically impact neurodegeneration in a mouse model of tauopathy (Shi et al., 2017). Similar to the order of effect of ApoE isoform on Aβ pathology, increased neurodegeneration was observed with ApoE4 compared to ApoE3 and ApoE2 while the absence of ApoE is neuroprotective. Due to its pleiotropic effects, ApoE may influence AD in other ways, including effects on the blood-brain-barrier, synaptic plasticity, and possibly other mechanisms (Bell et al., 2012; Liu et al., 2013). In addition to AD, ApoE has also been reported to have an impact on dementia and synucleinopathy in Parkinson’s Disease dementia and Dementia with Lewy Bodies as well as in mouse models of synucleinopathy (Davis et al., 2020; Dickson et al., 2018; Tsuang et al., 2013; Zhao et al., 2020).
The dramatic effect of ApoE isoform on AD, neurodegeneration, and other neuropathologies is likely the consequence of both the direct interaction of ApoE with specific proteins such as Aβ and ApoE receptors, as well as indirectly through regulation of ApoE levels by the interaction of ApoE with its receptors and other molecules. Some early studies have suggested that ApoE may directly interact with Aβ to induce Aβ fibrillization (Aleshkov et al., 1997; LaDu et al., 1994; Ma et al., 1994; Strittmatter et al., 1993b; Wisniewski et al., 1993). In fact, our lab also recently discovered that the ApoE in Aβ plaques exists in a unique conformation that is not lipidated that may promote Aβ plaque formation (Liao et al., 2018). On the other hand, ApoE2 may be protective due to deficiencies in ApoE-LDLR binding (Hui et al., 1984; Rall et al., 1983) or via other effects. Modulation of LDL receptors, including LDLR and LRP1, has been demonstrated to affect Aβ clearance by different groups (Cam et al., 2005; Castellano et al., 2012; Kim et al., 2009). To better understand and develop treatment for AD and other neurodegenerative disorders it will be critical to investigate the molecular interactions that occur between ApoE, pathological proteins, ApoE receptors, and other interacting molecules. Indeed, much progress has been made in the last 40 years to elucidate the molecular behavior of ApoE and its impact on neurodegeneration. Advances in understanding the structure of ApoE have yielded insights into how seemingly small changes at the structural level manifest themselves as dramatic changes in ApoE receptor binding and AD pathology. In this review, we discuss the historical progress in understanding ApoE structure and how these insights can be used to better understand AD neuropathology and pathogenesis. We will also discuss new directions that will be necessary to further advance our understanding of ApoE structure/function relationships.
Historical Progress in Understanding ApoE Structure
Soon after the discovery of apolipoprotein E (Shore and Shore, 1973), structural predictions were proposed based on the primary sequence of ApoE (Baker et al. 1975; Weisgraber 1994). Biochemists and evolutionary biologists have focused on conservative internal repeats of the primary amino acid sequences that exist in most apolipoproteins (Huebbe and Rimbach, 2017; Li et al., 1988). These internal repeats exhibit features that are favorable for forming amphiphilic α-helical structures according to the primary amino acid sequence (Li et al., 1988; Weisgraber 1994). Such amphiphilic helices supported the amphiphilic theory that charged residues create electrostatic association with lipid head groups, and their adjacent hydrophobic residues further stabilize the neck of those phospholipids on lipoprotein particles. The predicted helical secondary structures were experimentally verified by circular dichroism spectra (Weisgraber, 1994). More interestingly, it was also shown that the heparin-binding domain of apolipoprotein E expanded the helical portion in the presence of heparin or other glycosaminoglycans (Cardin et al., 1991; Mahley et al., 1979), strengthening the belief that the helical structures in the apolipoprotein family are required to interact with lipids on lipoprotein particles (Jackson et al., 1975).
Thrombolytic cleavage of the ApoE molecule yields two unique fragments, the N-terminal domain and C-terminal domain, where the N-terminal domain is responsible for LDLR binding, but only weakly interacts with lipids, and the C-terminal is considered to bind to the surface of lipoproteins and is not involved in LDLR recognition (Fig. 1A&B) (Bradley et al., 1982; Innerarity et al., 1983; Weisgraber, 1990). However, while point mutagenesis and antibody competition experiments have unveiled that residues 136 to 150 are required for LDLR binding to ApoE (Innerarity et al., 1983; Lalazar et al., 1988; Weisgraber et al., 1983; Wilson et al., 1991), the single residue replacement of arginine by cysteine at position 158 on ApoE2 (Fig. 1A&C), seemingly distant from the LDLR binding site, dramatically reduces the binding of ApoE2 to LDLR. In addition, while DMPC (1,2-dimyristoyl-sn-glycero-3-phosphocholine) associated full length ApoE2 has a significant reduction in affinity for LDLR (Weisgraber et al., 1982), the N-terminal truncated form of ApoE2 regains partial binding ability when bound to DMPC (Yamamoto et al., 2007). The ApoE4 amino acid change of Cys112 to Arg112 (Fig. 1A&C), that is also not part of the LDLR binding site (136-150), is strongly associated with increased cholesterol and LDL in plasma (Eto et al., 1987, 1991; Saito et al., 2004). These observations contributed to delineate a critical question that has defined the field: how do these single amino acid substitutions dramatically change the characteristics of ApoE-containing lipoprotein particles and their binding properties to other molecules, consequently impacting on AD pathology?
This question was first addressed by the crystal structure of a recombinant N-terminal portion of human ApoE3 expressed in E. coli (Wilson et al., 1991). While building one of the first vertebrate apolipoprotein structures (Wilson et al., 1991; Breiter et al., 1991), Wilson et al. achieved a density map at 2.5 Å resolution for de novo modeling generated from dimethyl mercury labeling and combined heavy atom phase refinement. According to their structural model, the major N-terminal portion of human ApoE3 exhibits an elongated four-antiparallel-helix bundle. This helix bundle buries most of the hydrophobic residues inside a compact core and exposes the hydrophilic residues on the surface, where the repeating leucine residues stack with each other with leucine side chains from adjacent antiparallel helices and further “zipper” the helical bundle. Moreover, seven salt bridges were observed to form between pairs of helices, further stabilizing the super-secondary structure. On the other hand, supporting previous determinations of the LDLR binding site (Weisgraber, 1994), the structure described by Wilson et al. revealed that many basic residues, such as Arg134, Arg136, His140, Arg142, Lys143 Arg145, Lys146, Arg147, and Arg150, are mostly accessible on the surface and free from intramolecular salt bridges, forming a strong basic site for LDLR binding (Fig. 2A). It also explains why none of the naturally occurring variants or single point mutagenesis in that region could completely eliminate the ApoE-LDLR interaction, because all these basic residues may cooperatively interact with LDLR and foster a high binding affinity (Innerarity et al., 1983; Lalazar et al., 1988; Landschulz et al., 1996; Mann et al., 1989).
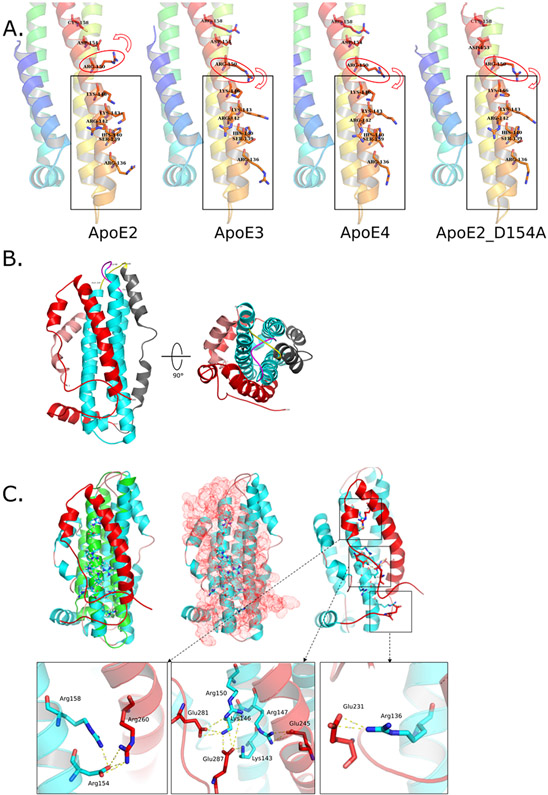
Figure 2. Structural analysis focusing on LDLR binding regions on ApoE.
A) Putative explanation of isoform differences in affecting LDLR binding region from crystal structures. From left to right: ApoE2, ApoE3, ApoE4 and ApoE2_D154A. Red ellipses highlight the key Arg150 only captured by Asp154 in ApoE2 that affects LDLR binding site. Red arrows indicate different orientations of Arg150 in those structures. Grey rectangles show the LDLR binding sites on ApoE structures. PDB files from left to right: 1le2, 1ple, 1le4, 1nfo.
B) NMR structure of full-length ApoE3 (PDB: 2l7b). Salmon: N-terminal residues (1-24); Cyan: N-terminal α-helix bundle (25-163); Yellow: free loop (164-168); Grey: hinge region; Red: C-terminal. Cyan structure shows the α-helix bundle consistent with previous X-ray crystal structures. Yellow loop highlights the “lock” of N-terminal α-helix bundle to unfold. The grey hinge region and red C-terminal domain cover N-terminal α-helix bundle that was assumed solvent exposed.
C) NMR structure of full-length ApoE3 showed that LDLR binding region is completed covered in non-lipidated form. Top-left panel shows the superposition of crystal structure of ApoE3 N-terminal domain (Green, PDB: 1lpe) to NMR structure (Cyan and Red, PDB: 2l7b), key residues involved in LDLR binding are highlighted with hydrogen atoms (white); Top-middle panel shows that LDLR binding site on N-terminal domain of ApoE3 is completely capped by C-terminal (Red dots represent the atomic surface of C-terminal domain); Top-right panel highlights key interactions between acidic residues on C-terminal domain and basic residues in LDLR binding, enlarged views of three interaction cores between N-terminal and C-terminal domains are presented at the bottom. NOTE: Q279 and E287 are mutations generated by the authors.
Prompted by this pioneering work, the similar N-terminal fragment of ApoE4 and ApoE2 were resolved by X-ray crystallography (Dong et al., 1994; Wilson et al., 1994). Combining all these N-terminal crystal structures, an intriguing structural model was proposed by Weisgraber and colleagues to explain the differences in ApoE isoforms (Fig. 3A) (Weisgraber, 1994). This model consists of a N-terminal four helix bundle and an undetermined α-helical C-terminal region connected by a hinge domain (Fig. 3A). ApoE possesses many Lys and Arg residues in the Helix 4 in the N-terminal helical bundle, creating a positively charged pocket to interact with acidic residues in the lipoprotein binding site on LDLR protein family members. Meanwhile, lipid association with ApoE would induce a conformational change of the N-terminal and hinge region, especially recruiting Arg172 in the hinge region to fully activate the LDLR binding affinity of ApoE (Morrow et al., 2000). Such a conformational change induced by lipid binding is thought to initiate in the C-terminal domain, because, unlike the C-terminal domain, the isolated N-terminal domain showed little lipid binding ability (Bradley et al., 1982; Innerarity et al., 1983; Weisgraber, 1990). On the other hand, compared with ApoE3, the single residue substitution on ApoE2 – R158C – releases the Asp154 to form a new salt bridge with Arg150, and therefore disrupts the initiation of LDLR-ApoE interaction starting with Arg150 and the adjacent basic residues (Fig. 2A). Weisgraber’s idea was supported by a point mutagenesis that switched Asp154 to Ala and successfully restored the LDLR binding affinity of mutant ApoE2 (Fig. 2A) (Dong and Weisgraber, 1996). On the other hand, the ApoE4 change – C112R – was proposed to be involved in a putative interaction with Glu255 in the C-terminal region (Fig. 3A). The first evidence for this came from the N-terminal structure resolved by the Weisgraber and Agard group, in which the substitution C112R pushed Arg 61 away from the helix bundle while in ApoE3 Arg61 was buried between two helices (Dong et al., 1994). The second evidence was from a mutagenesis study that demonstrated a single mutation E255A changed ApoE4’s preference in plasma LDL binding to HDL like ApoE2 and ApoE3 (Saito, 2003). To address these observations, Weisgraber and colleagues hypothesized that there is an interaction between Arg61 and Glu255 caused by Arg112 in ApoE4 that locks ApoE4 in a less open conformation compared to ApoE2 and ApoE3, rendering a much tighter interaction between the N-terminal domain and C-terminal domain (Fig. 3A) (Hatters et al., 2005; Xu et al., 2004). Mutagenesis of Glu109 in ApoE3 and Arg61 in ApoE4 also supported the idea that the position of Arg61 in ApoE4 is critical in determining ApoE4 plasma VLDL binding preference (Dong et al., 1994; Dong and Weisgraber, 1996). The relevance of such a lipoprotein preference based on this model might be relevant in the plasma where large lipoproteins such as LDL and VLDL exist. Although only HDL-like lipoparticles exitst in the CNS, these findings may also be important in understanding size differences in ApoE-lipoprotein particles secreted from different cell types. Of note, the other rearrangements of the salt bridges on helix 3 in ApoE2 may not completely explain its lack of binding to LDLR (Weisgraber, 1994; Wilson et al., 1991), and these N-terminal structures also provided no information of other key residues for ApoE-LDLR interaction such as arginine 172 (Morrow et al., 2000). The lack of the integral view on full-length ApoE molecular structure has hampered the interpretations of increasing pathological observations that linked ApoE to AD pathology for years.
