Abstract
The p63 gene encodes at least 10 isoforms, which can be classified into TA and ∆N isotypes (TAp63 and ∆Np63 proteins) according to their differences at the N termini. TAp63γ is an important transcription factor. We previously reported that peptidyl‐prolyl isomerase (PPI) Pin1 directly binds to TAp63γ protein and identified that serine 12 (S12) in the transactivation domain (TAD) of TAp63γ is required for regulation of its transcriptional activity. In the present study, we report that Pin1 stimulates transcriptional and pro‐apoptotic activities of TAp63γ; this Pin1‐mediated stimulation may depend on phosphorylation of S12 mediated by JNK1 and results in striking activation of TAp63γ. JNK1 represses transactivity of TAp63γ in cells without abundant Pin1 proteins and enhances it in the presence of sufficient levels of Pin1. Collectively, our data suggest a novel mechanism for regulation of TAp63γ transactivity: TAp63γ with unphosphorylated S12 is moderately active, phosphorylation at this residue (pS12) makes it hypoactive, and Pin1 binds to the pS12‐P13 motif and makes TAp63γ hyperactive. Our findings will aid in the elucidation of the mechanism underlying modulation of TAp63γ.
Keywords: JNK1, Pin1, TAp63γ, transactivity
When the serine 12 residue (S12) in the transactivation domain of TAp63γ is unphosphorylated, TAp63γ is moderately active to transactivate its downstream genes. Phosphorylation at this residue (pS12) can be mediated by kinase JNK1, leading to a repression of TAp63γ transactivity; isomerization of the pS12‐P13 motif can be mediated by Pin1, which makes TAp63γ hyperactive.
Abbreviations
- A
alanine
- CL‐PARP1
cleaved PARP1
- CoIP
co‐immunoprecipitation
- IB
immunoblotting
- IP
immunoprecipitation
- MTT
3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyl‐tetrazolium bromide
- P
proline
- PPI
peptidyl‐prolyl isomerase
- pS
phosphorylated serine
- S
serine
- SAM
sterile alpha motif
- T
threonine
- TAD
transactivation domain
- TID
transinhibition domain
- Y
tyrosine
The p63 gene belongs to the p53 family and encodes at least 10 isoforms, which can be classified into TA and ∆N isotypes (TAp63 and ∆Np63 proteins) according to their differences at the N termini. TAp63s contain the full transactivation domain (TAD) at the N termini, while ∆Np63 isotypes have an incomplete TAD with a weaker transactivity. After transcription, both TA and ∆N isotypes can be spliced into mRNAs with different 3’ termini, generating at least 5 different C termini, α, β, γ, δ, and ε. Among them, the γ types miss the sterile alpha motif (SAM) and the transinhibition domain (TID) at their C termini compared with the α isoform of p63 proteins [1, 2, 3]. TAp63 proteins express at relatively lower levels in somatic cells. However, like p53, these TA isoforms of p63 play key roles in cell cycle arrest and apoptotic cell death via transactivating pro‐apoptotic factors such as p21, Puma, Bax, and Noxa [4, 5, 6]. Thus, TAp63s function as quality control factors in the female germline upon genotoxic stress [7, 8, 9, 10]. Studies with mouse models demonstrate that specific knockout of TAp63 can cause premature aging [11, 12] and metabolic syndrome [13]. These TAp63‐null mice are also highly tumor prone and develop metastatic diseases [11, 14], reaffirming the tumor suppressor functions of TAp63 proteins. Data from Ernesto Bruno group suggest that TAp63 suppresses recurrence of nasal polyps [15]. According to reports from group of Esther H. Chang, miR‐130b and TAp63 form a feed‐forward loop, and this miR‐130b/TAp63 axis is a druggable pathway that has the potential to uncover broad‐spectrum therapeutic options for the majority of p53‐altered cancers [16]. It has been reported that TAp63 may also function as a repressor of transcription [17]. Recently, Suenaga Y and Nakagawara A et al found that TAp63 restrains neuroblastoma growth via repressing MYCN/NCYM bidirectional transcription [18]. As a short isoform of TAp63, TAp63γ is assumed to have a high activity to mediate transcription and apoptosis, since it lacks TID and SAM at the C terminus [1]. Some recent reports demonstrate that TAp63γ promotes myogenic differentiation, osteoblastic differentiation, and cartilage development [19, 20, 21].
