Abstract
TP53 is the most frequently mutated gene in breast cancer, but its role in survival is confounded by different studies concluding that TP53 mutations are associated with negative, neutral, or positive outcomes. Closer examination showed that many studies were limited by factors such as imprecise methods to detect TP53 mutations and small cohorts that combined patients treated with drugs having very different mechanisms of action. When only studies of patients receiving the same treatment(s) were compared, they tended to agree. These analyses reveal a role for TP53 in response to different treatments as complex as its different biological activities. We discuss studies that have assessed the role of TP53 mutations in breast cancer treatment and limitations in interpreting reported results.
TP53 Mutation Status and Clinical Outcomes in Breast Cancer
TP53 is the most frequently mutated gene in cancer [1]. TP53 codes for the transcription factor p53, which initiates transcription of genes involved in processes such as cell cycle arrest, apoptosis, metabolism, DNA repair, and cellular senescence in response to stressors like radiation and chemotherapy [2,3]. TP53 mutations occur predominantly in the DNA binding domain, thus rendering the mutant p53 protein transcriptionally inactive in response to stress [4]. While the degree of p53 induction and the specific targets transactivated by p53 vary by tumor and cell type, wild type p53 activity is generally thought to confer a favorable prognosis in cancer [2,5].
In breast cancer, TP53 is mutated in nearly 30% of all cases and is the most frequently mutated gene [1]. As every molecular subtype of breast cancer contains tumors with mutant TP53 [1], many preclinical and clinical studies have attempted to discern the role of p53 in breast cancer. However, the clinical relevance of TP53 mutation status in breast cancers has been confounded by different studies showing that TP53 mutations are associated with negative, neutral, or positive outcomes (e.g., [6–9]).
In this review, we examine these past studies and why they differed so markedly in their conclusions, revealing that many have been limited by various factors. These include using imprecise methods to determine tumor TP53 status and using different clinical endpoints to assess predictive or prognostic significance of TP53 mutations. Perhaps the most confounding factor, however, has been the use of relatively small sample sizes of patients that fail to properly account for heterogeneity in subtype and, importantly, treatment regimens.
Treatment Strategies for Management of Breast Cancer
Breast cancer is a heterogeneous disease with many different treatment options that can include combinations of surgery, chemotherapies, hormonal therapies (herein termed ‘HRT’), therapies targeting human epidermal growth factor receptor 2 (HER2), and no treatment [10,11]. Different options exist within each of these categories as well. The primary determinant of optimal treatment is the presence or absence of molecular markers for steroid hormone receptors (HR) [estrogen receptor (ER) or progesterone receptor], and HER2 [11].
Chemotherapies routinely used in clinic for breast cancer include DNA-damaging anthracycline- and nonanthracycline-based drugs. Anthracycline-based therapies, such as doxorubicin and epirubicin, exert cytotoxic effects by intercalating into the DNA-topoisomerase II complex and interfering with DNA synthesis [12,13]. Nonanthracycline-based DNA-damaging therapies are broader in mechanism of action and include DNA-alkylating agents like cyclophosphamide [14] and antimetabolites like 5-fluorouracil that interfere with DNA synthesis [15]. Similarly, radiation treatment causes DNA damage by generating strand breaks [16]. Breast cancer is also treated with non-DNA damaging taxanes, which interfere with mitotic cell division through disruption of microtubule function [17]. p53 activity is critical in determining the fate of cells with stress caused by DNA damage [2,18] or mitotic failure [19].
Chemotherapy can be administered either as a neoadjuvant or adjuvant treatment [20]. Adjuvant treatments are therapies delivered after the primary treatment (i.e., surgical resection of tumor). Neoadjuvant treatments are therapies delivered prior to the primary treatment. This allows pathologists to determine how effective the treatment was in shrinking the tumor by evaluating the presence or absence of residual disease, according to the following criteria: (i) complete response (CR), (ii) partial response, (iii) stable disease, and (iv) progressive disease (PD) [21].
HR-positive breast cancers are routinely treated with HRTs that interfere with HR-signaling pathways that drive these cancers. HRTs are comprised of several classes of drugs, including selective ER modulators, selective ER degraders, and aromatase inhibitors (reviewed in [22–25]).
In addition to biological factors, breast cancer treatment can vary from patient to patient due to patient and physician preferences, institutional guidelines, access to healthcare, insurance coverage [11], and reimbursement incentives [26–28]. Simply put, breast cancer is a complex disease that is managed using a variety of treatment strategies, depending on a multitude of biological and nonbiological factors.
Methods for Detection of TP53 Mutation Status in Breast Cancer
Because of the clearly important role for p53 in cancer development and progression, including in breast cancer, scientists and clinicians have examined how TP53 mutation correlates with clinical outcomes. These studies have used several techniques to determine TP53 mutation status, including DNA sequencing, immunohistochemical (IHC) staining, and yeast-based functional assays.
IHC Staining
Many earlier studies utilized positive IHC staining as a surrogate marker for p53 mutation. Missense mutations in the TP53 DNA-binding domain, which account for the majority of mutations in the gene, often cause hyperstabilization of the p53 protein that can be detected by strong nuclear IHC staining [29]. However, some point mutations, splice variants, frameshift mutations, and all truncating alterations in TP53 will fail to result in a stable protein; thus, using IHC to determine p53 status in these cases will incorrectly designate these tumors TP53 wild type [29,30]. This is significant because recent studies using more sensitive methods of detection show ~40% of TP53 mutations in breast cancer will not produce a stable protein [31]. Moreover, wild type p53 protein can stain positively by IHC in tumor samples [7,32]. In fact, it is estimated that correlation between p53 accumulation measured by IHC and TP53 mutations determined through DNA sequencing is less than 75% in breast cancer [33]. Thus, due to these limitations of the technique, some patients who are inferred to have TP53 wild type tumors based on IHC may in fact have TP53 mutant tumors and some designated as mutant because of positive IHC staining may be wild type. This inaccuracy likely confounds survival analysis on mutation status determined by IHC [7].
Yeast-Based Functional Assay
TP53 mutation status can also be determined through a yeast-based functional assay. In this assay, RNA is extracted from patient tumor biopsies and reverse-transcribed into cDNA. TP53 transcripts are then amplified by PCR and transfected into yeast with an integrated p53 transcription reporter plasmid. Yeast colonies that are transfected with wild type p53 will form white colonies and colonies transfected with mutant and thus transcriptionally inactive p53 will form red colonies [34].
DNA-Sequencing
TP53 mutations are also detected in breast cancer by DNA sequencing [35]. Early studies showed that the majority of mutations in TP53 occur in exons 5–8 of the gene [36]. In turn, this is the region that has been most frequently, and often exclusively, sequenced [37]. Studies that only assess mutations in this region will designate patients with TP53 mutation in other exons as wild type. Next generation sequencing methods have allowed for detection of TP53 mutations outside of the exon 5–8 region [35]. DNA sequencing is regarded as the gold standard for identifying TP53 mutations [38].