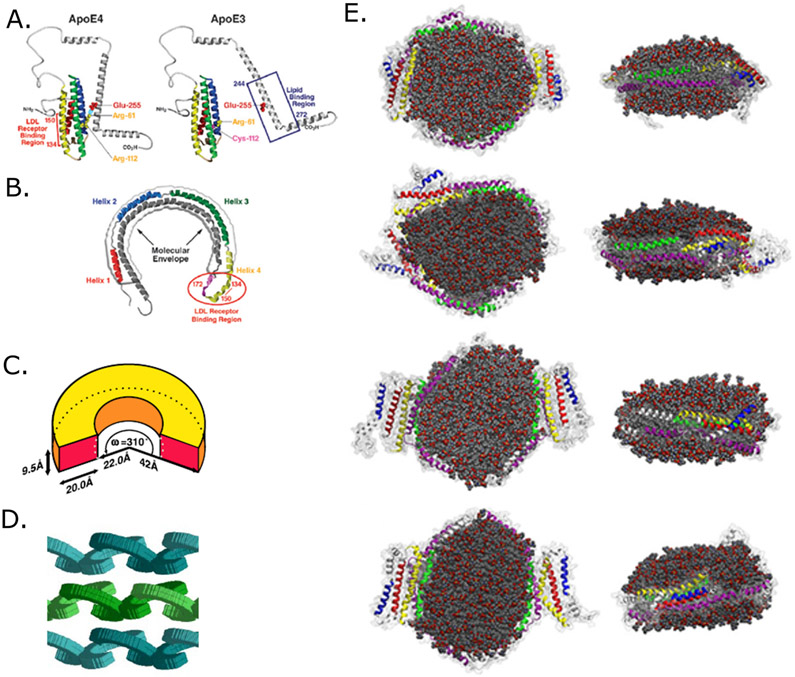
Figure 3. Two structural models of lipidated ApoE lipoprotein particle (adapted from Mahley et al., 2009; Peters-Libeu et al., 2006; Henry et al., 2018).
A) Early ApoE lipidation and explanation of isoform differences in lipidation, adapted from Mahley et al., 2009. Left panel shows the impact of Arg112 substitution in ApoE4 on strengthening N/C-terminal domain interaction (Arg61-Glu255) comparing to ApoE3.
B) Figure shows the “horse-shoe” structural model fit into low-resolution electronic density map from X-ray diffraction data by Weisgraber’s group. LDLR binding region is circled in red. Adapted from Mahley et al., 2009.
C) Cartoon adapted from Peters-Libeu et al., 2006, presents a cartoon model how the “horse-shoe” ApoE monomer curves on lipoparticles based on the X-ray crystallography data.
D) Cartoon adapted from Peters-Libeu et al., 2006, showing a dimeric packing of two ApoE molecules on each repeat unit of ApoE-DPPC.
E) Molecular dynamic simulation results of ApoE4 dimers on a lipid disc, adapted from Henry et al., 2018. From top to bottom show different possible configurations of two ApoE4 molecules wrapping around a lipid disc.
It wasn’t until 2011, that the first full-length apolipoprotein E structure was resolved by the group of Jiajun Wang by NMR, though this was done with multiple mutations (R215C/R217S/F257A/W264R/V269A/L279Q/V287E) needed to stabilize ApoE3 in a monomeric form (Zhao et al., 2008; Chen et al., 2011). Their structural model successfully extended the N-terminal models from X-ray crystallography and previous NMR structure of the C-terminal domain, in which the N-terminal domain (residues 1–167) and C-terminal domain (residues 206–299) are shown to link together by a long hinge domain (residues 168–205) (Fig. 2B). This NMR structure showed that the folding and unfolding process of ApoE could potentially be much more complicated than suggested by the Weisgraber model (Fig. 2C). According to their full-length NMR structure of human ApoE3, the N-terminal four helix-bundle possesses similar configurations compared to previous N-terminal truncated structures, except for some minor changes: the existence of the C-terminal domain attracts the helix 1’ (45–52) to slide down and results in helices 3 (89–125) and 4 (131–164) subtly tilted towards the C-terminal domain. The long hinge domain forms two more helices that tightly cover the hydrophobic surface on helix 2 and 3 of the N-terminal domain, regulating the interaction between N- and C-terminal domains. The C-terminal domain ultimately turns to fold into one curved long helix and two short helical structures that covers the other side of N-terminal domain helix bundle (Fig. 2B). Indeed, the general folding of the N-terminal and C-terminal helices are consistent with previous structures (Fig. 2B). However, if the mutated residues do not significantly alter ApoE structure, then this full-length ApoE3 structure would change our understanding of molecular characteristics of ApoE and its interaction with lipids or even receptors, such as LDLR or its family members because of its unique topology. First, the hinge region forms two helices instead of completely random coils that partially cover the surface of the N-terminal helix bundle. Second, the enshrouding superposition by the loop between hinge helix and helix 4 on top of the loop between helix 2 and 3 forces ApoE3 to undergo a unique unfolding process compared to what was previously proposed (Weisgraber, 1994). Last but not least, the major region of LDLR binding site on in the N-terminal domain is in fact shielded by the C-terminal domain containing helices and intrinsic disordered regions instead of simply G helices (Fig. 2C). The key arginine 150 may not be free from interactions with multiple residues in the C-terminal domain including Gln253, Glu281 and Glu287. Moreover, the proposed original salt bridges become more complicated as residues Asp154 and Arg158 also interact with Lys157, Arg260, Gln275 and Gln279 (Fig. 2C). Also, the sole substitution in ApoE2 – C112R – may alter the unfolding process or affinity since Arg158 is predicted to be involved in dissociation of helix 3 and helix 4 and the final dimerization between two stretched ApoE3 monomers (Chen et al., 2011). Wang’s group also pointed out that the previously predicted domain interaction through Arg61 and Glu255 was not present in their model. Rather, the interaction between the N-terminal and C-terminal domain is proposed to occur between Lys95 and Glu255. However, Gln279 and Glu287 were in fact induced for NMR restraints and turned out to be strongly involved in those interactions (Fig. 2C). Whether the covering of the C-terminal domain onto N-terminal helical bundle comes from an artifact is never clearly discussed. Moreover, since physiological ApoE is lipidated it is unclear whether the shielding of the N-terminal domain by the C-terminal domain is physiologically significant.
This structure eventually leads to the most comprehensive and rigorous model for ApoE folding and lipidation process proposed. Carl Frieden group’s data from hydrogen – deuterium exchange coupled to mass spectrometry (HDX-MS) experiments unveiled that all peptides that behaved differently between ApoE3 and ApoE4 (except 148–159) are close in proximity to residue 112, where the single residue substitution is, providing a more straightforward explanation of isoform structural difference between ApoE3 and E4 (Frieden et al., 2017). It is quite interesting that the major difference between full-length ApoE3 and ApoE4 that Frieden et al. observed appear to be in peptides 27–37, 60–64, and 102–108, which surprisingly are all in the N-terminal helix bundle. These observations were all performed at 10 μM, conditions in which the protein is in tetrameric form. Thus, part of the signal might result from oligomeric interactions. Nonetheless, this difference in flexibility in those regions may account for the distinct preference of ApoE4 to plasma VLDL because of the curvature accommodation. Whether this is relevant to lipid binding in HDL-like particles in the CNS is not clear. The Frieden group also completed the ApoE lipid binding model concluding with a discussion of isoform differences (Frieden et al., 2017). They postulated that there is a domain-domain separation between N- and C-terminal domains as the first step in lipid binding, where charged lipid molecules replace and break the original salt bridges. This finalized model is consistent with the structural differences between ApoE3 and ApoE4 and also succeeded in explaining the difference in receptor binding between lipidated ApoE and non-lipidated ApoE due to the opening up of the C-terminal domain, which is required for accessing the LDLR binding site on the shielded N-terminal domain. Indeed, it is possible that the Weisgraber model still matches with the situation where ApoE gets fully lipidated and adopts an extended helical conformation. However, experimental data is lacking to answer whether molecular structures of ApoE2 and ApoE4 fit with Wang and Frieden’s model or if ApoE2 and ApoE4 in fact adopt different conformations, leading to a mystery whether these two single residue substitutions could induce non-trivial conformational changes that might alter the current model. It is also unknown if ApoE2 and ApoE4 would undergo the same lipidation process or have different steps of conformational changes as suggested before (Morrow et al., 2002). In addition, fluorescent measurements of ApoE lipid binding suggested that ApoE4 has a more disordered C-terminal domain, which lead to ApoE4’s higher affinity for lipid and preferential association with plasma VLDL (Saito et al., 2003). This is opposite to Frieden’s HDX results, yet again, those experiments were performed at 10 μM where ApoE is tetrameric. Nonetheless, there are some interesting results from molecular dynamic simulations that supported Wang and Frieden’s model and provided pure computational models focusing on ApoE4 (Ray et al., 2017; Williams et al., 2015). Importantly, none of the ApoE structures above were resolved in their native, physiological lipidated forms present in vivo. However, these structures indeed provide vital information for us to investigate the molecular features controlling folding, lipid interaction, and receptor binding of ApoE, whereas the empirical evidence supports aspects of these structural models to this day.
Early assessments of lipidated ApoE started with C-terminal fragments when the group of Robert J. Cushley managed to mimic the lipid environment of lipid-associated C-terminal regions of ApoE and ApoAI and restored their secondary structure by applying micelles of SDS or dodecylphosphocholine (Shaw et al., 1997; Wang et al., 1996a). Further, Guangshun Wang and colleagues determined the first soluble structure of the partial ApoE C-terminal domain by circular dichroism spectra and 2D nuclear magnetic resonance (NMR) (Wang et al., 1996b). The structure of both constructs demonstrated that the C-terminal domain of ApoE mostly forms helical structures in the lipid-associated environments, while it collapses into random structures in the absence of dosium dodecyl sulfate (SDS), according to the CD spectra results. Although previous helical wheel projections had already led to a prediction that the C-terminal domain of ApoE forms an atypical class of helix called G amphipathic helix (Segrest et al., 1992; Sparrow et al., 1992), it was shown for the first time that the C-terminal domain would fold into such an amphipathic helix, creating a hydrophobic gulf for DMPC interaction through Trp264, Phe265, Leu268, and Val269. While this domain is not folded in an ideal way – Val269 was not in the hydrophobic face in construct 267-289, the NMR structures confirmed that Trp276 is vital for lipid-association as its side chain is consistently motion-restricted due to its insertion into the micelle interior (Wang et al., 1996b). However, such a lipidated form of ApoE was restricted to only ~ 22 amino acids in the C-terminal domain, leaving it still unclear as to how full-length ApoE present in a lipoprotein particle behaves.
To further address this question, the first and sole structure of lipidated ApoE was determined by Karl Weisgraber’s group in 2006 via X-ray crystallography (Peters-Libeu et al., 2006). They crystallized ApoE4 with dipalmitoylphosphatidylcholine (DPPC) and analyzed the unique diffuse scattering pattern in their diffraction data. Surprisingly, the crystallized ApoE4-DPPC particle did not fall into a lipid bilayer with phospholipid molecules as previous suggested (Raussens et al., 1998). Rather, ApoE4-DPPC formed a spheroidal shape without a complete ring of diffuse scattering centered on 4.2 Å which was supported by the non-crystallographic 2-fold rotation axis (Peters-Libeu et al., 2006). Indeed, it is most likely that ApoE could have flexibility to accommodate different shapes and sizes of the lipoprotein particles since other methods showed that ApoE particles could form disc-like shapes with different concentrations of cholate and lipid as does ApoE particles secreted by astrocytes (DeMattos et al., 2001; Hatters et al., 2009; Mahley et al., 2009). Alternatively, the spheroidal shape of the particles might be more favorable for the molecular packing under the crystallization condition they optimized. Both forms of ApoE lipoprotein particles could exist in different physiological environments. Meanwhile, their final model suggested a stoichiometry of two ApoE4 molecules per particle, each bending into a horseshoe-like shape, circumscribed around the lipid spheroid with a circular cross-section in the equatorial plane as a dimer (Fig. 3B&C) (Peters-Libeu et al., 2006). One question that remains open is whether the structural properties of ApoE when bound to ApoE-lipid particles depend on the stoichiometry of ApoE per ApoE-lipid particle or is intrinsic to the protein. Although the overall resolution of this data set was limited at 10 Å, the height and width of the density map does fit with a side-by-side pair of helices model. It suggests that the ApoE protein backbone can possess some short helices and connecting loops (or bends) to form the semicircular arc and match with the curvature of a lipoprotein particle surface (Fig. 3B&C). Upon ApoE lipidation, the Weisgraber group hypothesized that there are three steps: 1) the N-terminal four helix bundle stretches out and exposes its hydrophobic core, while the C-terminal domain dissociates from the original compact conformation; 2) the N-terminal domain further opens up and extends into a line where C-terminal domain in an anti-parallel fashion sits on top of the hydrophobic residues exposed on N-terminal helices and together forms a belt-like configuration, but unlike the naive belt model, hydrophobic residues were thought to contribute to hairpin interactions between two helices, instead of interacting with lipid tails; 3) two ApoE “belts” eventually dimerize on the edge of the spheroidal shaped lipid core with an stagger angle at ~42°, in which the each ApoE protein stabilizes the lipoprotein particle by enveloping half of the polar phospholipid headgroups (Fig. 3A-D) (Peters-Libeu et al., 2006; Hatters et al., 2006). The Weisgraber model also explains how esterified cholesterol is incorporated into the HDL core as the ApoE horseshoe dimer can provide enough flexibility to accommodate expansion of the size and the changes of the curvature. Yet, the Weisgraber model is based substantially on the crystal structures with the N-terminal truncated forms of the ApoE isoforms and full-length ApoE-DPPC structure was based on a prediction according to the low-resolution electron density map. (Hatters et al., 2006; Weisgraber, 1994). It is undeniable that the missing portion of ApoE protein might influence the conformations of the known motifs if even a single residue substitution can make a huge impact on the protein structure and functionality. Without a higher resolution structure, it remains unclear if the details of this structure are correct.