Due to their key roles in cell cycle control, both expression levels and activities of p63 proteins are tightly regulated in cells [2]. According to data from our group and other laboratories, p63 proteins undergo various post‐translational modifications including phosphorylation, ubiquitination, and isomerization [2, 22, 23, 24, 25, 26, 27, 28]. Particularly, we previously reported that peptidyl‐prolyl isomerase (PPI) Pin1 physically interacts with several protein isoforms of p63, including TAp63α, ∆Np63α, and TAp63γ; Pin1 specifically binds to the T‐P‐P‐P‐P‐Y motif in the SAM of p63α proteins and inhibits the proteasomal degradation of them [22]. However, γ isoforms lack the T‐P‐P‐P‐P‐Y motif and SAM. Therefore, the binding sites and effects of Pin1 on TAp63γ remain obscure. In another study, we found that c‐Jun N‐terminal kinase 1 (JNK1) may phosphorylate TAp63γ at serine 12 and impair its transactivity and pro‐apoptotic activity [27]. In the present work, we find that Pin1 stimulates transcriptional and pro‐apoptotic activities of TAp63γ; S12A mutation in TAp63γ impairs its physical interaction with Pin1 and deprives Pin1‐mediated stimulation of TAp63γ; we further find that Pin1 strikingly reverses JNK1‐repressed transactivity of TAp63γ and makes it hyperactive. Our findings are helpful to elucidate how transactivity of TAp63γ is modulated.
Materials and methods
Cell culture, transfection, and plasmids
Saos‐2, Hela, and H1299 cells were cultured in Modified McCoy's 5a Medium (BI) supplemented with 10% FBS (BI) and 1% penicillin G/streptomycin (Hyclone, Logan, UT, USA) at 37 °C in a humidified 5% CO2 incubator. Transient transfection was performed with Entranster™‐H4000 (Engreen Biosystem, Beijing, China), and total amounts of plasmid DNA were balanced with corresponding vectors for each transfection. Constructs of pcDNA3.1‐HA‐TAp63γ, pcDNA3.1‐HA‐TAp63γ(S12A), pcDNA3.1‐Pin1, pcDNA3.1‐Pin1(W34A), and pcDNA3.1‐JNK1 were previously described [22, 27, 29]. JNK1 siRNA and scrambled control were purchased from Santa Cruz Biotechnology (Dallas, TX, USA). Cell viabilities were determined by 3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyl‐tetrazolium bromide (MTT; Promega, Madison, WI, USA) as described in the instruction.
Immunoprecipitation and immunoblotting analysis
Immunoprecipitation (IP) and immunoblotting (IB) analyses were performed as previously described [22, 27]. Antibodies used were specific for Pin1 (rabbit polyclonal antibody; Cell Signaling Technology, Beverly, MA, USA; 1 : 1000), JNK1 (rabbit polyclonal antibody; Abcam, Cambridge, MA, USA; 1 : 1000), HA (mouse monoclonal antibody; Millipore, Billerica, CA, USA;1 : 500), p63 (rabbit polyclonal antibody; Zen‐bio, Chengdu, Sichuan, China; 1 : 1000), PARP1 (rabbit polyclonal antibody; Zen‐bio, Chengdu, Sichuan, China; 1 : 2000), and GAPDH (rabbit polyclonal antibody; Zen‐bio, Chengdu, Sichuan, China; 1 : 1000). Blots were detected using an ECL system (GE Amersham Pharmacia Biotech, Boston, MA, USA).
Luciferase reporter assay
Luciferase assays were performed as described previously [22, 27]. Saos‐2 cells were transfected with a mixture of Bax‐Luc and pRL‐TK‐Renilla plus indicated plasmids or siRNAs. Total amount of DNAs or RNAs was balanced with control vectors or scramble control RNAs. Cells were harvested at 48 h post‐transfection and lysed in Passive Lysis Buffer (Promega). Lysates were analyzed for firefly and Renilla luciferase activities using the Dual Luciferase Reagent Assay Kit (Promega). Luminescence was measured in a luminometer. Relative luciferase activity was determined by normalizing luciferase activity with Renilla.
Statistical analysis
All experiments were carried out in triplicate. Two‐tailed t‐test was used for comparison between two groups. P < 0.05 was considered statistically significant. All the error bars indicate SD.