TP53 Mutation Status and Survival of Breast Cancer Patients: Studies That Combine all Treatments and Subtypes
When TP53 status is taken into account in large heterogeneous populations that combine all treatments and subtypes, many studies demonstrate that patients with TP53 mutant tumors have worse survival than patients with TP53 wild type tumors.
One of the earliest studies of TP53 mutation status and breast cancer survival was performed by Ostrowski et al. [39]. They analyzed the correlation of p53 IHC expression and survival of 90 breast cancer patients. They determined that there was no significant difference in survival between patients with p53 positive and negative tumors. In another study of p53 expression and survival in 31 breast cancer patients, p53 positivity by IHC was associated with worse overall survival (OS) (P <0.01) [40]. However, the number of patients with p53 positive tumors was relatively small (n = 5) and patient treatment exposures were not reported. In a later study of 73 patients with invasive ductal carcinoma, p53 overexpression by IHC was associated with worse postrelapse survival (P <0.0001) [41]. In this study, while all patients received HRT, some patients received chemotherapy while others did not. Also, survival analysis for clinical endpoints like OS and recurrence-free survival (RFS) were not reported. In all of these studies, TP53 status was inferred based on IHC expression of p53, which is now regarded as an inaccurate method of mutation detection (discussed above).
Studies that employed more accurate sequencing methodology have generally agreed with studies using IHC. Bergh et al. found that patients with TP53 mutant tumors had worse OS (P = 0.001) and RFS (P = 0.002) than patients with wild type tumors [42]. This study included patients that were treated with very different therapies, including radiotherapy, HRT, and/or different chemotherapies. Shiao et al. assessed the role of TP53 mutation status in breast cancer survival in black and white patients [43]. While TP53 mutations were associated with worse survival in black patients (n = 45, P = 0.012), there was no survival difference between white patients with or without mutations in TP53 (n = 47, P = 0.670). These patients were treated with different therapies, including chemotherapy, radiation therapy, and HRT. In a later study of 90 patients with breast cancer, Blaszyk et al. determined that patients with TP53 mutant tumors had worse OS (P = 0.0001) and disease-free survival (DFS) (P = 0.003) compared with patients that have TP53 wild type tumors [44]. These patients were treated with a variety of therapies, including systemic chemotherapy for positive lymph nodes and/or HRT if tumors were positive for ER. Andersson et al. determined that in a cohort of 396 breast cancer patients, those with TP53 mutant tumors had worse OS (P = 0.0005) compared with patients that have wild type tumors [45].
In this study, however, treatments included different combinations of therapies like HRT and chemotherapy. Rossner, Jr. et al. found in a study of 859 patients with breast cancer that TP53 mutations were associated with poor breast cancer-specific and all-cause mortality [46]. While this study included a large sample size of patients, it was noted that follow-up information like treatment history was missing for many of the patients. Olivier et al. noted in a study of 1794 breast cancer patients that those with tumors harboring TP53 mutations in exons 5–8 of the gene had a worse risk of dying of breast cancer within 10 years following surgery (P <0.0001) [47]. However, treatment history for patients included in this study was not provided. Meric-Bernstam et al. reported that amongst 165 patients with HR+ metastatic breast cancer, patients with TP53 mutant tumors had worse OS (P = 0.003) [48]. However, some of these patients were treated with chemotherapy, while others were not.
While these studies suggest TP53 mutations are correlated with worse survival in breast cancer, as noted, many report differences in key elements such as patient treatment stratifications, clinical endpoints, and TP53 mutation detection methods. Specifically, many of these studies used IHC to infer p53 status, performed analysis on heterogeneously treated patients, and were powered by small sample sizes. In turn, it is difficult to compare these studies to clarify the role of TP53 mutations in breast cancer survival.
The METABRIC Dataset Allows Stratification of Different Treatments
As a result of the complexity of breast cancer treatment, cross-comparisons and meta-analyses of studies on the role of TP53 mutations in breast cancer have been exceedingly difficult. Individual studies, however, have not generally been powered by a sufficient sample size to comprehensively analyze TP53 mutations and survival across different clinical and treatment groups. To resolve the deficiencies found in previous reports, our group performed an analysis [49] of the large METABRIC dataseti of primary breast cancer patients [50].
This study was powered by a large sample size of patients (n = 1979), treatment histories were properly annotated, and all patient tumors were sequenced, allowing for reliable stratification of patients based on TP53 mutation status [50,51]. When OS was compared between patients stratified by TP53 mutation status across multiple clinical features (all patients, regardless of treatment) patients with TP53 mutant tumors had shorter median OS compared with patients that have TP53 wild type tumors (P <0.0001) [49] (Figure 1A).
Figure 1. Breast Cancer Patient Survival after Stratification for TP53 Status and Treatment.
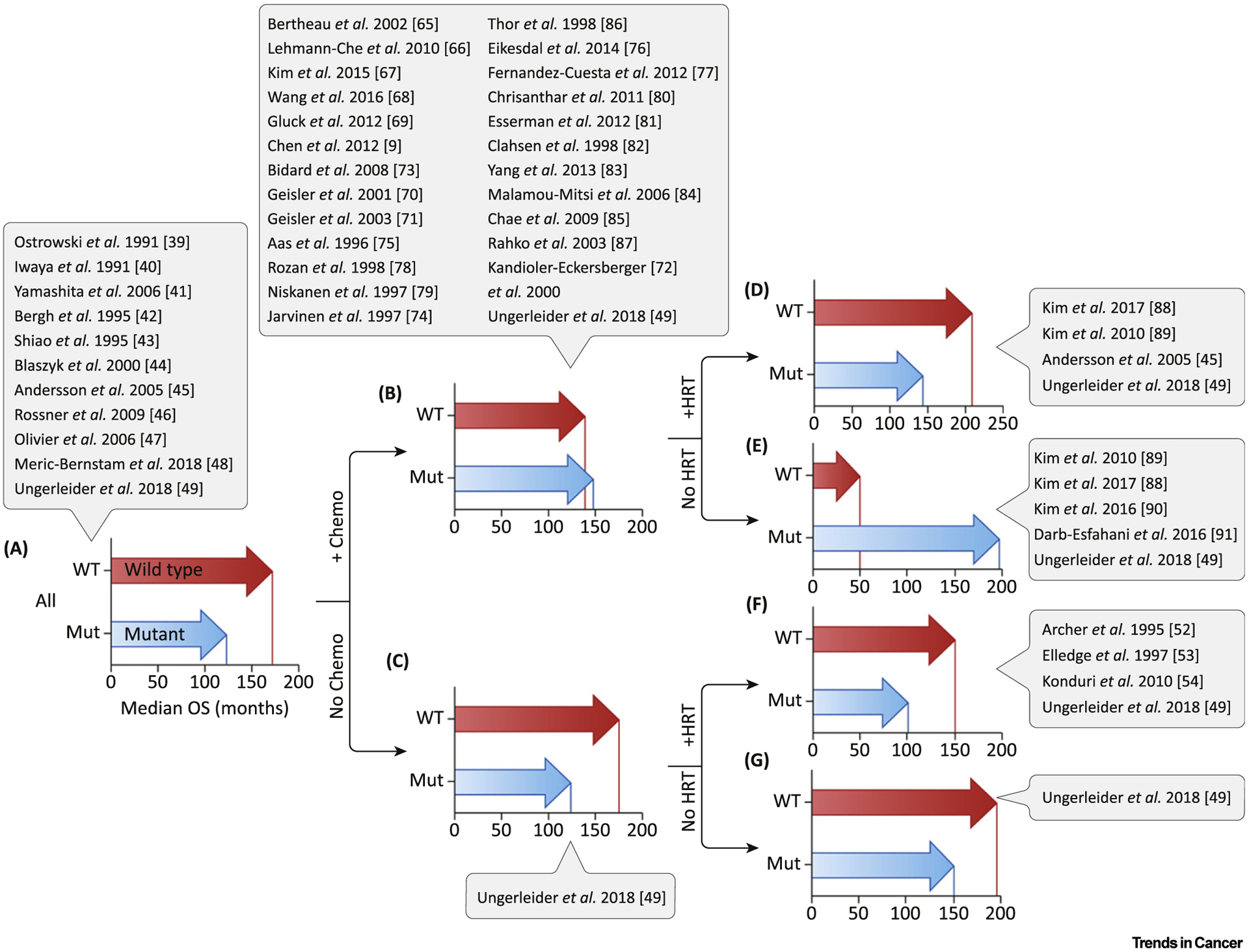