Recently, a computational model of full-length, lipidated ApoE4 was presented by Henry et al. with support from cross-linking mass spectrometry (MS) data (Henry et al., 2018). They achieved a relatively homogenous population of lipidated human recombinant ApoE4 by mixing ApoE4 and POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocoline) at a molar ratio of 1:110. After size-exclusion chromatography purification, they treated the ApoE4-containing lipid particle with an equimolar mixture of light DSS (sodium trimethylsilylpropanesulfonate) (DSS-H12) and heavy DSS (DSS-D12) to crosslink spatially nearby Lys residue pairs, from which the special information of adjacent secondary structures would be recorded by MS and applied for molecular dynamic simulations. Their data suggested that each monomeric ApoE4 may fold into two major configurations: 1) an open hairpin structure where the hairpin bends at N-terminal helices 3 and 4 in juxtaposition to the C-terminal domain; 2) a compact hairpin that adopted a previously proposed N-terminal four helix bundle (Fig. 3E). Then two ApoE4 monomers dimerize into either head-to-head or head-to-tail conformations wrapping around the edge of POPC lipid-disc, protecting the hydrophobic surface of packed POPC tails. Their molecular dynamics simulation indicated that ApoE4 secondary structures could stretch or shrink between helices or bend to accommodate the size changes of different lipoprotein particles. Interestingly, their data also suggested that the end of N-terminal helix 4 could change from original random coil to an α-helical conformation with the existence of POPC disc and caused reorientation of Arg172 – a previously reported key residue involved in ApoE-LDLR interaction, but is not in the LDLR binding site (Morrow et al., 2000). However, because DSS conjugation only applies for Lys pairs, helices without Lys residues such as helix 1 on the N-terminal domain could not be applied to their crosslinking MS (XL-MS) method. In addition, in the view of multiple configurations of ApoE protein on the lipid-containing particles, their XL-MS results would result from an ensemble of all the ApoE molecules after the DSS treatment, instead of only one or two conformations. Although the authors 1) distinguished two different conformations of ApoE4 on the lipid-containing particles to satisfy the XL-MS data; and 2) applied a reasonable initial model for simulation based on previous literature, it is still possible that these two structural models may not be able to completely represent the true nature of native ApoE lipoprotein particles. Moreover, the helical bundle presented both in their open hairpin and compact hairpin models was not clearly observed in their and in other groups initial cryoEM images (Henry et al., 2018; Zhang et al., 2010). Additionally, some of the XL-MS Lys pairs might come from less lipidated ApoE particles or even delipidated ones, which was not considered in the simulation. Nevertheless, their model raises a question whether ApoE does have different organizations on lipid nanodiscs and whether such a difference would have resulted from interchangeable, but distinguishable folding/unfolding steps or reflects a broader conformational heterogeneity of the protein. It is also unknown as to whether what they observed from ApoE4 could be applied to the other two isoforms or explain the isoform differences for AD pathology in a novel way.
In addition to these hallmark structures of the ApoE molecule, there are many other structural determinations worth mentioning which provide vital supplementary information for our understanding of ApoE and related interactions. For example, Guttman et al. characterized interactions between ApoE and LRP1 by solving the complex structure of ApoE fragment (130–149) fused with one conserved complement repeat (CR17) from LRP1 by NMR (Guttman et al., 2010). Their model suggests a universal mechanism how CR-containing receptors such as LRP1 and LDLR interact, presumably through a side chain of tryptophan and some acidic residues surrounding one calcium ion to attract the Lys143 and Lys146 on the ApoE helix. The interaction between ApoE and heparin has been determined in 2001, showing that Arg142 and Arg145 are critical to the 6-Osulfo group of glucosamine (Dong et al., 2001). Ren’s group also pioneered the possibility of applying novel techniques such as CryoEM in lipoprotein structure determination, including ApoE (Zhang et al., 2010, 2013). Going forward, the use of CryoEM to obtain high resolution structural features may ultimately prove to be critical in determining the structure of lipidated forms of ApoE produced by different cell types to answer some of these unresolved issues.
Physiological form of ApoE and Neuropathology
In its native form, ApoE is secreted as HDL-like lipoprotein particles in the CNS (Fig. 4A). This alters its ability to bind ApoE receptors and its interactions with pathological proteins, such as Aβ (Garai et al., 2014; Strittmatter et al., 1993b) as well as structures such as normal and damaged synapses. The lipidation status of ApoE has also been postulated to contribute to the progression of AD as the size and components of ApoE-containing lipoproteins have been associated with disease progression in AD (Grimm et al., 2017). The lipidation process of ApoE in vivo is not well understood, nor is the conformation of lipidated ApoE. As discussed, only two structural models of lipidated recombinant ApoE have been assessed (Hatters et al., 2006; Henry et al., 2018). While the crystal structure has unveiled a single, low-resolution model of lipidated ApoE on a spherical DPPC lipid-containing particle, the later simulation model assumes ApoE is bound to a discoidal lipoparticle. Both structural models suggest that two ApoE molecules form a dimeric ring to stabilize lipids in the center in its final state, while the lipoprotein particle might be disc-shaped or spheroidal, depending on certain lipidation conditions. However, neither of those two structures could provide detailed information of the molecular configurations of ApoE protein on the lipid-containing particle at even near-atomic resolution, which has limited our understanding of characteristics of native ApoE in vivo.
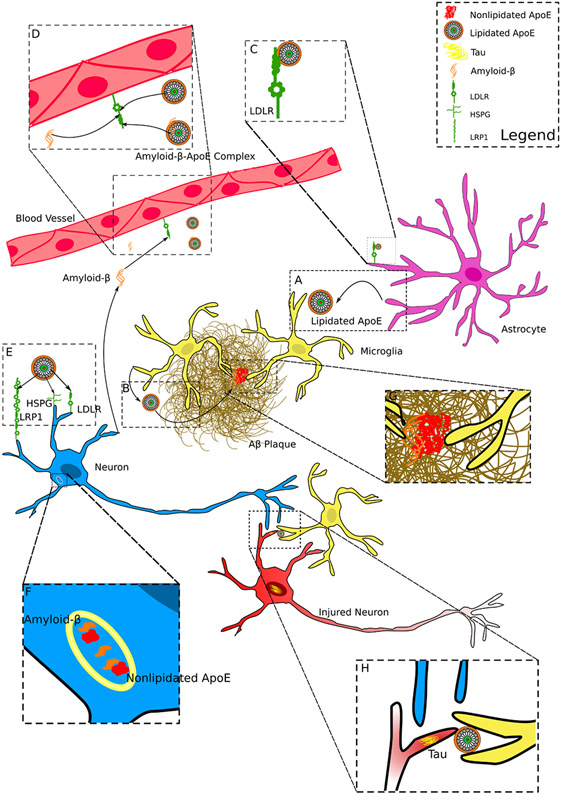
Figure 4. Model of ApoE and where it may impact AD Neuropathology.
A) Lipidated ApoE is secreted primarily by astrocytes in the CNS.
B) Microglia secrete ApoE in disease conditions and microglial ApoE can be integrated into existing amyloid-β plaques.
C) ApoE is uptaken by LDLR present on astrocytes and other cell types, such as endothelial cells (see D) and neurons (see E).
D) Amyloid-β and lipidated ApoE may compete for clearance by binding to LDLR receptors. ApoE might also form complexes with amyloid-β which may promote clearance of amyloid-β.
E) ApoE is uptaken by neurons through receptors such as LDLR, LRP1, and HSPG.
F) Recycling endosomes or lysosomes in neurons are potential sites where non-lipidated or partially lipidated ApoE interacts with amyloid-β inducing amyloid-β fibrilization.
G) ApoE in plaques is presumably required for microglial clustering around amyloid-β plaques.
H) ApoE may promote the engulfment of dying or damaged synapses/neurons with fibrillar tau accumulation.
Moreover, more subtle isoform specific size differences are also observed in the CNS. Observational studies of lipoprotein particle size in human CSF showed that ApoE4 was present in smaller HDL-like particles, whereas ApoE2 was associated with larger HDL-like particles (Heinsinger et al., 2016). In mouse models expressing different human ApoE isoforms, ApoE4 expression results in smaller particles while ApoE2 forms larger particles compared to ApoE3 (Hu et al., 2015). Consistency in the size preference between ApoE isoforms across species indicates that these differences are independent from the human/mouse brain environment. Rather these preferences result from intrinsic properties of the ApoE protein. This is supported by experiments using hydrogen-deuterium exchange coupled to MS experiments that showed that ApoE4 was more flexible at several residues (27-37, 60-64, and 102-108) than ApoE3, which may explain the prevalence of ApoE4 in VLDL (Frieden et al., 2017). Isoform specific differences may due to the ability of the N-terminal and C-terminal domains to separate. However, it is still not clear how and why ApoE is present as discoidal particles secreted by astrocytes (Fagan et al., 2002). This may be driven by the ability of ApoE to become lipidated in the CNS, which is a property both of the protein glycosylation and secretory apparatus of particular cell types. The lipidation of ApoE lipoparticles in the CNS may have a strong association with AD pathology. It has been found that the lipid efflux protein ABCA1 is required for ApoE to become lipidated by astrocytes (Hirsch-Reinshagen et al., 2004; Wahrle et al., 2004). Further, the lack of ABCA1 in Aβ-depositing mice results in increased amyloidosis while overexpression of ABCA1 increases ApoE-lipidation in the brain and decreases amyloidosis (Hirsch-Reinshagen et al., 2005; Koldamova et al., 2005; Wahrle et al., 2005, 2008). In addition, LXR agonists which increase ABCA1, among other effects, have been shown to reduce Aβ pathology in mice (Fitz et al., 2010). However, current studies do not suggest isoform specific differences in ABCA1 binding, nor the ultimate lipidation states after interacting with ABCA1 (Krimbou et al., 2004). Nonetheless, these studies suggest that the degree of ApoE lipidation influences its effect on Aβ even though the impact on the conformation of ApoE is not completely understood.
The lipidation of ApoE and isoform specific differences in stability may further contribute to differences in AD neuropathogenesis. The overall protein stability and lipid binding preferences appear to be linked. This is predicted to be driven by interactions between the N-terminal and C-terminal domains (Hatters et al., 2005; Xu et al., 2004). This was supported by a mutagenesis study where mutation of Glu255 to Ala255 altered the lipid binding preference of ApoE4 from plasma VLDL to HDL, similar to ApoE3 and ApoE2 (Weisgraber, 1990). Meanwhile, ApoE lipidation and its impact on AD pathology may be linked with secretion by different cell types. For example, while astrocytes produce and secrete most of the ApoE containing HDL-like lipoproteins in the normal CNS, following CNS damage and in the setting of neurodegeneration, microglia strongly upregulate ApoE levels (Krasemann et al., 2017). The function of ApoE produced by microglia has been proposed to be involved in the microglial activation response (Krasemann et al., 2017). It has recently been shown the microglia secrete ApoE in very poorly lipidated lipid particle as compared to astrocytes (Huynh et al., 2019). This is of particular interest as the deposition of ApoE into Aβ plaques appears to be driven primarily by ApoE produced from microglia (Parhizkar et al., 2019) (Fig. 4B). It has also been reported that neurons may upregulate ApoE in disease conditions; in the context of ApoE4 this may result in neurotoxcicity (Mahley and Huang, 2012). Moreover, it has been reported that the overexpression of ApoE4 from astrocytes is neuroprotective while the overexpression of ApoE4 by neurons in not (Buttini et. al., 2010). Whether there is a unique lipidation status and conformation of ApoE produced in neurons is currently unknown. Further research is needed to better understand the conformation of ApoE on differentially lipidated lipoprotein particles and how such differences influence ApoE structure and the effect of ApoE on AD neuropathology, both in an isoform specific and cell specific manner.