Results
Pin1 enhances TAp63γ‐induced transcription and apoptosis
In a previous study, we performed a pull‐down experiment and found that TAp63γ protein forms a complex with PPI Pin1; mutation on tryptophan 34 to alanine (W34A) in Pin1, which was reported to disrupt the binding of this isomerase to its substrates, significantly impairs its physical interaction with TAp63γ [22]. To confirm this interaction in mammalian cells, we transiently overexpressed HA‐tagged TAp63γ (HA‐TAp63γ), along with wild‐type Pin1 or its W34A mutant, in human osteosarcoma cell Saos‐2, and performed a co‐immunoprecipitation (CoIP) assay. The results demonstrate that Pin1 can form a stable complex with TAp63γ, while W34A mutation in Pin1 significantly impairs this interaction (Fig. 1A). Bax is a downstream gene of TAp63; luciferase reporter driven by Bax promoter (Bax‐Luc) can be used to measure the transactivity of TAp63 proteins [22]. To further investigate whether Pin1 modulates transactivity of TAp63γ, we performed a luciferase reporter assay. The results demonstrate that the wild‐type Pin1, but not its W34A mutant (M), significantly enhances TAp63γ‐mediated expression of Bax‐Luc (Fig. 1B). On the other hand, we used MBC1‐4‐Luc reporter as a nonresponsive promoter control and found that neither TAp63γ nor Pin1 can activate its expression (data not shown) [30], indicating the specific regulation of both proteins on Bax‐Luc expression. The IB analysis results reveal that neither wild‐type Pin1 nor its W34A M affects the expression level of TAp63γ; wild‐type Pin1, but not the mutant, significantly increases the level of cleaved PARP1 (CL‐PARP1), which is a molecular marker of cell apoptosis and can be induced by TAp63γ (Fig. 1B). These effects of Pin1 and TAp63γ are consistent with the results of cell survival/proliferation assay: wild‐type Pin1, but not its W34A mutant, significantly aggravates cell proliferation/survival inhibition of TAp63γ (Fig. 1C). Further, we found that Pin1 stimulates TAp63γ‐mediated expression of Bax‐Luc in a dose‐dependent manner (Fig. 1D). These results suggest that Pin1 stimulates transcriptional and pro‐apoptotic activities of TAp63γ.
Fig. 1.
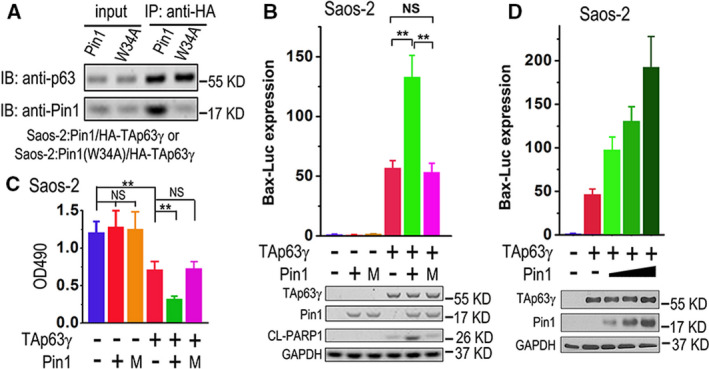
Pin1 enhances TAp63γ‐induced transcription and apoptosis. (A) Saos‐2 cells transfected with HA‐TAp63γ, plus Pin1 or its W34A mutant, were lysed and subjected to IP with anti‐HA. The cell lysates (inputs) or IP products were subjected to immunoblot (IB) analysis with indicated primary antibodies. (B) Saos‐2 cells were transfected with a mixture of Bax‐Luc and TK‐Renilla plus indicated plasmids. M, W34A mutant Pin1. Firefly and Renilla luciferase activities were measured, while IB analyses were performed to detect indicated proteins. The Bax‐Luc activity was normalized to Renilla activity and presented as Bax‐Luc expression level with SD (n = 3). Bax‐Luc expression in cells transfected with Bax‐Luc/TK‐Renilla mixture alone was set as 1. Two‐tailed t‐test was used for comparison between two groups; **P < 0.01; NS, nonsignificant. (C) Saos‐2 cells transfected with indicated plasmids were subjected to cell survival measurement with 3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyl‐tetrazolium bromide (MTT). Cell viabilities were presented as optical density values at the wavelength of 490 nm (OD490) with SD (n = 3). Two‐tailed t‐test was used for comparison between two groups; **P < 0.01; NS, nonsignificant. (D) Saos‐2 cells were transfected with a mixture of Bax‐Luc and TK‐Renilla plus HA‐TAp63γ and increasing amounts of Pin1 plasmid as indicated. Bax‐Luc expression levels were measured and presented as mentioned above, while IB analyses were performed to detect indicated proteins. The error bars indicate SD (n = 3).
Serine 12 in the transactivation domain of TAp63γ is crucial to Pin1‐mediated stimulation
In another previous report from our group, we found that serine 12 (S12) is crucial to transactivity of TAp63γ [27]. S12 is followed by a proline residue (P13), composing a putative Pin1 modification site [22]. It is well known that phosphorylation of the serine or threonine followed by proline is essential for the binding of Pin1 [31]. As a PPI, Pin1 mediates isomerization of proline, which is prevented by phosphorylation of the adjacent serine or threonine residue (pS‐P or pT‐P) [32]. This isomerization offers a molecular switch for recruitment of protein binding or post‐translational modification and modulates transactivity of multiple transcription factors [33, 34, 35]. To investigate whether this pS12‐P13 site is involved in Pin1‐mediated stimulation of TAp63γ (Fig. 1B,D), we tested the effect of Pin1 on expression of Bax‐Luc mediated by S12A mutant TAp63γ, which loses phosphorylation at this site. The results demonstrate that though S12A mutation enhances transactivity of TAp63γ, the expression of Bax‐Luc mediated by the mutant cannot be stimulated by Pin1 (Fig. 2A). The results of CoIP show that TAp63γ readily binds to Pin1 and this physical interaction can be significantly impaired by S12A mutation (Fig. 2B). These results reveal that serine 12 in the TAD of TAp63γ is crucial to its interaction with Pin1 and Pin1‐mediated stimulation.