Median survival was determined for patients from the METABRIC database, stratified by TP53 mutation status and treatment exposure. Studies that analyzed patients under similar treatment conditions are listed in adjacent captions. (A) All patients (P <0.0001). (B) All chemotherapy-treated patients (P = ns). (C) All patients who did not receive chemotherapy (P <0.0001). (D) All patients treated with chemotherapy + HRT (P <0.05). (E) All patients treated with chemotherapy alone (P <0.05). (F) All patients treated with HRT only (P <0.0001). (G) All patients who did not receive chemotherapy or HRT (P = ns). Abbreviations: HRT, hormonal therapies; mut, mutant; ns, not significant; OS, overall survival; WT, wild type. (See [9,39–49,52–54,65–91].)
Stratifying by Different Treatment Types: TP53 Status and HRT Response in Breast Cancer
TP53 is mutated in approximately 20% of HR-positive breast cancers [49]. Because of this prevalence, studies have aimed to determine the role of TP53 mutation status in response to HRT. For example, Archer et al. found that in 92 patients treated with first-line HRT, those patients with p53-positive and negative tumors by IHC had no difference in survival (P = 0.71) [52]. Elledge et al. found in a study of 205 patients with ER-positive, metastatic breast cancer that patients with higher p53 IHC expression had worse survival after HRT (P = 0.008) [53]. Konduri et al. determined that amongst 35 ER-positive breast cancer patients treated with tamoxifen, patients with p53-positive tumors by IHC had superior survival compared with patients that have p53-negative tumors (P = 0.007) [54]. In all of these studies, however, it is unclear whether patients received treatments in addition to HRT. Also, p53 status was determined via IHC, which may have incorrectly designated some TP53 mutation status (discussed above).
Studies on the role of TP53 mutation and response to HRT are limited and those identified just above may include some patients treated with therapies in addition to HRT. Our analysis of METABRIC for survival in patients that received only HRT, with no confounding additional therapies, demonstrated that patients with TP53 mutant tumors had worse OS compared with patients that have wild type tumors (P <0.0001) (Figure 1F).
TP53 Status and Chemotherapy Response in Breast Cancer
Chemotherapy is frequently used in the management of breast cancer. In fact, patients with triple-negative breast cancers (TNBCs) lack targeted therapy options and are primarily treated with a standard chemotherapy regimen.
p53 and the Apoptotic Response to Cell Stress
Chemotherapy and radiation treatments inflict DNA damage on tumor cells. One of the first functions identified for p53 was the induction of apoptosis after DNA damage. In classic studies, mice that lack p53 failed to undergo apoptosis in radiosensitive organs after exposure to irradiation [55]. Subsequent studies using mouse fibrosarcoma models showed p53 activation of apoptosis was critical for the response to DNA damaging chemotherapy [56]. p53 is also activated in response to mitotic stress caused by agents such as taxanes that disrupt microtubules [57].
Subsequent research demonstrated that p53 is a potent transcription factor [58] that, in response to stress, turns on genes such as PUMA, NOXA, PERP, and BAX that directly mediate apoptosis [59]. Because Lowe et al. [56] and other studies demonstrated that p53 triggers apoptosis when a cell incurs significant DNA damage, it was often presumed that functional p53 activity is necessary for a robust response to DNA-damaging chemotherapy [60]. However, p53 also transactivates the CDKN1A gene, coding for the potent cell cycle inhibitor p21 [61–63] and thus can halt the cell cycle and, in some contexts, activate a program of cellular senescence instead of apoptosis [64]. Because of the critical role p53 plays in cellular responses to chemotherapy, many groups have expanded p53 studies into other tumor types, including breast cancer. Interestingly, TP53 is mutant in 30% of all breast cancers and nearly 80% of TNBCs [1]. Thus, many breast cancer patients who will receive first-line chemotherapy treatment have tumors harboring TP53 mutations and many studies have attempted to clarify the role of these mutations in chemotherapy response, as research has seemingly produced mixed results.
TP53 Mutated Tumors Are More Likely to Undergo CR after Chemotherapy
Two frequently used endpoints for evaluating chemotherapy efficacy are: (i) the extent of residual disease present at the time of surgery following a chemotherapy regimen; (ii) survival (overall or relapse/disease-free). Contrary to a role for wild type p53 in activating apoptotic cell death that results in tumor regression after chemotherapy, numerous studies have now shown that TP53 mutant breast cancers have significantly higher rates of pathologic complete response (pCR) compared with wild type breast cancers.
Perhaps the first report to suggest TP53 mutation might actually be beneficial to chemotherapy response in breast cancer came from Bertheau et al. [65]. The authors showed that out of 50 non-inflammatory locally advanced breast cancers treated with high doses of epirubicin and cyclophosphamide, all of the pCRs occurred in TP53-mutated tumors (TP53 wild type: 0/36, TP53 mutant: 8/14; P <0.0001). A follow-up study by the same group showed that in a cohort of similarly treated breast cancer patients, 36.5% (23/63) of patients with TP53 mutated tumors underwent pCR compared with 0% (0/65) of patients that have wild type tumors [66]. Both studies utilized a sensitive yeast-based functional assay to determine mutation status of TP53. Numerous other studies and meta-analyses have confirmed these initial findings.