ApoE Receptors and Neuropathology
In 1970s, it was found that ApoE was a primary ligand for LDLR (Fig. 4C) and that ApoE2 was involved in familial type III hyperlipoproteinemia due to the dramatic reduction in binding to LDLR (Innerarity and Mahley, 1978; Innerarity et al., 1979; Wardell et al., 1982; Weisgraber et al., 1982). A classic theory suggested that the interaction between ApoE and LDLR was dependent on the arrangement of salt bridges in the LDLR binding site of ApoE (Dong and Weisgraber, 1996). However, the full-length structure of ApoE3 shows that the C-terminal domain potentially shields the LDLR receptor binding region that was assumed to be exposed in the Weisgraber model (Chen et al., 2011). Certainly, once ApoE is lipidated, the salt bridges between Arg150 and Asp154 may exist in the fully extended helical conformation of lipidated ApoE2. Therefore, it remains an important question as to whether the impact of Cys158 on ApoE-LDLR binding is due to its effect on the initial lipid binding stage of ApoE leading to an inability to adopt a specific conformation on the lipid surface in order to bind LDLR or if it is due to salt bridge rearrangement on lipidated ApoE that was predicted based on the structure of the N-terminus. As such, lipidation of ApoE is indispensable to understand ApoE’s ability to bind to LDLR. This interaction may be especially important as ApoE2 shows a dramatic reduction in the ability to bind LDLR and decreases AD risk (Innerarity et al., 1984; Lalazar et al., 1988).
In fact, the lipidation state of ApoE and interactions with the C-terminal domain were known from early on to play a critical role in the binding of ApoE to LDLR. ApoE2 bound to DPMC has <1% binding activity to LDLR as does plasma VLDL isolated from subjects with ApoE2/2 genotype (Innerarity et al., 1984). However, the N-terminal fragment of ApoE2 has 12-fold greater binding to LDLR, and Cys-modification with cysteamine and binding to DPMC restores its binding activity to that of ApoE3-DPMC (Innerarity et al., 1984). This strongly suggests that the reduction in ApoE2 binding to LDLR is due to conformational changes only present in the full length lipidated structure of ApoE2. FRET studies have investigated the conformational changes that occur when ApoE binds to lipid, with these studies suggesting that N-terminal domain of ApoE adopts an open conformation upon lipid binding (Fisher et al., 2000). These results are further supported by an assay using soluble LDLR where only lipidated full length ApoE3 was found to bind LDLR compared to lipid-free ApoE3 (Yamamoto et al., 2007). Lipidation of ApoE is also critical to expose this N-terminal region to LDLR as the C-terminal domain shields this region based on the NMR full length ApoE3 strucuture (Chen et al., 2011). The exposure of the LDLR binding site may be further regulated by the conformation of ApoE on lipoprotein particles. It has been proposed that there are two different major conformations of ApoE that co-exist – open hairpin and compact hairpin – which may undergo different lipidation processes (Henry et al., 2018).
Answering this question of how ApoE isoform and lipidation affect receptor binding is critical to understanding the impact of ApoE on processes related to neurodegeneration. ApoE is known to bind to LDLR, LRP1, and other receptors that are LDLR family members (Holtzman and Herz, 2012). This is highlighted not only by the differential risk for late-onset AD conferred by ApoE4>ApoE3>ApoE2, but also by the recent data from an individual with autosomal dominant AD who is also homozygous for ApoE3 with the Christchurch mutation (R136S). This individual has a strongly delayed onset of cognitive decline and tau-related pathology (Arboleda-Velasquez et al., 2019). The Christchurch mutation on ApoE3 leads to strongly decreased LDLR binding and also decreased binding to heparin (Lalazar et al., 1988; Arboleda-Velasquez et al., 2019). On the other hand, ApoE4 binds to heparin sulfate and dermatan sulfate more strongly than ApoE2 and ApoE3 as measured by surface plasmon resonance (SPR) (Yamauchi et al., 2008). The binding constants of ApoE-heparin binding also differ among isoforms (Futamura et al., 2005). These results are interesting in light of the fact that the individual with delayed dementia with autosomal dominant AD had a lot of amyloid deposition in the brain but much less tau pathology and neurodegeneration than would be expected (Arboleda-Velasquez et al., 2019). As data in a mouse model of tauopathy shows that ApoE strongly increases not only tauopathy, but tau-mediated neurodegeneration via microglia (Shi et al., 2017, 2019), perhaps ApoE binding to heparin linked with other receptors on microglia is important in this process. The related structures and interactions that mediate these types of effects are completely unknown.
Likely as a consequence of receptor binding, ApoE isoform is known to affect the amount of ApoE in the CNS. ApoE4 has the lowest protein levels and ApoE2 has the highest protein levels (Bales et al., 2009; Huynh et al., 2019). Since the binding of ApoE2 to LDLR is dramatically reduced, these data suggest that the protein levels of ApoE in the CNS are a direct consequence of the ability of ApoE to bind LDLR. Global knockout of LDLR significantly increased ApoE4 and ApoE3 levels while not affecting ApoE2 levels (Fryer et al., 2005). This is of particular interest as not only the isoform but also the levels of ApoE determine the amount of Aβ deposition that develops in the brain (Bien-Ly et al., 2012; Kim et al., 2011). In fact, it has been suggested that ApoE and Aβ compete for the same receptors for clearance (Kanekiyo et al., 2013; Verghese et al., 2013) (Fig. 4D). Further, LDLR has been demonstrated to directly bind Aβ, contributing to cellular uptake of Aβ (Basak et al., 2012), although the exact binding site is still unknown. Interestingly, overexpression of LDLR leads to a reduction in Aβ pathology (Kim et al., 2009). Similarly, knockout of LRP1 in astrocytes also leads to greater accumulation of Aβ pathology (Liu et al., 2017) (Fig. 4E). However, these affects appear to be cell type dependent as knockout of LRP1 in neurons decreased Aβ pathology in APPPS1/ApoE4 mice back to the levels seen in ApoE3/APPPS1 mice (Tachibana et al., 2019). These results may be neuron specific as neuronal synapses are postulated to be a site of ApoE-Aβ interaction that initiates Aβ aggregation (Bilousova et al., 2019) (Fig. 4F)
ApoE and Aβ Fibrilization: Consequences for Neuropathology
Due to the strong association of ApoE isoform with AD risk and the presence of ApoE in Aβ plaques, there has been much interest in the biochemical interaction between ApoE and Aβ. Early studies using plasma purified, but de-lipidated ApoE, found that ApoE4 binds to Aβ more quickly than ApoE3 (Strittmatter et al., 1993b). These initial findings were supported by studies where it was found that de-lipidated ApoE4 isolated from human plasma associated more rapidly with Aβ than ApoE3 and formed a denser matrix of monofibrils than from co-incubation of ApoE3 with Aβ (Sanan et al., 1994). In both cases immunogold labeling showed ApoE uniformly distributed across the surface of Aβ monofibrils. Using a thioflavin-T assay, ApoE was found to increase the rate of Aβ fibrillogenesis and total amount of fibrillar Aβ (Castano et al., 1995). In contrast, using lipidated ApoE showed a different trend where ApoE2 was found to associate most strongly with Aβ-peptide followed by ApoE3 then by ApoE4 (Aleshkov et al., 1997). It was also found using lipidated ApoE, that ApoE3 rather than ApoE4 formed more SDS-stable complexes with Aβ (LaDu et al., 1994, 1995). Together these findings began to demonstrate that the binding mechanism of ApoE to Aβ is dependent on differences in the conformation of non-lipidated ApoE and lipidated ApoE (Aleshkov et al., 1997). Moreover, there might also be a thermal and chemical stability tier in the order ApoE4<ApoE3<ApoE2 with the support of reducing folding intermediate data from ApoE4 to ApoE3 to ApoE2. Such a difference in stability was also considered to affect the ApoE-Aβ interaction (Segrest et al., 1992). The less stable ApoE4 is also thought to adopt a more molten-globule-like conformation with Aβ-bound that may break down the lysosomal membrane (Ji et al., 2002, 2006).
More studies have been undertaken in order to try to understand the impact of ApoE on the aggregation of Aβ with new techniques. Studies looking at how ApoE influences the rate of Aβ aggregation have found that ApoE actually slows Aβ aggregation (Garai et al., 2014; Raulin et al., 2019). Non-lipidated ApoE4 was found to slow aggregation the most compared to ApoE3 and ApoE2 slowed aggregation the least. These experiments were carried out with ApoE concentrations of 100 μM where ApoE is likely oligomeric. However, DMPC lipidated ApoE showed the same trend with ApoE4-DMPC slowing Aβ aggregation more than ApoE2 and ApoE3. ApoE derived from astrocytes also slowed Aβ aggregation, but did not show an isoform specific difference, highlighting the importance of the lipid environment in understanding ApoE-Aβ interaction (Fig. 4G&H). However, these studies do not completely address the pathological consequences of ApoE on Aβ aggregation, due to the known association of ApoE with Aβ plaques in vivo. ApoE isoforms show differences in the rate of self-assembly and in their propensity to form fibrillar structures. Whether such fibrillization occurs in vivo is not clear, yet in vitro, ApoE4 was found to rapidly self-associate and form fibrils faster than ApoE3; ApoE2 showed the slowest rate of self-aggregation and fibrillization (Raulin et al., 2019). On the other hand, a study applying fluorescence cross-correlation spectroscopy (FCCS) combined with alternating laser excitation (ALEX) characterized this interaction in solution, overcoming the immobilization restraint in SPR and noise signal from self-association in EPR assay (Ly et al., 2013). Their data clearly suggests that Aβ oligomers bind to ApoE4 slower than ApoE3, where both interactions prevented Aβ aggregation by itself. Further, the ApoE-Aβ complex showed limited dissociation, indicating a much tighter complex molecule formed, which was consistent with the SDS-resistant feature reported before (Bentley et al., 2002; LaDu et al., 1994). The delay of Aβ aggregation by ApoE binding has been further demonstrated by FRET recently (Ghosh et al., 2019). However, the new data still led to a conclusion, not observed in vivo, that ApoE4 binds faster than ApoE3 and is more efficient in preventing Aβ aggregation. Also, full-length ApoE has a higher affinity binding to Aβ comparing with either the N- or C-terminal domains. Meanwhile, DMPC-associated ApoE4 seemed to fail to prevent Aβ aggregation but DMPC-associated ApoE3 retained that ability in this study, which is contradictory compared to earlier results (Garai et al., 2014; Raulin et al., 2019). Indeed, lipidated ApoE seems to behave much differently from non-lipidated ApoE in its interaction with Aβ. Presumably, the interaction between Aβ and ApoE will substantially and kinetically depend on the lipidation state of ApoE.
Domain studies of ApoE-Aβ binding also yielded different conclusions. Some early studies on the interaction of ApoE and Aβ revealed that the lipid binding region of ApoE (244-272) is critical for ApoE-Aβ binding (Strittmatter et al., 1993b). This is further supported by the full-length NMR model of ApoE3, where the ApoE C-terminal domain contains multiple hydrophobic residues that are hypothesized to initiate Aβ binding. Although, other studies have also identified residues in the NT domain to be involved (Winkler et al., 1999). This suggests that Aβ may compete with ApoE-containing lipoproteins for binding to LDLR and may also explain differences in ApoE isoform affinity for Aβ that is dependent on the lipidation state of ApoE. A recent study found that N-and C-terminal domains and full-length ApoE may have similar binding affinity at sub-micromolar range (where ApoE monomeric and dimeric forms are prevalent) to Aβ fibrils (Gunzburg et al., 2007). Their FRET results suggested that the ApoE N-terminal domain could undergo similar conformational changes to adapt the binding to different amyloid fibril structures such as ApoC-II fibrils and Aβ fibrils. Interestingly, R90, C112 and R119 were predicted to interact with E22 on the Aβ fibril, where C112 is the key residue to distinguish ApoE2 from ApoE3 and ApoE4.
While the true biochemical interactions between ApoE and Aβ are still under debate, in vivo it is possible that ApoE has two distinct impacts on Aβ pathology depending on its state. The impact of ApoE isoform on Aβ pathology is dramatic in humans and animal models and is one of the best studied consequences of ApoE on AD pathology (Morris et al., 2010; Reiman et al., 2009). Expression of human ApoE in various mouse models of AD-like Aβ amyloidosis has found significant isoform dependent effects where ApoE4 increases the amount of Aβ plaques and ApoE2 decreases the amount of Aβ pathology relative to ApoE3 (Bales et al., 2009; Holtzman et al., 2000; Huynh et al., 2017, 2019; Liu et al., 2017). There is a lot of evidence that ApoE influences Aβ accumulation via effects on Aβ clearance and seeding as reviewed below. In addition, one study reported that ApoE directly altered Aβ production in embryonic stem cell derived neurons (Huang et al., 2017). Studies using either antisense oligonucleotides to decrease ApoE or tamoxifen inducible expression to increase ApoE have shown that the impact of ApoE isoform on Aβ pathology is time dependent (Huynh et al., 2017; Liu et al., 2017). Reduction of ApoE early prior to the onset of Aβ deposition reduces the amount of Aβ pathology while reduction after the initial seeding stage of amyloid pathology had no major effect (Huynh et al., 2017). Similarly, induction of ApoE4 expression before Aβ deposition increases Aβ pathology, but when done after Aβ deposition there was no effect (Liu et al., 2017). Together, these results suggest that the early stage of Aβ pathology where Aβ first misfolds into a β-pleated sheet and starts to form plaques “seeding” is impacted by ApoE isoform. In fact, knockout of ApoE results in a huge reduction of Aβ fibril formation into the β-pleated sheet structures in amyloid plaques (Bales et al., 1999; Holtzman et al., 2000; Ulrich et al., 2018). Furthermore, the presence of ApoE in plaques affects the microglial response to Aβ plaques and may ultimately impact tau-mediated neurodegeneration (Shi et al., 2017; Ulrich et al., 2018) (Fig. 4I). Lipidated ApoE likely competes for clearance with Aβ and its impact on pathology is driven by isoform specific differences in receptor binding. In contrast, the self-association and aggregation of ApoE may promote the formation/seeding of Aβ plaques and also be involved in Aβ pathology in an isoform specific manner. Interestingly, some hydrophobic residues (Phe257, Phe265, Trp276, Val283, Leu279, Val287) in the C-terminal domain of ApoE remain solvent exposed and might form a strong hydrophobic pocket which may contribute to initial lipid binding (PDB: 2l7p). Such a hydrophobic surface may also explain how ApoE can attract the Aβ peptide and eventually incorporate into the dense core of amyloid plaques in an aggregated form (Liao et al., 2018). Also, the interaction of ApoE and Aβ may be mediated by cleavage of the ApoE protein as an increased number of N-terminal ApoE fragments have been reported in ApoE4 plaques (Jones et al., 2011). On the other hand, Aβ has numerous biophysical forms that might potentially interact with ApoE (Frieden et al., 2017). It is still not clear as to which form or forms of ApoE initiate and propagate ApoE-Aβ interactions that contribute to pathology. This highlights the importance of knowing the structure of different forms of ApoE.