Fig. 2.
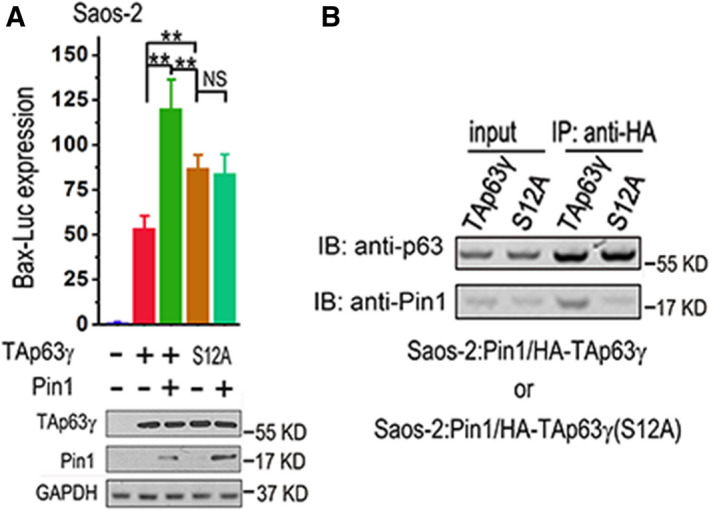
Serine 12 in the TAD of TAp63γ is crucial to Pin1‐mediated stimulation. (A) Saos‐2 cells were transfected with a mixture of Bax‐Luc and TK‐Renilla plus indicated plasmids. S12A, S12A mutant TAp63γ. Bax‐Luc expression levels were measured and presented as mentioned above (n = 3), while IB analyses were performed to detect indicated proteins. The error bars indicate SD. Two‐tailed t‐test was used for comparison between two groups; **P < 0.01; NS, nonsignificant. (B) Saos‐2 cells transfected with HA‐TAp63γ, plus Pin1 or its W34A mutant, were lysed and subjected to IP with anti‐HA. The cell lysates (inputs) or IP products were subjected to IB analysis with indicated primary antibodies.
Pin1 strikingly reverses JNK1‐repressed transcriptional and pro‐apoptotic activities of TAp63γ and makes it hyperactive
In our previous report mentioned above, we found that JNK1 can phosphorylate TAp63γ at serine 12, resulting in a repression of its transcriptional and pro‐apoptotic activities [27]. To further investigate the effects of JNK1 and Pin1 on TAp63γ, we transfected JNK1 and (or) Pin1 along with TAp63γ into Saos‐2 cells. The results of luciferase reporter assay show that TAp63γ‐mediated Bax‐luc expression is repressed by JNK1 but boosted by Pin1; unexpectedly, simultaneous overexpression of JNK1 can further enhance Pin1‐mediated activation of TAp63γ (Fig. 3A). The IB analysis reveals that overexpression of JNK1 or Pin1 has no significant effects on the protein level of TAp63γ; JNK1 significantly impairs the production of CL‐PARP1 induced by TAp63γ; on the contrary, Pin1 obviously promotes TAp63γ‐induced CL‐PARP1; intriguingly, simultaneous overexpression of Pin1 and JNK1 can strikingly exacerbate cleavage of PARP1 induced by TAp63γ (Fig. 3A). In line with the PARP1 cleavage results, TAp63γ‐induced inhibition of cell survival/proliferation is rescued by JNK1 and intensified by Pin1, while further exacerbated by simultaneous overexpression of Pin1 and JNK1 (Fig. 3B). Next, we knocked down endogenous JNK1 with siRNA used previously [27] and tested the Pin1‐mediated activation of TAp63γ. The results of IB analysis show that the specific siRNA can effectively ablate endogenous JNK1 in Saos‐2 cells; TAp63γ induces the production of CL‐PARP1, which can be further increased by the ablation of JNK1; overexpression of both TAp63γ and Pin1 makes an even higher CL‐PARP1 level, while simultaneous knockdown of JNK1 impairs the effect of Pin1 on TAp63γ‐induced production of CL‐PARP1 (Fig. 3C). The luciferase reporter assay demonstrates that ablation of JNK1 significantly increases TAp63γ‐mediated expression of Bax‐Luc; ablation of JNK1 abrogates the effect of Pin1 on TAp63γ‐mediated expression of Bax‐Luc (Fig. 3C). These results suggest that Pin1 strikingly reverses JNK1‐repressed transcriptional and pro‐apoptotic activities of TAp63γ and makes it hyperactive.