In a study of 198 TNBC patients treated with neoadjuvant chemotherapy (NAC), p53 expression detected by IHC was predictive of pCR after NAC [67]. Wang et al. found in a study of primary breast cancer patients treated with NAC that patients with TP53 mutant tumors had higher rates of pCR compared with patients that have wild type tumors [68]. Gluck et al. similarly determined that patients with TP53 mutated breast cancers had a superior response rate to NAC compared with wild type patients (TP53 mutant: 30%, TP53 wild type: 10%; P = 0.0032) [69].
Geisler et al. suggest that certain TP53 mutations can portend poor response to chemotherapy [70]. In their study of 91 breast cancer patients treated with doxorubicin, only patients with tumors that were mutated for the TP53 L2 or L3 loop domains responded poorly to treatment (P = 0.063 for all TP53 mutations and P = 0.008 for L2/L3 mutations). Notably, 12 of the patients included in this study had metastases at diagnosis. In a later study of 35 patients treated with 5-fluorouracil/mitomycin, Geisler et al. similarly reported that patients with TP53 L2/L3 mutated tumors had worse response to treatment (P = 0.177 for all mutations and P = 0.006 for L2/L3 mutations) [71].
One study found that TP53 gene mutations were associated with poor response to treatment with a neoadjuvant regimen of 5-fluorouracil/epirubicin/cyclophosphamide (P = 0.0029) [72]. However, there were only seven patients with TP53 mutated tumors treated with this regimen, as compared with 28 patients that have TP53 wild type tumors.
Two studies have reported no role for TP53 mutation and response; however, these utilized IHC as the method of mutation detection and/or had very small sample sizes [73,74].
While there is some conflict in the conclusions of these studies, in a meta-analysis of 3476 breast cancer cases across 26 different studies, Chen et al. reported that mutant TP53 status (assigned using IHC and/or sequencing) was associated with increased rates of pCR following NAC treatment [9].
As multiple studies and a meta-analysis have shown the superior rates of pCR for TP53 mutant tumors in response to chemotherapy, one might surmise that these patients would have superior survival. However, studies have conflicted and only recently have explanations emerged.
TP53 Mutation Status and Survival Studies That Combine Patients That Received Chemotherapy and Chemotherapy plus HRT
A number of studies suggest that, despite having significantly better rates of pCR, TP53 mutant breast cancers have worse survival after chemotherapy compared with their wild type counterparts. However, conclusions about p53 and chemotherapy response reached by these reports are confounded for multiple reasons, making comparisons difficult.
The overwhelming majority of reports of TP53 status and survival after chemotherapy include patients treated with therapies in addition to chemotherapy. For example, in a study of 63 patients with locally advanced breast cancer, four out of the six patients with PD following anthracycline-based (doxorubicin) treatment had TP53 mutant tumors [75]. However, 59 out of 63 of the total patients included in this study were determined to be HR-positive by IHC and were thus likely treated with HRT in addition to chemotherapy. In a second publication based on an extended survival study of these patients treated with doxorubicin, patients with TP53 mutant tumors experienced significantly reduced RFS and OS compared with patients that have TP53 wild type tumors (RFS: 14 vs 83 months, P <0.001; OS: 35 versus 90 months, P <0.001) [76]. Similar to their earlier study, this study included primarily HR-positive patients who were stated to have received adjuvant HRT in addition to chemotherapy. Breast cancer patients with TP53 mutant tumors also had worse survival after a regimen of 5-fluorouracil/mitomycin compared with patients that have wild type tumors (OS: 22 vs 80 months, P = 0.03) [76]. Likewise, the patients included in this study were primarily HR-positive patients who were treated with adjuvant HRT.
Fernandez-Cuesta et al. reported that in node-positive breast cancer patients treated with adjuvant doxorubicin +/− docetaxel, DFS and OS was comparable between patients with TP53 wild type and mutant tumors (n = 520; DFS: P = 0.577, OS: P = 0.367) [77]. Further stratification of patients by TP53 mutation class (missense vs truncating mutations) showed truncating mutations in TP53 were associated with a significant reduction in both DFS and OS (P <0.001). In this study, over 70% of patients also received HRT. Two other studies using IHC to infer TP53 mutation status showed that p53 expression had no prognostic value in the response to chemotherapy [78,79].
In a study of stage III breast cancer, patients with TP53 mutations were shown to have worse DFS following treatment with paclitaxel (P = 0.007) [80]. Nearly half the patients included in this analysis were ER-positive and received some form of HRT. Further, only differences in DFS were reported. In the I-SPY-1 clinical trial, Esserman et al. determined from a cohort of 120 breast cancer patients who were treated with NAC that while patients with TP53 mutant tumors had higher rates of pCR (P <0.001), patients with TP53 wild type tumors had better RFS (P <0.05) [81]. In this clinical trial, while all patients received NAC, many patients received HRT.
Studies using IHC to infer TP53 mutation status also showed that patients with p53-positive tumors had worse OS and/or DFS compared with patients that have p53-negative tumors after chemotherapy +/− HRT [82–86]. Also, one study using IHC showed that patients with p53-positive tumors had higher rates of PD after chemotherapy [87]. In each of these studies [82–84,86,87], a varying percentage of patients also received HRT.
In sum, while the majority of studies above correlate TP53 mutations to worse survival after chemotherapy, they each include a mix of patients that received chemotherapy alone and chemotherapy plus HRT and ratios that vary markedly. Further, many of these studies were powered by small sample sizes and differ in methodology such as TP53 mutation detection. These differences confound comparisons between studies, which may lead to conflicting conclusions regarding TP53 mutations and response/survival after chemotherapy.
Our analysis of the METABRIC dataset on all chemotherapy-treated primary breast cancer patients revealed survival was not significantly different between patients with TP53 wild type and mutant tumors (Figure 1B). This cohort that included chemotherapy-treated and chemotherapy plus HRT-treated patients is in agreement with larger studies in the previous section [77–79].
We next identified studies that analyzed cohorts with patients who were all treated with either chemotherapy and HRT or chemotherapy alone.
TP53 Mutation Status and Survival Studies of Patients That Only Received Chemotherapy plus HRT
Studies reviewed above suggest that chemotherapy-treated patients with TP53 mutant breast tumors have worse or similar survival, despite having much higher rates of pCR than patients with TP53 wild type tumors. As noted above, nearly all of these studies combined ER-negative patients with ER-positive patients that were also treated with HRT. Taken together, the seemingly contradictory findings in studies examining pCR rate and survival suggest the possibility that HRT may affect survival of patients with TP53 mutant and wild type tumors differently. Most studies, however, failed to separate ER positive and negative patients in survival analysis due to relatively small sample sizes.
We have identified studies that analyzed the survival of only those patients that received both chemotherapy and HRT and excluded patients receiving chemotherapy or HRT alone, untreated patients, etc. Kim et al. found in a study of 17 breast cancer patients that those with TP53 wild type tumors treated with chemotherapy and HRT had superior survival compared with patients that have TP53 mutant tumors [88]. Patients (n = 1964) with p53 IHC negative tumors who received chemotherapy followed by HRT had RFS comparable with patients that have p53-positive tumors (P = 0.128) [89]. Andersson et al. showed patients treated with cyclophosphamide/methotrexate/5-fluorouracil (CMF) and HRT had no difference in OS, whether the tumor was TP53 wild type or mutant (P = 0.27) [45].