It is challenging to analyze and interpret biochemical data addressing interactions between ApoE and Aβ due to the self-association of each molecule. Non-lipidated ApoE exists in at least three forms – monomer, dimer, and tetramer – sometimes with higher order oligomeric forms. Further, Aβ is known to strongly self-aggregate under specific conditions. To date, it is still unclear when and how the interaction between ApoE and Aβ occurs, although ApoE has been observed in amyloid plaques for years. Since ApoE is secreted in a lipidated form in vivo and a non-lipidated or poorly lipidated form of ApoE is observed in Aβ plaques (Liao et al., 2018), one hypothesis is that the interaction of Aβ starts with lipidated ApoE and the ApoE found in plaques is in a specific conformation resulting from its partial or complete de-lipidation. The models of lipidated ApoE4 proposed by Weisgraber or Wang/Frieden both suggest that residues in the C-terminal domain of ApoE are bound to phospholipid, and it is possible that Aβ binding to this site may result in de-lipidation of ApoE. Indeed, using ELISA to measure soluble ApoE-Aβ complexes, it was found that lower levels of ApoE-Aβ soluble complexes were detectable in hippocampal homogenates from ApoE4 expressing mice compared to ApoE2 and ApoE3 expressing mice (Tai et al., 2013). This may explain earlier results where ApoE binding was measured by examining SDS-stable complexes that likely represent complexes of aggregated ApoE-Aβ (Strittmatter et al., 1993b). Additionally, soluble Aβ oligomers in the brain were higher in ApoE4 carriers compared to ApoE3 with ApoE2 having the lowest levels (Hashimoto et al., 2012).
Of importance in future work will be to characterize 1. How different lipidation states of ApoE influence its structure and ability regulate its ability to bind Aβ; 2. Whether ApoE-Aβ affects the binding of ApoE to the lipoprotein and its structural conformation; 3. Whether ApoE-Aβ interaction occurs prior to or is concomitant with amyloid fibrillization; 4. If this interaction leads to or prevents fibrillization of Aβ or even plaque formation; 5. Molecular details from future structural determinations of complex between lipidated and non-lipidated ApoE and monomeric vs. oligomeric Aβ.
Conclusion
ApoE plays a critical role in the pathogenesis of several neurodegenerative diseases; including its dramatic impact on AD risk. Resolving the molecular function and interactions of ApoE under physiological conditions and in the setting of disease will be critical to understanding disease pathogenesis, but also in designing therapeutics to combat these diseases. Our current understanding of the role of ApoE in these diseases is limited by incomplete structural information. ApoE2 and ApoE4 differ from ApoE3 by a single amino acid substitution, which results in dramatically different impacts on disease risk and outcomes. Changes in the interaction of ApoE with its receptors or with proteins involved in disease pathogenesis are correlated with either slowing down or accelerating disease progression. In addition, ApoE has been shown to directly interact with Aβ, tau, and α-synuclein; likely directly contributing to the formation of protein aggregates or the response of the brain to these aggregates in various diseases (Davis et al., 2020; Strittmatter et al., 1993a; Zhao et al., 2018, 2020). The molecular details of these interactions have not yet been elucidated, impeding progress in the development of effective therapeutics targeting these interactions. To advance our understanding of ApoE and its role in AD, diseases of the CNS, and in normal physiology, it is also essential to solve, at high-resolution, structure of lipidated ApoE in the CNS – its physiological form. Lipidation of ApoE and the conformational change that occurs in ApoE on the lipid surface is required for binding of ApoE to ApoE receptors that are members of the LDLR family. In addition, interdomain interactions within ApoE may drive isoform specific differences in the binding of ApoE to its receptors, lipids, and pathological proteins, such as Aβ. Moreover, some biochemical results indicated that non-lipidated ApoE undergoes dimerization and then tetramerization at high concentration, while the model that monomeric ApoE binds to lipids fit best with binding curves (Frieden et al., 2017; Garai et al., 2011). Current methods are insufficient for elucidating these interactions within ApoE. However, solving isoform-specific structures of non-lipidated ApoE and lipidated ApoE complex both alone and with the binding partners would allow for the design of drugs to directly modulate ApoE-receptor and ApoE-protein interactions at a molecular level. Moreover, structure correctors also showed some potential to alleviate toxic effects from ApoE4 (Wang et al., 2018). Indeed, there are current studies that postulate modulating the structure of ApoE4 can reverse its negative effects on Aβ pathology, but we currently lack methodologies to confirm the occurrence and extent of such modulations.
To summarize, our review has focused on the biology of ApoE that relates our evolving understanding of ApoE structure over the last 40 years to the pathophysiology of AD. Three structural models of ApoE have yielded insights that explain apparently contradictory observations in the molecular features and interactions of ApoE. Despite this, each model appears to explain some specific, but not all, aspects of the molecule. Although current models of ApoE have managed to explain different functions and interactions of this molecule, we still lack critical experimental data that would complete the models, such as a full-length, high resolution structure of ApoE2 and ApoE4, including all isoforms when lipidated. Even, the isoform-specific differences between non-lipidated forms of ApoE2, ApoE3, and ApoE4 have not been completely resolved. Further studies utilizing single molecule FRET, or in vivo FRET, based on emerging models would be informative. Moreover, novel drug targets that might leverage ApoE or related interactions still await determination of ApoE-Aβ and ApoE-receptor complexes. For example, the complex structure of a reported interaction between ApoE and TREM2 has never been determined (Atagi et al., 2015). As new structural models generate new hypotheses, future studies that apply emerging techniques such as small molecule inhibitors of ApoE-receptor and ApoE-protein interactions, molecular modulators, and cryo-EM will pave the way for therapeutic development for AD and other neurodegenerative diseases based on knowledge of the relationship between the structure and function of ApoE.
Acknowledgments:
This work was supported by NIH grants NS090934 (DMH), AG047644 (DMH), AG062837 (AS), AG58518-1 (MRS), NSF grant DGE-1745038 (MRS), the Cure Alzheimer’s Fund (DMH), JPB Foundation (DMH), Ruth K. Broad Biomedical Research Foundation (AS), and the Alzheimer's Association (AS).
Footnotes
Declaration of Interests
D.M.H. is listed as an inventor on a US patent application #20190270794 entitled “Anti-APOE antibodies” from Washington University. D.M.H. co-founded and is on the scientific advisory board of C2N Diagnostics. D.M.H. is on the scientific advisory board of Denali and consults for Genentech, Merck, and Idorsia. Washington University (D.M.H.) has a sponsored research agreement to work on apoE antibodies from NextCure. All other authors have no competing interests.
References
- Aggerbeck LP, Wetterau JR, Weisgraber KH, Mahley RW, and Agard DA (1988). Crystallization and preliminary X-ray diffraction studies on the amino-terminal (receptor-binding) domain of human apolipoprotein E3 from serum very low density lipoproteins. Journal of Molecular Biology 202, 179–181. [DOI] [PubMed] [Google Scholar]
- Aleshkov S, Abraham CR, and Zannis VI (1997). Interaction of nascent apoe2, apoe3, and apoe4 isoforms expressed in mammalian cells with amyloid peptide β (1-40). Relevance to Alzheimer’s disease. Biochemistry 36, 10571–10580. [DOI] [PubMed] [Google Scholar]
- Arboleda-Velasquez JF, Lopera F, O’Hare M, Delgado-Tirado S, Marino C, Chmielewska N, Saez-Torres KL, Amarnani D, Schultz AP, Sperling RA, et al. (2019). Resistance to autosomal dominant Alzheimer’s disease in an APOE3 Christchurch homozygote a case report. Nature Medicine 25, 1680–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atagi Y, Liu C-C, Painter MM, Chen X-F, Verbeeck C, Zheng H, Li X, Rademakers R, Kang SS, Xu H, et al. (2015). Apolipoprotein E Is a Ligand for Triggering Receptor Expressed on Myeloid Cells 2 (TREM2). The Journal of Biological Chemistry 290, 26043–26050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker HN, Gotto AM, and Jackson RL (1975). The primary structure of human plasma high density apolipoprotein glutamine I (ApoA-I). II. The amino acid sequence and alignment of cyanogen bromide fragments IV, III, and I. Journal of Biological Chemistry 250, 2725–2738. [PubMed] [Google Scholar]
- Bales KR, Verina T, Dodel RC, Du Y, Altstiel L, Bender M, Hyslop P, Johnstone EM Little SP, Cummins DJ, et al. (1997). Lack of apolipoprotein E dramatically reduces amyloid beta-peptide deposition. Nature Genetics 17, 263–264. [DOI] [PubMed] [Google Scholar]
- Bales KR, Verina T, Cummins DJ, Du Y, Dodel RC, Saura J, Fishman CE, DeLong CA, Piccardo P, Petegnief V, et al. (1999). Apolipoprotein E is essential for amyloid deposition in the APP(V717F) transgenic mouse model of Alzheimer’s disease. Proceedings of the National Academy of Sciences of the United States of America 96, 15233–15238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bales KR, Liu F, Wu S, Lin S, Koger D, DeLong C, Hansen JC, Sullivan PM, and Paul SM (2009). Human APOE isoform-dependent effects on brain β-amyloid levels in PDAPP transgenic mice. Journal of Neuroscience 29, 6771–6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basak JM, Verghese PB, Yoon H, Kim J, and Holtzman DM (2012). Low-density lipoprotein receptor represents an apolipoprotein E-independent pathway of Aβ uptake and degradation by astrocytes. Journal of Biological Chemistry 287, 13959–13971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell RD, Winkler EA, Singh I, Sagare AP, Deane R, Wu Z, Holtzman DM, Betsholtz C, Armulik A, Sallstrom J, et al. (2012). Apolipoprotein E controls cerebrovascular integrity via cyclophilin A. Nature 485, 512–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley NM, Ladu MJ, Rajan C, Getz GS, and Reardon CA (2002). Apolipoprotein E structural requirements for the formation of SDS-stable complexes with beta-amyloid-(1-40): the role of salt bridges. The Biochemical Journal 366, 273–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bien-Ly N, Gillespie AK, Walker D, Yoon SY, and Huang Y (2012). Reducing human apolipoprotein E levels attenuates age-dependent Aβ accumulation in mutant human amyloid precursor protein transgenic mice. The Journal of Neuroscience : The Official Journal of the Society for Neuroscience 32, 4803–4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilousova T, Melnik M, Miyoshi E, Gonzalez BL, Poon WW, Vinters HV, Miller CA, Corrada MM, Kawas C, Hatami A, et al. (2019). Apolipoprotein E/Amyloid-β Complex Accumulates in Alzheimer Disease Cortical Synapses via Apolipoprotein E Receptors and Is Enhanced by APOE4. The American Journal of Pathology 189, 1621–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blue ML, Williams DL, Zucker S, Khan SA, and Blum CB (1983). Apolipoprotein E synthesis in human kidney, adrenal gland, and liver. Proceedings of the National Academy of Sciences of the United States of America 80, 283–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyles JK, Pitas RE, Wilson E, Mahley RW, and Taylor JM (1985). Apolipoprotein E associated with astrocytic glia of the central nervous system and with nonmyelinating glia of the peripheral nervous system. Journal of Clinical Investigation 76, 1501–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley WA, Gilliam EB, Gotto AM, and Gianturco SH (1982). Apolipoprotein-E degradation in human very low density lipoproteins by plasma protease(s): Chemical and biological consequences. Biochemical and Biophysical Research Communications 109, 1360–1367. [DOI] [PubMed] [Google Scholar]