Fig. 3.
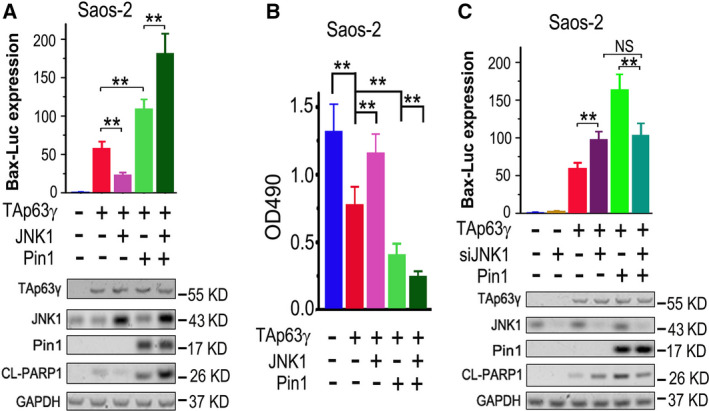
Pin1 strikingly reverses JNK1‐repressed activity of TAp63γ and makes it hyperactive. (A) Saos‐2 cells were transfected with a mixture of Bax‐Luc and TK‐Renilla plus indicated plasmids. Bax‐Luc expression levels were measured and presented as mentioned above (n = 3), while IB analyses were performed to detect indicated proteins. Two‐tailed t‐test was used for comparison between two groups; **P < 0.01. (B) Saos‐2 cells transfected with indicated plasmids were subjected to cell survival measurement with MTT. Cell viabilities were presented as mentioned above (n = 3). Two‐tailed t‐test was used for comparison between two groups; **P < 0.01. (C) Saos‐2 cells were transfected with a mixture of Bax‐Luc and TK‐Renilla plus indicated plasmids or siRNAs. Bax‐Luc expression levels were measured and presented as mentioned above, while IB analyses were performed to detect indicated proteins. Two‐tailed t‐test was used for comparison between two groups; **P < 0.01; NS, nonsignificant. The error bars (A–C) indicate SD (n = 3).
JNK1 may repress or promote transactivity of TAp63γ depending on Pin1 level
As shown in Fig. 4A, there is a high level of endogenous Pin1 in Hela cells, while a moderate level in H1299. shRNA‐based knockdown of Pin1 can significantly impair TAp63γ‐mediated expression of Bax‐Luc in Hela cells (Fig. 4B). On the other hand, overexpression of JNK1 enhances TAp63γ‐mediated expression of Bax‐Luc in a dose‐dependent manner in Hela cells (Fig. 4C). This is contrary to our previous study in H1299 cells [27], as well as the results in Saos‐2 cells in the present study (Fig. 3). Intriguingly, in Hela cells ablated with Pin1, overexpression of JNK1 represses TAp63γ‐mediated expression of Bax‐Luc in a dose‐dependent manner (Fig. 4D). In H1299 cells, overexpression of Pin1 strikingly reverses effects of JNK1 on TAp63γ transactivity (Fig. 4E), just like it does in Saos‐2 cells (Fig. 3A).
Fig. 4.
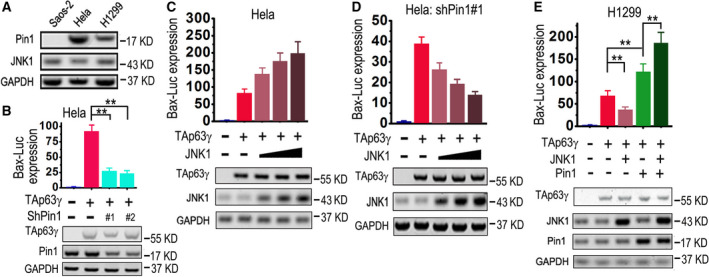
JNK1 may repress or promote transactivity of TAp63γ depending on Pin1 level. (A) Saos‐2, Hela and H1299 cells were lysed and indicated proteins were detected by means of IB analysis. (B) Hela cells were transfected with a mixture of Bax‐Luc and TK‐Renilla plus indicated plasmids. Bax‐Luc expression levels were measured and presented as mentioned above (n = 3), while IB analyses were performed to detect indicated proteins. Two‐tailed t‐test was used for comparison between two groups; **P < 0.01. (C, D) Hela cells, or Hela cells stably ablated with Pin1, were transfected with a mixture of Bax‐Luc and TK‐Renilla, plus HA‐TAp63γ and increasing amounts of JNK1 plasmid as indicated. Bax‐Luc expression levels were measured and presented as mentioned above (n = 3), while IB analyses were performed to detect indicated proteins. (E) H1299 cells were transfected with a mixture of Bax‐Luc and TK‐Renilla plus indicated plasmids. Bax‐Luc expression levels were measured and presented as mentioned above (n = 3), while IB analyses were performed to detect indicated proteins. Two‐tailed t‐test was used for comparison between two groups; **P < 0.01. The error bars (B–E) indicate SD.