Our analysis of the METABRIC dataset showed patients with TP53 wild type tumors had marginally better survival (significant in univariate analysis only) compared with patients that have TP53 mutant tumors when chemotherapy plus HRT was administered (Figure 1D).
TP53 Mutation Status and Survival Studies of Patients That Received Only Chemotherapy
As discussed, p53 mediates the cell response to stress, such as that caused by DNA damage or aberrant mitoses. Thus, a close examination of patients that received only chemotherapy is warranted.
In one study of TP53 mutation in 1275 breast cancer patients treated with chemotherapy, the authors showed patients with p53-positive and -negative tumors by IHC had comparable RFS (P = 0.359) [89]. However, it was observed that amongst patients age >50 years (n = 427), those with p53-positive tumors had better RFS than patients with p53-negative tumors (P = 0.027). In another study of 37 ER-negative patients with metastatic breast cancer, patients with TP53 mutant tumors had better OS compared with patients that have wild type tumors (P = 0.026) [88]. However, some patients in this cohort harbored HER2+ tumors and received targeted treatment and, though statistically significant, the number of TP53 wild type patients was small (n = 2). Despite the limitations of these studies, they suggest a survival benefit for patients with TP53 mutant tumors.
In our analysis of chemotherapy-only treated patients in the METABRIC dataset, we showed that patients with TP53 mutant tumors had significantly longer survival than patients with wild type tumors (TP53 mutant tumors: median survival 195 months, TP53 wild type tumors: median survival 45 months, P = 0.0072) (Figure 1E). This finding was true in all subtypes, including basal-like or triple negative tumors. For example, in TNBC patients, median survival for those with TP53 mutant tumors was 263 months and for TP53 wild type, 45 months (P = 0.0083) [49]. These data from METABRIC are consistent with the high rate of pCR observed in previous neoadjuvant studies.
Some studies with very short follow up, however, have not found a survival advantage associated with TP53 mutation. In 174 TNBC patients treated with adjuvant chemotherapy after curative surgery, TP53 mutation status had no impact on patient survival [90]. In this study, only 49% of TNBC were determined by sequencing to be TP53 mutant and distant RFS was at least 80% in all treatment arms. Darb-Esfahani et al. examined TP53 mutation rates in 450 breast cancers treated with NAC and while their study confirmed high rates of mutations in TNBC, mutation status did not predict survival rates [91]. Of note, the cohort had only been followed for 50 months and ~80–90% of patients in all arms were still alive.
Interesting Case of TP53 Mutation Status and Response to Chemotherapy Alone versus Chemotherapy plus HRT
Both individual smaller studies and analysis of the large METABRIC dataset showed that patients with TP53 mutant tumors had superior survival after treatment with chemotherapy alone (Figure 1E). However, when chemotherapy plus HRT are given, the advantage to having a TP53 mutation disappears (Figure 1D). A closer examination revealed, in fact, that TP53 mutant tumors responded very well to chemotherapy plus HRT. Addition of HRT actually failed to extend survival at all in these patients [49]. The patients with TP53 wild type tumors, however, derived an enormous benefit from addition of HRT, extending survival from 46 months (Figure 1E) to 206 months (Figure 1D). In this way, HRT appears to be an ‘equalizer’ in survival between patients with TP53 wild type and mutant tumors that are treated with chemotherapy.
In studies of heterogeneously treated patients discussed above, a greater number of TP53 wild type patients that are able to receive HRT will dramatically skew the curve toward longer survival. If a large fraction of TP53 wild type patients are ER negative, then median survival will skew much shorter. In future studies this should be accounted for, or, more optimally, populations of patients getting different treatments should be segregated into separate survival curves.
Why Do TP53 Mutant Tumors Respond Differently to Treatment with Chemotherapy and/or HRT?
Many studies assessed the role of TP53 mutation status in survival after chemotherapy in patients treated with chemotherapy and HRT. Combining these patient populations makes it difficult to reach any conclusions regarding the role of TP53 mutations in the response to chemotherapy for many reasons.
First, while it is well-established that p53 plays a central role in cellular response to DNA-damaging agents like chemotherapy, the role of p53 in cellular responses to HRT is much less clear. Some preclinical studies suggest that p53 interacts with ER, leading to speculation that p53 may play a role in cancer sensitivity to HRT [54,92]. However, even preclinical studies have conflicted on the result of p53 and ER interactions, with some studies showing that ER represses p53 activity [93] and others showing that ER augments p53 activity [94]. Preclinical data suggest that breast cancer cell lines with mutated p53 are less responsive to HRT [95,96]. These data support the survival studies on HRT-treated patients discussed above. Thus, how p53 regulates the cellular response to HRT remains largely unclear and is likely not related to how p53 responds to DNA damage or mitotic stress. More research is warranted and this includes studies of patients that receive no chemotherapy (Figure 1C) or no treatment (Figure 1G).
The question of how wild type p53 mediates a poor response to chemotherapy treatment has been addressed in several studies. As mentioned, p53 directs cells to undergo apoptosis in response to chemotherapy in many cell and tumor types. This well-characterized function is counter to the observations in breast cancer: p53 wild type tumors are less likely to undergo massive cell death and a pCR and have worse survival after chemotherapy treatment alone. Evidence from breast cancer models and tumors have shown p53 directs cells to undergo cell cycle arrest and senescence [97–99]. Cells of the senescent tumors secrete chemokines and cytokines of the senescence-associated secretory phenotype [100] that can drive protumor phenotypes such as survival, metastasis, and proliferation [101]. One mechanism that contributes to senescent cell survival and persistence in the residual disease after chemotherapy treatment is the engulfing and cannibalizing of neighboring cells [102]. Less well understood is how the senescent cells avoid immune detection and clearance [101].
Concluding Remarks
Our analysis revealed the importance of looking beyond the headlines and titles of manuscripts to understand the role TP53 plays in breast cancer treatment response. The patient characteristics, treatment regimens, and methodologies behind declarations of ‘TP53 mutation predicts better/worse/no different survival’ should be examined carefully. p53 activation and the cellular outcomes it dictates are complicated and context dependent. It is unsurprising that TP53 mutation plays very different roles in outcomes based on the very different biological consequences caused by treatments as disparate as DNA damage and hormone deprivation. Functional p53 directs breast cancer cells to undergo cell cycle arrest and senescence in response to chemotherapy, leaving residual disease that is highly refractory and resistant to immune clearance. In these treatment settings where wild type p53 is disadvantageous, additional pharmacological intervention may be warranted (see Outstanding Questions).
Outstanding Questions.
Can therapies that target residual senescent cells improve outcomes for chemotherapy-treated TP53 wild type breast cancer patients?