- Buttini M, Masliah E, Yu G-Q, Palop JJ, Chang S, Bernardo A, Lin C, Wyss-Coray T, Huang Y, and Mucke L (2010). Cellular source of apolipoprotein E4 determines neuronal susceptibility to excitotoxic injury in transgenic mice. The American Journal of Pathology 177, 563–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam JA, Zerbinatti CV, Li Y, and Bu G (2005). Rapid Endocytosis of the Low Density Lipoprotein Receptor-related Protein Modulates Cell Surface Distribution and Processing of the β-Amyloid Precursor Protein. Journal of Biological Chemistry 280, 15464–15470. [DOI] [PubMed] [Google Scholar]
- Cardin AD, Demeter DA, Weintraub HJ, and Jackson RL (1991). Molecular design and modeling of protein-heparin interactions. Methods in Enzymology 203, 556–583. [DOI] [PubMed] [Google Scholar]
- Castaño EM, Prelli F, Pras M, and Frangione B (1995). Apolipoprotein E carboxyl-terminal fragments are complexed to amyloids A and L. Implications for amyloidogenesis and Alzheimer’s disease. The Journal of Biological Chemistry 270, 17610–17615. [DOI] [PubMed] [Google Scholar]
- Castellano JM, Kim J, Stewart FR, Jiang H, DeMattos RB, Patterson BW, Fagan AM, Morris JC, Mawuenyega KG, Cruchaga C, et al. (2011). Human apoE isoforms differentially regulate brain amyloid-β peptide clearance. Science Translational Medicine 3, 89ra57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano JM, Deane R, Gottesdiener AJ, Verghese PB, Stewart FR, West T, Paoletti AC, Kasper TR, DeMattos RB, Zlokovic BV, et al. (2012). Low-density lipoprotein receptor overexpression enhances the rate of brain-to-blood Aβ clearance in a mouse model of β-amyloidosis. Proceedings of the National Academy of Sciences of the United States of America 109, 15502–15507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Li Q, and Wang J (2011). Topology of human apolipoprotein E3 uniquely regulates its diverse biological functions. Proceedings of the National Academy of Sciences of the United States of America 108, 14813–14818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, and Pericak-Vance MA (1993). Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science (New York, N.Y.) 261, 921–923. [DOI] [PubMed] [Google Scholar]
- Davignon J, Bouthillier D, Nestruck AC, and Sing CF (1988). Apolipoprotein E polymorphism and atherosclerosis: insight from a study in octogenarians. Transactions of the American Clinical and Climatological Association 99, 100–110. [PMC free article] [PubMed] [Google Scholar]
- Davis AA, Inman CE, Wargel ZM, Dube U, Freeberg BM, Galluppi A, Haines JN, Dhavale DD, Miller R, Choudhury FA, et al. (2020). APOE genotype regulates pathology and disease progression in synucleinopathy. Science Translational Medicine 12, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMattos RB, Curtiss LK, and Williams DL (1998). A minimally lipidated form of cell-derived apolipoprotein E exhibits isoform-specific stimulation of neurite outgrowth in the absence of exogenous lipids or lipoproteins. Journal of Biological Chemistry 273, 4206–4212. [DOI] [PubMed] [Google Scholar]
- DeMattos RB, Rudel LL, and Williams DL (2001). Biochemical analysis of cell-derived apoE3 particles active in stimulating neurite outgrowth. Journal of Lipid Research 42, 976–987. [PubMed] [Google Scholar]
- Dickson DW, Heckman MG, Murray ME, Soto AI, Walton RL, Diehl NN, Gerpen J.A. van, Uitti RJ, Wszolek ZK, Ertekin-Taner N, et al. (2018). APOE ϵ4 is associated with severity of Lewy body pathology independent of Alzheimer pathology. Neurology 91, e1182–e1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L-M, and Weisgraber KH (1996). Human Apolipoprotein E4 Domain Interaction. Journal of Biological Chemistry 271, 19053–19057. [DOI] [PubMed] [Google Scholar]
- Dong J, Peters-Libeu CA, Weisgraber KH, Segelke BW, Rupp B, Capila I, Hernáiz MJ, LeBrun LA, and Linhardt RJ (2001). Interaction of the N-terminal domain of apolipoprotein E4 with heparin. Biochemistry 40, 2826–2834. [DOI] [PubMed] [Google Scholar]
- Dong LM, Wilson C, Wardell MR, Simmons T, Mahley RW, Weisgraber KH, and Agard DA (1994). Human apolipoprotein E. Role of arginine 61 in mediating the lipoprotein preferences of the E3 and E4 isoforms. Journal of Biological Chemistry 269, 22358–22365. [PubMed] [Google Scholar]
- Eto M, Watanabe K, Iwashima Y, Morikawa A, Chonan N, Oshima E, Sekiguchi M, and Ishii K (1987). Increased frequency of apolipoprotein 4 allele in type II diabetes with hypercholesterolemia. Diabetes 36, 1301–1306. [DOI] [PubMed] [Google Scholar]
- Eto M, Watanabe K, and Makino I (1991). Relationship of metabolic control and body weight to hypercholesterolemia in NIDDM patients with apolipoprotein E4. Diabetes Care 14, 127–129. [DOI] [PubMed] [Google Scholar]
- Fagan AM, Holtzman DM, Munson G, Mathur T, Schneider D, Chang LK, Getz GS, Reardon CA, Lukens J, Shah JA, et al. (1999). Unique lipoproteins secreted by primary astrocytes from wild type, apoE (−/−), and human apoE transgenic mice. Journal of Biological Chemistry 274, 30001–30007. [DOI] [PubMed] [Google Scholar]
- Fagan AM, Watson M, Parsadanian M, Bales KR, Paul SM, and Holtzman DM (2002). Human and murine ApoE markedly alters A beta metabolism before and after plaque formation in a mouse model of Alzheimer’s disease. Neurobiology of Disease 9, 305–318. [DOI] [PubMed] [Google Scholar]
- Fisher CA, Narayanaswami V, and Ryan RO (2000). The lipid-associated conformation of the low density lipoprotein receptor binding domain of human apolipoprotein E*. Journal of Biological Chemistry 275, 33601–33606. [DOI] [PubMed] [Google Scholar]
- Fitz NF, Cronican A, Pham T, Fogg A, Fauq AH, Chapman R, Lefterov I, and Koldamova R (2010). Liver X receptor agonist treatment ameliorates amyloid pathology and memory deficits caused by high-fat diet in APP23 mice. The Journal of Neuroscience : The Official Journal of the Society for Neuroscience 30, 6862–6872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieden C, Wang H, and Ho CM (2017). A mechanism for lipid binding to apoE and the role of intrinsically disordered regions coupled to domain-domain interactions. Proceedings of the National Academy of Sciences of the United States of America 114, 6292–6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer JD, DeMattos RB, McCormick LM, O’Dell MA, Spinner ML, Bales KR, Paul SM, Sullivan PM, Parsadanian M, Bu G, et al. (2005). The low density lipoprotein receptor regulates the level of central nervous system human and murine apolipoprotein E but does not modify amyloid plaque pathology in PDAPP mice. Journal of Biological Chemistry 280, 25754–25759. [DOI] [PubMed] [Google Scholar]
- Futamura M, Dhanasekaran P, Handa T, Phillips MC, Lund-Katz S, and Saito H (2005). Two-step mechanism of binding of apolipoprotein E to heparin: Implications for the kinetics of apolipoprotein E-heparan sulfate proteoglycan complex formation on cell surfaces. Journal of Biological Chemistry 280, 5414–5422. [DOI] [PubMed] [Google Scholar]
- Garai K, Baban B, and Frieden C (2011). Dissociation of apolipoprotein E oligomers to monomer is required for high-affinity binding to phospholipid vesicles. Biochemistry 50, 2550–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garai K, Verghese PB, Baban B, Holtzman DM, and Frieden C (2014). The binding of apolipoprotein e to oligomers and fibrils of amyloid-β alters the kinetics of amyloid aggregation. Biochemistry 53, 6323–6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Sil TB, Dolai S, and Garai K (2019). High-affinity multivalent interactions between apolipoprotein E and the oligomers of amyloid-β. FEBS Journal 286, 4737–4753. [DOI] [PubMed] [Google Scholar]
- Goldstein JL, and Brown MS (1976). The LDL pathway in human fibroblasts: a receptor-mediated mechanism for the regulation of cholesterol metabolism. Current Topics in Cellular Regulation 11, 147–181. [DOI] [PubMed] [Google Scholar]
- Grimm MO, Michaelson DM, and Hartmann T (2017). Omega-3 fatty acids, lipids, and apoE lipidation in Alzheimer’s disease: A rationale for multi-nutrient dementia prevention. Journal of Lipid Research 58, 2083–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunzburg MJ, Perugini MA, and Howlett GJ (2007). Structural Basis for the Recognition and Cross-linking of Amyloid Fibrils by Human Apolipoprotein E. Journal of Biological Chemistry 282, 35831–35841. [DOI] [PubMed] [Google Scholar]
- Guttman M, Prieto JH, Handel TM, Domaille PJ, and Komives EA (2010). Structure of the minimal interface between ApoE and LRP. Journal of Molecular Biology 398, 306–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T, Serrano-Pozo A, Hori Y, Adams KW, Takeda S, Banerji AO, Mitani A, Joyner D, Thyssen DH, Bacskai BJ, et al. (2012). Apolipoprotein E, especially apolipoprotein E4, increases the oligomerization of amyloid β peptide. The Journal of Neuroscience : The Official Journal of the Society for Neuroscience 32, 15181–15192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatters DM, Peters-Libeu CA, and Weisgraber KH (2005). Engineering conformational destabilization into mouse apolipoprotein E: A model for a unique property of human apolipoprotein E4. Journal of Biological Chemistry 280, 26477–26482. [DOI] [PubMed] [Google Scholar]
- Hatters DM, Peters-Libeu CA, and Weisgraber KH (2006). Apolipoprotein E structure: insights into function. Trends in Biochemical Sciences 31, 445–454. [DOI] [PubMed] [Google Scholar]
- Hatters DM, Voss JC, Budamagunta MS, Newhouse YN, and Weisgraber KH (2009). Insight on the Molecular Envelope of Lipid-Bound Apolipoprotein E from Electron Paramagnetic Resonance Spectroscopy. Journal of Molecular Biology 386, 261–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinsinger NM, Gachechiladze MA, and Rebeck GW (2016). Apolipoprotein E Genotype Affects Size of ApoE Complexes in Cerebrospinal Fluid. Journal of Neuropathology and Experimental Neurology 75, 918–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry N, Krammer E-M, Stengel F, Adams Q, Van Liefferinge F, Hubin E, Chaves R, Efremov R, Aebersold R, Vandenbussche G, et al. (2018). Lipidated apolipoprotein E4 structure and its receptor binding mechanism determined by a combined cross-linking coupled to mass spectrometry and molecular dynamics approach. PLOS Computational Biology 14, e1006165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch-Reinshagen V, Zhou S, Burgess BL, Bernier L, McIsaac SA, Chan JY, Tansley GH, Cohn JS, Hayden MR, and Wellington CL (2004). Deficiency of ABCA1 impairs apolipoprotein E metabolism in brain. The Journal of Biological Chemistry 279, 41197–41207. [DOI] [PubMed] [Google Scholar]
- Hirsch-Reinshagen V, Maia LF, Burgess BL, Blain JF, Naus KE, McIsaac SA, Parkinson PF, Chan JY, Tansley GH, Hayden MR, et al. (2005). The absence of ABCA1 decreases soluble ApoE levels but does not diminish amyloid deposition in two murine models of Alzheimer disease. Journal of Biological Chemistry 280, 43243–43256. [DOI] [PubMed] [Google Scholar]
- Holtzman D, and Herz J (2012). Apolipoprotein E and apolipoprotein receptors: normal biology and roles in Alzheimer’s disease. Cold Spring Harbor Perspectives in Medicine 2, a006312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman DM, Bales KR, Wu S, Bhat P, Parsadanian M, Fagan AM, Chang LK, Sun Y, and Paul SM (1999). Expression of human apolipoprotein E reduces amyloid-beta deposition in a mouse model of Alzheimer’s disease. The Journal of Clinical Investigation 103, R15–R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman DM, Bales KR, Tenkova T, Fagan AM, Parsadanian M, Sartorius LJ, Mackey B, Olney J, McKeel D, Wozniak D, et al. (2000). Apolipoprotein E isoform-dependent amyloid deposition and neuritic degeneration in a mouse model of Alzheimer’s disease. Proceedings of the National Academy of Sciences of the United States of America 97, 2892–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Liu C-C, Chen X-F, Zhang Y-W, Xu H, and Bu G (2015). Opposing effects of viral mediated brain expression of apolipoprotein E2 (apoE2) and apoE4 on apoE lipidation and Aβ metabolism in apoE4-targeted replacement mice. Molecular Neurodegeneration 10, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y-WA, Zhou B, Wernig M, and Sudhof TC (2017). ApoE2, ApoE3, and ApoE4 Differentially Stimulate APP Transcription and Aβ Secretion. Cell 168, 427–441.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebbe P, and Rimbach G (2017). Evolution of human apolipoprotein E (APOE) isoforms: Gene structure, protein function and interaction with dietary factors. Ageing Research Reviews 37, 146–161. [DOI] [PubMed] [Google Scholar]