These results suggest that JNK1‐mediated phosphorylation of TAp63γ at serine 12 can repress its transactivity, in the absence of abundant Pin1 (e.g., in Saos‐2 and H1299 cells, or Hela cells ablated with Pin1); in cells rich in Pin1 (e.g., Hela and Saos‐2 or H1299 ectopically overexpressing Pin1), the peptidyl‐prolyl isomerization of this phosphoserine‐proline (pS12‐P13) motif in the TAD of TAp63γ can strikingly activate its transcriptional activity (depicted as Graphical abstract figure).
Discussion
The p63 gene encodes multiple transcription factors [3]. Despite its low expression, TAp63γ plays key roles in quality control of germline cells, tumorigenesis, and aging, via its potent transactivity [7, 8, 9, 10, 11, 12, 13, 14]. We previously reported that Pin1 physically interacts with several isoforms of p63, including TAp63γ; Pin1 stabilizes TAp63α and ∆Np63α via mediating the isomerization of pT‐P‐P‐P‐P‐Y motif in the SAM and consequently impairing their affinity to E3 ligase WWP1 at this motif; however, the effect of this protein–protein interaction between TAp63γ and Pin1 was unknown [22]. In the present study, we find that Pin1 enhances transcriptional and pro‐apoptotic activities of TAp63γ (Fig. 1). On the other hand, we and others previously found that serine 12 (S12) in the TAD is critical to regulation of TAp63γ transactivity [24, 27]. S12 and the adjacent residue, proline 13 (P13), compose a potential Pin1‐binding site, which is supposed to lose the putative interaction by S12A mutation. We find that S12A mutant TAp63γ cannot be stimulated by Pin1 (Fig. 2A). Our further data show that this point mutation in TAp63γ significantly impairs its interaction with Pin1; the residual interaction between TAp63γ(S12A) and Pin1 indicates other binding sites of Pin1 than S12 in TAp63γ (Fig. 2B). Together, these results suggest that Pin1 promotes transactivity via binding to S12‐P13 in the TAD of TAp63γ. Since TAp63α and TAp63β also have this site, we speculate that Pin1 and JNK1 may regulate them in the same way. However, this regulation may not exist in △Np63 proteins, because they do not have the S12‐P13 motif in their truncated TAD [1].
S12 in TAp63γ is phosphorylated by IKKβ or JNK1, leading to an impairment of its transactivity [24, 27]. In our present study, we find that this inhibition of transactivity mediated by phosphorylation at this residue can be strikingly reversed by Pin1; in combination with JNK1, Pin1 can even enhance the transcriptional and pro‐apoptotic activities of TAp63γ to an extent that is higher than that in the absence of JNK1 (Fig. 3). JNK1 exhibits negative effects on TAp63γ activity in cells lacking abundant Pin1 proteins, while stimulates TAp63γ in cells rich in Pin1 (Fig. 4). Based on these results, we propose the following model to interpret the regulation of TAp63γ transactivity (as shown in Graphical abstract figure): TAp63γ with S12 unphosphorylated is moderately active; phosphorylation at this residue (pS12) mediated by IKKβ or JNK1 can repress its activity; in the presence of Pin1, isomerization of this pS12‐P13 motif makes TAp63γ hyperactive. Our data are helpful to elucidate the regulation of TAp63γ, which is an important transcription factor in tumorigenesis and germline quality control, as well as a potential therapeutic target against p53‐altered tumors [10, 16].
Conflict of interest
The authors declare no conflict of interest.
Author contributions
CL devised the hypothesis. XF, WH, KH, HC, LC, and SF designed and performed the experiments. W H and CL analyzed the data and wrote the manuscript.
Acknowledgements
This work was supported by National Natural Science Foundation of China (#91749121 and #32070747), and the Fundamental Research Funds for the Central Universities (SCU2019D013).
Xueying Fan and Wei He contributed equally to this work
[Correction added on 25 February, 2021, after first online publication: Sichuan University has been added to the first affiliation and correspondence address]
Data Accessibility
We state that our data will be available from the corresponding author upon reasonable request.