Can immunotherapy be sequenced following chemotherapy to promote clearance of residual disease?
Should TP53 mutation status be taken into consideration in determining whether patients are candidates for HRT?
Highlights.
TP53 is mutant in 30% of all breast cancers and is the most frequently mutated gene. The role of TP53 in the management of breast cancer remains unclear.
Recent analysis reveals that mutant p53 can be detrimental or beneficial to clinical outcome, depending on treatments given.
Because p53 has diverse activities in response to different treatments, each treatment should ideally have its own survival arm.
Acknowledgments
The authors apologize for omitting any of our colleague’s work due to space restrictions. J.G.J. is supported by a Department of Defense Breast Cancer Research Program grant W81XWH-14-1-0216. A.S. is supported by Award Number TL1TR003106 from the National Center for Advancing Translational Sciences. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Advancing Translational Sciences.
Footnotes
References
- 1.TCGA-Cancer Genome Atlas Network (2012) Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kastenhuber ER and Lowe SW (2017) Putting p53 in context. Cell 170, 1062–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tonnessen-Murray C et al. (2016) The p53 protein: from cell regulation to cancer. In The p53 Protein: From Cell Regulation to Cancer (Lozano G and Levine AJ, eds), pp. 173–186, Cold Spring Harbor Laboratory Press [Google Scholar]
- 4.Soussi T (2011) TP53 mutations in human cancer: database reassessment and prospects for the next decade. Adv. Cancer Res 110, 107–139 [DOI] [PubMed] [Google Scholar]
- 5.Soussi T and Beroud C (2001) Assessing TP53 status in human tumours to evaluate clinical outcome. Nat. Rev. Cancer 1, 233–240 [DOI] [PubMed] [Google Scholar]
- 6.Berns EM et al. (2000) Complete sequencing of TP53 predicts poor response to systemic therapy of advanced breast cancer. Cancer Res 60, 2155–2162 [PubMed] [Google Scholar]
- 7.Bertheau P et al. (2008) TP53 status and response to chemotherapy in breast cancer. Pathobiology 75, 132–139 [DOI] [PubMed] [Google Scholar]
- 8.Silwal-Pandit L et al. (2014) TP53 mutation spectrum in breast cancer is subtype specific and has distinct prognostic relevance. Clin. Cancer Res 20, 3569–3580 [DOI] [PubMed] [Google Scholar]
- 9.Chen MB et al. (2012) Value of TP53 status for predicting response to neoadjuvant chemotherapy in breast cancer: a meta-analysis. PLoS One 7, e39655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalez-Angulo AM et al. (2007) Overview of resistance to systemic therapy in patients with breast cancer. Adv. Exp. Med. Biol 608, 1–22 [DOI] [PubMed] [Google Scholar]
- 11.Waks AG and Winer EP (2019) Breast cancer treatment: a review. JAMA 321, 288–300 [DOI] [PubMed] [Google Scholar]
- 12.Roninson IB et al. (2001) If not apoptosis, then what? Treatment-induced senescence and mitotic catastrophe in tumor cells. Drug Resist. Updat 4, 303–313 [DOI] [PubMed] [Google Scholar]
- 13.Gianni L et al. (2009) Role of anthracyclines in the treatment of early breast cancer. J. Clin. Oncol 27, 4798–4808 [DOI] [PubMed] [Google Scholar]
- 14.Fu D et al. (2012) Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat. Rev. Cancer 12, 104–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Longley DB et al. (2003) 5-fluorouracil: mechanisms of action and clinical strategies. Nat. Rev. Cancer 3, 330–338 [DOI] [PubMed] [Google Scholar]
- 16.Vignard J et al. (2013) Ionizing-radiation induced DNA double-strand breaks: a direct and indirect lighting up. Radiother. Oncol 108, 362–369 [DOI] [PubMed] [Google Scholar]
- 17.Sparreboom A et al. (1998) Preclinical pharmacokinetics of paclitaxel and docetaxel. Anti-Cancer Drugs 9, 1–17 [DOI] [PubMed] [Google Scholar]
- 18.Gudkov AV and Komarova EA (2003) The role of p53 in determining sensitivity to radiotherapy. Nat. Rev. Cancer 3, 117–129 [DOI] [PubMed] [Google Scholar]
- 19.Meek DW (2000) The role of p53 in the response to mitotic spindle damage. Pathol. Biol. (Paris) 48, 246–254 [PubMed] [Google Scholar]
- 20.Buchholz TA et al. (2003) Neoadjuvant chemotherapy for breast carcinoma: multidisciplinary considerations of benefits and risks. Cancer 98, 1150–1160 [DOI] [PubMed] [Google Scholar]
- 21.Eisenhauer E et al. (2008) New response evaluation criteria in solid tumors: revised RECIST guideline version 1.1. EJC Suppl. 6, 13. [DOI] [PubMed] [Google Scholar]
- 22.Osborne CK et al. (2000) Selective estrogen receptor modulators: structure, function, and clinical use. J. Clin. Oncol 18, 3172–3186 [DOI] [PubMed] [Google Scholar]
- 23.Osborne CK and Schiff R (2005) Aromatase inhibitors: future directions. J. Steroid Biochem. Mol. Biol 95, 183–187 [DOI] [PubMed] [Google Scholar]
- 24.Nasrazadani A et al. (2018) Precision medicine in hormone receptor-positive breast cancer. Front. Oncol 8, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fabian CJ (2007) The what, why and how of aromatase inhibitors: hormonal agents for treatment and prevention of breast cancer. Int. J. Clin. Pract 61, 2051–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitchell AP et al. (2019) Association between reimbursement incentives and physician practice in oncology: a systematic review. JAMA Oncol. 5, 893–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Epstein AJ and Johnson SJ (2012) Physician response to financial incentives when choosing drugs to treat breast cancer. Int. J. Health Care Finance Econ 12, 285–302 [DOI] [PubMed] [Google Scholar]
- 28.Hadley J et al. (2003) Medicare breast surgery fees and treatment received by older women with localized breast cancer. Health Serv. Res 38, 553–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murnyak B and Hortobagyi T (2016) Immunohistochemical correlates of TP53 somatic mutations in cancer. Oncotarget 7, 64910–64920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Slebos RJ et al. (1996) Clinical and pathological associations with p53 tumour-suppressor gene mutations and expression of p21WAF1/Cip1 in colorectal carcinoma. Br. J. Cancer 74, 165–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shahbandi A and Jackson JG (2019) Analysis across multiple tumor types provides no evidence that mutant p53 exerts dominant negative activity. NPJ Precis. Oncol 3, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valentin-Vega YA et al. (2007) High levels of the p53 inhibitor MDM4 in head and neck squamous carcinomas. Hum. Pathol 38, 1553–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Norberg T et al. (1998) Comparison between p53 protein measurements using the luminometric immunoassay and immunohistochemistry with detection of p53 gene mutations using cDNA sequencing in human breast tumors. Int. J. Cancer 79, 376–383 [DOI] [PubMed] [Google Scholar]