- Hui DY, Innerarity TL, and Mahley RW (1984). Defective hepatic lipoprotein receptor binding of β-very low density lipoproteins from type III hyperlipoproteinemic patients. Importance of apolipoprotein E. Journal of Biological Chemistry 259, 860–869. [PubMed] [Google Scholar]
- Huynh T-PV, Liao F, Francis CM, Robinson GO, Serrano JR, Jiang H, Roh J, Finn MB, Sullivan PM, Esparza TJ, et al. (2017). Age-Dependent Effects of apoE Reduction Using Antisense Oligonucleotides in a Model of β-amyloidosis. Neuron 96, 1013–1023.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh T-PV, Wang C, Tran AC, Tabor GT, Mahan TE, Francis CM, Finn MB, Spellman R, Manis M, Tanzi RE, et al. (2019). Lack of hepatic apoE does not influence early Aβ deposition: observations from a new APOE knock-in model. Molecular Neurodegeneration 14, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innerarity TL, and Mahley RW (1978). Enhanced binding by cultured human fibroblasts of apo-E-containing lipoproteins as compared with low density lipoproteins. Biochemistry 17, 1440–1447. [DOI] [PubMed] [Google Scholar]
- Innerarity TL, Pitas RE, and Mahley RW (1979). Binding of arginine-rich (E) apoprotein after recombination with phospholipid vesicles to the low density lipoprotein receptors of fibroblasts. The Journal of Biological Chemistry 254, 4186–4190. [PubMed] [Google Scholar]
- Innerarity TL, Friedlander EJ, Rall SC, Weisgraber KH, and Mahley RW (1983). The receptor-binding domain of human apolipoprotein E. Binding of apolipoprotein E fragments. Journal of Biological Chemistry 258, 12341–12347. [PubMed] [Google Scholar]
- Innerarity TL, Weisgraber KH, Arnold KS, Rall SCJ, and Mahley RW (1984). Normalization of receptor binding of apolipoprotein E2. Evidence for modulation of the binding site conformation. The Journal of Biological Chemistry 259, 7261–7267. [PubMed] [Google Scholar]
- Jackson RL, Morrisett JD, Gotto AM, and Segrest JP (1975). The mechanism of lipid-binding by plasma lipoproteins. Molecular and Cellular Biochemistry 6, 43–50. [DOI] [PubMed] [Google Scholar]
- Ji Z-S, Miranda RD, Newhouse YM, Weisgraber KH, Huang Y, and Mahley RW (2002). Apolipoprotein E4 potentiates amyloid beta peptide-induced lysosomal leakage and apoptosis in neuronal cells. The Journal of Biological Chemistry 277, 21821–21828. [DOI] [PubMed] [Google Scholar]
- Ji Z-S, Mullendorff K, Cheng IH, Miranda RD, Huang Y, and Mahley RW (2006). Reactivity of apolipoprotein E4 and amyloid beta peptide: lysosomal stability and neurodegeneration. The Journal of Biological Chemistry 281, 2683–2692. [DOI] [PubMed] [Google Scholar]
- Jones PB, Adams KW, Rozkalne A, Spires-Jones TL, Hshieh TT, Hashimoto T, Armin C.A. von, Mielke M, Bacskai BJ, and Hyman BT (2011). Apolipoprotein E: Isoform specific differences in tertiary structure and interaction with amyloid-β in human Alzheimer brain. PLoS ONE 6, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanekiyo T, Cirrito JR, Liu C-C, Shinohara M, Li J, Schuler DR, Shinohara M, Holtzman DM, and Bu G (2013). Neuronal clearance of amyloid-β by endocytic receptor LRP1. The Journal of Neuroscience : The Official Journal of the Society for Neuroscience 33, 19276–19283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Castellano JM, Jiang H, Basak JM, Parsadanian M, Pham V, Mason SM, Paul SM, and Holtzman DM (2009). Overexpression of low-density lipoprotein receptor in the brain markedly inhibits amyloid deposition and increases extracellular A beta clearance. Neuron 64, 632–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Jiang H, Park S, Eltorai AEM, Stewart FR, Yoon H, Basak JM, Finn MB, and Holtzman DM (2011). Haploinsufficiency of human APOE reduces amyloid deposition in a mouse model of amyloid-β amyloidosis. The Journal of Neuroscience : The Official Journal of the Society for Neuroscience 31, 18007–18012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koldamova R, Staufenbiel M, and Lefterov I (2005). Lack of ABCA1 considerably decreases brain ApoE level and increases amyloid deposition in APP23 mice. The Journal of Biological Chemistry 280, 43224–43235. [DOI] [PubMed] [Google Scholar]
- Krasemann S, Madore C, Cialic R, Baufeld C, Calcagno N, El Fatimy R, Beckers L, O’Loughlin E, Xu Y, Fanek Z, et al. (2017). The TREM2-APOE Pathway Drives the Transcriptional Phenotype of Dysfunctional Microglia in Neurodegenerative Diseases. Immunity 47, 566–581.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimbou L, Denis M, Haidar B, Carrier M, Marcil M, and Genest J (2004). Molecular interactions between apoE and ABCA1: Impact on apoE lipidation. Journal of Lipid Research 45, 839–848. [DOI] [PubMed] [Google Scholar]
- Kuszczyk MA, Sanchez S, Pankiewicz J, Kim J, Duszczyk M, Guridi M, Asuni AA, Sullivan PM, Holtzman DM, and Sadowski MJ (2013). Blocking the interaction between apolipoprotein E and Aβ reduces intraneuronal accumulation of Aβ and inhibits synaptic degeneration. The American Journal of Pathology 182, 1750–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaDu MJ, Falduto MT, Manelli AM, Reardon CA, Getz GS, and Frail DE (1994). Isoform-specific binding of apolipoprotein E to β-amyloid. Journal of Biological Chemistry 269, 23403–23406. [PubMed] [Google Scholar]
- LaDu MJ, Pederson TM, Frail DE, Reardon CA, Getz GS, and Falduto MT (1995). Purification of apolipoprotein E attenuates isoform-specific binding to beta-amyloid. The Journal of Biological Chemistry 270, 9039–9042. [DOI] [PubMed] [Google Scholar]
- Lalazar A, Weisgraber KH, Rall SC, Giladi H, Innerarity TL, Levanon AZ, Boyles JK, Amit B, Gorecki M, Mahley RW, et al. (1988). Site-specific mutagenesis of human apolipoprotein E. Receptor binding activity of variants with single amino acid substitutions. Journal of Biological Chemistry 263, 3542–3545. [PubMed] [Google Scholar]
- Landschulz KT, Pathak RK, Rigotti A, Krieger M, and Hobbs HH (1996). Regulation of scavenger receptor, class B, type I, a high density lipoprotein receptor, in liver and steroidogenic tissues of the rat. Journal of Clinical Investigation 98, 984–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.-h., Tanimura M, Luo C.-c., Datta S, and Chant L (1988). The apo ipoprotein multigene fam ly : biosynthesis , structure , structure-function relationships , Journal of Lipid Research 29, 245–271. [PubMed] [Google Scholar]
- Liao F, Li A, Xiong M, Bien-Ly N, Jiang H, Zhang Y, Finn MB, Hoyle R, Keyser J, Lefton KB, et al. (2018). Targeting of nonlipidated, aggregated apoE with antibodies inhibits amyloid accumulation. Journal of Clinical Investigation 128, 2144–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C-C, Liu C-C, Kanekiyo T, Xu H, and Bu G (2013). Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nature Reviews. Neurology 9, 106–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C-C, Hu J, Zhao N, Wang J, Wang N, Cirrito JR, Kanekiyo T, Holtzman DM, and Bu G (2017). Astrocytic LRP1 Mediates Brain Aβ Clearance and Impacts Amyloid Deposition. The Journal of Neuroscience 37, 4023–4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly S, Altman R, Petrlova J, Lin Y, Hilt S, Huser T, Laurence TA, and Voss JC (2013). Binding of apolipoprotein E inhibits the oligomer growth of amyloid-β peptide in solution as determined by fluorescence cross-correlation spectroscopy. Journal of Biological Chemistry 288, 11628–11635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Yee A, Brewer HBJ, Das S, and Potter H (1994). Amyloid-associated proteins alpha 1-antichymotrypsin and apolipoprotein E promote assembly of Alzheimer beta-protein into filaments. Nature 372, 92–94. [DOI] [PubMed] [Google Scholar]
- Mahley RW (1988). Apolipoprotein E : Cholesterol Transport. Science 240, 622–630. [DOI] [PubMed] [Google Scholar]
- Mahley RW, Weisgraber KH, and Innerarity TL (1979). Interaction of plasma lipoproteins containing apolipoproteins B and E with heparin and cell surface receptors. Biochimica et Biophysica Acta (BBA)/Lipids and Lipid Metabolism 575, 81–91. [DOI] [PubMed] [Google Scholar]
- Mahley RW, Weisgraber KH, and Huang Y (2009). Apolipoprotein E: structure determines function, from atherosclerosis to Alzheimer’s disease to AIDS. Journal of Lipid Research 50, S183–S188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahley RW, and Huang Y (2012). Apolipoprotein e sets the stage: response to injury triggers neuropathology. Neuron 76, 871–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann WA, Gregg RE, Sprecher DL, and Brewer HB (1989). Apolipoprotein E-1Harrisburg: a new variant of apolipoprotein E dominantly associated with type III hyperlipoproteinemia. Biochimica et Biophysica Acta 1005, 239–244. [DOI] [PubMed] [Google Scholar]
- McPherson A (2020). Crystal structure of a proteolytically cleaved, amino terminal domain of apolipoprotein E3. Biochemical and Biophysical Research Communications 525, 57–60. [DOI] [PubMed] [Google Scholar]
- Morris JC, Roe CM, Xiong C, Fagan AM, Goate AM, Holtzman DM, and Mintun MA (2010). APOE predicts amyloid-beta but not tau Alzheimer pathology in cognitively normal aging. Annals of Neurology 67, 122–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow JA, Arnold KS, Dong J, Balestra ME, Innerarity TL, and Weisgraber KH (2000). Effect of arginine 172 on the binding of apolipoprotein E to the low density lipoprotein receptor. Journal of Biological Chemistry 275, 2576–2580. [DOI] [PubMed] [Google Scholar]
- Morrow JA, Hatters DM, Lu B, Hochtl P, Oberg KA, Rupp B, and Weisgraber KH (2002). Apolipoprotein E4 forms a molten globule. A potential basis for its association with disease. The Journal of Biological Chemistry 277, 50380–50385. [DOI] [PubMed] [Google Scholar]
- Näslund J, Thyberg J, Tjernberg LO, Wernstedt C, Karlström AR, Bogdanovic N, Gandy SE, Lannfelt L, Terenius L, and Nordstedt C (1995). Characterization of stable complexes involving apolipoprotein E and the amyloid beta peptide in Alzheimer’s disease brain. Neuron 15, 219–228. [DOI] [PubMed] [Google Scholar]
- Newman TC, Dawson PA, Rudel LL, and Williams DL (1985). Quantitation of apolipoprotein E mRNA in the liver and peripheral tissues of nonhuman primates. Journal of Biological Chemistry 260, 2452–2457. [PubMed] [Google Scholar]
- Parhizkar S, Arzberger T, Brendel M, Kleinberger G, Deussing M, Focke C, Nuscher B, Xiong M, Ghasemigharagoz A, Katzmarski N, et al. (2019). Loss of TREM2 function increases amyloid seeding but reduces plaque-associated ApoE. Nature Neuroscience 22, 191–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters-Libeu CA, Newhouse Y, Hatters DM, and Weisgraber KH (2006). Model of biologically active apolipoprotein E bound to dipalmitoylphosphatidylcholine. Journal of Biological Chemistry 281, 1073–1079. [DOI] [PubMed] [Google Scholar]
- Pitas RE, Boyles JK, Lee SH, Foss D, and Mahley RW (1987). Astrocytes synthesize apolipoprotein E and metabolize apolipoprotein E-containing lipoproteins. Biochimica et Biophysica Acta 917, 148–161. [DOI] [PubMed] [Google Scholar]
- Rall SC, Weisgraber KH, Innerarity TL, and Mahley RW (1983). Identical structural and receptor binding defects in apolipoprotein E2 in hypo-, normo-, and hypercholesterolemic dysbetalipoproteinemia. Journal of Clinical Investigation 71, 1023–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raulin AC, Kraft L, Al-Hilaly YK, Xue WF, McGeehan JE, Atack JR, and Serpell L (2019). The Molecular Basis for Apolipoprotein E4 as the Major Risk Factor for Late-Onset Alzheimer’s Disease. Journal of Molecular Biology 431, 2248–2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raussens V, Fisher CA, Goormaghtigh E, Ryan RO, and Ruysschaert JM (1998). The low density lipoprotein receptor active conformation of apolipoprotein E: Helix organization in N-terminal domain-phospholipid disc particles. Journal of Biological Chemistry 273, 25825–25830. [DOI] [PubMed] [Google Scholar]
- Ray A, Ahalawat N, and Mondal J (2017). Atomistic Insights into Structural Differences between E3 and E4 Isoforms of Apolipoprotein E. Biophysical Journal 113, 2682–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiman EM, Chen K, Liu X, Bandy D, Yu M, Lee W, Ayutyanont N, Keppler J, Reeder SA, Langbaum JBS, et al. (2009). Fibrillar amyloid-beta burden in cognitively normal people at 3 levels of genetic risk for Alzheimer’s disease. Proceedings of the National Academy of Sciences of the United States of America 106, 6820–6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H, Dhanasekaran P, Nguyen D, Holvoet P, Lund-Katz S, and Phillips MC (2003). Domain structure and lipid interaction in human apolipoproteins A-I and E, a general model. The Journal of Biological Chemistry 278, 23227–23232. [DOI] [PubMed] [Google Scholar]