References
- 1. Chen Y, Peng Y, Fan S, Li Y, Xiao ZX and Li C (2018) A double dealing tale of p63: an oncogene or a tumor suppressor. Cell Mol Life Sci 75, 965–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Li C and Xiao ZX (2014) Regulation of p63 protein stability via ubiquitin‐proteasome pathway. Biomed Res Int 2014, 175721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mangiulli M, Valletti A, Caratozzolo MF, Tullo A, Sbisa E, Pesole G and D'Erchia AM (2009) Identification and functional characterization of two new transcriptional variants of the human p63 gene. Nucleic Acids Res 37, 6092–6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gressner O, Schilling T, Lorenz K, Schulze Schleithoff E, Koch A, Schulze‐Bergkamen H, Lena AM, Candi E, Terrinoni A, Catani MV et al. (2005) TAp63alpha induces apoptosis by activating signaling via death receptors and mitochondria. EMBO J 24, 2458–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Osada M, Ohba M, Kawahara C, Ishioka C, Kanamaru R, Katoh I, Ikawa Y, Nimura Y, Nakagawara A, Obinata M et al. (1998) Cloning and functional analysis of human p51, which structurally and functionally resembles p53. Nat Med 4, 839–843. [DOI] [PubMed] [Google Scholar]
- 6. Wu G, Nomoto S, Hoque MO, Dracheva T, Osada M, Lee CC, Dong SM, Guo Z, Benoit N, Cohen Y et al. (2003) DeltaNp63alpha and TAp63alpha regulate transcription of genes with distinct biological functions in cancer and development. Cancer Res 63, 2351–2357. [PubMed] [Google Scholar]
- 7. Gonfloni S, Di Tella L, Caldarola S, Cannata SM, Klinger FG, Di Bartolomeo C, Mattei M, Candi E, De Felici M, Melino G et al. (2009) Inhibition of the c‐Abl‐TAp63 pathway protects mouse oocytes from chemotherapy‐induced death. Nat Med 15, 1179–1185. [DOI] [PubMed] [Google Scholar]
- 8. Deutsch GB, Zielonka EM, Coutandin D and Dotsch V (2011) Quality control in oocytes: domain‐domain interactions regulate the activity of p63. Cell Cycle 10, 1884–1885. [DOI] [PubMed] [Google Scholar]
- 9. Deutsch GB, Zielonka EM, Coutandin D, Weber TA, Schafer B, Hannewald J, Luh LM, Durst FG, Ibrahim M, Hoffmann J et al. (2011) DNA damage in oocytes induces a switch of the quality control factor TAp63alpha from dimer to tetramer. Cell 144, 566–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bildik G, Acilan C, Sahin GN, Karahuseyinoglu S and Oktem O (2018) C‐Abl is not activated in DNA damage‐induced and Tap63‐mediated oocyte apoptosis in human ovary. Cell Death Dis 9, 943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Guo X, Keyes WM, Papazoglu C, Zuber J, Li W, Lowe SW, Vogel H and Mills AA (2009) TAp63 induces senescence and suppresses tumorigenesis in vivo . Nat Cell Biol 11, 1451–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Su X, Paris M, Gi YJ, Tsai KY, Cho MS, Lin YL, Biernaskie JA, Sinha S, Prives C, Pevny LH et al. (2009) TAp63 prevents premature aging by promoting adult stem cell maintenance. Cell Stem Cell 5, 64–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Su X, Gi YJ, Chakravarti D, Chan IL, Zhang A, Xia X, Tsai KY and Flores ER (2012) TAp63 is a master transcriptional regulator of lipid and glucose metabolism. Cell Metab 16, 511–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Su X, Chakravarti D, Cho MS, Liu L, Gi YJ, Lin YL, Leung ML, El‐Naggar A, Creighton CJ, Suraokar MB et al. (2010) TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature 467, 986–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Terrinoni A, Palombo R, Pitolli C, Caporali S, De Berardinis R, Ciccarone S, Lanzillotta A, Mauramati S, Porta G, Minieri M et al. (2019) Role of the TAp63 isoform in recurrent nasal polyps. Folia Biol 65, 170–180. [DOI] [PubMed] [Google Scholar]
- 16. Gunaratne PH, Pan Y, Rao AK, Lin C, Hernandez‐Herrera A, Liang K, Rait AS, Venkatanarayan A, Benham AL, Rubab F et al. (2019) Activating p53 family member TAp63: a novel therapeutic strategy for targeting p53‐altered tumors. Cancer 125, 2409–2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yamaki T, Suenaga Y, Iuchi T, Alagu J, Takatori A, Itami M, Araki A, Ohira M, Inoue M, Kageyama H et al. (2013) Temozolomide suppresses MYC via activation of TAp63 to inhibit progression of human glioblastoma. Sci Rep 3, 1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Suenaga Y, Yamamoto M, Sakuma T, Sasada M, Fukai F, Ohira M, Yamaguchi Y, Yamamoto T, Ando K, Ozaki T et al. (2019) TAp63 represses transcription of MYCN/NCYM gene and its high levels of expression are associated with favorable outcome in neuroblastoma. Biochem Biophys Res Commun 518, 311–318. [DOI] [PubMed] [Google Scholar]