- 34.Flaman JM et al. (1995) A simple p53 functional assay for screening cell lines, blood, and tumors. Proc. Natl. Acad. Sci. U. S. A 92, 3963–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hainaut P and Pfeifer GP (2016) Somatic TP53 mutations in the era of genome sequencing. Cold Spring Harb. Perspect. Med 6, a026179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hollstein M et al. (1999) New approaches to understanding p53 gene tumor mutation spectra. Mutat. Res 431, 199–209 [DOI] [PubMed] [Google Scholar]
- 37.Hartmann A et al. (1995) p53 gene mutations inside and outside of exons 5–8: the patterns differ in breast and other cancers. Oncogene 10, 681–688 [PubMed] [Google Scholar]
- 38.Robles AI and Harris CC (2010) Clinical outcomes and correlates of TP53 mutations and cancer. Cold Spring Harb. Perspect. Biol 2, a001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ostrowski JL et al. (1991) p53 expression in human breast cancer related to survival and prognostic factors: an immunohistochemical study. J. Pathol 164, 75–81 [DOI] [PubMed] [Google Scholar]
- 40.Iwaya K et al. (1991) Nuclear p53 immunoreaction associated with poor prognosis of breast cancer. Jpn. J. Cancer Res 82, 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamashita H et al. (2006) p53 protein accumulation predicts resistance to endocrine therapy and decreased post-relapse survival in metastatic breast cancer. Breast Cancer Res 8, R48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bergh J et al. (1995) Complete sequencing of the p53 gene provides prognostic information in breast cancer patients, particularly in relation to adjuvant systemic therapy and radiotherapy. Nat. Med 1, 1029–1034 [DOI] [PubMed] [Google Scholar]
- 43.Shiao YH et al. (1995) Racial disparity in the association of p53 gene alterations with breast cancer survival. Cancer Res 55, 1485–1490 [PubMed] [Google Scholar]
- 44.Blaszyk H et al. (2000) A prospective trial of midwest breast cancer patients: a p53 gene mutation is the most important predictor of adverse outcome. Int. J. Cancer 89, 32–38 [DOI] [PubMed] [Google Scholar]
- 45.Andersson J et al. (2005) Worse survival for TP53 (p53)-mutated breast cancer patients receiving adjuvant CMF. Ann. Oncol 16, 743–748 [DOI] [PubMed] [Google Scholar]
- 46.Rossner P Jr. et al. (2009) Mutations in p53, p53 protein overexpression and breast cancer survival. J. Cell. Mol. Med 13, 3847–3857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Olivier M et al. (2006) The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clin. Cancer Res 12, 1157–1167 [DOI] [PubMed] [Google Scholar]
- 48.Meric-Bernstam F et al. (2018) Survival outcomes by TP53 mutation status in metastatic breast cancer. JCO Precis. Oncol Published online April 25, 2018. 10.1200/PO.17.00245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ungerleider NA et al. (2018) Breast cancer survival predicted by TP53 mutation status differs markedly depending on treatment. Breast Cancer Res. 20, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Curtis C et al. (2012) The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pereira B et al. (2016) The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat. Commun 7, 11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Archer SG et al. (1995) Expression of ras p21, p53 and cerbB-2 in advanced breast cancer and response to first line hormonal therapy. Br. J. Cancer 72, 1259–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Elledge RM et al. (1997) bcl-2, p53, and response to tamoxifen in estrogen receptor-positive metastatic breast cancer: a Southwest Oncology Group study. J. Clin. Oncol 15, 1916–1922 [DOI] [PubMed] [Google Scholar]
- 54.Konduri SD et al. (2010) Mechanisms of estrogen receptor antagonism toward p53 and its implications in breast cancer therapeutic response and stem cell regulation. Proc. Natl. Acad. Sci. U. S. A 107, 15081–15086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lowe SW et al. (1993) p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature 362, 847–849 [DOI] [PubMed] [Google Scholar]
- 56.Lowe SW et al. (1993) p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell 74, 957–967 [DOI] [PubMed] [Google Scholar]
- 57.Sablina AA et al. (2001) p53 activation in response to microtubule disruption is mediated by integrin-Erk signaling. Oncogene 20, 899–909 [DOI] [PubMed] [Google Scholar]
- 58.Raycroft L et al. (1990) Transcriptional activation by wild-type but not transforming mutants of the p53 anti-oncogene. Science 249, 1049–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vousden KH and Lu X (2002) Live or let die: the cell’s response to p53. Nat. Rev. Cancer 2, 594–604 [DOI] [PubMed] [Google Scholar]
- 60.Brown JM and Wouters BG (1999) Apoptosis, p53, and tumor cell sensitivity to anticancer agents. Cancer Res. 59, 1391–1399 [PubMed] [Google Scholar]
- 61.Noda A et al. (1994) Cloning of senescent cell-derived inhibitors of DNA synthesis using an expression screen. Exp. Cell Res 211, 90–98 [DOI] [PubMed] [Google Scholar]
- 62.Serrano M et al. (1993) A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature 366, 704–707 [DOI] [PubMed] [Google Scholar]
- 63.el-Deiry WS et al. (1993) WAF1, a potential mediator of p53 tumor suppression. Cell 75, 817–825 [DOI] [PubMed] [Google Scholar]
- 64.Chang BD et al. (1999) Role of p53 and p21waf1/cip1 in senescence-like terminal proliferation arrest induced in human tumor cells by chemotherapeutic drugs. Oncogene 18, 4808–4818 [DOI] [PubMed] [Google Scholar]
- 65.Bertheau P et al. (2002) Effect of mutated TP53 on response of advanced breast cancers to high-dose chemotherapy. Lancet 360, 852–854 [DOI] [PubMed] [Google Scholar]
- 66.Lehmann-Che J et al. (2010) Cyclophosphamide dose intensification may circumvent anthracycline resistance of p53 mutant breast cancers. Oncologist 15, 246–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim T et al. (2015) Predictive significance of p53, Ki-67, and Bcl-2 expression for pathologic complete response after neoadjuvant chemotherapy for triple-negative breast cancer. J. Breast Cancer 18, 16–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Y et al. (2016) TP53 mutations are associated with higher rates of pathologic complete response to anthracycline/cyclophosphamide-based neoadjuvant chemotherapy in operable primary breast cancer. Int. J. Cancer 138, 489–496 [DOI] [PubMed] [Google Scholar]
- 69.Gluck S et al. (2012) TP53 genomics predict higher clinical and pathologic tumor response in operable early-stage breast cancer treated with docetaxel-capecitabine +/− trastuzumab. Breast Cancer Res. Treat 132, 781–791 [DOI] [PubMed] [Google Scholar]
- 70.Geisler S et al. (2001) Influence of TP53 gene alterations and c-erbB-2 expression on the response to treatment with doxorubicin in locally advanced breast cancer. Cancer Res. 61, 2505–2512 [PubMed] [Google Scholar]