- Saito M, Eto M, Nitta H, Kanda Y, Shigeto M, Nakayama K, Tawaramoto K, Kawasaki F, Kamei S, Kohara K, et al. (2004). “Effect of apoüpoprotein E4 alíele on plasma LDLcholesterol response to diet therapy in type 2 diabetic patients”. Apunts. Medicina de L’Esport 39, 41–42. [DOI] [PubMed] [Google Scholar]
- Sanan DA, Weisgraber KH, Russell SJ, Mahley RW, Huang D, Saunders A, Schmechel D, Wisniewski T, Frangione B, Roses AD, et al. (1994). Apolipoprotein E associates with β amyloid peptide of Alzheimer’s disease to form novel monofibrils. Isoform ApoE4 associates more efficiently than ApoE3. Journal of Clinical Investigation 94, 860–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segrest JP, Jones MK, De Loof H, Brouillette CG, Venkatachalapathi YV, and Anantharamaiah GM (1992). The amphipathic helix in the exchangeable apolipoproteins: a review of secondary structure and function. Journal of Lipid Research 33, 141–166. [PubMed] [Google Scholar]
- Shaw RA, Buchko GW, Wang G, Rozek A, Treleaven WD, Mantsch HH, and Cushley RJ (1997). Infrared spectroscopy of human apolipoprotein fragments in SDS/D2O: Relative lipid-binding affinities and a novel amide I assignment. Biochemistry 36, 14531–14538. [DOI] [PubMed] [Google Scholar]
- Shi Y, Yamada K, Liddelow SA, Smith ST, Zhao L, Luo W, Tsai RM, Spina S, Grinberg LT, Rojas JC, et al. (2017). ApoE4 markedly exacerbates tau-mediated neurodegeneration in a mouse model of tauopathy. Nature 549, 523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Manis M, Long J, Wang K, Sullivan PM, Remolina Serrano J, Hoyle R, and Holtzman DM (2019). Microglia drive APOE-dependent neurodegeneration in a tauopathy mouse model. The Journal of Experimental Medicine 216, 2546–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore VG, and Shore B (1973). Heterogeneity of human plasma very low density lipoproteins. Separation of species differing in protein components. Biochemistry 12, 502–507. [DOI] [PubMed] [Google Scholar]
- Song Y, Stampfer MJ, and Liu S (2004). Meta-analysis: Apolipoprotein E genotypes and risk for coronary heart disease. Annals of Internal Medicine 141, 137–147. [DOI] [PubMed] [Google Scholar]
- Sparrow JT, Sparrow DA, Fernando G, Culwell AR, Kovar M, and Gotto AM (1992). Apolipoprotein E: Phospholipid Binding Studies with Synthetic Peptides from the Carboxyl Terminus. Biochemistry 31, 1065–1068. [DOI] [PubMed] [Google Scholar]
- Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, and Roses AD (1993a). Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proceedings of the National Academy of Sciences of the United States of America 90, 1977–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strittmatter WJ, Weisgraber KH, Huang DY, Dong LM, Salvesen GS, Pericak-Vance M, Schmechel D, Saunders AM, Goldgaber D, and Roses AD (1993b). Binding of human apolipoprotein E to synthetic amyloid beta peptide: isoform-specific effects and implications for late-onset Alzheimer disease. Proceedings of the National Academy of Sciences of the United States of America 90, 8098–8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Holm M-L, Liu C-C, Shinohara M, Aikawa T, Oue H, Yamazaki Y, Martens YA, Murray ME, Sullivan PM, et al. (2019). APOE4-mediated amyloid-β pathology depends on its neuronal receptor LRP1. The Journal of Clinical Investigation 129, 1272–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai LM, Bilousova T, Jungbauer L, Roeske SK, Youmans KL, Yu C, Poon WW, Cornwell LB, Miller CA, Vinters HV, et al. (2013). Levels of soluble apolipoprotein E/amyloid-β (Aβ) complex are reduced and oligomeric Aβ increased with APOE4 and Alzheimer disease in a transgenic mouse model and human samples. The Journal of Biological Chemistry 288, 5914–5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuang D, Leverenz JB, Lopez OL, Hamilton RL, Bennett DA, Schneider JA, Buchman AS, Larson EB, Crane PK, Kaye JA, et al. (2013). APOE ϵ4 increases risk for dementia in pure synucleinopathies. JAMA Neurology 70, 223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich JD, Ulland TK, Mahan TE, Nyström S, Nilsson KP, Song WM, Zhou Y, Reinartz M, Choi S, Jiang H, et al. (2018). ApoE facilitates the microglial response to amyloid plaque pathology. The Journal of Experimental Medicine 215, 1047–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utermann G (1975). Isolation and Partial Characterization of an Arginine-Rich Apolipoprotein from Human Plasma Very-Low-Density Lipoproteins: Apolipoprotein E. Hoppe-Seyler′s Zeitschrift Für Physiologische Chemie 356, 1113–1122. [DOI] [PubMed] [Google Scholar]
- Verghese PB, Castellano JM, Garai K, Wang Y, Jiang H, Shah A, Bu G, Frieden C, and Holtzman DM (2013). ApoE influences amyloid-β (Aβ) clearance despite minimal apoE/Aβ association in physiological conditions. Proceedings of the National Academy of Sciences of the United States of America 110, E1807–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahrle SE, Jiang H, Parsadanian M, Legleiter J, Han X, Fryer JD, Kowalewski T, and Holtzman DM (2004). ABCA1 is required for normal central nervous system ApoE levels and for lipidation of astrocyte-secreted apoE. The Journal of Biological Chemistry 279, 40987–40993. [DOI] [PubMed] [Google Scholar]
- Wahrle SE, Jiang H, Parsadanian M, Hartman RE, Bales KR, Paul SM, and Holtzman DM (2005). Deletion of Abca1 increases Abeta deposition in the PDAPP transgenic mouse model of Alzheimer disease. The Journal of Biological Chemistry 280, 43236–43242. [DOI] [PubMed] [Google Scholar]
- Wahrle SE, Jiang H, Parsadanian M, Kim J, Li A, Knoten A, Jain S, Hirsch-Reinshagen V, Wellington CL, Bales KR, et al. (2008). Overexpression of ABCA1 reduces amyloid deposition in the PDAPP mouse model of Alzheimer disease. The Journal of Clinical Investigation 118, 671–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Najm R, Xu Q, Jeong DE, Walker D, Balestra ME, Yoon SY, Yuan H, Li G, Miller ZA, et al. (2018). Gain of toxic apolipoprotein E4 effects in human iPSC-derived neurons is ameliorated by a small-molecule structure corrector article. Nature Medicine 24, 647–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Treleaven WD, and Cushley RJ (1996a). Conformation of human serum apolipoprotein A-I(166-185) in the presence of sodium dodecyl sulfate or dodecylphosphocholine by 1H-NMR and CD. Evidence for specific peptide-SDS interactions. Biochimica et Biophysica Acta - Lipids and Lipid Metabolism 1301, 174–184. [DOI] [PubMed] [Google Scholar]
- Wang G, Pierens GK, Treleaven WD, Sparrow JT, and Cushley RJ (1996). Conformations of Human Apolipoprotein E(263–286) and E(267–289) in Aqueous Solutions of Sodium Dodecyl Sulfate by CD and 1H NMR. Biochemistry 35, 10358–10366. [DOI] [PubMed] [Google Scholar]
- Wardell MR, Suckling PA, and Janus ED (1982). Genetic variation in human apolipoprotein E. Journal of Lipid Research 23, 1174–1182. [PubMed] [Google Scholar]
- Weisgraber KH (1990). Apolipoprotein E distribution among human plasma lipoproteins: Role of the cysteine-arginine interchange at residue 112. Journal of Lipid Research 31, 1503–1511. [PubMed] [Google Scholar]
- Weisgraber KH (1994). Apolipoprotein E: Structure-Function Relationships. In Advances in Protein Chemistry, pp. 249–302. [DOI] [PubMed] [Google Scholar]
- Weisgraber KH, Innerarity TL, and Mahley RW (1982). Abnormal lipoprotein receptor-binding activity of the human E apoprotein due to cysteine-arginine interchange at a single site. Journal of Biological Chemistry 257, 2518–2521. [PubMed] [Google Scholar]
- Weisgraber KH, Rall SCJ, Bersot TP, Mahley RW, Franceschini G, and Sirtori CR (1983). Apolipoprotein A-IMilano. Detection of normal A-I in affected subjects and evidence for a cysteine for arginine substitution in the variant A-I. The Journal of Biological Chemistry 258, 2508–2513. [PubMed] [Google Scholar]
- Wernette-Hammond ME, Lauer SJ, Corsini A, Walker D, Taylor JM, and Rall SC (1989). Glycosylation of human apolipoprotein E. The carbohydrate attachment site is threonine 194. Journal of Biological Chemistry 264, 9094–9101. [PubMed] [Google Scholar]
- Williams B2, Convertino M, Das J, and Dokholyan NV (2015). ApoE4-specific Misfolded Intermediate Identified by Molecular Dynamics Simulations. PLoS Computational Biology 11, e1004359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DL, Dawson PA, Newman TC, and Rudel LL (1985). Apolipoprotein E synthesis in peripheral tissues of nonhuman primates. Journal of Biological Chemistry 260, 2444–2451. [PubMed] [Google Scholar]
- Wilson C, Wardell M, Weisgraber K, Mahley R, and Agard D (1991). Three-dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science 252, 1817–1822. [DOI] [PubMed] [Google Scholar]
- Wilson C, Mau T, Weisgraber KH, Wardell MR, Mahley RW, and Agard DA (1994). Salt bridge relay triggers defective LDL receptor binding by a mutant apolipoprotein. Structure 2, 713–718. [DOI] [PubMed] [Google Scholar]
- Winkler K, Scharnagl H, Tisljar U, Hoschützky H, Friedrich I, Hoffmann MM, Hüttinger M, Wieland H, and März W (1999). Competition of Abeta amyloid peptide and apolipoprotein E for receptor-mediated endocytosis. Journal of Lipid Research 40, 447–455. [PubMed] [Google Scholar]
- Wisniewski T, and Frangione B (1992). Apolipoprotein E: A pathological chaperone protein in patients with cerebral and systemic amyloid. Neuroscience Letters 135, 235–238. [DOI] [PubMed] [Google Scholar]
- Wisniewski T, Golabek A, Matsubara E, Ghiso J, and Frangione B (1993). Apolipoprotein E: Binding to Soluble Alzheimer′s β-Amyloid. Biochemical and Biophysical Research Communications 192, 359–365. [DOI] [PubMed] [Google Scholar]
- Xu Q, Brecht WJ, Weisgraber KH, Mahley RW, and Huang Y (2004). Apolipoprotein E4 domain interaction occurs in living neuronal cells as determined by fluorescence resonance energy transfer. The Journal of Biological Chemistry 279, 25511–25516. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Kiyota T, Horiba M, Buescher JL, Walsh SM, Gendelman HE, and Ikezu T (2007). Interferon-γ and tumor necrosis factor-α regulate amyloid-β plaque deposition and β-secretase expression in Swedish mutant APP transgenic mice. American Journal of Pathology 170, 680–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Choi HW, and Ryan RO (2008). Apolipoprotein E isoform-specific binding to the low-density lipoprotein receptor. Analytical Biochemistry 372, 222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y, Deguchi N, Takagi C, Tanaka M, Dhanasekaran P, Nakano M, Handa T, Phillips MC, Lund-Katz S, and Saito H (2008). Role of the N- and C-terminal domains in binding of apolipoprotein E isoforms to heparan sulfate and dermatan sulfate: a surface plasmon resonance study. Biochemistry 47, 6702–6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youmans KL, Tai LM, Nwabuisi-Heath E, Jungbauer L, Kanekiyo T, Gan M, Kim J, Eimer WA, Estus S, Rebeck GW, et al. (2012). APOE4-specific changes in Aβ accumulation in a new transgenic mouse model of Alzheimer disease. The Journal of Biological Chemistry 287, 41774–41786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Song J, Newhouse Y, Zhang S, Weisgraber KH, and Ren G (2010). An optimized negative-staining protocol of electron microscopy for apoE4•POPC lipoprotein. Journal of Lipid Research 51, 1228–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Tong H, Garewal M, and Ren G (2013). Optimized negative-staining electron microscopy for lipoprotein studies. Biochimica et Biophysica Acta - General Subjects 1830, 2150–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao N, Liu C-C, Van Ingelgom AJ, Linares C, Kurti A, Knight JA, Heckman MG, Diehl NN, Shinohara M, Martens YA, et al. (2018). APOE ϵ2 is associated with increased tau pathology in primary tauopathy. Nature Communications 9, 4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao N, Attrebi ON, Ren Y, Qiao W, Sonustun B, Martens YA, Meneses AD, Li F, Shue F, Zheng J, et al. (2020). APOE4 exacerbates α-synuclein pathology and related toxicity independent of amyloid. Science Translational Medicine 12, eaay1809. [DOI] [PMC free article] [PubMed] [Google Scholar]