- 19. Cefalu S, Lena AM, Vojtesek B, Musaro A, Rossi A, Melino G and Candi E (2015) TAp63gamma is required for the late stages of myogenesis. Cell Cycle 14, 894–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Curtis KM, Aenlle KK, Frisch RN and Howard GA (2015) TAp63gamma and DeltaNp63beta promote osteoblastic differentiation of human mesenchymal stem cells: regulation by vitamin D3 Metabolites. PLoS One 10, e0123642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang Q, Li N, Chen F, Hei R, Gu J, Lu Y, Sun L and Zheng Q (2020) TAp63gamma influences mouse cartilage development. Aging 12, 8669–8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li C, Chang DL, Yang Z, Qi J, Liu R, He H, Li D and Xiao ZX (2013) Pin1 modulates p63alpha protein stability in regulation of cell survival, proliferation and tumor formation. Cell Death Dis 4, e943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li Y, Zhou Z and Chen C (2008) WW domain‐containing E3 ubiquitin protein ligase 1 targets p63 transcription factor for ubiquitin‐mediated proteasomal degradation and regulates apoptosis. Cell Death Differ 15, 1941–1951. [DOI] [PubMed] [Google Scholar]
- 24. Liao JM, Zhang Y, Liao W, Zeng SX, Su X, Flores ER and Lu H (2013) IkappaB kinase beta (IKKbeta) inhibits p63 isoform gamma (TAp63gamma) transcriptional activity. J Biol Chem 288, 18184–18193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. He H, Peng Y, Fan S, Chen Y, Zheng X and Li C (2018) Cullin3/KCTD5 induces monoubiquitination of DeltaNp63alpha and impairs its activity. FEBS Lett 592, 2334–2340. [DOI] [PubMed] [Google Scholar]
- 26. Chen J, Shi H, Chen Y, Fan S, Liu D and Li C (2017) DNA damage induces expression of WWP1 to target DeltaNp63alpha to degradation. PLoS One 12, e0176142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen J, Shi H, Qi J, Liu D, Yang Z and Li C (2015) JNK1 inhibits transcriptional and pro‐apoptotic activity of TAp63gamma. FEBS Lett 589, 3686–3690. [DOI] [PubMed] [Google Scholar]
- 28. Li D, Li C, Wu M, Chen Q, Wang Q, Ren J and Zhang Y (2015) PKCdelta stabilizes TAp63 to promote cell apoptosis. FEBS Lett 589, 2094–2099. [DOI] [PubMed] [Google Scholar]
- 29. Ying H, Chang DL, Zheng H, McKeon F and Xiao ZX (2005) DNA‐binding and transactivation activities are essential for TAp63 protein degradation. Mol Cell Biol 25, 6154–6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Han A, Li J, Li Y, Wang Y, Bergholz J, Zhang Y, Li C and Xiao ZhX (2016) p63alpha modulates c‐Myc activity via direct interaction and regulation of MM1 protein stability. Oncotarget 7, 44277–44287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lu KP, Liou YC and Zhou XZ (2002) Pinning down proline‐directed phosphorylation signaling. Trends Cell Biol 12, 164–172. [DOI] [PubMed] [Google Scholar]
- 32. Lu KP and Zhou XZ (2007) The prolyl isomerase PIN1: a pivotal new twist in phosphorylation signalling and disease. Nat Rev Mol Cell Biol 8, 904–916. [DOI] [PubMed] [Google Scholar]
- 33. Huang GL, Liao D, Chen H, Lu Y, Chen L, Li H, Li B, Liu W, Ye C, Li T et al. (2016) The protein level and transcription activity of activating transcription factor 1 is regulated by prolyl isomerase Pin1 in nasopharyngeal carcinoma progression. Cell Death Dis 7, e2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Poolman TM, Farrow SN, Matthews L, Loudon AS and Ray DW (2013) Pin1 promotes GR transactivation by enhancing recruitment to target genes. Nucleic Acids Res 41, 8515–8525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Werwein E, Cibis H, Hess D and Klempnauer KH (2019) Activation of the oncogenic transcription factor B‐Myb via multisite phosphorylation and prolyl cis/trans isomerization. Nucleic Acids Res 47, 103–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
We state that our data will be available from the corresponding author upon reasonable request.