- 71.Geisler S et al. (2003) TP53 gene mutations predict the response to neoadjuvant treatment with 5-fluorouracil and mitomycin in locally advanced breast cancer. Clin. Cancer Res 9, 5582–5588 [PubMed] [Google Scholar]
- 72.Kandioler-Eckersberger D et al. (2000) TP53 mutation and p53 overexpression for prediction of response to neoadjuvant treatment in breast cancer patients. Clin. Cancer Res 6, 50–56 [PubMed] [Google Scholar]
- 73.Bidard FC et al. (2008) p53 status and efficacy of primary anthracyclines/alkylating agent-based regimen according to breast cancer molecular classes. Ann. Oncol 19, 1261–1265 [DOI] [PubMed] [Google Scholar]
- 74.Jarvinen TA et al. (1998) Predictive value of topoisomerase IIalpha and other prognostic factors for epirubicin chemotherapy in advanced breast cancer. Br. J. Cancer 77, 2267–2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Aas T et al. (1996) Specific P53 mutations are associated with de novo resistance to doxorubicin in breast cancer patients. Nat. Med 2, 811–814 [DOI] [PubMed] [Google Scholar]
- 76.Eikesdal HP et al. (2014) TP53 status predicts long-term survival in locally advanced breast cancer after primary chemotherapy. Acta Oncol. 53, 1347–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fernandez-Cuesta L et al. (2012) Prognostic and predictive value of TP53 mutations in node-positive breast cancer patients treated with anthracycline- or anthracycline/taxane-based adjuvant therapy: results from the BIG 02–98 phase III trial. Breast Cancer Res. 14, R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rozan S et al. (1998) No significant predictive value of c-erbB-2 or p53 expression regarding sensitivity to primary chemotherapy or radiotherapy in breast cancer. Int. J. Cancer 79, 27–33 [DOI] [PubMed] [Google Scholar]
- 79.Niskanen E et al. (1997) Predictive value of c-erbB-2, p53, cathepsin-D and histology of the primary tumour in metastatic breast cancer. Br. J. Cancer 76, 917–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chrisanthar R et al. (2011) Predictive and prognostic impact of TP53 mutations and MDM2 promoter genotype in primary breast cancer patients treated with epirubicin or paclitaxel. PLoS One 6, e19249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Esserman LJ et al. (2012) Chemotherapy response and recurrence-free survival in neoadjuvant breast cancer depends on biomarker profiles: results from the I-SPY 1 TRIAL (CALGB 150007/150012; ACRIN 6657). Breast Cancer Res. Treat 132, 1049–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Clahsen PC et al. (1998) p53 protein accumulation and response to adjuvant chemotherapy in premenopausal women with node-negative early breast cancer. J. Clin. Oncol 16, 470–479 [DOI] [PubMed] [Google Scholar]
- 83.Yang P et al. (2013) The impact of p53 in predicting clinical outcome of breast cancer patients with visceral metastasis. Sci. Rep 3, 2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Malamou-Mitsi V et al. (2006) Evaluation of the prognostic and predictive value of p53 and Bcl-2 in breast cancer patients participating in a randomized study with dose-dense sequential adjuvant chemotherapy. Ann. Oncol 17, 1504–1511 [DOI] [PubMed] [Google Scholar]
- 85.Chae BJ et al. (2009) p53 as a specific prognostic factor in triple-negative breast cancer. Jpn. J. Clin. Oncol 39, 217–224 [DOI] [PubMed] [Google Scholar]
- 86.Thor AD et al. (1998) erbB-2, p53, and efficacy of adjuvant therapy in lymph node-positive breast cancer. J. Natl. Cancer Inst 90, 1346–1360 [DOI] [PubMed] [Google Scholar]
- 87.Rahko E et al. (2003) A mutant TP53 gene status is associated with a poor prognosis and anthracycline-resistance in breast cancer patients. Eur. J. Cancer 39, 447–453 [DOI] [PubMed] [Google Scholar]
- 88.Kim JY et al. (2017) Clinical implications of genomic profiles in metastatic breast cancer with a focus on TP53 and PIK3CA, the most frequently mutated genes. Oncotarget 8, 27997–28007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kim HS et al. (2010) Overexpression of p53 is correlated with poor outcome in premenopausal women with breast cancer treated with tamoxifen after chemotherapy. Breast Cancer Res. Treat 121, 777–788 [DOI] [PubMed] [Google Scholar]
- 90.Kim JY et al. (2016) Association between mutation and expression of TP53 as a potential prognostic marker of triple-negative breast cancer. Cancer Res. Treat 48, 1338–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Darb-Esfahani S et al. (2016) Role of TP53 mutations in triple negative and HER2-positive breast cancer treated with neoadjuvant anthracycline/taxane-based chemotherapy. Oncotarget 7, 67686–67698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bailey ST et al. (2012) Estrogen receptor prevents p53-dependent apoptosis in breast cancer. Proc. Natl. Acad. Sci. U. S. A 44, 18060–18065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu W et al. (2006) Estrogen receptor-alpha binds p53 tumor suppressor protein directly and represses its function. J. Biol. Chem 281, 9837–9840 [DOI] [PubMed] [Google Scholar]
- 94.Berger CE et al. (2012) p53, a target of estrogen receptor (ER) alpha, modulates DNA damage-induced growth suppression in ER-positive breast cancer cells. J. Biol. Chem 287, 30117–30127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fernandez-Cuesta L et al. (2011) Estrogen levels act as a rheostat on p53 levels and modulate p53-dependent responses in breast cancer cell lines. Breast Cancer Res. Treat 125, 35–42 [DOI] [PubMed] [Google Scholar]
- 96.Fernandez-Cuesta L et al. (2011) p53 status influences response to tamoxifen but not to fulvestrant in breast cancer cell lines. Int. J. Cancer 128, 1813–1821 [DOI] [PubMed] [Google Scholar]
- 97.Jackson JG et al. (2012) p53-Mediated senescence impairs the apoptotic response to chemotherapy and clinical outcome in breast cancer. Cancer Cell 21, 793–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.te Poele RH et al. (2002) DNA damage is able to induce senescence in tumor cells in vitro and in vivo. Cancer Res. 62, 1876–1883 [PubMed] [Google Scholar]
- 99.Tonnessen-Murray C et al. (2018) p53 Mediates vast gene expression changes that contribute to poor chemotherapeutic response in a mouse model of breast cancer. Transl. Oncol 11, 930–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tchkonia T et al. (2013) Cellular senescence and the senescent secretory phenotype: therapeutic opportunities. J. Clin. Invest 123, 966–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rao SG and Jackson JG (2016) SASP: tumor suppressor or promoter? Yes! Trends Cancer 2, 676–687 [DOI] [PubMed] [Google Scholar]
- 102.Tonnessen-Murray CA et al. (2019) Chemotherapy-induced senescent cancer cells engulf other cells to enhance their survival. J. Cell Biol 218, 3827–3844 [DOI] [PMC free article] [PubMed] [Google Scholar]


